Abstract
To understand the molecular epidemiology of circulating dengue viruses (DENV) in Upper Myanmar, DENV isolation was attempted by inoculating the sera of a panel of 110 serum samples onto a C6/36 mosquito cell line. The samples were collected from dengue (DEN) patients admitted at Mandalay Children’s Hospital in 2006. Infected culture fluids were subjected to a RT-PCR to detect the DENV genome. Three DENV strains were isolated. This was the first DENV isolation performed either in Mandalay or in Upper Myanmar. One strain belonged to DENV serotype-3 (DENV-3), and two other strains belonged to DENV serotype-4 (DEN-4). The sequence data for the envelope gene of these strains were used in a phylogenetic comparison of DENV-3 and DENV-4 from various countries. Phylogenetic analyses revealed that this DENV-3 strain was clustered within genotype II, and the two DENV-4 strains were clustered within genotype I in each serotype. The Myanmar strains were closely related to strains from the neighboring countries of Thailand and Bangladesh. These results are important for elucidating the trends of recent and future DEN outbreaks in Myanmar.
Keywords: Dengue virus, Molecular epidemiology, Upper Myanmar
Introduction
The dengue virus (DENV) belongs to the genus Flavivirus of the family Flaviviridae, which exists as four serotypes (DENV-1,-2,-3, and -4) [1]. DENV infection is the most important of the mosquito-borne viral diseases, and it affects mainly tropical and subtropical countries [2]. It is well documented that all four serotypes of DENV co-circulate in Asian countries including Myanmar [3, 4]. The first major epidemic of dengue hemorrhagic fever (DHF) occurred in Myanmar in 1970 [5]. Currently, DHF occurs throughout the country, with the notable exception of the Chin State. Almost 80% of cases are reported from three divisions (Yangon, Bago and Mandalay) and one state (Mon), with more than 50% of cases recorded exclusively from the Yangon Division [5]. DEN outbreaks have been recorded in Upper Myanmar, especially in Mandalay, the largest city in the region. However, an extensive study has never been accomplished due to insufficient laboratory facilities. The present study focused on highlighting current DENV infections in Upper Myanmar with a special emphasis on molecular epidemiology.
Methods
Patients
In total, 110 serum samples were obtained from 110 patients (≤ 12 years old) who were clinically suspected for DEN according to World Health Organization [6] criteria and who were admitted to the 550-bed Mandalay Children’s Hospital (MCH), Mandalay City, Upper Myanmar in 2006 with the informed consent of parents or legal guardians. The study protocol was reviewed and approved by the Ethical Committee on Medical Research Involving Human Subjects, Department of Medical Research (Upper Myanmar), Pyin Oo Lwin, Myanmar. The sera were stored at −70°C until further use.
Methods
Both IgM- and IgG-capture ELISAs were performed using Dengue Duo IgM-capture and IgG-capture ELISA Kits (PANBIO, Brisbane, Australia) to determine primary and secondary DENV infections. All the commercial kits in the present study were used following the manufacturer’s instructions.
The frozen sera were transferred to Japan, and the virus culture was conducted in the Department of Virology, Institute of Tropical Medicine, Nagasaki University, Japan. Each serum sample was inoculated onto Aedes albopictus clone C6/36 mosquito cells and incubated at 28°C for 7 days [7]. The presence of DENV in the infected culture fluid (ICF) was verified by in-house Flavivirus antigen detection ELISA (Ag-ELISA) [8] and RT-PCR. RNA extraction from ICF was performed using a viral RNA Mini Kit (QIAGEN, Hilden, Germany). DENV serotyping was done using 4 sets of serotype-specific primers [9–11] employing the PrimeScriptTM One Step RT-PCR Kit (Takara Bio Inc., Shiga, Japan).
The desired DNA bands were excised from the agarose gel, and were extracted and purified using QIAEX® II Gel Extraction Kit (QIAGEN, Hilden, Germany). The primer extension dideoxy chain termination method was used for direct sequencing of the PCR product. DNA sequencing analysis was performed with BigDye® Terminator version 3.1 Cycle Sequencing Ready Reaction Mixture (Applied Biosystems, Foster City, USA) following the thermal cycle sequencing parameters described previously [12]. The reaction mixture was then purified using an AGENCOURT® CLEANSEQ® Sequencing Reaction Clean-up system (Agencourt Bioscience Corp., Massachusetts, USA). The final product was loaded on an ABI PrismTM Capillary Sequencer 3100-Avant Genetic Analyzer (Applied Biosystems, Foster City, USA). Nucleotide sequences were edited and homology searches and comparisons of the sequences done using DNASIS (Mac version 3.6 Software system; Hitachi, Tokyo, Japan). Nucleotide sequence alignments were carried out using CLUSTAL X, version 2.0 [13], and the phylogenetic analysis was performed using either the heuristic or the branch and bound algorithm of PAUP version 4.0b10 (Altivec) software [14]. The neighbor-joining method was used to construct the phylogenetic tree with a bootstrap analysis of 1,000 replicates [15].
The Genbank accession numbers, EU478408, EU478409 and EU478410, for the three Myanmar isolates used in the present study, and the accession numbers of all the other strains used for the phylogenetic analysis, are listed in Tables 1 and 2 with the geographic origin and the year of isolation.
Table 1.
Clinical information of the 110 patients suspected with dengue virus infection and from whom blood samples were collected
| Parameters | Confirmed dengue casesa (%) | Non dengue casesb (%) |
|---|---|---|
| number of cases | 96 (87) | 14 (13) |
| Mean age in years (± SD) | 5.5 (± 3.2) | 5.6 (± 4.0) |
| Male/Female | 49/47 | 6/8 |
| DHF I | 26 (27) | 5 (36) |
| DHF II | 32 (33) | 5 (36) |
| DHF III | 21 (22) | 2 (14) |
| DHF IV | 1 (1) | 0 (0) |
| DSS | 16 (17) | 2 (14) |
Confirmed dengue cases were positive for dengue IgM capture ELISA. Non-dengue cases were negative for dengue IgM capture ELISA.
DHF I, dengue hemorrhagic fever grade I; DHF II, dengue hemorrhagic fever grade II; DHF III, dengue hemorrhagic fever grade III; DHF IV, dengue hemorrhagic fever grade IV; DSS, dengue shock syndrome. DHF grading were classified according to WHO criteria (WHO, 1997).
Table 2.
DENV-3 strains used for phylogenetic analysis
| Strain | Code in tree | Geographic origin | Year of isolation | Accession number |
|---|---|---|---|---|
| BDH 02-01 | Bdesh 0201 | Bangladesh | 2002 | AY 496871 |
| BDH 02-07 | Bdesh 0207 | Bangladesh | 2002 | AY 496877 |
| 114 | Bdesh 00114 | Bangladesh | 2000 | AY656669 |
| 165 | Bdesh 00165 | Bangladesh | 2000 | AY656671 |
| 058 | Bdesh 00058 | Bangladesh | 2000 | AY656674 |
| Jacob | Bdesh 2001 | Bangladesh | 2001 | AY656673 |
| 68784 | Brazil 2000 | Brazil | 2000 | AY038605 |
| 80-2 | China 80 | China | 1980 | AF317645 |
| Cuba-21/02 | Cuba 02 | Cuba | 2002 | AY702031 |
| 29472 | Fiji 92 | Fiji | 1992 | L11422 |
| 1416 | India 84 | India | 1984 | L11424 |
| 228761 | Indon 73 | Indonesia | 1973 | L11425 |
| 1280 | Indon 78 | Indonesia | 1978 | L11426 |
| 85-159 | Indon 85 | Indonesia | 1985 | L11428 |
| 1300 | Malay 74 | Malaysia | 1974 | L11429 |
| 29586 | Malay 81 | Malaysia | 1981 | L11427 |
| LN 5547 | Malay 92 | Malaysia | 1992 | AF147457 |
| LN 1746 | Malay 93 | Malaysia | 1993 | AF147458 |
| LN 6083 | Malay 94 | Malaysia | 1994 | AF147460 |
| D3/H/IMTSSA-MART/2001/2012 | Martiniq 01 | Martinique | 2001 | AY099340 |
| MEX6097 | Mexico 95 | Mexico | 1995 | AY146763 |
| 1559 | Mozambiq 85 | Mozambique | 1985 | L11430 |
| 31985 KLA | Myan 98 | Myanmar | 1998 | AY145712 |
| DV3/Myanmar/0508aTw/ 2005 | Myan 05 | Myanmar | 2005 | DQ518666 |
| DV3/Mandalay.MYA/ H58/2006* | Myan 06 | Myanmar | 2006 | EU478409 |
| 24/94 | Nicaragu 94 | Nicaragua | 1994 | AY702033 |
| D3 PY/A59/03 | Paraguay 03 | Paraguay | 2003 | DQ 118885 |
| H 87 | Philip 56 | Philippines | 1956 | L 11423 |
| 168-AP-2 | Philip 83 | Philippines | 1983 | L11432 |
| PhMH-J1-97 | Philip 97 | Philippines | 1997 | AY 496879 |
| PR6 | PRico 63 | Puerto Rico | 1963 | L11433 |
| 1340 | PRico 77 | Puerto Rico | 1977 | L11434 |
| 1696 | Samoa 86 | Samoa | 1986 | L11435 |
| 1326 | SLanka 81 | Sri Lanka | 1981 | L11431 |
| 1594 | SLanka 85 | Sri Lanka | 1985 | L11436 |
| 260698 | SLanka 89 | Sri Lanka | 1989 | L11437 |
| 2783 | SLanka 91 | Sri Lanka | 1991 | L11438 |
| D3/Srilanka 9912aTw/1999 | SLanka 99 | Sri Lanka | 1999 | DQ 518679 |
| 2167 | Tahiti 89 | Tahiti | 1989 | L11619 |
| D3/Taiwan/813KH9408a/1994 | Taiwan 94 | Taiwan | 1994 | DQ 518667 |
| D3/Taiwan/701TN9811a/1998 | Taiwan 98 | Taiwan | 1998 | DQ 518662 |
| D3/Taiwan/807KH0509a/2005 | Taiwan 05 | Taiwan | 2005 | DQ 518659 |
| 5987 | Thai 62 | Thailand | 1962 | L11440 |
| CH3489D73-1 | Thai 73 | Thailand | 1973 | L11620 |
| D86-007 | Thai 86 | Thailand | 1986 | L11441 |
| MK315 | Thai 87 | Thailand | 1987 | L11442 |
| D88-303 | Thai 88 | Thailand | 1988 | AY145714 |
| D89-273 | Thai 89 | Thailand | 1989 | AY145715 |
| D91-393 | Thai 91 | Thailand | 1991 | AY145716 |
| D92-431 | Thai 92 | Thailand | 1992 | AY145719 |
| D92-423 | Thai 92 | Thailand | 1992 | AY145718 |
| D93-044 | Thai 93 | Thailand | 1993 | AY145720 |
| D94-283 | Thai 94 | Thailand | 1994 | AY145723 |
| D95-0400 | Thai 95 | Thailand | 1995 | AY145725 |
| D 96-313 | Thai 96 | Thailand | 1996 | AY145726 |
| D 97-0291 | Thai 97 | Thailand | 1997 | AY145730 |
| 00-27-1 Hu NIID | NIID 2000 | Thailand/Bangladesh | 2000 | AB111080 |
| LARD 5990 | Venezu 2000 | Venezuela | 2000 | AY146764 |
| LARD 6668 | Venezu 2001 | Venezuela | 2001 | AY146774 |
| D3/Vietnam/9609aTw/1996 | Vietnam 96 | Vietnam | 1996 | DQ518655 |
| D3/Vietnam/0409aTw/2004 | Vietnam 04 | Vietnam | 2004 | DQ518656 |
| D3/Vietnam/0507aTw/2005 | Vietnam 05 | Vietnam | 2005 | DQ518658 |
* New strain from Myanmar presented in this study
Results
Among 110 clinically diagnosed DEN patients, 70 (64%) were positive for both IgM and IgG by DEN IgM capture and DEN IgG capture ELISA and were confirmed as secondary DENV infections. Primary DENV infection was confirmed in 26 (24%) patients who were positive only for IgM. The remaining 14 (13%) patients were not confirmed to have DENV infections. Among the 96 dengue-confirmed patients, dengue virus strains were sucessfully isolated from three patients whose sera were collected within 7 days from the onset of fever. One isolate was DEN-3 from a patient having a primary DENV infection with DHF grade (I), and two isolates were DEN-4 from a patient having primary DENV infection with DSS. The other patient had secondary DENV infection with DHF grade (I). The clinical information of the 110 patients is shown in Table 1.
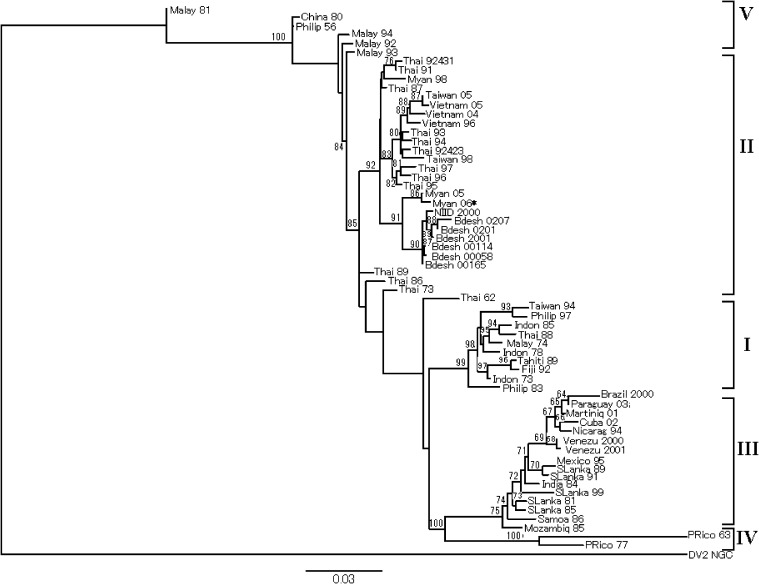
The nucleotide sequence of the E gene of the newly isolated DENV-3 strain from Upper Myanmar, designated as the DV3/Mandalay.MYA/H58/2006 strain (Myan 06, code in tree), was compared with other published sequences of 61 DENV-3 strains originating from various geographic regions (Table 2). The phylogenetic tree constructed for the 62 DENV-3 strains, employing the DENV-2 New Guinea C strain as an out-group strain, is shown in Fig. 1. The tree reveals that the newly isolated DENV-3 strain from Upper Myanmar was grouped together with previously published strains from Lower Myanmar, as well as the strains from Thailand, Bangladesh, Malaysia, Vietnam and Taiwan in a well-defined genotype II.
Fig. 1.
Phylogenetic tree of DENV-3 strains (n = 62). The tree is rooted by the DENV-2-NGC (New Guinea C, Accession No. M29095) strain (Table 2). All horizontal branch lengths are drawn to a scale of nucleotide substitutions per site. Bootstrap support values are shown and the genotypes of DENV-3 are indicated. For simplicity, each strain name was replaced by a code that consists of the country and the year of isolation. The DENV-3 isolate presented in the present study is indicated by a asterisk (*).
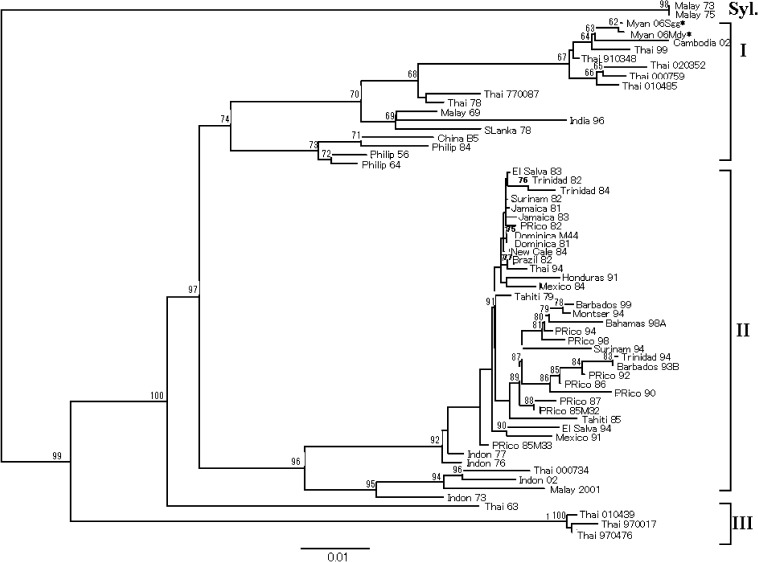
Similarly, the nucleotide sequences of the E gene of two newly isolated DENV-4 strains from Upper Myanmar, designated as the DV4/Sagaing.MYA/H27/2006 strain (Myan 06 Sgg, code in tree) and the DV4/Mandalay.MYA/H64/2006 strain (Myan 06 Mdy, code in tree), were compared with other published sequences of 59 DENV-4 strains originating from various geographic regions (Table 3). The phylogenetic tree constructed for a total of 61 DENV-4 strains is shown in Fig. 2. The tree reveals that the two new strains from Upper Myanmar were grouped together with those from Thailand, Cambodia, Malaysia, India, Sri Lanka, China and the Philippines in the Asian genotype I.
Table 3.
DENV-4 strains used for phylogenetic analysis
| Strain | Code in tree | Geographic origin | Year of isolation | Accession number |
|---|---|---|---|---|
| Bahamas A/98 | Bahamas 98A | Bahamas | 1998 | AY 152364 |
| Barbados B/93 | Barbados 93B | Barbados | 1993 | AY 152376 |
| Barbados/99 | Barbados 99 | Barbados | 1999 | AY 152368 |
| 1385/82 | Brazil 82 | Brazil | 1982 | U 18425 |
| 02-21-1 Hu NIID | Cambodia 02 | Cambodia | 2002 | AB 111089 |
| China.GuangzhoB5 | China B5 | China | NA | AF 289029 |
| 814669/81 | Dominica 81 | Dominica | 1981 | AF 326573 |
| M 44/81 | Dominica M44 | Dominica | 1981 | AY 152360 |
| 1411/83 | El Salva 83 | El Salvador | 1983 | U 18426 |
| BC 6494/94 | El Salva 94 | El Salvador | 1994 | U 18427 |
| Honduras/91 | Honduras 91 | Honduras | 1991 | AY 152379 |
| 96-33-1 Hu NIID | India 96 | India | 1996 | AB 111086 |
| 30153/73 | Indon 73 | Indonesia | 1973 | U 18428 |
| 1036/76 | Indon 76 | Indonesia | 1976 | U 18429 |
| 1132/77 | Indon 77 | Indonesia | 1977 | U 18430 |
| 02-12-1 Hu NIID | Indon 02 | Indonesia | 2002 | AB 111088 |
| Jamaica/81 | Jamaica 81 | Jamaica | 1981 | AY 152389 |
| Jamaica/83 | Jamaica 83 | Jamaica | 1983 | AY 152384 |
| P7-1006 | Malay 69 | Malaysia | 1969 | AF 231722 |
| P73-1120 | Malay 73 | Malaysia | 1973 | AF 231724 |
| P75-514 | Malay 75 | Malaysia | 1975 | AF 231723 |
| MY 01-23096 | Malay 2001 | Malaysia | 2001 | AJ 428557 |
| 1492/84 | Mexico 84 | Mexico | 1984 | U 18431 |
| Mexico/91 | Mexico 91 | Mexico | 1991 | AY 152378 |
| Montserrat-A/94 | Montser 94 | Montserrat | 1994 | AY 152369 |
| DV4/Sagaing.MYA/H27/2006* | Myan 06 Sgg | Myanmar | 2006 | EU 478410 |
| DV4/Mandalay.MYA/H64/2006* | Myan 06 Mdy | Myanmar | 2006 | EU 478408 |
| 5489/84 | New Cale 84 | New Caledonia | 1984 | U 18432 |
| H241/56 | Philip 56 | Philippines | 1956 | U 18433 |
| 16589/64 | Philip 64 | Philippines | 1964 | U 18434 |
| 12123/84 | Philip 84 | Philippines | 1984 | U 18435 |
| M5/82 | PRico 82 | Puerto Rico | 1982 | AY 152336 |
| M32/85 | PRico 85 M32 | Puerto Rico | 1985 | AY 152856 |
| M33/85 | PRico 85 M33 | Puerto Rico | 1985 | AY 152857 |
| 1650/86 | PRico 86 | Puerto Rico | 1986 | U 18436 |
| 69/87 | PRico 87 | Puerto Rico | 1987 | AY 152252 |
| 96/90 | PRico 90 | Puerto Rico | 1990 | AY 152855 |
| 28/92 | PRico 92 | Puerto Rico | 1992 | AY 152196 |
| 84/94 | PRico 94 | Puerto Rico | 1994 | AY 152084 |
| 17/98 | PRico 98 | Puerto Rico | 1998 | AY 152056 |
| S-44750/78 | SLanka 78 | Sri Lanka | 1978 | U 18437 |
| B/82 | Surinam 82 | Surinam | 1982 | AY 152386 |
| A/94 | Surinam 94 | Surinam | 1994 | AY 152372 |
| S-44754/79 | Tahiti 79 | Tahiti | 1979 | U 18438 |
| 114-094-85/85 | Tahiti 85 | Tahiti | 1985 | U 18439 |
| TC 2443/63 | Thai 63 | Thailand | 1963 | U 18440 |
| Thai D4-0087/77 | Thai 770087 | Thailand | 1977 | AY 618991 |
| Thailand/78 | Thai 78 | Thailand | 1978 | U 18441 |
| Thai D4-0348/91 | Thai 910348 | Thailand | 1991 | AY 618990 |
| 703-4/94 | Thai 94 | Thailand | 1994 | AF 231726 |
| Thai D4-0017/97 | Thai 970017 | Thailand | 1997 | AY 618978 |
| Thai D4-0476/97 | Thai 970476 | Thailand | 1997 | AY 618979 |
| 99-10-1 Hu NIID | Thai 99 | Thailand | 1999 | AB 111087 |
| Thai D4-0734/00 | Thai 000734 | Thailand | 2000 | AY 618993 |
| Thai D4-0759/00 | Thai 000759 | Thailand | 2000 | AY 618938 |
| Thai D4-0439/01 | Thai 010439 | Thailand | 2001 | AY 618940 |
| Thai D4-0485/01 | Thai 010485 | Thailand | 2001 | AY 618992 |
| Thai D4-0352/02 | Thai 020352 | Thailand | 2002 | AY 618945 |
| Trinidad A/82 | Trinidad 82 | Trinidad | 1982 | AY 152382 |
| Trinidad A/84 | Trinidad 84 | Trinidad | 1984 | AY 152380 |
| Trinidad/94 | Trinidad 94 | Trinidad | 1994 | AY 152377 |
* New strains from Myanmar presented in this study
Fig. 2.
Phylogenetic tree of DENV-4 strains (n = 61). The tree is rooted by the two sylvatic strains, Malay73 and Malay75 (Table 3). All horizontal branch lengths are drawn to a scale reflecting nucleotide substitutions per site. Bootstrap support values are shown and the genotypes of DENV-4 are indicated. For simplicity, each strain name was replaced by a code that consists of the country and the year of isolation. The DENV-4 isolates presented in the present study are indicated by a asterisk (*).
Discussion
The DENV-3 isolate from Upper Myanmar in the present study belonged to genotype II, like two previously published Lower Myanmar strains: the 31985 KLA strain (Myan 98, code in tree) and the DV3/Myanmar/0508aTw/2005 strain (Myan 05, code in tree). It clustered together with strains from Bangladesh in a well-defined sub-cluster. Further support came from the fact that three unique aminoacid (aa) changes, I140T, S447G and A489T, were found in this strain and were shared by Myan 05 and the Bangladesh strains. To examine the introduction of the DENV-3 genotype II to the country (although DENV-3 isolates from Myanmar are very few), we compared an older strain, Myan 98, to the two most recent ones: Myan 05 and Myan 06 [16]. In the phylogenetic tree, Myan 98 was clustered in a separate sub-cluster of genotype II together with earlier Thai strains. This clustering is supported by four aa changes, I140T, S447G, A489T and A479V, which are present in the two most recent Myanmar isolates, Myan 05 and Myan 06, but are not present in either the Myan 98 strain or in the Thai isolates that were clustered together with the latter strain. These results indicate that the genotype II of DENV-3 reached Myanmar most likely through independent entries from Thailand, a supposition supported by the appearance of the more recent lineage including the isolates from 2005 and 2006 (Fig. 1). The fact that the Bangladesh strains isolated from 2000 to 2002 showed little evidence of independent evolution suggests that genotype II was also introduced recently from neighboring countries. Our results support Podder et al.’s (2006) suggestion that recent DEN outbreaks in Bangladesh (2000 and 2001) were associated with the introduction of DENV-3 from eastern countries, rather than the evolution of a virulent strain in situ [17]. In addition, Islam et al. (2006) speculated that DENV-3 circulating in Bangladesh in 2002 might have entered from neighboring countries [12]. Recently, it was reported that seven DENV-3 strains isolated from Yangon (Lower Myanmar) in 2007 belonged to genotype III [18]. Therefore, it appears that more than one DENV-3 genotype is circulating in the country. It would be interesting to analyze the time and route of introduction.
The two newly isolated DENV-4 strains from Upper Myanmar in the present study were clustered together with other Asian strains in genotype I being the closest related strains from Thailand and Cambodia [9], but V238M and L489P aa changes were unique to Myan 06 Mdy and Myan 06 Sgg strains, respectively. Although the existence of DENV-1 and DENV-2 among the circulating viruses in Upper Myanmar could not be ruled out, the present study demonstrated that DENV-3 and DENV-4 were co-circulating in the area in 2006. This is the first report on the molecular analysis of DENV-4 strains in Myanmar, particularly those circulating in the upper part of the country. Therefore, if DENV-3 is currently regarded as the prevailing serotype for recent outbreaks, then DENV-4 might be in the pipeline to take the lead in future outbreaks in Myanmar.
Acknowledgements
The authors are grateful to Dr. Myint-Myint Thein, Medical Superintendent of Mandalay Children’s Hospital, for the collection of samples from the hospitalized patients, and to the WHO Country Office in Myanmar for providing the biennium budget for the years 2006/2007 and for conducting the ELISA experiments. The molecular epidemiological study was supported by a Grant-in-Aid for Scientific Research (No. 18406017 and 21256004) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and the Program of Founding Research Centers for Emerging and Re-emerging Infectious Diseases MEXT Japan, the Global COE Program, the 21st century COE Program MEXT Japan, and the JSPS Core University Program.
The authors also appreciate the valuable scientific suggestions of Corazon Cerilla Buerano and the support of the members of the Department of Molecular Epidemiology and the Department of Virology, Institute of Tropical Medicine, Nagasaki University, Nagasaki City, Japan.
Potential Conflict of Interest
We declare no conflicts of interest.
References
- 1.Henchal EA, Putnak JR. The dengue viruses. Clin Microbiol Rev 1990; 3: 376–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monath TP. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci U S A 1994; 91: 2395–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guzman MG, Halstead SB, Artsob H, et al. Dengue: a continuing global threat. Nat Rev Microbiol 2010; 8: S7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thu HM, Lowry K, Myint TT, et al. Myanmar dengue outbreak associated with displacement of serotypes 2, 3, and 4 by dengue 1. Emerg Infect Dis 2004; 10: 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thein S. Dengue haemorrhagic fever in Myanmar. DMR Bull 1991; 5: 1–14. [Google Scholar]
- 6.World Health Organization. Dengue Haemorrhagic fever. Diagnosis, Treatment, Prevention and Control. 2nd ed. Geneva: WHO; 1997. [Google Scholar]
- 7.Igarashi A. Isolation of a Singh’s Aedes albopictus cell clone sensitive to Dengue and Chikungunya viruses. J Gen Virol 1978; 40: 531–544. [DOI] [PubMed] [Google Scholar]
- 8.Igarashi A, Mohamed H, Yusof A, et al. Production of type 2 dengue (D2) monoclonal antibody and cell culture derived D2 antigen for use in dengue IgM capture ELISA. Trop Med 1995; 37: 165–173. [Google Scholar]
- 9.Klungthong C, Zhang C, Mammen MP Jr, et al. The molecular epidemiology of dengue virus serotype 4 in Bangkok, Thailand. Virology 2004; 329: 168–179. [DOI] [PubMed] [Google Scholar]
- 10.Lanciotti RS, Calisher CH, Gubler DJ, et al. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol 1992; 30: 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morita K, Tanaka M, Igarashi A. Rapid identification of dengue virus serotypes by using polymerase chain reaction. J Clin Microbiol 1991; 29: 2107–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Islam MA, Ahmed MU, Begum N, et al. Molecular characterization and clinical evaluation of dengue outbreak in 2002 in Bangladesh. Jpn J Infect Dis 2006; 59: 85–91. [PubMed] [Google Scholar]
- 13.Thompson JD, Gibson TJ, Plewniak F, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997; 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (* and other methods). Version 4.0b. Sunderland, Massachusetts: Sinauer Associates, 1998. [Google Scholar]
- 15.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 16.Wittke V, Robb TE, Thu HM, et al. Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology 2002; 301: 148–156. [DOI] [PubMed] [Google Scholar]
- 17.Podder G, Breiman RF, Azim T, et al. Origin of dengue type 3 viruses associated with the dengue outbreak in Dhaka, Bangladesh, in 2000 and 2001. Am J Trop Med Hyg 2006; 74: 263–265. [PubMed] [Google Scholar]
- 18.Thu HM JA, Han AM, Aye KM, et al. Molecular epidemiology of dengue 3 viruses in Myanmar (abstract). In: Myanmar Health Research Congress. Yangon, 2008. [Google Scholar]