Abstract
Precise regulation of Notch signaling is essential for normal vertebrate development. Mind bomb (Mib) is a ubiquitin ligase that is required for activation of Notch by Notch's ligand, Delta. Sorting Nexin 5 (SNX5) co-localizes with Mib and Delta complexes and has been shown to directly bind to Mib. We show that microRNA-216a (miR-216a) is expressed in the retina during early development and regulates snx5 to precisely regulate Notch signaling. miR-216a and snx5 have complementary expression patterns. Knocking down miR-216a and/or overexpression of snx5 resulted in increased Notch activation. Conversely, knocking down snx5 and/or miR-216a overexpression caused a decrease in Notch activation. We propose a model in which SNX5, precisely controlled by miR-216a, is a vital partner of Mib in promoting endocytosis of Delta and subsequent activation of Notch signaling.
Keywords: microRNA, Notch signaling, miR-216a, sorting nexin 5, zebrafish, retina
Introduction
Since their discovery as regulators of C. elegans developmental timing in 1993 (Lee et al., 1993; Wightman et al., 1993), miRNAs have been shown to be involved in diverse aspects of development. miRNAs are 21-23 nucleotide (nt) non-coding RNAs that regulate gene expression by binding to complementary sequences in the 3’UTR of messenger RNAs (Bartel, 2004; Fabian et al., 2010; He and Hannon, 2004; Huntzinger and Izaurralde, 2011; Liu et al., 2012). This results in the recruitment of the RNA Induced Silencing Complex (RISC), the effector complex that mediates translation repression, deadenylation, and decay of target mRNAs (Bazzini et al., 2012; Djuranovic et al., 2012; Giraldez et al., 2006). We and others have identified developmental roles for several individual miRNAs in zebrafish (Flynt et al., 2007; Flynt et al., 2009; Giraldez et al., 2005; Li et al., 2008; Li et al., 2011; Mishima et al., 2009; Stahlhut et al., 2012; Wei et al., 2013). However, the exact roles and mRNA targets for most miRNAs that function during development are still unknown.
Notch signaling regulates many processes during vertebrate development, from vasculogenesis to segmentation (Fortini, 2009; Lawson et al., 2001; Wright et al., 2011). It is especially important during neurogenesis (Louvi and Artavanis-Tsakonas, 2006), is instructive for gliogenesis in the zebrafish retina (Scheer et al., 2001), and has been shown to be essential for zebrafish retinal development (Bernardos et al., 2005). Notch is a transmembrane receptor that mediates interaction with adjacent cells through membrane bound ligands, such as Delta, that trigger proteolytic cleavage of Notch and release of an intracellular domain that travels to the nucleus to alter gene expression (Louvi and Artavanis-Tsakonas, 2006). Mind bomb is a ubiquitin ligase that ubiquitylates Delta, thereby facilitating its endocytosis, which is essential for cleavage of Notch and subsequent activation of signaling (Itoh et al., 2003). Mutants in Mind bomb have disorganized retinal architecture and do not have Müller glia (Bernardos et al., 2005).
Sorting Nexin 5 (SNX5) is part of the large sorting nexin protein family, members of which have been previously shown to bind phosphoinositides through a specialized phoxhomology (PX) domain (Cullen, 2008; Cullen and Korswagen, 2012). SNX5 is part of a select group of sorting nexins that also contain a carboxy-terminal BAR (Bin, amphiphysin, Rvs) domain, thought to facilitate binding to and/or induce membrane curvature, possibly functioning in endocytosis or vesicle budding (Cullen, 2008). The sorting nexins function in diverse cellular trafficking processes, including developmental signaling cascades as in the case of SNX3, which has been shown to be required for Wnt secretion (Harterink et al., 2011) and SNX17 which functions in integrin recycling (Steinberg et al., 2012). SNX5 was previously shown to co-localize with Mib and Delta (Yoo et al., 2006). Knockdown of SNX5 using morpholinos in zebrafish causes defects in vascular development (Eckfeldt et al., 2005; Yoo et al., 2006). Accumulating evidence, therefore, suggests that SNX5 could play a role in modulating Notch signaling.
In this study, we show for the first time that miR-216a, a miRNA that is expressed in the developing zebrafish retina, regulates snx5. Results using reporter fish show that miR-216a regulates snx5 to modulate Notch signaling during eye development.
Materials and Methods
Zebrafish Lines and Maintenance
Wildtype (AB) (Walker, 1999), albino (University of Oregon, Eugene, OR), Tg(gfap:GFP) (Bernardos and Raymond, 2006), Tg(her4:dRFP) (Yeo et al., 2007) Tg(flk1:GFP) (Choi et al., 2007) and Tg(Tp1bglob:eGFP) (Parsons et al., 2009) lines were maintained at 28.5°C on a 14:10 hour light:dark cycle. Embryos were raised in egg water (0.03% Instant Ocean) at 28.5°C and staged according to morphology (Kimmel et al., 1995) and hours post fertilization (hpf). All experiments were performed with the approval of the Vanderbilt University Institutional Animal Care and Use Committee (M/09/398).
Microarrays of developing eyes
Developing eyes were dissected at 2 and 5 days post fertilization (dpf), homogenized in Trizol and total RNA was extracted. Small RNAs were enriched and arrays were performed and normalized as previously described (Thatcher et al., 2007). Fold changes were calculated compared to a negative control consisting of probes for Pseudomonas aeruginosa dehydrogenase (Thatcher et al., 2007). Microarray data was analyzed using GeneSpring software, and paired t-tests were performed using Prism (GraphPad) to determine p values.
Molecular Cloning
Potential target mRNA 3’UTRs were amplified by RT-PCR using the primers below. Each 3’ UTR was cloned into pCS2+ downstream of the coding sequence of GFP (Flynt et al., 2007).
| Gene | Forward Primer (5’-3’) | Reverse Primer (5’-3’) |
|---|---|---|
| snx5 (NM_214769) | ACCTGATCGAGATGACTGAG | TTATCTTCGCTGAGTTGCAC |
| her4.2 (NM_131090.3) | AGTCACATCTGGAGACCCTG | GCTTCAACACACAAACAAGTCC |
| notch1b (NM_131302.2) | GTCACAAATCGGACACATGC | CACAAATCGTTTCAATCGGATG |
| heyl (NM_181736.1) | GGGCTTTGAGTTCCTCCAG | TCTCCTCAAGCACTTCAATCTC |
| numb (NM_001040406.1) | CGCTCCATCACCCACAAACC | GACGAGTCGTTCCCTGTATGG |
| hey2 (NM_131622.2) | AGTAAACCATACCGACCGTG | GGTTACATCTTACAGAGGGTGG |
miRNA recognition elements (MREs) were deleted from the snx5 3’UTR with PCR. For MRE 1, forward (5’-TGCAGACACATAAAGTACCACTATG-3’) and reverse (5’-GCTAATATTTGCATAACTTGGAATATG-3’) primers and for MRE 2, forward (5’-GTCCGAATGCATTACTCTGCATTACAGAT-3’) and reverse (5’-TATTAGGAGGAAAGATATCTGAAGCATTACA-3’) primers were designed to exclude each MRE. snx5 mRNA was amplified by RT-PCR using forward (5’-GCCGAGGGATCCTGAGGAACGAGCTTGCTGCTGGAA-3’) and reverse (5’-GCCGAGCTCGAGCAACTGGGGACATCAGTCAGTCCTT-3’) primers and cloned into pCS2+ (Rupp et al., 1994). snx5 mRNA without its 3’UTR was amplified by RT-PCR using forward (5’-GCCGAGGGATCCTGAGGAACGAGCTTGCTGCTGGAA-3’) and reverse (5’-GCCGAGCTCGAGGTCATCATCGTGTGGGTC-3’) primers and cloned into pCS2+. All clones and MRE deletions were verified by Sanger sequencing in the Vanderbilt DNA Sequencing Core.
Microinjection
All injections were performed in fertilized 1-cell zebrafish embryos. Phenol red dye (0.05%) was used in each injection solution and alone as an injection control. Capped snx5 RNA (from the pCS2+ vector containing the snx5 mRNA without 3’UTR) or GFP-snx5 3’UTR RNA (from the pCS2+ vector containing the coding sequence of GFP and either the full length snx5 3’UTR or the snx5 3’UTR with both MREs deleted) were prepared using an Sp6 mMessage Machine Kit (Ambion). snx5 RNA was injected at 100 pg/embryo concentration for functional experiments and 50 pg/embryo for rescue experiments. GFP RNA was injected at 25 pg/embryo concentration. Synthetic miR-216a duplexes (Dharmacon) were injected at 50 pg/embryo concentration in functional experiments and 25 pg/embryo in GFP reporter experiments. Two different morpholinos against miR-216a (one against the mature miR-216a: 5’-TCACAGTTCCCAGCTGAGATTA-3’ and a second against the loop of pre-miR-216a: 5’-GCAGCGCCTGTGAGAGGGATGAAAA-3’), a morpholino against the snx5 start site: 5’-ACGTCATGTTCAGGAGATATTTCGC-3’ (Eckfeldt et al., 2005), and an exon 4 splice donor morpholino: 5’-CAGAGTTAGACTCACGCCTCAAGTT-3’ (Yoo et al., 2006), and a p53 morpholino (5’-GCGCCATTGCTTTGCAAGAATTG-3’) were from Gene Tools. Two different miR-216a morpholinos were injected together at 150 pg each/embryo for functional experiments and a morpholino targeting just the mature form of miR-216a was used at 100 pg/embryo for GFP reporter experiments. snx5 morpholinos were injected together at 100 pg each/embryo for all experiments. The p53 morpholino was injected at 150 pg/embryo. All injection amounts were experimentally determined to be the lowest effective dose.
In situ hybridization
Staged albino zebrafish embryos were fixed in 4% paraformaldehyde (PFA) in 1X PBS (pH 7.4) at 4°C overnight on a 3D rocker. Whole-mount mRNA in situ hybridization was performed as described (Thisse and Thisse, 2008) using a digoxigenin (DIG)-labeled snx5 RNA probe generated with Roche Applied Science reagents and pCS2+ vector containing the full length snx5 mRNA sequence. Whole-mount miRNA in situ hybridization was performed as described (Lagendijk et al., 2012) using a miRCURY 5’- and 3’-DIG labeled hsa-miR-216a LNA probe (Exiqon).
Immunoblotting
Embryos were deyolked at 1 dpf (day post fertilization) and placed in lysis buffer [25 mM HEPES (pH 7.5), 5 mM MgCl2, 300 mM NaCl, 1 mM EDTA, 0.2 mM EGTA, 1 mM DTT, 10% glycerol, 1.0% Triton X-100, 1 mM PMSF] for protein extraction. Total proteins were separated on 10% SDS-PAGE gels and transferred to PVDF-plus membranes (GE Osmonics). Membranes were incubated with rabbit polyclonal antibodies against SNX5 (1:2000, Aviva Systems Biology) and α-tubulin (1:500, Abcam). Anti-rabbit HRP-conjugated secondary antibodies (1:5000, GE Healthcare) were used for visualization with ECL reagents (Perkin Elmer). Using ImageJ, SNX5 levels were normalized to α-tubulin control levels, after which the ratio of SNX5 under varying injection conditions was determined. One-way ANOVA using Bonferroni's correction to adjust for multiple comparisons was performed using StatPlus (AnalystSoft).
Staining and Imaging
Live embryos, either Tg(flk1:GFP) at 3-4 dpf or those injected with GFP reporter transcripts were briefly anesthetized with 0.02% tricaine for imaging on a Zeiss Discovery V8 stereo microscope and photographed using an Axiocam MRM black and white camera and Axiovision software (Zeiss). Live embryos that were staged and fixed in 4% PFA in 1X PBS (pH 7.4) at room temperature for 2-3 hours or embryos upon which in situ hybridization had been performed were embedded in 1.5% agarose/5% sucrose in egg water. The resulting blocks were cryoprotected in 30% sucrose overnight, frozen, and sectioned on a Leica CM1850 cryostat (10-15μm sections). The resulting transverse sections were mounted on VistaVision Histobond slides (VWR). Tg(her4:dRFP) sections were stained with Alexa Fluor 488-conjugated phalloidin (1:100, Molecular Probes) and Hoescht (1:3000, Molecular Probes), and Tg(gfap:GFP)sections were stained with the mouse monoclonal antibody zpr-1 (1:1000, Zebrafish International Research Center), HuC/D (1:1000, Invitrogen), and/or TOPRO-3 (1:1000, Molecular Probes). TUNEL labeling was performed using an in situ Cell Death Detection Kit, TMR red (Roche). Fluorescent sample slides were mounted with Vectashield (Vector Laboratories) and in situ sample slides were mounted in 100% glycerol. In situ and Tg(her4:dRFP) samples were imaged on a Leica DM6000B microscope or Leica LSM 510 confocal (inverted) microscope with a 40× objective. Tg(gfap:GFP) samples were imaged on a Leica LSM 510 confocal (inverted) microscope with a 20x or 40x objective in the Vanderbilt Cell Imaging Shared Resource. Images were processed using ImageJ and Adobe Photoshop, and one-way ANOVA was calculated as described for immunoblotting.
Results
miRNA expression analysis in developing eyes
In order to examine the role of miRNAs during vertebrate eye development, we dissected developing eyes from zebrafish at 2 and 5 dpf and isolated RNA for miRNA expression profiling. We detected 12 miRNAs expressed at levels above background at 2 dpf and 23 miRNAs detected at 5 dpf (Table 1). From in situ localization experiments, only three of these miRNAs (miR-9, miR-124, and miR-216a) are expressed specifically in the developing eye at these times, the remainder are expressed ubiquitously (Ason et al., 2006; Kapsimali et al., 2007; Wienholds et al., 2005; Wienholds and Plasterk, 2005). Because miR-9 and miR-124 have been extensively studied during neural development (Gao, 2010), we decided to focus on the role of miR-216a in zebrafish eye development.
Table 1.
miRNA expression profiling in developing zebrafish eyes.
| 2 dpf | Fold Difference | p-value |
|---|---|---|
| miR-9 | 4.1791 | 0.0002 |
| miR-17-5p | 7.7904 | 0.0002 |
| miR-19a | 3.6866 | 0.0069 |
| miR-20 | 4.6253 | 0.0018 |
| miR-25 | 2.6127 | < 0.0001 |
| miR-31 | 3.3801 | 0.0008 |
| miR-93 | 3.7530 | 0.0002 |
| miR-108 | 4.1121 | 0.0033 |
| miR-124a | 7.0932 | < 0.0001 |
| miR-152 | 4.9246 | 0.0017 |
| miR-210 | 2.8556 | 0.0076 |
| miR-216 | 3.9684 | 0.0016 |
| 5 dpf | Fold Difference | p-value |
|---|---|---|
| miR-9 | 5.4529 | < 0.0001 |
| miR-17-5p | 7.1188 | < 0.0001 |
| miR-18 | 3.8517 | 0.0002 |
| miR-19a | 6.6342 | < 0.0001 |
| miR-19b | 3.9508 | < 0.0001 |
| miR-20 | 6.0296 | < 0.0001 |
| miR-22 | 5.6745 | < 0.0001 |
| miR-25 | 5.3233 | < 0.0001 |
| miR-31 | 3.4258 | 0.0001 |
| miR-93 | 5.2311 | < 0.0001 |
| miR-108 | 5.1436 | 0.001 |
| miR-124a | 7.3811 | < 0.0001 |
| miR-125b | 6.0221 | < 0.0001 |
| miR-152 | 4.0968 | < 0.0001 |
| miR-181a | 4.4360 | 0.0001 |
| miR-181b | 4.6483 | < 0.0001 |
| miR-182 | 5.4448 | < 0.0001 |
| miR-183 | 6.3569 | < 0.0001 |
| miR-204 | 7.9874 | < 0.0001 |
| miR-210 | 8.0629 | < 0.0001 |
| miR-213 | 3.8558 | < 0.0001 |
| miR-216 | 8.8500 | < 0.0001 |
| miR-217 | 6.8886 | 0.0002 |
Microarrays containing probes for 346 zebrafish miRNAs were performed on tissue from developing zebrafish retinas at 2 and 5 days post fertilization (dpf). Fold differences were calculated by dividing the normalized expression values by negative control signals derived from probes against a Pseudomonas aeruginsa dehydrogenase. All p-values were calculated based on paired t-tests.
*Fold difference is calculated by dividing the normalized miRNA expression by the negative control
**All p-values are calculated based on a paired t-test
Expression of miR-216a in developing eyes is temporally and spatially specific
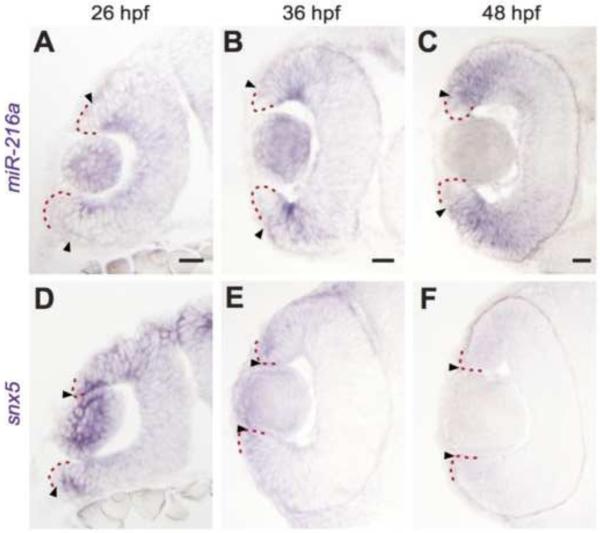
To determine the expression of miR-216a over the course of eye development, we performed whole mount LNA in situ hybridization for miR-216a on zebrafish embryos, which were then sectioned and visualized (Figure 1). miR-216a is robustly and widely expressed throughout the eye cup at 22 hpf (Supplemental Fig. 1), but its localization then progressively changes as development proceeds (Fig 1A-C). From 26 to 48 hpf, miR-216a expression shifts from the central retina to an increasingly restricted marginal region that will become the Circumferential Germinal Zone (CGZ) or Ciliary Marginal Zone (CMZ)(Hitchcock and Raymond, 2004). Given the role that miRNAs play in regulating the expression of target mRNAs, we conclude that the temporal and spatial specificity of the expression of miR-216a suggest that it plays a role in patterning the developing retina.
Figure 1. miR-216a and snx5 have complementary expression patterns during development.
Transverse sections of whole mount in situ hybridizations for miR-216a and snx5 at 26 (A,D), 36 (B,E), and 48 (C,F) hours post fertilization (hpf). miR-216a expression spreads from the center of the developing retina toward the periphery. snx5 is detected in a complementary pattern becoming increasingly restricted over time to a small number of cells at the far periphery of the developing retina. Arrowheads indicate the extent of signal, the red dashed line indicates the lateral edge of the optic cup. Scale bar: 20μm.
miR-216a targets snx5
MicroCosm and TargetScan online target prediction algorithms (Griffiths-Jones et al., 2008; Lewis et al., 2005) were used to identify potential targets of miR-216a. Concurrently, we conducted a series of miR-216a gain- and loss-of-function experiments in developing zebrafish embryos. We observed vascular defects upon altered expression of miR-216a that were remarkably similar to previous reports demonstrating an involvement of Notch signaling and a requirement for SNX5 in vascular development (Supplemental Figs. 2,3)(Lawson et al., 2001; Yoo et al., 2006). Thus, we focused our target search on Notch pathway related genes and SNX5. Several Notch related genes contain one predicted miRNA recognition element (MRE) for miR-216a in their 3’ UTRs, including her4.2, heyl, notch1b, hey2, and numb. In contrast, snx5 contains two MREs in its 3’ UTR (Fig. 2A). Based on the involvement of Notch signaling in retinogenesis (Bernardos et al., 2005; Scheer et al., 2001), we assessed whether these predicted targets of miR-216a were true targets using GFP reporter assays.
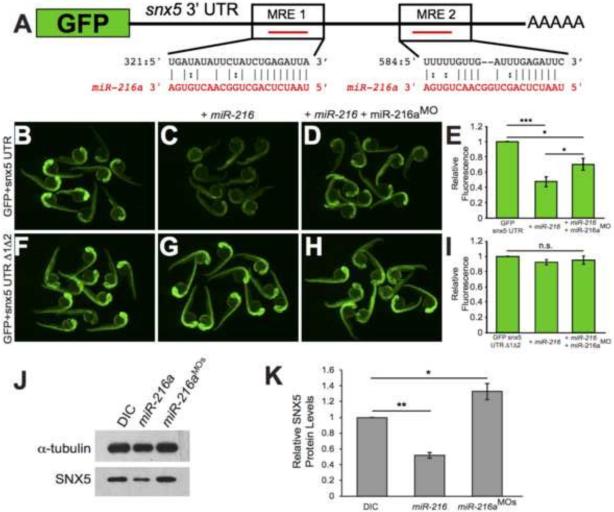
Figure 2. snx5 is a target of miR-216a.
The coding sequence of GFP was fused to the 3’UTR of snx5. Predicted pairing of each MRE in the 3’ UTR (black) and miR-216a (red) are pictured. (B) Embryos injected at the 1-cell stage with GFP-snx5 3’ UTR reporter mRNA alone, with miR-216a (C), or with miR-216a and miR-216aMO (D) were imaged using a fluorescence dissecting scope at 1 dpf. (F) Both MREs were deleted from the GFP-snx5 3’ UTR reporter. Embryos injected at the 1-cell stage with this mRNA alone, with miR-216a (G), or with miR-216a and miR-216aMO (H) were imaged at 1 dpf using a fluorescence dissecting scope. (E, I) Relative fluorescence was quantified using ImageJ, and comparisons were made using one-way ANOVA with Bonferroni's correction. (J) Western blots for SNX5 and alpha-tubulin were performed on protein lysates from 1 dpf zebrafish injected at 1-cell stage with dye control (DIC), miR-216a, or miR-216aMOs. (K) Band density was quantified using ImageJ, and comparisons were made using one-way ANOVA with Bonferroni's correction. *, p<0.05; **, p<0.01; ***, p<0.001. Error bars show SEM.)
The full-length 3’ UTR of each of these predicted targets was fused to the coding sequence of GFP. mRNA transcripts were then generated from these reporter constructs and injected into single cell zebrafish embryos in the presence or absence of co-injected, exogenous miR-216a. The effect of miR-216a was determined by measuring GFP fluorescence at 24 hpf. Fluorescence levels of the her4.2, heyl, notch1b, hey2, and numb 3’UTR reporters were comparable with or without co-injection of miR-216a, suggesting that these genes are not targeted by miR-216a (Supplemental Fig. 4). However, for snx5, we observed a robust decrease in GFP fluorescence upon co-injection with miR-216a (Fig. 2B,C,E). Importantly, the effect of miR-216a could be partially suppressed by co-injection of a morpholino targeting the mature sequence of miR-216a, indicating specific suppression of snx5 by miR-216a (Fig. 2D,E). To further test for specificity, we deleted each of the two predicted MREs in the snx5 3’UTR. No differences were observed in GFP fluorescence among fish injected with the mutated reporter transcripts compared to co-injection with miR-216a (Fig. 2F,G,I). As an additional test of specificity, co-injection of both miR-216a and miR-216aMO with the GFP reporter containing a mutated snx5 3’ UTR resulted in no change in fluorescence (Fig. 2H,I). These results indicate that miR-216a can regulate snx5 via two MREs located in its 3’ UTR.
To address whether endogenous snx5 is targeted by miR-216a, we isolated protein from 1 dpf embryos injected at the one cell stage with either a dye control (DIC), miR-216a, or two morpholinos targeted to miR-216a, one complementary to the mature sequence of miR-216a and one targeted to the Dicer cleavage site of the miR-216a precursor (miR-216aMOs). We initially performed the experiments with just one of the morpholinos but combining the two allowed us to use a lower dose of each, reducing the chances of off target effects. We then performed western blots using an antibody against SNX5 protein and α-tubulin as a control. Injection of miR-216a significantly decreased endogenous levels of SNX5, while injection of miR-216aMOs led to a significant increase in endogenous SNX5 (Fig. 2J,K). Taken together, these results indicate that miR-216a targets endogenous snx5 via two MREs in its 3’UTR.
miR-216a spatially and temporally restricts expression of snx5 in the eye
Because we observed specific spatial and temporal expression of miR-216a over the course of early eye development (Fig. 1A-C), we were interested to examine the expression of snx5 at corresponding time points. We thus performed in situ hybridization using snx5 riboprobes on whole mount zebrafish embryos, which were then sectioned and imaged (Fig. 1D-F). Expression of miR-216a was largely complementary to that observed for snx5. As miR-216a expression moved toward the future CGZ at 36 and 48 hpf (Fig. 1B,C), localization of snx5 became increasingly restricted (Fig. 1E,F) until snx5 expression was virtually undetectable from all cells of the developing retina except for a limited number of cells at the very margins of the future CGZ. The complementary expression patterns of miR-216a and snx5 suggest that miR-216a restricts temporal and spatial expression of snx5 in the developing eye.
Notch-Delta signaling and miR-216a-snx5 interaction
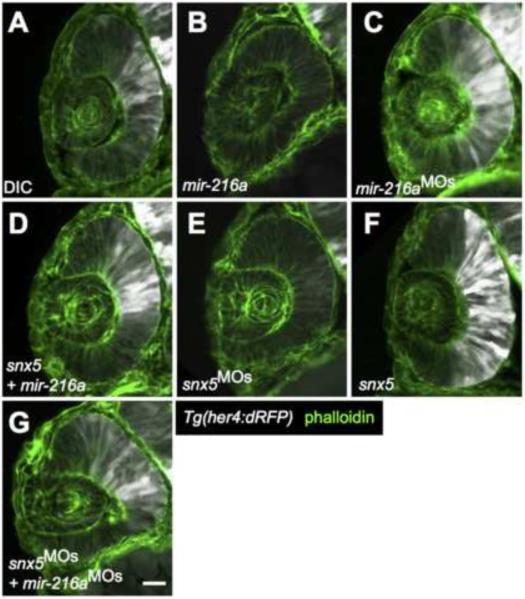
Previous experiments have demonstrated interaction between SNX5 with MIB, co-localization with MIB and Delta (Yoo et al., 2006), and a role for MIB and Notch-Delta signaling in gliogenesis (Bernardos et al., 2005; Scheer et al., 2001). However, the exact effects of snx5 on Notch-Delta signaling have not been characterized nor has there been any previous work investigating the regulation of snx5 during early retina development. We therefore used a Notch reporter zebrafish line (Tg(her4:dRFP)) which expresses dRFP under the control of the her4 Notch-responsive element (Takke et al., 1999; Yeo et al., 2007). We injected Tg(her4:dRFP) single cell embryos with either dye control, synthetic miR-216a duplexes, miR-216aMOs, snx5MOs, or snx5 mRNA, and then fixed the embryos at 30 hpf and sectioned to examine Notch activation in the developing retina. Strikingly, overexpression of miR-216a, or knockdown of snx5, resulted in a marked decrease in Notch activation compared to DICs, as reported by the loss of Tg(her4:dRFP) fluorescent protein expression (Fig. 3A,B,E). Conversely, knockdown of miR-216a, or overexpression of snx5, mRNA resulted in expansion of the zone of Tg(her4:dRFP) fluorescence and presumptive Notch activation compared to DICs (Fig. 3C,F). Co-injection of snx5 lacking its 3’UTR with miR-216a restored the zone of Tg(her4:dRFP) activation (Fig. 3D), as did co-injection of snx5MOs and miR-216aMOs (Fig. 3G). These data indicate that snx5 is a positive regulator of Notch-Delta signaling and that miR-216a negatively regulates Notch-Delta signaling via its interaction with snx5. Consistent with this hypothesis, we used a second zebrafish Notch reporter line (Tg(Tp1bglob:eGFP)) and observed repression of Notch activation by increasing amounts of miR-216a or knockdown of snx5 (Supplemental Fig. 5)(Parsons et al. 2009).
Figure 3. miR-216a and snx5 regulate Notch activation.
Transverse sections of developing retinas from 30 hours post fertilization (hpf) Tg(her4:dRFP) embryos were injected with dye control (DIC; A), miR-216a (B), miR-216aMOs (C), snx5MOs (E), or snx5 mRNA (F). Reporter expression (white) indicates changes in the zone of Notch activation. Partial rescue of Notch activity is shown in (D) and (G) where embryos were co-injected with combinations of either snx5 and miR-216a (D) or snx5MOs and miR-216aMOs (G). Sections were stained with Alexa Fluor 488-conjugated phalloidin (green) to visualize cell boundaries. Scale bar: 20μm.
Because it was formally possible that the effects we observed might be due to morpholino-induced apoptosis as opposed to regulation of snx5 by miR-216a, we conducted TUNEL staining. Previous work has illustrated potential pitfalls with the use of morpholinos, including increased levels of apoptosis due to activation of p53 (Gerety and Wilkinson, 2011). To ensure that the effects we observed were specific to knockdown of miR-216a or snx5, we injected morpholinos in the presence and absence of p53 and found no change in the levels of TdT-mediated incorporation of dUTP (Supplemental Fig. 6). Combined with our suppression/rescue experiments, these results demonstrate that the effects of miR-216a and snx5 knockdown are specific and that the changes in Notch activation we observe are due to regulation of snx5 by miR-216a.
Disruption of Müller glia
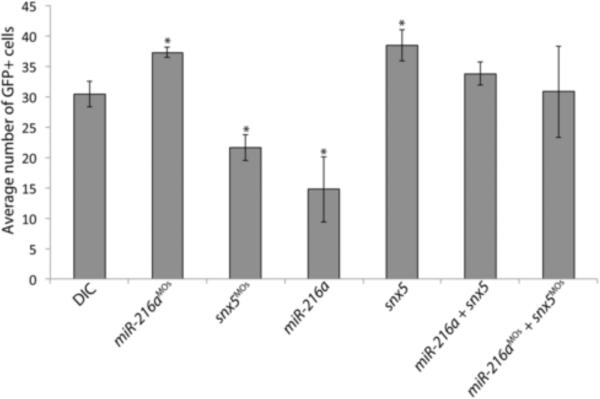
Notch signaling is required for gliogenesis (Bernardos et al., 2005; Scheer et al., 2001) and the prediction is that early alteration in Notch signaling by miR-216a and snx5 should affect the subsequent number of Müller glia during retinal development. To assess the functional consequences of disrupting miR-216a and snx5 expression, we injected miR-216a, miR-216aMOs, snx5MOs, or snx5 mRNA into single cell Tg(gfap:GFP) zebrafish embryos and examined fluorescence levels during early development. These animals express GFP under the control of the glial-specific GFAP promoter (Bernardos and Raymond, 2006). We initially examined retinas from embryos at 30 hpf to coincide with the her4 reporter experiments. Fluoresence was detectable at this time but the levels were not robust, consistent with the timing of Müller glia specification (Easter and Malicki, 2002). Since it has been reported that Müller glia are specified by 65 hpf (Bernardos et al., 2005), and because we observed Notch activation in Müller glia at 65 hours using the her4 reporter fish (Fig. 4), we counted GFP+ cells at this time. Upon overexpression of miR-216a, a significant decrease in GFP+ cells was observed compared to DICs (Fig. 5). In contrast, knocking down miR-216a with morpholinos resulted in an increase in GFP+ numbers (Fig. 5). Correspondingly, knockdown of snx5 resulted in significantly decreased numbers of GFP+ cells whereas overexpression of snx5 led to an increase in GFP+ cells (Fig. 5). These results are consistent with regulation of snx5 by miR-216a. To further test this hypothesis, we conducted co-injection rescue/suppression experiments. The prediction is that the decreased numbers of GFP+ cells caused by knockdown of snx5 should be suppressed by co-injection of miR-216aMOs. Similarly, the effects of overexpression of miR-216a should be suppressed by co-injection of snx5. In both cases, we observed rescue of GFP+ cell numbers indicating that Müller glia numbers were largely restored (Fig. 5). Taken together, these data are consistent with the hypothesis that miR-216a modulates gliogenesis via its interaction with snx5.
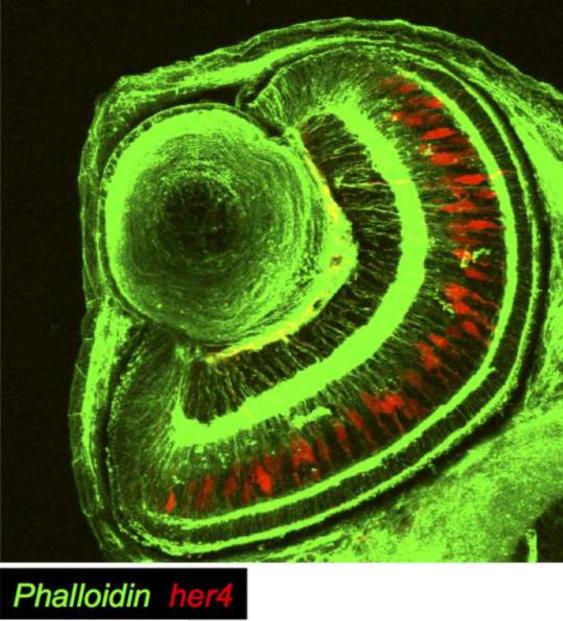
Figure 4. Notch activation in Müller glia at 65 hpf.
In a cross section of Tg(her4:dRFP) fish at 65 hpf, Notch activation (in red) was detected primarily in Müller glia. Cell membranes are labeled with phalloidin, here visualized in green.
Figure 5. miR-216a and snx5 regulate Müller glia cell numbers.
Tg(gfap:GFP) transgenic zebrafish were injected as indicated, grown to 65 hpf, and GFP+ cell numbers were counted. Compared to DICs, injection of miR-216aMOs or snx5 caused a significant increase in GFP+ cells (p<0.05). Injections with miR-216a or snx5MOs caused a significant decrease in GFP+ cells (p<0.05). Partial rescue of GFP+ cell counts was observed in embryos co-injected with combinations of either snx5 and miR-216a, or snx5MOs and miR-216aMOs. Error bars=SEM.
Effects of Müller glia specification on cone photoreceptor differentiation
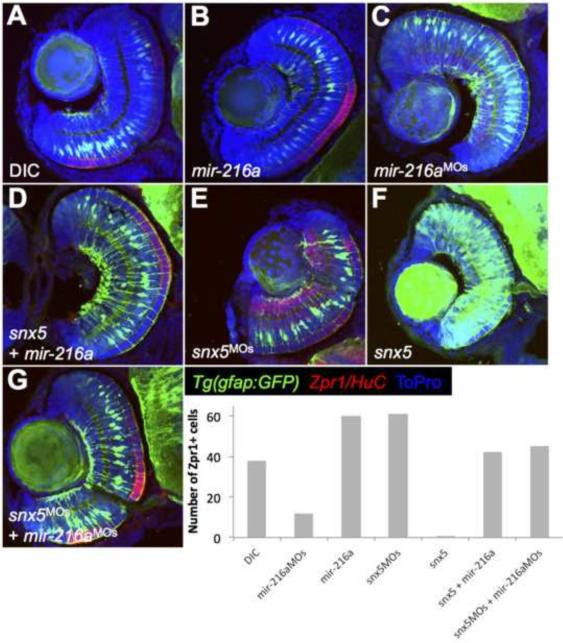
A prediction of the effects of altered gliogenesis is that other retinal neuronal cell types would be altered after either loss or gain of Müller glia. For these experiments we used Tg(gfap:GFP) embryos fixed at 65 hpf and stained transverse retinal sections using antibodies that mark cone photoreceptors (Zpr-1). As shown in Fig. 6, alteration in Müller glia number was accompanied by complementary changes in the extent of Zpr-1 staining in the outer nuclear layer. Overexpression of snx5 or knockdown of miR-216a led to increased Müller glia and decreased Zpr-1 staining while overexpression of miR-216a or knockdown of snx5 led to decreased Müller glia and increased Zpr-1 staining. These results are consistent with the model that altered gliogenesis can in turn affect neuronal differentiation.
Figure 6. Inverse correlation between MG numbers and cone photoreceptor staining.
Tg(gfap:gfp) embryos were injected with dye control (DIC; A), miR-216a (B), miR-216aMOs (C), snx5MOs (E), or snx5 mRNA (F) at the 1-cell stage, fixed at 65 hpf, and transverse sections of developing retinas were obtained. Immunohistochemistry was performed using antibodies to identify cone photoreceptors in the outer nuclear layer (Zpr-1) or amacrine/ganglion cells in the inner nuclear layer and the ganglion cell layer (HuC). Changes in Müller glia cell numbers led to consistent changes in cone photoreceptor numbers. Zpr-1 staining increased in embryos injected with mir-216a or snx5MOs and decreased in embryos injected with mir-216aMOs or snx5 compared to embryos injected with dye. Partial rescue of Zpr-1 levels is shown in (D) and (G) where embryos were co-injected with combinations of either snx5 and miR-216a (D) or snx5MOs and miR-216aMOs (G). Amacrine and ganglion cell numbers demonstrated similar, though less striking and less consistent changes compared to cone photoreceptors. Nuclei were marked by staining with To-Pro.
Discussion
We used expression profiling experiments to identify several candidate miRNA regulators of zebrafish eye development. As demonstrated by snx5 and miR-216a expression, GFP reporter assays, and SNX5 immunoblotting, we show that miR-216a regulates snx5. Based on the expression of miR-216a and snx5 in the retinal neuroepithelium, it appears that miR-216a plays a role in both spatial and temporal control of snx5 expression and, in turn, Notch signaling.
miR-216a regulates Notch signaling via snx5
SNX5 binds Mib and knocking down SNX5 leads to vascular defects (Eckfeldt et al., 2005; Yoo et al., 2006). The role of Notch signaling in vascular development is also well established (Lawson et al., 2001). In addition to changes in fluorescent protein expression in Tg(her4:dRFP) fish, we also observed defects in vascular patterning upon knockdown and overexpression of miR-216a and snx5 (Supplemental Figs. 2,3). This suggests that miR-216a and snx5 also play a role in Notch signaling in zebrafish vascular development. We also show that perturbing expression of miR-216a and snx5 causes changes in Notch activation, as reported by altered zones of fluorescent protein expression in the retinas of Tg(her4:dRFP) embryos.
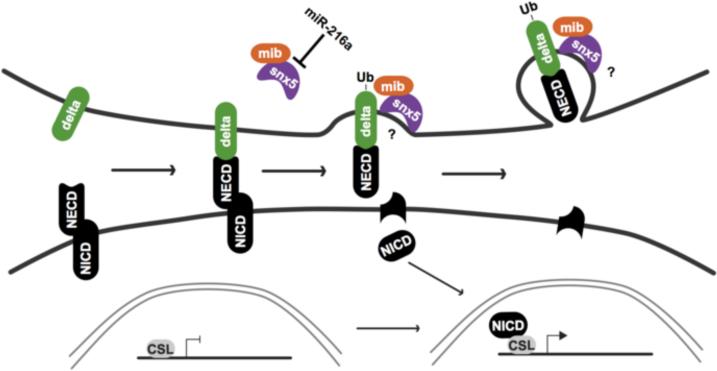
Based on prior work about SNX5 and Mib and our experiments, we propose a model where miR-216a regulates Notch-Delta signaling via regulation of snx5 (Fig. 7). We hypothesize that SNX5 (bound to Mib) moves to the site of Delta activation where it binds to the membrane as Mib ubiquitylates Delta. SNX5 then facilitates membrane curvature through its BAR domain with subsequent Delta endocytosis, which is required for Notch activation and neuronal development (Parks et al., 2000) (Louvi and Artavanis-Tsakonas, 2006).
Figure 7. Model for the role of SNX5 and miR-216a in Notch signaling.
SNX5 (bound to Mib) moves to the site of Delta activation, where it binds to the membrane as Mib ubiquitylates Delta. SNX5 then facilitates membrane curvature and Delta endocytosis, which is required for cleavage of the Notch extracellular domain (NECD). Cleavage of the NECD frees the Notch intracellular domain (NICD) which is translocated to the nucleus to co-activate downstream target genes with the CSL transcription factor.
While our experiments show a role for snx5/miR-216a in controlling Notch activity during retinal development, it is likely that overall control of Notch involves multiple factors and control points during cell fate specification and development. Focusing just on the retina, we show that early changes in Notch signaling manifest themselves at later time points by altering neuronal cell fate. However, several other Notch components, including Delta, are likely to be subject to additional temporal regulation as the wave of differentiation spreads from the central retinal to the periphery. Despite the fact that our morpholino knockdown experiments of miR-216a allow sufficient Notch activity to affect changes in cell fate, our experiments cannot preclude the role of additional Notch components and/or regulators during the dynamic processes occurring during retina development. This likely includes other miRNAs that might regulate other components of the Notch pathway.
miR-216a and snx5 modulate Müller glia cell numbers
The changes in Notch signaling in response to perturbation of snx5 and miR-216a expression that we observed are striking and consistent with previous experiments. Scheer et al. (2001) showed that expressing a constitutively active version of Notch1a resulted in a disruption of neurogenesis and an increase in gliogenesis (Scheer et al., 2001). Additionally, differentiation of Müller glia does not occur in mib mutant fish (Bernardos et al., 2005). These results suggest that Notch signaling is instructive for gliogenesis in the zebrafish retina. We observed that high Notch activation at 30 hpf, as reported by fluorescent protein expression in the Tg(her4:dRFP) zebrafish and induced by either miR-216a knockdown or snx5 overexpression, caused increased numbers of Müller glia at 65 dpf, as reported by Tg(gfap:GFP) fluorescence. Because high Notch signaling at 30 hpf, in the case of miR-216a knockdown or snx5 overexpression, translates to increased numbers of Müller glia, we hypothesize that the snx5-miR-216a interaction may directly impact Notch signaling, and therefore gliogenesis, in the developing retina. Of note, we observed Notch activation in Müller glia at 65hpf (detected by Tg(her4:dRFP; Fig. 4).
It has been suggested that SNX5 is localized to a distinct domain of the early endosome, a cellular location where it could be playing multiple, as yet unknown, roles in cellular trafficking (Yoo et al., 2006). Furthermore, miR-216a and snx5 are each expressed throughout the developing optic cup and retinal neuroepithelium in early development (Supplemental Fig. 1). By knocking down or overexpressing both miR-216a and snx5 globally at early stages, we have likely disrupted functions that manifest themselves later in development leading to a disruption in Notch activation and correspondingly, specification of Müller glia. It has been shown that the interaction of different Delta ligands with Notch can result in different outcomes for Delta activation in neural tissue (Matsuda and Chitnis, 2009).
We also found that altered gliogenesis impacts neuronal differentiation. We show that MG numbers show an inverse correlation with the staining of a marker of cone photoreceptor differentiation. This suggests that overall specification of cell types in the developing retina are coordinately regulated.
miRNAs regulate developmental signaling
We have previously shown that miRNAs play regulatory roles in Hedgehog signaling (Flynt et al., 2007), the development of endoderm and left-right asymmetry (Li et al., 2011), and synaptogenesis (Wei et al., 2013). miRNA regulation of Notch signaling is important during Drosophila follicle development (Poulton et al., 2011) and bone development in mice (Bae et al., 2012). Additionally, Notch signaling has been shown to regulate the expression of miR-9, a miRNA that we detected in our eye-field microarray and is involved in multiple aspects of neural development (Coolen et al., 2012). The finding that miR-216a regulates snx5 adds to the mounting evidence for the importance of miRNAs in regulating developmental processes in vertebrates.
Supplementary Material
Highlights.
miR-216a regulates SNX5 in the developing zebrafish retina.
Perturbation of miR-216a and SNX5 expression leads to changes in Notch activation.
Altering miR-216a and SNX5 expression causes changes in Müller glia number.
Acknowledgements
We thank Qiang Guan for zebrafish maintenance at the Vanderbilt University Stevenson Center Fish Facility. Transgenic zebrafish lines were shared by Ajay Chitnis (Tg(her4:dRFP)) and Michael Parsons (Tg(Tp1bglob:eGFP)). This study was supported in part by grants from the National Eye Institute–National Institutes of Health to J.G.P (R21 EY 019759) and Vanderbilt University. AFO and MBR are part of the Program in Developmental Biology at Vanderbilt and were supported in part by the Gisela Mosig fund in the Department of Biological Sciences. AFO was supported by T32HD007502.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The authors declare no financial or non-financial competing interests.
References
- Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RHA. From the Cover: Differences in vertebrate microRNA expression. PNAS. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T, Dawson BC, Munivez E, Tao J, Lee BH. miRNA-34c regulates Notch signaling during bone development. Human Molecular Genetics. 2012;21:2991–3000. doi: 10.1093/hmg/dds129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardos RL, Lentz SI, Wolfe MS, Raymond PA. Notch-Delta signaling is required for spatial patterning and Muller glia differentiation in the zebrafish retina. Dev Biol. 2005;278:381–395. doi: 10.1016/j.ydbio.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Raymond PA. GFAP transgenic zebrafish. Gene Expr Patterns. 2006;6:1007–1013. doi: 10.1016/j.modgep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Choi J, Dong L, Ahn J, Dao D, Hammerschmidt M, Chen JN. FoxH1 negatively modulates flk1 gene expression and vascular formation in zebrafish. Dev Biol. 2007;304:735–744. doi: 10.1016/j.ydbio.2007.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen M, Thieffry D, Drivenes O, Becker TS, Bally-Cuif L. miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Dev Cell. 2012;22:1052–1064. doi: 10.1016/j.devcel.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Cullen PJ. Endosomal sorting and signalling: an emerging role for sorting nexins. Nat Rev Mol Cell Biol. 2008;9:574–582. doi: 10.1038/nrm2427. [DOI] [PubMed] [Google Scholar]
- Cullen PJ, Korswagen HC. Sorting nexins provide diversity for retromer-dependent trafficking events. Nat Cell Biol. 2012;14:29–37. doi: 10.1038/ncb2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easter SS, Jr., Malicki JJ. The zebrafish eye: developmental and genetic analysis. Results and Problems in Cell Differentiation. 2002;40:346–370. doi: 10.1007/978-3-540-46041-1_17. [DOI] [PubMed] [Google Scholar]
- Eckfeldt CE, Mendenhall EM, Flynn CM, Wang TF, Pickart MA, Grindle SM, Ekker SC, Verfaillie CM. Functional analysis of human hematopoietic stem cell gene expression using zebrafish. PLoS Biol. 2005;3:e254. doi: 10.1371/journal.pbio.0030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Flynt A, Li N, Thatcher E, Solnica-Krezel L, Patton J. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt A, Thatcher E, Burkewitz K, Li N, Liu Y, Patton J. miR-8 microRNAs regulate the response to osmotic stress in zebrafish embryos. J. Cell Biol. 2009;185:115–127. doi: 10.1083/jcb.200807026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Gao FB. Context-dependent functions of specific microRNAs in neuronal development. Neural Dev. 2010;5:25. doi: 10.1186/1749-8104-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerety SS, Wilkinson DG. Morpholino artifacts provide pitfalls and reveal a novel role for pro-apoptotic genes in hindbrain boundary development. Dev Biol. 2011;350:279–289. doi: 10.1016/j.ydbio.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs Regulate Brain Morphogenesis in Zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 Promotes Deadenylation and Clearance of Maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Research. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harterink M, Port F, Lorenowicz MJ, McGough IJ, Silhankova M, Betist MC, van Weering JR, van Heesbeen RG, Middelkoop TC, Basler K, Cullen PJ, Korswagen HC. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat Cell Biol. 2011;13:914–923. doi: 10.1038/ncb2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hitchcock PF, Raymond PA. The teleost retina as a model for developmental and regeneration biology. Zebrafish. 2004;1:257–271. doi: 10.1089/zeb.2004.1.257. [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Kapsimali M, Kloosterman WP, de Bruijn E, Rosa F, Plasterk RH, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Developmental Dynamics. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Lagendijk AK, Moulton JD, Bakkers J. Revealing details: whole mount microRNA in situ hybridization protocol for zebrafish embryos and adult tissues. Biology Open. 2012;1:566–569. doi: 10.1242/bio.2012810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Scheer N, Pham VN, Kim CH, Chitnis AB, Campos-Ortega JA, Weinstein BM. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128:3675–3683. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li N, Flynt A, Kim HR, Solnica-Krezel L, Patton J. Dispatched Homolog 2 is targeted by miR-214 through a combination of three weak microRNA recognition sites. Nucleic Acids Res. 2008;36:4277–4285. doi: 10.1093/nar/gkn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Wei C, Olena AF, Patton JG. Regulation of endoderm formation and left-right asymmetry by miR-92 during early zebrafish development. Development. 2011;138:1817–1826. doi: 10.1242/dev.056697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jin DY, McManus MT, Mourelatos Z. Precursor microRNA-programmed silencing complex assembly pathways in mammals. Mol Cell. 2012;46:507–517. doi: 10.1016/j.molcel.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Chitnis AB. Interaction with Notch determines endocytosis of specific Delta ligands in zebrafish neural tissue. Development. 2009;136:197–206. doi: 10.1242/dev.027938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, Abreu-Goodger C, Staton AA, Stahlhut C, Shou C, Cheng C, Gerstein M, Enright AJ, Giraldez AJ. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23:619–632. doi: 10.1101/gad.1760209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- Parsons MJ, Pisharath H, Yusuff S, Moore JC, Siekmann AF, Lawson ND, Leach SD. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mechanisms of Development. 2009;126:898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton JS, Huang YC, Smith L, Sun J, Leake N, Schleede J, Stevens LM, Deng WM. The microRNA pathway regulates the temporal pattern of Notch signaling in Drosophila follicle cells. Development. 2011;138:1737–1745. doi: 10.1242/dev.059352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp RA, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes and Development. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- Scheer N, Groth A, Hans S, Campos-Ortega JA. An instructive function for Notch in promoting gliogenesis in the zebrafish retina. Development. 2001;128:1099–1107. doi: 10.1242/dev.128.7.1099. [DOI] [PubMed] [Google Scholar]
- Stahlhut C, Suarez Y, Lu J, Mishima Y, Giraldez AJ. miR-1 and miR-206 regulate angiogenesis by modulating VegfA expression in zebrafish. Development. 2012;139:4356–4364. doi: 10.1242/dev.083774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg F, Heesom KJ, Bass MD, Cullen PJ. SNX17 protects integrins from degradation by sorting between lysosomal and recycling pathways. J Cell Biol. 2012;197:219–230. doi: 10.1083/jcb.201111121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takke C, Dornseifer P, v Weizsacker E, Campos-Ortega JA. her4, a zebrafish homologue of the Drosophila neurogenic gene E(spl), is a target of NOTCH signalling. Development. 1999;126:1811–1821. doi: 10.1242/dev.126.9.1811. [DOI] [PubMed] [Google Scholar]
- Thatcher E, Flynt A, Li N, Patton J, Patton J. MiRNA expression analysis during normal zebrafish development and following inhibition of the Hedgehog and Notch signaling pathways. Dev Dyn. 2007;236:2172–2180. doi: 10.1002/dvdy.21211. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Walker C. Haploid screens and gamma-ray mutagenesis. Methods Cell Biol. 1999;60:43–70. doi: 10.1016/s0091-679x(08)61893-2. [DOI] [PubMed] [Google Scholar]
- Wei C, Thatcher EJ, Olena AF, Cha DJ, Perdigoto AL, Marshall A, Carter BD, Broadie K, Patton JG. miR-153 regulates SNAP-25, synaptic transmission, and neuronal development. PloS One. 2013;8:e57080. doi: 10.1371/journal.pone.0057080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RHA. MicroRNA Expression in Zebrafish Embryonic Development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wright GJ, Giudicelli F, Soza-Ried C, Hanisch A, Ariza-McNaughton L, Lewis J. DeltaC and DeltaD interact as Notch ligands in the zebrafish segmentation clock. Development. 2011;138:2947–2956. doi: 10.1242/dev.066654. [DOI] [PubMed] [Google Scholar]
- Yeo SY, Kim M, Kim HS, Huh TL, Chitnis AB. Fluorescent protein expression driven by her4 regulatory elements reveals the spatiotemporal pattern of Notch signaling in the nervous system of zebrafish embryos. Dev Biol. 2007;301:555–567. doi: 10.1016/j.ydbio.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Yoo KW, Kim EH, Jung SH, Rhee M, Koo BK, Yoon KJ, Kong YY, Kim CH. Snx5, as a Mind bomb-binding protein, is expressed in hematopoietic and endothelial precursor cells in zebrafish. FEBS Lett. 2006;580:4409–4416. doi: 10.1016/j.febslet.2006.07.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.