Abstract
Background: Polycystic ovaries and irregular menstruation/anovulation are important diagnostic criteria along with hyperandrogenism as per the Androgen Excess Society–2006 criteria for polycystic ovarian syndrome (PCOS). In the etiopathogenesis of PCOS, one of the candidate genes causing ovarian failure is the luteinizing hormone (LH) chorionic gonadotropin hormone receptor (LHCGR). Our aim was to study the association of LHCGR polymorphism (rs2293275) with PCOS in our study population. Materials and Methods: Genetic case–control study from multiple gynecological centers from Hyderabad, a cosmopolitan city in South India. The study involved 204 women with PCOS and 204 healthy, sex-, and age-matched controls. Anthropometric and biochemical profiles were taken in a well-designed pro forma. Isolation of deoxyribonucleic acid (DNA) and genotype analysis were done for the entire study population using the polymerase chain reaction–restriction fragment length polymorphism method followed by 12% polyacrylamide gel electrophoresis. Results: In this study, we have demonstrated an association between LHCGR (rs2293275) polymorphism and PCOS. The frequency of the G allele was 0.60 in PCOS and 0.49 in controls (odds ratio [OR] 1.531, confidence interval [CI] 1.16–2.01, and p-value=0.0026), which indicates that the G allele is associated with PCOS in our population. The GG genotype conferred a significant risk of developing PCOS (OR 3.36, CI 1.96–5.75, and p-value<0.0001). We found a significant association of the GG allele with body–mass index, waist to hip ratio, insulin resistance, LH, and LH/follicle-stimulating hormone (FSH) ratio in PCOS when compared with controls. The AA allele showed high basal FSH levels. Conclusions: This study suggests that LHCGR (rs2293275) polymorphism is associated with PCOS and could be used as a relevant molecular marker to identify women with the risk of developing PCOS in our population and may provide an understanding about the etiology of PCOS.
Background
Polycystic ovarian syndrome (PCOS) is one of the most common reproductive endocrine disorders of women, with a prevalence of ∼5–10% worldwide (Azziz et al., 2006). As per the newer diagnostic criteria of PCOS (Azziz et al., 2006, 2009), irregular menstrual cycles/anovulation and polycystic ovaries form important features along with hyperandrogenism. An altered hypothalamic–pituitary–gonadotropin (HPG) axis plays a key role in causing ovarian failure. Many candidate genes have been studied along the HPG axis like follicle-stimulating hormone (FSH), FSH receptor, luteinizing hormone (LH), LH receptor (LHCGR), androgen receptors, and sex-hormone binding globulin (Zhao and Chen, 2013). The genome-wide association studies of PCOS in the Han Chinese population have identified three susceptible loci 2p16.3, 2p21, and 9q33.3 (Chen et al., 2011).
LHCGR is expressed in theca cells of the ovary and in Leydig cells of the testicles, as well as in adipose tissue, and it encodes for the receptor mediating the actions of both LH and human chorion gonadotropin on steroid biosynthesis. LHCGR encodes for a transmembrane protein, belonging to the G-protein-coupled receptor family, comprising 11 exons (Capalbo et al., 2012). The LH receptor gene carries a large number of single-nucleotide polymorphisms (SNPs) (Themmen, 2005). According to the SNP per website (http://SNPper.chip.org), 300 SNPs are present in the LH receptor gene, resulting in an average distance between SNPs of 306 base pairs. The most frequent LH receptor polymorphisms are an insertion at position 18 in exon 1 (insLQ) and two variable amino acids at position 291 and 312, respectively, located in exon 10. Our aim in the present study was to assess the association of LHCGR (rs2293275) polymorphism with PCOS, which is located in exon 10.
Materials and Methods
Subjects
This study was approved by the institutional ethics committee, and informed written consent was obtained from all subjects. In this prospective genetic case–control study, we included 204 consecutive PCOS patients from different obstetrics and gynecology centers and the general population from July, 2011, to January, 2013. Subjects ranged in age from 17 to 35 years and were diagnosed using the 2006-Androgen Excess Society (AES) criteria: (1) hyperandrogenism, clinical or biochemical and either, (2) oligo-anovulation, or (3) polycystic ovarian morphology (Balen et al., 2003). All subjects underwent a transvaginal ultrasound or transabdominal ultrasound in the follicular phase to evaluate ovarian morphology and any lesions in the pelvic area.
Exclusion criteria
Women excluded from the study were those with inherited disorders like congenital adrenal hyperplasia, androgen secreting neoplasms, androgenic/anabolic drug use or abuse, Cushing's syndrome, syndromes of severe insulin resistance, thyroid dysfunction, and hyperprolactinemia.
There were also 204 controls included in this study over the same period. They visited the healthcare center in a super specialty hospital as a part of group check up for work or an individual need for annual comprehensive medical checkup with no specific health problems. Subjects ranged from 17 to 35 years and did not show hirsutism, acne, or male-type alopecia. All of them had regular menstrual cycles and none of them satisfied any of the 2006 AES criteria. All control subjects also underwent an ultrasonographic examination, and women who had any pathologic findings like polycystic ovaries were excluded from the study.
Sampling
Two milliliters of peripheral blood was collected in EDTA from all the patients and controls along with clinical data, personal history, and family history. Five milliliters of blood was collected in a plain vial for serum.
Anthropometric measurements like body–mass index (BMI) using height and weight and waist to hip ratio using waist circumference and hip circumference were calculated.
Biochemical Analysis: serum samples were immediately processed in a refrigerated centrifuge upon sampling and stored at −20°C until assay. Biochemical analysis was performed in all subjects: fasting insulin, FSH, LH, insulin, LH/FSH, and homeostatic model assessment (HOMA) score for insulin resistance. Fasting plasma glucose (enzymatic colorimetric method), serum FSH (Hitachi Analyzer), LH (Omega Diagnostics), and insulin (DRG) were measured by ELISA in both patients and controls. Laboratory controls were used to check the accuracy and precision of the analyzer, reagents, and assay results.
Isolation of DNA and genotype analysis
Genomic DNA was isolated from the peripheral blood of subjects according to the method routinely used in our laboratory (Govindan et al., 2007; Movva et al., 2007). The DNA was stored at −20°C until processing. Genotyping for the LHCGR polymorphism (rs2293275) was performed by polymerase chain reaction (PCR) with specific published primers (Capalbo et al., 2012): forward primer: 5′-CCTCTTCTCTTTCAGACAGA-3′; reverse primer: 5′-CATGCAAATACTTACAG TGTTTTGGTA-3′ synthesized from Sigma-Aldrich Chemical Pvt Limited, followed by restriction fragment length polymorphism (RFLP) analysis using RsaI. A three-step PCR was performed using an XP thermal cycler, as described by us earlier (Kodati et al., 2010). Briefly, the PCR conditions included an initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 58°C for 30 s, extension at 72°C for 45 s, and a final extension at 72°C for 5 min. The 111 bp amplified PCR product was digested with RsaI at 37°C for 2 h and electrophoresed on 12% polyacrylamide gel electrophoresis (PAGE) at 200 V for 2 h. Bands of 111 bp (undigested) were observed in case of AA (homozygous wild) genotype, 111/86/25 bp in AG (heterozygous) genotype, and 86/25 bp in GG (homozygous mutant) genotype.
Data and statistics
BMI=weight/height2 (kg/m2) and insulin resistance (homeostatic model assessment [HOMA] score) were calculated by using the following formula: fasting serum insulin (μU/mL)×fasting plasma glucose (mg/dL)/405 (Liang et al., 2011).
Statistical analysis was performed using Medcalc statistical software. The chi square test, odds ratio (OR), and 95% confidence interval (CI) were done to assess the association between the groups. One-way analysis of variance with post t-test was performed using GraphPad Insta3 software. A p-value of <0.05 was considered statistically significant.
Results
A PCR product of 111 bp was obtained, which on digestion with a restriction enzyme gave fragments of 111 bp indicating AA genotype, 111/86/25 bp indicating AG genotype, and 86/25 bp indicating GG genotype (Fig. 1 and Table 1). The homozygous GG genotype was seen in 28.92% of patients with PCOS when compared with 10.78% of healthy controls (Table 2). Data showed that a recessive (GG vs. AG+AA) genotype pattern of inheritance exhibited a significant association with PCOS (Table 3). The GG genotype was associated with PCOS (OR, 3.36; 95% CI 1.96–5.75, p<0.0001). The frequency of the G allele was 0.60 in PCOS and 0.49 in controls (p=0.0026), (OR 1.531, 95% CI 1.16–2.01) (Table 2). There is significant elevation (p<0.0001) of BMI, waist to hip ratio (WHR), fasting insulin, HOMA score, LH, and LH/FSH in PCOS compared to controls (Table 4). Similarly, we have observed significant elevation of height, weight, BMI, WHR, fasting insulin, HOMA score, LH, and LH/FSH with the GG genotype of PCOS (p<0.0001) rather than AG and AA genotypes when compared with age-matched healthy controls (Table 5). The genotype and allele frequencies for the 204 controls (22 G/G, 155 A/G, and 27 A/A) were in Hardy–Weinberg equilibrium (Rodriguez et al., 2009).
FIG. 1.
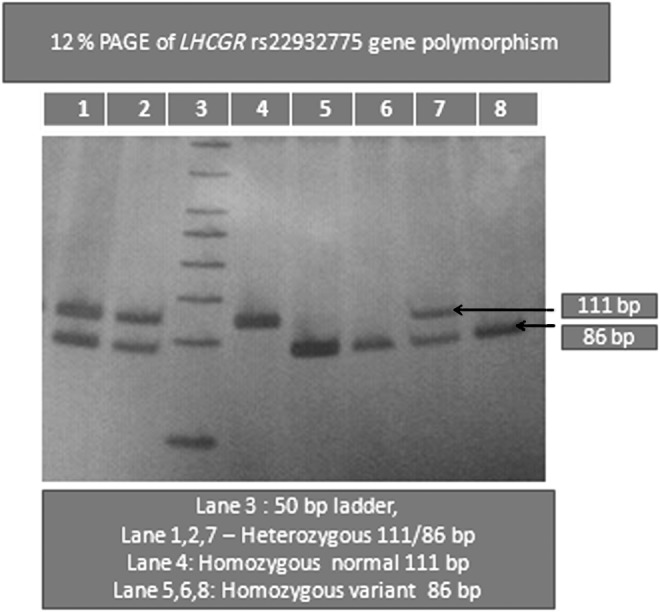
Twelve percent PAGE of LHCGR (rs2293275) gene polymorphism. PAGE, polyacrylamide gel electrophoresis.
Table 1.
Luteinizing Hormone Chorionic Gonadotropin Receptor Gene (rs2293275) Primer Sequence
| LHCGR 86 bp (v) | Primers | PCR product | AA | 111 | |
| RsaI | Forward | CCTCTTCTCTTTCAGACAGA | 111 bp | AG | 111/86/25 |
| rs2293275 | Reverse | CATGCAAATACTTACAGTGTTTTGGTA | GG | 86/25 |
PCR, polymerase chain reaction.
Table 2.
Genotype and Allele Frequencies of LHCGR Polymorphism Identified in the Study
| LHCGR | AA | AG | GG | A allele | G allele |
|---|---|---|---|---|---|
| Patients | 21 (10.29%) | 124 (60.78%) | 59 (28.92%) | 166 (0.40) | 242 (0.60) |
| Controls | 27 (13.23%) | 155 (75.98%) | 22 (10.78%) | 209 (0.51) | 199 (0.49) |
Allele frequency odds ratio 1.5311; 95% CI 1.16–2.01; p-value=0.0026.
CI, confidence interval.
Table 3.
Statistical Analysis of Genotypes of LHCGR Polymorphism Identified in the Study
| Genotype | PCOS | Controls | Odds ratio | 95% CI | p-Value |
|---|---|---|---|---|---|
| GG vs. AG+AA | 59/145 | 22/182 | 3.36 | 1.96–5.75 | <0.0001 |
| GA vs. GG+AA | 124/80 | 155/49 | 0.49 | 0.319–0.75 | 0.0001 |
| GG+GA vs. AA | 183/21 | 177/27 | 1.3293 | 0.724–2.43 | 0.3577 |
| AA+AG vs. GG | 145/59 | 182/22 | 0.2971 | 0.1738–0.3878 | <0.0001 |
| AA vs. GG+GA | 21/183 | 27/177 | 0.7523 | 0.4102–1.37 | 0.352 |
p, significance; PCOS, polycystic ovarian syndrome.
Table 4.
Comparison of Mean Values of Anthropometric and Biochemical Characteristics in Polycystic Ovarian Syndrome Patients and Controls with Their Mean and Standard Deviation
| S.No | Parameter | PCOS (n=204) | Controls (n=204) | p-Value |
|---|---|---|---|---|
| 1 | Age, years | 28±3.6 | 28±5.1 | 1.0000 |
| 2a | BMI, kg/m2 | 27.12±4.93 | 23.4±3.2 | <0.0001 |
| 3a | W/H | 0.93±0.04 | 0.79±0.05 | <0.0001 |
| 4 | Fasting glucose, gm/dL | 88±8.6 | 86.85±7.1 | 0.0678 |
| 5a | Fasting insulin, μIU/mL | 16.94±7.26 | 6.66±3.19 | <0.0001 |
| 6a | HOMA score | 3.73±3.8 | 1.44±0.75 | <0.0001 |
| 7a | LH, mIU/mL | 11.97±6.08 | 7.9±5.46 | <0.0001 |
| 8 | FSH, mIU/mL | 5.48±1.98 | 6.47±3.16 | 0.0002 |
| 9a | LH/FSH | 2.62±1.2 | 1.5±1.2 | <0.0001 |
Significant values (p<0.0001).
BMI, body–mass index; FSH, follicle-stimulating hormone; HOMA, homeostatic model assessment; LH, luteinizing hormone.
Table 5.
Distribution of LHCGR Genotypes, Age, BMI, W/H, FG, FI, HOMA Score, LH, FSH, LH/FSH in Patients and Controls and Their Comparison with Mean and Standard Deviation
| A/A genotype | A/G genotype | G/G genotype | |||||
|---|---|---|---|---|---|---|---|
| Parameters | PCOS | Controls | PCOS | Controls | PCOS | Controls | p-Value |
| Age, years | 27.76±5.6 | 27.29±6.1 | 27.95±3.73 | 28.5±4.8 | 29±3.5 | 27.27±5.7 | 0.0020 |
| BMI (Kg/m2) | 24.02±3.9 | 22.5±3.4 | 26.8±5.4 | 23.6±3.12 | 28.7±3.3 | 23.18±3.5 | <0.0001 |
| W/H | 0.9±0.03 | 0.78±0.05 | 0.93±0.04 | 0.78±0.05 | 0.94±0.04 | 0.8±0.04 | <0.0001 |
| Fasting glucose, gm/dL | 83.04±11.24 | 91.29±18.4 | 88.68±10 | 88.2±10 | 89.32±11.5 | 90±12 | <0.0001 |
| Fating insulin, μIU/mL | 12.09±7.69 | 6.3±2.9 | 11.07±6.45 | 6.8±3.4 | 11.9±8 | 6.59±2.73 | <0.0001 |
| HOMA score | 2.4±1.5 | 1.5±0.9 | 2.54±1.6 | 1.44±0.78 | 3.13±2.3 | 1.45±0.6 | <0.0001 |
| LH, mIU/mL | 9.9±4.3 | 7.28±4.8 | 11.69±6.3 | 8.18±5.4 | 13.3±5.8 | 6.64±3.72 | <0.0001 |
| FSH, mIU/mL | 5.9±2.5 | 5.4±3.6 | 5.37±2 | 5.7±3.2 | 5.54±1.86 | 5.7±2.67 | 0.8331 |
| LH/FSH | 2.38±1.47 | 1.4±0.9 | 2.32±1.2 | 1.55±1.2 | 2.5±0.8 | 1.3 8±1.05 | <0.0001 |
Data are shown as mean±SD.
p-Values were evaluated by one-way analysis of variance with post-test.
Significant values (p<0.05).
W/H, waist to hip ratio.
Discussion
The principal features of PCOS include hyperandrogenism, obesity, polycystic ovaries, and irregular menstrual cycles/anovulation (Azziz et al., 2006, 2009). Many candidate genes have been proposed as important contributors in PCOS (Legro, 1995; Urbanek et al., 1999) In the genetic association studies for PCOS, many researchers selected the genes involving steroidogenesis, the HPG axis, obesity, and chronic inflammatory pathways (Zhao and Chen, 2013).
In the present study, we have evaluated the role of polymorphism of LHCGR rs2293275 as a risk factor for PCOS in a prospective genetic case–control study of South Indian women. In our cohort of PCOS subjects, the prevalence of obesity was more than 70% when compared with controls and this increased the presence of obesity in our PCOS as per the Asian-Pacific definition of obesity (Chen et al., 2010), which can be explained by the Indian lifestyle and food habits. Our PCOS subjects had shown elevated levels of insulin resistance, supported by the increased HOMA score and increased LH and LH/FSH values. All the subjects had shown a majority of polycystic ovaries being bilateral (Balen et al., 2003). Eighty-nine percent of PCOS showed irregular menstrual cycles/anovulation. To assess the role of LHCGR polymorphism in the altered HPG axis resulting in the manifestations of PCOS, we studied LHCGR polymorphism (rs2293275) in 204 PCOS patients and 204 controls. We noticed a significant difference in the distribution of alleles and genotypes between PCOS and controls (Table 2). A strong correlation identified between the PCOS phenotype and S312N variant was similar to the Sardinian population (Capalbo et al., 2012). In case of the homozygous variant, the risk of PCOS in our population is 3.36-fold compared to other genotypes, which was higher than the risk of the Sardinian population (2.7-folds). The S312N variant investigated by Valkenburg et al. (2009) did not find an increase in PCOS risk. The presence of a higher risk in our population can be explained by the strict inclusion of PCOS patients as per the AES Criteria–2006 (Azziz et al., 2006, 2009) in our study and the different geographic regions of our cases and controls.
When we tried to assess the extent to which the genotypic variants of LHCGR influence the severity of clinical features of PCOS, we were able to demonstrate a significant association with LHCGR polymorphism. The homozygous variant of LHCGR polymorphism (GG) of PCOS patients in our population had shown increased values of BMI, waist to hip ratio, insulin resistance, and LH and LH/FSH values when compared with other genotypes, which highlights the contribution of this polymorphism to the phenotype of PCOS and helps in better understanding the role of genetic variation in the HPG axis.
To the best of our knowledge, this is the first study in the South Indian population that examined the association of LHCGR polymorphism (rs2293275) with PCOS.
Conclusion
To conclude, LHCGR variants were strongly associated with disease risk (3.36-fold), and LHCGR polymorphism contributes to a significant difference in the phenotype of PCOS suggesting that the LHCGR (rs2293275) polymorphism could be used as a relevant molecular marker to identify women at risk of developing PCOS in our population.
Acknowledgments
The authors thank the Women Scientist Scheme-A, Department of Science and Technology, Ministry of Science and Technology, Government of India (New Delhi, India) for granting funds. Grant Number: SR/LS-91/2011/WOS-A.
The authors also thank Dr. Anuradha, Anu Fertility Centre, Somajiguda, Hyderabad, and the Management of Kamineni Academy of Medical Sciences and Research Centre, Hyderabad, for their support in sample collection.
Authors' Contributions
Sujatha Thathapudi: study design, literature search, laboratory work, data acquisition, data analysis, statistical analysis, and manuscript preparation. Vijayalakshmi Kodati: study design and manuscript review. Jayashankar Erukkambattu: literature search, data analysis, statistical analysis, and manuscript preparation. Uma Addepally: laboratory work and statistical analysis. Qurratulain Hasan: study design, definition of intellectual content, data analysis, manuscript editing, and manuscript review.
Author Disclosure Statement
No competing financial interests exist.
References
- Azziz R, Carmina E, Dewailly D, et al. (2006) Position statement: criteria for defining PCOS as a predominantly hyperandrogenic syndrome: an androgen excess society guideline. J Clin Endocrinol Metab 91:4237–4245 [DOI] [PubMed] [Google Scholar]
- Azziz R, Carmina E, Dewailly D, et al. (2009) The androgen excess and PCOS society criteria for the Polycystic ovarian syndrome: the complete task force report. Fertil Steril 91:456–488 [DOI] [PubMed] [Google Scholar]
- Balen AH, Laven JSE, Tan SL, Dewailly D. (2003) Ultrasound assessment of Polycystic ovary: international consensus definitions. Hum Reprod Update 9:505–514 [DOI] [PubMed] [Google Scholar]
- Capalbo A, Saqnella F, Apa R, et al. (2012) The 312N variant of the luteinizing hormone/choriogonadotropin receptor gene (LHCGR) confers up to 2.7-fold increased risk of polycystic ovary syndrome in a Sardinian population. Clin Endocrinol (Oxf) 77:113–119 [DOI] [PubMed] [Google Scholar]
- Chen X, Ni R, Mo Y, et al. (2010) Appropriate BMI levels for PCOS patients in Southern China. Hum Reprod 25:1295–1302 [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Zhao H, He L, et al. (2011) Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21, and 9q33.3. Nat Genet 43:55–59 [DOI] [PubMed] [Google Scholar]
- Govindan S, Ahamad SN, Vedicherla B, et al. (2007) Association of progesterone receptor gene polymorphism (PROGINS) with endometriosis, uterine fibroids and breast cancer. Cancer Biomark 3:73–78 [DOI] [PubMed] [Google Scholar]
- Kodati VL, Shetty P, Vattam K, et al. (2010) Tumor necrosis factor alpha-C850T polymorphism is significantly associated with endometriosis in Asian Indian women. Fertil Steril 94:453–456 [DOI] [PubMed] [Google Scholar]
- Legro RS. (1995) The genetics of polycystic ovary syndrome. Am J Med 98Suppl 1 A:165. [DOI] [PubMed] [Google Scholar]
- Liang SJ, Hsu CS, Tzeng CR, et al. (2011) Clinical and Biochemical parameters of Polycystic ovary syndrome between the age of 20 and 40. Hum Reprod 12:3443–3449 [DOI] [PubMed] [Google Scholar]
- Movva S, Alluri R, Komandur S, et al. (2007) Relationship of angiotensin-converting gene polymorphism with nephropathy associated with type 2 diabetes mellitus in Asian Indians. J Diabetes Complications 21:237–241 [DOI] [PubMed] [Google Scholar]
- Rodriguez S, Gaunt TR, Day IN. (2009) Hardy-Weinberg equilibrium testing of biological ascertainment for mendelian randomization studies. Am J Epidemiol 169:505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Themmen APN. (2005) Focus on gonadotropin signalling. An update of the pathophysiology of human gonadotrophin subunit and receptor gene mutations and polymorphisms. Reproduction 130:263–274 [DOI] [PubMed] [Google Scholar]
- Urbanek M, Legro RS, Driscoll DA, et al. (1999) Thirty-seven candidate genes for polycystic ovary syndrome: strongest evidence for linkage is with follistatin. Proc Natl Acad Sci U S A 96:8573–8578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkenburg O, Uitterlinden AG, Piersma D, et al. (2009) Genetic polymorphisms of GnRH and gonadotrophic hormone receptors affect the phenotype of polycystic ovary syndrome. Hum Reprod 24:2014–2022 [DOI] [PubMed] [Google Scholar]
- Zhao H, Chen ZJ. (2013) Genetic association studies in female reproduction: from candidate gene approaches to genome-wide mapping. Mol Hum Reprod 19:644–654 [DOI] [PubMed] [Google Scholar]


