Abstract
MicroRNAs (miRNAs) are endogenous, single-stranded small RNAs that have important regulatory functions at the post-transcriptional level. Here, we characterize miRNAs in two divergent mosquito species, Aedes aegypti and Anopheles stephensi, through deep sequencing of small RNAs spanning all developmental stages. We discovered eight novel miRNAs in Ae. aegypti and 20 novel miRNAs in An. stephensi, which enabled the first systematic analysis of miRNA evolution in mosquitos. We traced the phylogenetic history of all miRNAs in both species and report a rate of 0.055–0.13 miRNA net gain per million years. Most novel miRNAs originate de novo. Duplications that produced miRNA clusters and families are more common in Ae. aegypti than in An. stephensi. We also identified arm-switch as a source of new miRNAs. Expression profile analysis identified mosquito-specific miRNAs that showed strong stage-specific expression in one or both lineages. For example, the aae-miR-2941/2946 family represents the most abundant maternally-deposited and zygotically transcribed miRNAs in Ae. aegypti. miR-2943 is a highly expressed zygotic miRNA in both Ae. aegypti and An. stephensi. Such information provides the basis to study the function of these miRNAs in biology common to all mosquitos or unique to one particular lineage.
Introduction
MicroRNAs (miRNAs) are endogenous, single-stranded small RNAs that are ~21–25 nucleotides in length. They modulate gene expression at the post-transcriptional level and are widely distributed in eukaryotes and some viruses. In animals, mature miRNAs are usually processed from primary miRNAs (pri-miRNA). A pri-miRNA contains one or more hairpins, which are processed by the Drosha/Pasha complex to make precursor miRNA (pre-miRNA). The pre-miRNA hairpin is exported to the cytoplasm by Exportin-5 and further cleaved into the imperfect miRNA:miRNA* duplex by the RNAase III enzyme Dicer. The miRNA strand of the duplex is then incorporated into the RNA-induced silencing complex (RISC) and functions by interacting with the target mRNAs. In some cases, pre-miRNAs known as Mirtrons are spliced directly from introns, thus, bypassing the Drosha/Pasha complex. Once bound to a target, miRNA regulates target expression by either decreasing the mRNA stability or inhibiting its translation (reviewed in Bartel et al., 2004). However, certain miRNAs have also been shown to induce target gene expression (Place et al., 2008; Vasudevan et al., 2007). Many miRNAs play important roles in embryonic development, cell differentiation, neurogenesis, and apoptosis (reviewed in Bushati. et al. 2007). MiRNAs are also found to be involved in the pathogenesis of multiple cancers, cardiac disease, and neurological disorders (Greenberg et al., 2014; Harada et al., 2014; Kye and Gonçalves, 2014). There is evidence that mosquito miRNAs are important in regulating mosquito defense against parasite invasion (Winter et al., 2007), female reproduction (Bryant et al., 2010), and Wolbachia infection (Zhang et al., 2013).
Since the discovery of the first miRNA in Caenorhabditis elegans (Lee et al., 1993), over twenty thousand miRNAs have been predicted in 206 species and documented in miRBase (miRBase release20)(Kozomara and Griffiths-Jones, 2013). Several models have been proposed to explain the origin and expansion of miRNAs. Duplication and random hairpin formation are two main sources of new miRNAs (Allen et al., 2004; Axtell et al., 2011; Marco et al., 2013; Yuan et al., 2011), with duplication being further divided into inverted duplication of target genes, tandem duplication, and segmental duplication (Yuan et al., 2011). Some mammalian miRNAs were derived from repetitive sequences, mostly transposable elements, providing another source for new miRNAs (Piriyapongsa et al., 2007).
Aedes aegypti and Anopheles stephensi are important disease vectors that belong to two divergent mosquito subfamilies, Culicinae and Anophelinae, respectively. Ae. aegypti is an important vector for arboviruses such as the yellow fever, dengue fever, and chikungunya viruses. An. stephensi is the main vector for malaria in urban areas in India and the Middle East. These two mosquito subfamilies diverged between 145 and 200 million years (myr) ago (Krzywinski et al., 2006). Efforts have been made to identify both conserved and mosquito-specific miRNAs in Ae. aegypti (Akbari et al., 2013; Li et al., 2009), Ae. albopictus (Gu et al., 2013), and Culex quinquefasiatus (Skalsky et al., 2010) by small RNA sequencing. MiRNA genes were bioinformatically predicted in An. darlingi (Mendes et al., 2010) and only a small number of miRNAs were experimentally verified by a small scale cloning method in An. gambiae (Winter et al., 2007) and An. stephensi (Mead and Tu, 2008). Two recent studies significantly increased the number of experimentally verified miRNAs in An. stephensi (Jain et al., 2014) and An. gamibae (Biryukova et al., 2014) by small RNA sequencing of adult females before and after blood-feeding.
In this study, we performed small RNA sequencing of samples covering all major developmental stages of Ae. aegypti and An. stephensi using Illumina. We report the discovery of eight novel miRNAs in Ae. aegypti and the first comprehensive analysis of miRNAs in An. stephensi in all major developmental stages. Whole-genome analysis of miRNAs in Ae. aegypti and An. stephensi and comparisons to other insect species provide an opportunity for novel insights into the evolution of mosquito miRNAs. For example, the genome of Ae. aegypti (Nene et al., 2007) is approximately five-fold larger than the genome of An. stephensi (Jiang et al., 2014). Our analysis has enabled the comparison of the number of lineage-specific miRNAs and the mechanisms to generate novel miRNAs between two mosquitos of divergent genomes. The systematic characterization and expression analysis of mosquito miRNAs facilitates future studies of miRNA functions in mosquitos. Regulation by lineage-specific miRNA influences phenotypic divergence among animal species (Mor and Shomron, 2013). We are investigating mosquito-specific miRNAs because they may underlie mosquito-specific biological adaptations and could provide mosquito-specific targets for the control of mosquito-borne infectious diseases. We have shown that several mosquito-specific miRNAs are only expressed either in the embryo, or pupae, or adult male, indicating involvement in mosquito-specific functions during these developmental stages. The importance of miRNA in embryogenesis is well documented in model organisms (Bushati et al., 2008; Giraldez, 2010). There is indication that gene expression during the maternal-to-zygotic transition may vary significantly between Ae. aegypti and Drosophila melanogaster (Biedler and Tu, 2010; Biedler et al., 2012). Thus we also performed a detailed analysis of miRNAs during maternal-to-zygotic transition in Ae. aegypti.
Results
Discovery of conserved and novel miRNAs in Ae. aegypti and An. stephensi by small RNA sequencing
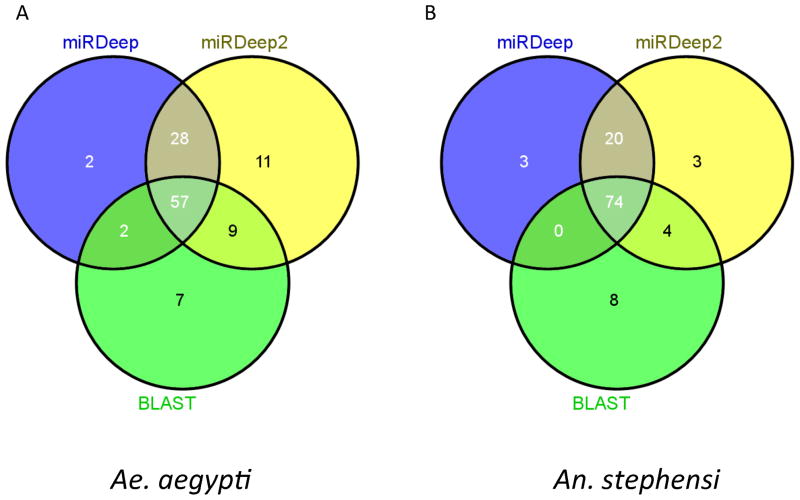
We performed small RNA sequencing on Ae. aegypti and An. stephensi samples from all life stages using the Illumina platform (NCBI PRJNA232374, PRJNA232180 and SRX116547; see Experimental Procedures). Small RNA reads with adapter removed were used to predict miRNAs de novo using the miRDeep (Friedländer et al., 2008) and miRDeep2 (Friedländer et al., 2012)(Figure S1) software packages, followed by two rounds of manual inspection to discard predictions that did not meet the stringent criteria set forth in previous publications (Axtell et al., 2011; Berezikov, 2011) (See Experimental Procedures). The first round of inspection removed sequences that did not have sufficient and homogeneous small RNA alignment to the predicted pre-miRNA stems. Sequences that passed the first round were regarded as miRNA candidates. Sequences that also passed the second round of inspection, which further considered other miRNA features (Table S4), were reported as predicted miRNAs. BLAST searches, using mature and precursor miRNAs in miRBase (v20) as queries, were also performed against the Ae. aegypti and An. stephensi small RNA databases and genomes to ensure coverage of conserved miRNAs. The numbers of miRNAs predicted by the three methods are indicated in Figure 1. The non-redundant miRNAs discovered by the three methods were taken as input to perform a reciprocal Mapmi search (Guerra-Assunção and Enright, 2010) in the two species.
Figure 1. Venn Diagrams showing the number of miRNAs predicted by miRDeep, miRDeep2 and BLAST methods in Aedes aegypti (A) and Anopheles stephensi (B).
The numbers represent miRNAs that passed the two rounds of manual inspection as described in the Experimental Procedures. Figures were generated by Venny (Oliveros, 2007).
In total, we discovered 120 miRNA loci in Ae. aegypti (Table S1) and 117 miRNA loci in An. stephensi (Table S2). The 117 miRNA loci in An. stephensi correspond to 108 unique mature miRNAs, 20 of which are novel. The 120 miRNA loci in Ae. aegypti correspond to 102 mature miRNAs, eight of which are novel. Akbari and colleagues (2013) recently predicted 36 novel miRNAs in Ae. aegypti based on small RNA sequencing results. Only seven of the 36 miRNAs overlap with our predictions. These seven include two insect-specific miRNAs (aae-miR-H-52, aae-miR-H104), two mosquito-specific miRNAs (aae-miR-H-65, aae-miR-H-73), and three Ae. aegypti specific miRNAs (aae-miR-H-62, aae-miR-H-115, aae-miR-H-85). Eighteen of the 36 mapped to more than five genomic loci and 10 did not meet the criteria set for a genuine miRNA upon manual inspection (Table S4). The only remaining miRNA predicted by Akbari et al. (aae-miR-H-88) had good secondary structure and unique genome location and were included in our subsequent analysis (Table S1). Jain et al. (2014) recently analyzed the change of miRNome before and after blood feeding and Plasmodium infection in An. stephensi (Jain et al., 2014). Except for two extremely lowly-expressed miRNAs (as-miR-2779 and as-miR-iab-8), we recovered all known An. stephensi miRNAs in Jain et al. (2014). We also discovered the mature sequence of three additional known miRNAs (ast-miR-2943, ast-miR-316, ast-miR-971) and the star reads of additional 71 miRNAs. Out of the 17 novel An. stephensi miRNA predictions described by Jain et al. (2014), five were found in the current work, nine did not pass the first round of inspection, one (as-nv-16) passed the first round and two (as-nv-3, as-nv-4) passed both rounds (Table S2, S3). As mentioned above, the analysis in the current study added eight novel miRNAs in Ae. aegypti (Table S1) and 20 novel miRNAs in An. stephensi (Table S2).
Clustered and intronic miRNAs in Ae. aegypti and An. stephensi
An miRNA cluster is often defined as a group of miRNAs that are within 10kb of each other on the same genomic strand (Marco et al., 2013). According to this definition, 40 miRNAs were found in 14 clusters in Ae. aegypti, and 41 miRNAs were grouped in 14 clusters in An. stephensi (Table 1). The proportion of clustered miRNAs in Ae. aegypti (33%) and An. stephensi (34.5%) are comparable to the proportions in other species (Olena and Patton, 2010). The majority of mosquito clusters are composed of two miRNAs, which is similar to the size of the D. melanogaster miRNA clusters (Marco et al., 2013) but smaller than the size of the Tribolium castaneum clusters (Marco et al., 2010). The largest miRNA cluster is the miR-2b/2a/13/2c/71 cluster in both Ae. aegypti and An. stephensi. The miRNA arrangements within homologous clusters are the same between Ae. aegypti and An. stephensi. Thirty-one Ae. aegypti miRNAs overlap with annotated protein coding genes, with 30 in introns and one in exon. The number of intragenic miRNAs is 22 in An. stephensi and all are in introns. The associations between intragenic miRNAs and their host genes are quite stable between the two divergent mosquitos. This positional conservation of intronic miRNAs is consistent with what has been observed in vertebrates (Hoeppner et al., 2009).
Table 1.
miRNA clusters in Ae. aegypti and An. stephensi
| Ae. aegypti | An. stephensi |
|---|---|
| aae-miR-1174/1175 | ast-miR-1174/1175 |
|
| |
| aae-miR-286a/2944b/2944a/309b | ast-miR-286a/2944b-1/2944a-1/309-1 |
| aae-miR-286b-1/2944c-1/2944d-1/309a-1 | ast-miR-286b/2944b-2/2944a-2/309-2 |
| aae-miR-286b-2/2944c-2/2944d-2/309a-2 | |
| aae-miR-275/305 | ast-miR-275/305 |
| aae-miR-277/34 | ast-miR-277/34 |
| aae-miR-2b/2a/13/2c/71 | ast-miR-2b/2a/13/2c/71 |
| aae-let-7/miR-125 | ast-miR-100/let-7/miR-125 |
| aae-miR-996/279 | ast-miR-996/279 |
| aae-miR-12/1889/283 | ast-miR-12/1889/283 |
| aae-miR-11/998 | ast-miR-11/998 |
| aae-miR-306/79/9b | ast-miR-9c/306/79/9b |
| aae-miR-2943-1/2943-2 | |
| aae-miR-2941-1/2941-2/2946 | |
| ast-miR-9a/new-5 | |
| ast-miR-new35/new36 | |
| ast-miR-new7/new11/new8/new9 | |
Gain and loss of miRNAs in Ae. aegypti and An. stephensi
To trace the evolution of miRNAs in Ae. aegypti and An. stephensi, we searched for homologs by performing BLAST using all Ae. aegypti and An. stephensi mature miRNAs and pre-miRNAs as queries against the miRBase (v20). The default e-value of 10 was used for the BLAST to include all possible candidates for further inspection. Because there are very limited miRNAs documented in miRBase for some species, we also performed Mapmi (Guerra-Assunção and Enright, 2010) using all Ae. aegypti and An. stephensi miRNAs as queries to search for homologous miRNAs in 15 species including 14 arthropods and humans. We re-constructed the phylogenetic history of Ae. aegypti and An. stephensi miRNAs following the parsimony principle. For instance, if a miRNA is present in lineage A and absent in its sister lineage B, it is considered to originate after the divergence of the two lineages unless there is evidence for the presence of the miRNA in their common ancestor. This is because such inference requires fewer evolutionary changes (Sperling and Peterson, 2009; Tarver et al., 2013). As shown in Figure 2, miRNAs were continually added through the evolution of mosquitos and occasionally lost in some lineages. A total number of 78 miRNAs (80 loci) were present before the divergence between Nematocera and Brachycera. Nine miRNAs were conserved in mosquitos but not present in other species, indicating their potential functions in mosquito-specific gene regulation. Five miRNA loci were duplicated or expanded in the mosquito lineage. Within the Nematocera lineage, there were four Culicinae-specific and two Anophelinae-specific miRNAs. It is noticeable that Ae. aegypti had more expansions or duplications of existing miRNAs than An. stephensi (13 expansions in Ae. aegypti vs. three expansions in An. stephensi), which may be explained at least partially by the larger and more repetitive genome of the former species. Assuming that the Culicinae and Anophelinae subfamilies diverged 145 to 200 myr ago (Krzywinski et al., 2006), the net gain of miRNA per myr was 0.055 to 0.076 for the Culicinae lineage and 0.095 to 0.13 for the Anophelinae lineage. This rate is slower than that in T. castaneum (0.18) and Drosophila (0.3 to 1) (Berezikov et al., 2010; Lu et al., 2008; Marco et al., 2010).
Figure 2. Gain and loss of miRNA in Aedes aegypti and Anopheles stephensi.
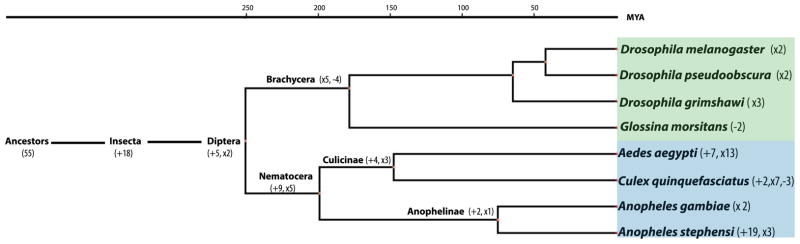
The gain and loss of miRNA in Ae. aegypti and An. stephensi were inferred based on the species distribution of miRNAs. Numbers in parentheses are the number of mosquito miRNAs that were gained (+) or lost (−) from that node; ‘x’ indicates expansion of existing miRNAs (i.e. miR-a/b and 1/2). The evolutionary time scale is adopted from (Liu et al., 2010). The names of specific miRNAs gained/lost/expanded are shown in Table S5.
Origin of new mosquito miRNA loci
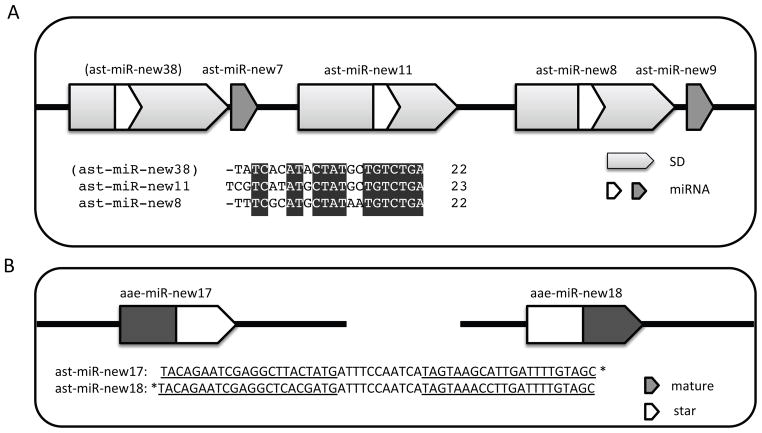
The majority of the mosquito-specific or lineage-specific miRNAs do not have an apparent homolog in their ancestral species (Figure 2) indicating that they either originated de novo or have evolved beyond recognition of its ancestral origin. However, a significant number of the mosquito-specific miRNAs and conserved miRNAs produced new miRNA loci by duplication (Table 2). We differentiated tandem duplication and segmental duplication as described in the Experimental Procedures and modified after Maher et al. (Maher et al., 2006). Seven out of 14 clusters in Ae. aegypti and five out of 14 clusters in An. stephensi had formed by tandem duplication (Tables 1 and 2). An interesting example of this is the An. stephensi-specific cluster ast-miR-new9/new8/new11/new7/new38 (Figure 3A), in which a ~200bp fragment duplicated at least twice and generated the miRNA family ast-new38/new11/new8. Ast-miR-new38 was regarded as a miRNA candidate because only mature reads had been recovered. Although the pre-miRNAs of the two miRNAs and one miRNA candidate are very similar (over 80% identity), mutations in seed regions have accumulated, providing the capacity to target different mRNAs. Segmental duplications involving a large flanking region are also common (Table 2).
Table 2.
Duplication events in Ae. aegypti and An. stephensi
| miRNA families | Estimated emergence time of the original miRNA | Estimated emergence time of the most recent duplication | |
|---|---|---|---|
|
Ae. aegypti
| |||
| segmental duplication | aae-miR-1000-1/1000-2 | after insect, before Diptera | after Ae. aegypti |
| aae-miR-137-1/-2 | before insect | after mosquito, before Culicinae | |
| aae-miR-1891-1/-2 | after mosquito, before Culicinae | after Ae. aegypti | |
| aae-miR-276-1/-2 | before insect | after Dipteran, before mosquito | |
| aae-miR-282-1/-2 | before insect | after Ae. aegypti | |
| aae-miR-317-1/-2 | before insect | after Culicinae, before Ae. aegypti | |
| aae-miR-929-1/-2 | after insect, before Diptera | after Ae. aegypti | |
| aae-miR-iab-4-5p-1/-2 | before insect | after Ae. aegypti | |
| aae-miR-new15-1/-2 | after mosquito, before Culicinae | after Ae. aegypti | |
| aae-miR-new5-1/-2 | after Ae. aegypti | after Ae. aegypti | |
| aae-miR-9a-1/-2 | before insect | after Ae. aegypti | |
| aae-miR-new19-1/-2 | after Culicinae, before Ae. aegypti | after Ae. aegypti | |
| aae-miR-new17/18 | after Ae. aegypti | after Ae. aegypti | |
|
| |||
| tandem duplication | aae-miR-2941-1/-2/2946 | after Culicinae, before Ae. aegypti | after Culicinae, before Ae. aegypti |
| aae-miR-2943-1/-2 | after mosquito, before Culicinae | after Culicinae, before Ae. aegypti | |
| aae-miR-2b/2a/13/2c | before insect | before insect | |
| aae-miR-92a/b | before insect | before insect | |
| aae-mIR-9b/79/9c | before insect | before insect | |
|
| |||
| tandem and segmental duplication | aae-miR-286a/2944b/2944a/309b | ||
| aae-miR-286b-1/2944c-1/2944d-1/309b-1 | |||
| aae-miR-286b-2/2944c-2/2944d-2/309b-2 | before insect | after Ae. aegypti | |
|
| |||
| other1 | aae-miR-263a/b | before insect | before insect |
| aae-miR-9a/9b,79,9c | before insect | before insect | |
|
| |||
|
An. stephensi
| |||
| segmental duplication | ast-miR-137-1/-2 | before insect | after mosquito, before Anophelinae |
| ast-mIR-1890-1/-2 | after mosquito, before Anophelinae | after An. stephensi | |
| ast-miR-92b-1/-2 | before insect | after An. stephensi | |
| ast-miR-965-1/-2 | before insect | after An. stephensi | |
| ast-miR-92a/b | before insect | before insect | |
| ast-miR-new12-1/-2 | after An. stephensi | after An. stephensi | |
|
| |||
| tandem duplication | ast-miR-276-1/-2 | before insect | after Diptera, after Anophelinae |
| ast-miR-2b/2a/13/2c | before insect | before insect | |
| ast-miR-9b/79/9c | before insect | before insect | |
| ast-mIR-new9/new8/new11/new7/(new38)2/new24 | after An. stephensi | after An. stephensi | |
|
| |||
| tandem and segmental duplication | ast-286a/2944b-1/2944a-1/309-1 | ||
| ast-286b/2944b-2/2944a-2/209-2 | before insect | after An. stephensi | |
|
| |||
| other1 | ast-miR-263a/b | before insect | before insect |
| ast-miR-9a/9b,79,9c | before insect | before insect | |
Notes:
“Other” indicates cases for which we cannot determine whether tandem or segmental duplications are responsible for the duplicated miRNAs.
Ast-miR-new38 in the parenthesis is a miRNA candidate.
Figure 3. Examples that illustrate the origin and evolution of mosquito miRNAs.
A) The ast-miR-(new38)/new7/nw11/new8/new9 cluster expanded by a series of tandem duplications. ast-miR-new38 is in parenthesis because it is an miRNA candidate B). Arm switch between aae-miR-new17 and new18. SD denotes segmental duplication. * denotes star sequence.
Another common scenario in miRNA evolution is arm switching. Eight homologous miRNA pairs between two mosquitos adopted different arms. Within Ae. aegypti, we detected one case of arm switching between the paralogous miRNAs aae-miR-new17 and aae-miR-new18, which evolved from a segmental duplication but used different arms as mature miRNA (Figure 3B).
Overall expression profile of Ae. aegypti and An. stephensi miRNAs
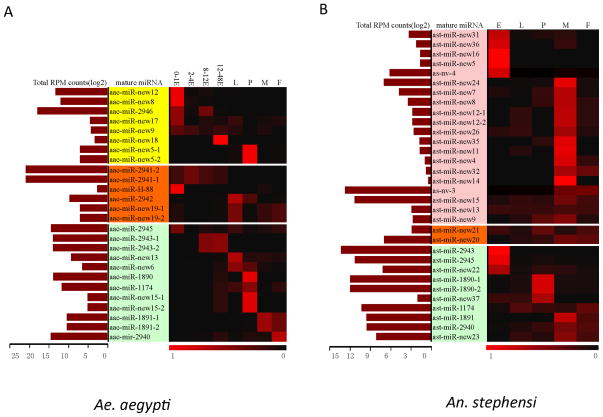
In both Ae. aegypti and An. stephensi, miR-1, miR-184, and miR-263 ranked among the most highly expressed miRNAs, which took up over half of the total hits in a majority of the developmental stages (Table S1, and S2). The expression profiles of miRNAs deduced from our small RNA-seq analysis (Tables S1 and S2) are in general agreement with published expression profiles of Aedes and Anopheles miRNAs based on northern blots (Mead and Tu, 2008; Li et al., 2009; Gu et al., 2013). Here, we focus our analysis on miRNAs that are found only in mosquitos (Figure 4) because they may be involved in mosquito adaptation and speciation. Except for aae-miR-new15 and its homolog ast-miR-new37, the 9 miRNAs that are conserved in all mosquitos showed high or moderate levels of expression. With a few exceptions, the expression patterns are consistent between Ae. aegypti and An. stephensi. In both species, miR-2943 and miR-2945 are highly expressed in the embryos; miR-1891 is most abundantly expressed in adult males and miR-1890 has peak expression in pupae (Figure 4). These mosquito-specific miRNAs are likely involved in important functions in both Aedes and Anopheles lineages. The most abundant miRNAs in Ae. aegypti embryos, the intronic aae-miR-2941-1/2941-2/2946 cluster, is specific to Culicinae (Figure 5, Table S1), indicating their important roles in embryogenesis specific to Culicinae. More than half of the Ae. aegypti-specific miRNAs showed embryo-biased expression while most of the An. stephensi-specific miRNAs showed male-biased expression (Figure 4).
Figure 4. Expression profiles of mosquito-specific miRNAs in Aedes aegypti (A) and Anopheles stephensi (B).
The maroon bars indicate the log2 values of the sum of reads per million (RPM) across all samples for each miRNA while the heatmap shows the relative expression of each miRNA throughout development. Green: mosquito-specific miRNAs found in both Ae. aegypti and An. stephensi; Orange: miRNAs that are restricted in Culicinae or Anophelinae; Yellow: miRNAs specific to Ae. aegypti; Pink: miRNAs specific to An. stephensi.
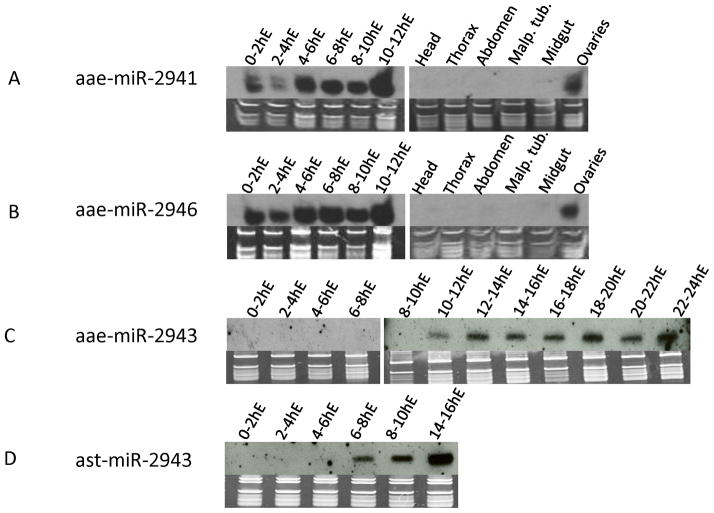
Figure 5. Northern blot of miRNAs that showed high embryonic expression.
Both aae-miR-2941 (A) and aae-miR-2946 (B) are expressed in ovaries, deposited into embryos and also zygotically expressed in early embryos. Both aae-miR-2943 (C) and ast-miR-2943 (D) are purely zygotic with expression starting from 10h post-oviposition in Ae. aegypti and 6h post-oviposition in An. stephensi, respectively.
Maternal and zygotic expression of Ae. aegypti and An. stephensi miRNAs
miRNAs found in 0–1h embryos in Ae. aegypti represent maternally deposited miRNAs as zygotic transcription does not commence until after ~2h post oviposition (Biedler and Tu, 2010; Biedler et al., 2012). We showed that 58 miRNAs have at least 5 raw counts, with 26 having more than 200 raw counts at the 0–1h time window (Table S1). We further investigated two miRNA families to gain insights into early embryonic development in mosquitos. As described earlier, the aae-miR-2941-1/2941-2/2946 miRNAs form a cluster within the intron of a potential transcription factor, AAEL009263. All three miRNAs share the same seed sequence while aae-miR-2941-1 and aae-miR-2941-2 have the same mature sequence with slightly different 3p variances (isomiR) (Morin et al., 2008) and miRNA*. At the four embryonic time points sequenced, the sum of miRNA-2941 and miR-2946 reads represents 38–93% of all identifiable miRNA reads (Table S1). Thus, this cluster is responsible for the most abundant miRNAs in a broad range of embryonic stages including 0–1h after egg-laying when only maternally deposited miRNAs are present (Biedler and Tu, 2010; Biedler et al., 2012). Strong expression in subsequent embryonic time points suggests that this cluster is also zygotically expressed. As shown in Figure S2, maps of the small RNA sequences demark the predicted miRNA and miRNA* boundaries, and the existence of unique miRNA* and 3p isomiRs suggests that both miR-2941 hairpins contribute to the production of miR-2941. Northern blot analysis of aae-miR-2941 and aae-miR-2946 (Figure 5A and 5B) is consistent with both maternal deposit, as indicated by strong signals in the ovary after blood-feeding and 0–2h embryos, and zygotic expression, as indicated by a decline in 2–4h embryos and increase afterwards. We also investigated the embryonic expression profile of miR-2943, a mosquito-specific miRNA, which has a single locus in An. stephensi and two loci in Aedes and Culex. Small RNA-seq data suggested that this miRNA had strong and stage-specific expression in the embryos in both species (Table S1, and S2). Northern blot analysis (Figure 5C and 5D) confirmed zygotic expression of miR-2943 in both species, starting at 10h post-oviposition in Ae. aegypti and 6h post-oviposition in An. stephensi. This time difference is consistent with the slower embryonic development in Ae. aegypti compared with An. stephensi (Juhn and James, 2006).
Discussion
In this study, we report sequencing of small RNAs across all developmental stages in two medically important and evolutionarily divergent mosquito species. We describe the first comprehensive comparative analysis of miRNAs in An. stephensi and Ae. aegypti spanning all developmental stages. We combined de novo prediction programs with homology-based searches to ensure inclusive coverage of miRNAs (Figure 1 and S1). Such comprehensive miRNA analysis in two divergent mosquitos provided a unique opportunity to glean genomic and evolutionary insights. MiRNAs tend to be continuously added into the genome during evolution and rarely lost once integrated into the regulation system (Sperling and Peterson, 2009; Tarver et al., 2013). We reconstructed the phylogenetic history of Ae. aegypti and An. stephensi miRNAs following the parsimony principle. As shown in Figure 2, each lineage has their own specific miRNA families, which suggests the involvement of these miRNAs in biological functions specific to the lineage. This analysis is based on all miRNAs in Ae. aegypti and An. stephensi and, thus, is only comprehensive for these two species. For example, we identified 7 and 19 miRNAs that are unique to Ae. aegypti and An. stephensi, respectively (Figure 2). No comprehensive statement can be made for miRNAs that are unique to An. gambiae or C. quinquefaciatus. However, our systematic analysis of miRNAs in Ae. aegypti, a species in the Culicinae subfamily, and An. stephensi, a species of the Anophelinae subfamily, allowed us to calculate a net gain rate of 0.055 to 0.076 miRNA per myr for Culicinae and 0.095 to 0.13 miRNA per myr for Anophelinae.
Genome-wide and comparative analysis also enabled insights into the evolutionary origin of new miRNAs or new miRNA loci in mosquitos. We found cases that support miRNA origins from de novo formation of hairpins, tandem duplication, segmental duplication, and arm switching. The majority of the mosquito-specific miRNAs does not have an apparent homolog indicating that they either originate de novo or have evolved beyond recognition of its ancestral origin. However, tandem duplications are important for miRNA cluster formation and expansion. In some cases, the evolution of miRNAs might be very complicated and involve multiple steps. One example, the evolution of miR309/2944a/2944b/286 cluster, was illustrated by Ninova et al. (2014).
We found that approximately 33% of the Ae. aegypti and 34.5% of the An. stephensi miRNAs are clustered within 10 kb intervals. The percentage of intragenic miRNAs, mostly in introns, is 25.6% in Ae. aegypti and 18.8% in An. stephensi. It is suggested that miRNA clusters often form by emergence of new miRNAs near an old miRNA and intragenic miRNAs often form by emergence of new miRNAs within host precursor mRNAs (Axtell et al., 2011; Marco et al., 2013). Our observation is consistent with the theory that the birth of miRNA is facilitated by locating near an extant transcript. This is not surprising since the established miRNA or mRNAs would enhance the chance of accessing transcription machinery or the Drosha/Parsha processing complex, thus, increase the probability of a newly formed hairpin to be transcribed and processed to miRNA precursors (Axtell et al., 2011; Marco et al., 2013).
The small RNA libraries in this study spanned the entire mosquito life cycle from embryo to adult. Despite lack of replicates, our data showed consistent profiles with northern blots of miRNAs in previous studies (Mead and Tu, 2008; Li et al., 2009; Gu et al., 2013). However, we need to be cautious when interpreting the expression profile analysis, especially of lowly transcribed miRNAs. There are several mosquito-specific miRNAs that showed highly embryo-specific, pupae-specific, or male-specific expressions in both species, indicating involvement in mosquito-specific functions at these stages (Figure 3). We further discuss three miRNA families to gain insights into early embryonic development in mosquitos. The miRNAs in the miR-2941/2946 cluster are maternally deposited to the embryo and are highly expressed at the early stage of embryogenesis (Figure 5A and 5B). This cluster is composed of two miR-2941 and one miR-2946 hairpins in Ae. aegypti and two miR-2941 and one miR-2952 hairpins in C. quinquefasiatus. MiR-2941/2946/2952 share the same seed sequence and likely have evolved by tandem duplication of the hairpins. The expansion of miR-2941/2946/2952 cluster in Culicinae and its extremely high expression in Ae. aegypti indicate that this cluster may have gained important and specific functions in Culicinae embryonic development. MiR-2943 is another mosquito-specific miRNA that is exclusively expressed in embryos. Unlike the miR-2941/2946 family, miR-2943 showed strong zygotic expression with no maternal deposition (Figure 5C and 5D). The expression of this miRNA starts at the beginning of germ band extension (Monnerat et al., 2002; Vital et al., 2010) and continues for at least 10 hours in both species. Another miRNA cluster that showed strong expression enriched in the embryonic stage is the miR-309/2944a/2944b/286 cluster, which contains miRNAs that are conserved in metazoan or insect species (Table S1 and S2). The expression patterns we observed in two mosquitos are consistent with those in flies (Leaman et al., 2005; Ninova et al., 2014), beetles (Marco et al., 2010), and moths (Wu et al., 2013). The miR-309 cluster has been shown to play roles in the degradation of maternal transcripts during maternal zygotic transition (MZT) in Drosophila (Bushati et al., 2008). The conservation of structure and expression of this cluster implies that its functions are conserved in embryogenesis among different insect species. Thus, it appears that mosquitos utilize highly conserved miRNAs as well as newly evolved miRNAs to control embryonic development to achieve common and lineage-specific functions.
The young and lineage specific miRNAs are generally expressed at lower levels compared with well-conserved miRNAs (Table S1, S2, Figure 4, S3). We noticed that a significant number of An. stephensi-specific miRNAs have their expression enriched in males (Figure 4A, S3), which is consistent with the observation that fast-evolving genes and miRNAs are testis biased (Levine et al., 2006; Marco, 2013). On the other hand, the majority of the Ae. aegypti specific miRNAs are enriched in 0–1h embryos (Figure 4B, S3), suggesting that the miRNA transcripts are maternally deposited. This observation may imply that novel miRNAs tend to have expression in the germ cell-enriched tissues (Wu and Sharp, 2013) and may play roles in mosquito reproduction or embryogenesis.
Within the aae-miR-2941/2946 cluster, there are two miR-2941 hairpins and one miR-2946 hairpin. The ratio of miR-2941 reads to miR-2946 reads varies from the expected ~2:1 to a highly biased ~60:1 in the four embryonic time points (Table S1). Although biased amplification could alter the miRNA ratio within a given sample, it is unlikely that this is the explanation here as the observed variation is between different samples. In addition, previous 454 sequencing of small RNAs of mixed embryos also showed the uneven expression among miR-2941 and miR-2946 (Table 1 in Li et al., 2009). Differential processing of the pre-miRNAs and differential stability of miRNAs are known mechanisms to control miRNA levels (Obernosterer et al., 2006; Bail et al., 2010), both of which could result in different levels of mature miRNAs from the same levels of pri-miRNAs. The small RNA northern blot shown in Figure 5 cannot be used to compare the miRNA-2941 and miR-2946 levels, as different probes are used and they are from different experiments. Quantitative measurement of the miRNAs in this cluster is needed to explain the variation in the miR-2941/miR-2946 ratios observed in the Illumina data. If confirmed, this will be the first case where differential processing or differential stability of miRNAs regulates miRNA expression in mosquitos.
We didn’t perform miRNA target prediction because the 3′ UTR annotations were not sufficiently informative in the two mosquito species. Moreover, in silico target prediction alone very often gives false positives since the target-miRNA recognition is relatively tolerant of mismatches in animals. Better annotations based on high throughput sequencing of full-length mRNAs will facilitate target analysis and shed new light on miRNA functions in mosquitos.
In conclusion, by performing small RNA sequencing across all developmental stages and by applying multiple analytical methods, we were able to obtain an inclusive annotation of miRNAs in two divergent mosquito species. We discovered eight novel miRNAs in Ae. aegypti and 20 novel miRNAs in An. stephensi. We also report genomic and evolutionary insights of mosquito miRNAs by performing systematic analysis of miRNA distribution and phylogeny. We found approximately 30% miRNAs are clustered and 20% miRNAs are in the introns in both mosquitos. We investigated the origin and evolution of miRNAs in Ae. aegypti and An. stephensi and showed that most lineage-specific miRNAs evolve de novo. Duplications that produced miRNA clusters and families are more common in Ae. aegypti than those in An. stephensi. Our analysis identifies a total number of nine mosquito-specific, four Culicinae-specific, and two Anopheles-specific miRNAs, which may provide the foundation for future analysis illustrating the biological differences between these different lineages. MiRNA profiles encompassing the entire mosquito life cycle shows miRNA enrichment in particular developmental stages. Several miRNAs, such as miR-2941/2946 and miR-2943, are particularly interesting because they are highly expressed in embryos and specific within mosquitos. Our expression analysis points to a direction for future miRNA functional studies.
Experimental Procedures
Sample collection and library preparation for small RNA sequencing
Small RNAs from different developmental stages of Ae. aegypti and An. stephensi were extracted, respectively, and subjected to Illumina sequencing. Ae. aegypti samples included 0–1h embryos, 2–4h embryos, 8–12h embryos, 12–48h embryos, mixed stage larvae, pupae, and adult males. An. stephensi samples were collected in the same manner except that embryos were collected at 0–2h and at 4h intervals from 2h to 42h post oviposition and were pooled to represent the entire embryonic stage. After removing the adapter sequence, identical reads within a single sequencing library were collapsed and different sequencing libraries were pooled together as the small RNA (smRNA) sequencing database. The collapsed Ae. aegypti smRNA database contained 6,836,662 reads and the collapsed An. stephensi smRNA database contained 3,027,738 reads. Those reads were non-redundant within the sequencing library, but may be the same across different libraries. Each read had a unique tag indicating its origin (sample name) and hit number (counts in a single library). All sequencing data can be found in NCBI SRA (PRJNA232374 and PRJNA232180) except the Ae. aegypti adult female sample, which was downloaded from SRA SRX116547 and treated the same way.
miRNA prediction by miRDeep, miRDeep2, BLAST and Mapmi
We followed the steps described in the miRDeep manual (Friedländer et al., 2008) to predict miRNA. The only exception was that we omitted the “filter by annotation” step because this step would only slightly increase the prediction accuracy and we performed manual inspection afterwards to filter false positives. The genome assembly of Ae. aegypti was downloaded from Vectorbase and the genome of An. stephensi was sequenced and assembled by our lab, which can be downloaded from NCBI (PRJNA168255). Briefly, the short reads in the smRNA database were mapped to their corresponding genome. For the purpose of this study, reads mapped to over 5 locations in genome were discarded as repeat sequences. The precursor miRNAs (pre-miRNAs) were retrieved from the genome after mapping and scored by the prediction programs. The output of miRDeep was manually inspected by an in-house script matchRNA. MatchRNA took the predicted pre-miRNA sequence, folded it with the embedded RNAfold program and aligned short sequencing reads onto the pre-miRNAs by BLAST (−e 0.0001, −b 200000). The output from matchRNA (Figure S4) allowed us to perform the first round of inspection which looked for miRNA candidates that have canonical structure and expression. A good miRNA candidate should satisfy that i. reasonable number of mature or star reads aligning to the pre-miRNA (we required a minimal of 5 reads that map to either mature or star sequence); ii. the 5′ and 3′ ends, especially the 5′ ends, are homogeneous; iii, most reads are mapped to one or two arms of the pri-miRNA(i.e. mature and star sequences) with few or no reads mapping to other places of pri-miRNA. The kept miRNA candidates from two species were then used as input for miRDeep2 prediction, which requires a reference file containing known miRNAs in the working species and another reference file containing known miRNAs in closely related species in addition to the genome and smRNA database. We again performed manual inspection upon all predicted miRNAs from miRDeep2. We also used BLAST to recover any conserved miRNAs. We first searched all mature sequences in miRBase (v20) against our smRNA database (−e 0.01) and then searched all precursor sequences in miRBase (v20) against our mosquito genome (−e 0.01). If the mature sequence of a particular miRNA was present in our smRNA database, its precursor sequence was then retrieved from the mosquito genome and inspected using matchRNA. Finally, we ran Mapmi (Guerra-Assunção and Enright, 2010) to detect whether any predicted novel Ae. aegypti miRNAs and miRNA candidates had homologs in An. stephensi, and vice versa. To be conservative, we allowed only one mismatch in mature sequences and verified all predicted miRNAs manually by matchRNA.
Newly-predicted miRNA candidates that passed the first round of inspection as described above were further divided into more confident “novel miRNAs” and less confident “novel miRNA candidates” after the second round of inspection. The factors we looked for during the second round of inspection included the presence of bulges or internal loops in the stem structure, the presence of miRNA star, 3′ 2nt overhang of miRNA/miRNA* duplex, conservation in other species, and the minimal free energy (MFE) of pre-miRNA structure (Table S4). All miRNAs that lack bulges or internal loops in their pre-miRNAs were put into miRNA candidate category because they might represent other small interfering RNAs. If one miRNA did not have reads mapping to the star arm, it must have homology support in other species to remain as a novel miRNA. If one miRNA overlapped with an exon, the 2 nt 3′ overhang was required to differentiate it from mRNA degradation, unless other evidence such as northern blot was available.
Infer phylogenetic history of mosquito miRNA
To infer the gain and loss of mosquito miRNAs during evolution, we investigated the distribution of the mosquito miRNA homologs in different species. We first ran Mapmi (Guerra-Assunção and Enright, 2010) using all Ae. aegypti and An. stephensi mature miRNAs as queries on 15 species with a maximum of one or three mismatches and 20 as the score cutoff. Using either one or three mismatch allowance produced the same final results. The 15 species were Ae. aegypti (Ensembl Metazoa release 9), C. quinquefasciatus (Ensembl Metazoa release 10), An. stephensi (NCBI PRJNA168255), An. gambiae (EnsemblMetazoa release 9), D. melanogaster (EnsemblMetazoa release 9), D. pseudoobscura (EnsemblMetazoa release 10), D. grimshaw (EnsemblMetazoa release 10), Glossina morsitans (VectorBase), Apis mellifera (Baylor College of Medicine Honey Bee Genome Project FTP), Bombyx mori (Genebank accession ID AADK00000000), Acyrthosiphon pisum (Ensembl Metazoa release 10), T. castaneum (Genebank accession ID AAJJ00000000), Ixodes scapularis (Ensembl Metazoa release 10), Pediculus humanus (Ensembl Metazoa release 10) and Homo sapiens (Ensembl release 62). We also searched miRBase (v20) by name as well as by BLAST using all Ae. aegypti and An. stephensi mature miRNAs and pre-miRNAs as queries (−e 10). Low similarity homologs were manually inspected for alignment. If a miRNA is absent in one species but present in its close relative, a further BLAST using the pre-miRNA in the relative species as query was conducted against the genome of the tested species under the e-value of 0.01. The gain of a miRNA refers to the first appearance of the miRNA in phylogeny. The loss of a miRNA was labeled when we failed to uncover the miRNA in one species but were able to infer its existence in the common ancestor.
Detection of tandem and segmental duplication
We defined and detected tandem and segmental duplication as described in Maher et al. (2006) with some modifications. Tandem duplicated miRNAs are contiguous miRNAs with the same or similar precursor sequences while segmental duplicated miRNAs are miRNA families derived from duplications of large DNA segments (from several hundred bp to several kb). To detect segmental duplication events, we looked at the similarity of flanking regions of miRNA pairs. Up to 25 kb flanking both sides of the Ae. aegypti miRNA pairs and 10 kb flanking each side of the An. stephensi miRNA pairs were retrieved and searched for homology by BLAST under the e-value of 0.0001. Sequences shorter than 25kb or 10kb might be retrieved if the scaffold was short or the gene was close to the end of the scaffold. If the genomic region where the paralog of this miRNA resides was recovered by BLAST under the e-value of 0.0001, the overall homologous region was larger than 1kb, and the same was observed in a reciprocal BLAST, we regarded these two miRNAs as candidates for segmental duplications. We then aligned the two homologous region by BLAST and the majority of them had e-value of 0, with the highest being 2E-84. Similarities between protein-coding genes flanking the miRNA pairs were also used to help infer homology.
Expression profile of mosquito miRNAs
The predicted mature miRNAs were used as queries in the BLAST search against the smRNA database (−e 0.0001, −b 200000). Total hits were counted based on different developmental stages and a tabulated table was generated in which each row contained raw counts of one miRNA. Bowtie (Langmead et al., 2009) mapping with no mismatch and BLAST search with the e-value of 0.01 were also performed to obtain read counts, which gave similar results with strong positive correlations (data not shown). To eliminate the effect of library size, the raw counts were normalized as reads per million (RPM) by dividing the total hairpin reads in a single library (Ruby et al., 2007). To visualize the expression pattern of each miRNA, the RPM normalized counts were further normalized by the total counts of that miRNA across all sequencing samples.
Small RNA northern blot analysis
All equipment was rinsed with DEPC-treated water to remove RNases. Samples were denatured at 95°C and run on a 15% polyacrylamide gel at 150V in 1x TBE buffer. 19- and 22-nucleotide oligomers were used as size markers. After staining the gel with ethidium bromide for visualization the RNAs were transferred to a Brightstar Plus membrane (Ambion, Life Technologies, Grand Island, NY) in 0.5x TBE at 4°C for 1.5 hours at 200mA. The membrane was then crosslinked with a SpectroLinker UV cross-linker at the optimal setting (120 mJ/cm2). The membrane was then prehybridized at 45°C while rotating with the ULTRAhyb -Oligo hybridization buffer (Ambion) for 30 minutes, after which 5′ Digoxigenin-labelled antisense LNA probe (Exiqon, Vedbaek, Denmark; miR-2941:5′-TCCGTGGAGTTCTAGCCGTACTA-3′, miR-2946:5′-TCCCCATATCTTTTCCGTACTA-3′, miR-2943: 5′-TTGCCTGCAAGTGCCTACTTAA) was added to a final concentration of 0.1 nM and left overnight. All subsequent procedures including washing, blocking, and incubation with the alkaline-phosphotase labelled anti- Digoxigenin antibody were as described in Mead and Tu (2008). Visualization of the membrane was achieved by 5 minutes of room temperature incubation with the CDP-star substrate followed by exposure to X-ray film.
Mature miRNA RT-PCR
Total RNAs were extracted from Ae. aegypti and An. stephensi mixed-age embryos, larvae, pupae, adult males and adult females. Approximately 500ng RNA was reverse transcribed to cDNA using Qiagen (Valencia, CA) miScript II RT kit under Hispec buffer. 1ul cDNA of each sample was used as template and amplified by rTaq polymerase using one universal primer and one miRNA specific primer. The PCR results were inspected using a 4% agarose gel.
Supplementary Material
Figure S1. Flowchart of miRNA prediction steps for Aedes aegypti and Anopheles stephensi. The inputs for each step were shown by arrows. The numbers of miRNAs that have been predicted before and after the first and second of round manual inspections were separated by “/” in parentheses.
Figure S2. Examples showing smRNA reads aligning to aae-miR-2941-1/2941-2 pre-miRNAs. Red color indicates unique miRNA* or isomiRs that can be used to indicate that both copies of the aae-miR-2941 are expressed.
Figure S3. RT-PCR showing the expression profiles of selected novel miRNAs in Aedes aegypti (A) and Anopheles stephensi (B). A). aae-miR-new8 and aae-miR-new 12 showed enriched expression in embryos; the conserved miRNA aae-miR-13 was used as a positive control. B) ast-miR-new24 and ast-miR-new7 showed enriched expression in males; ast-miR-new15 was present in all stages; ast-miR-13 was used as a positive control.
Figure S4. Examples of matchRNA output showing different patterns of smRNA alignment. The name right after the smRNA sequence (eg., Ste-E, Ste-P) indicates the sample origin of the aligned reads. The number after “x” is read count. 100% indicates the reads match the pre-miRNA sequence with 100% identity. A) A typical miRNA pattern with reads aligning to two arms of pre-miRNA hairpin. The boundaries of alignments are clear and there are no reads aligning to other places of pre-miRNA. B) An example where reads align to places other than arms of pre-miRNA. C) An example where the boundaries of alignment are not clear. Candidates as shown in B) and C) were discarded after the first round of manual inspection.
Table S1. Predicted novel and conserved miRNAs in Aedes aegypti
Table S2. Predicted novel and conserved miRNAs in Anopheles stephensi
Table S3. Predicted miRNA candidates in Aedes aegypti and Anopheles stephensi
Table S4. Criteria used in the second round of inspection to identify novel miRNAs from the miRNA candidates in Aedes aegypti and Anopheles stephensi
Table S5. The names of miRNAs that were gained and lost as described in Figure 2
Acknowledgments
We thank Randy Saunders for mosquito rearing and Janet Webster for critical reading and revision of the manuscript. This work was supported by NIH grants AI070854 and AI077680, FNIH Grant GC7 #316, and the Virginia Experimental Station.
References
- Akbari OS, Antoshechkin I, Amrhein H, Williams B, Diloreto R, Sandler J, Hay BA. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 (Bethesda) 2013;3:1493–509. doi: 10.1534/g3.113.006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–90. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Westholm JO, Lai EC. Vive la differénce: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Lee R, Feinbaum R. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function Genomics: The miRNA Genes. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12:846–60. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Liu N, Flynt AS, Hodges E, Rooks M, Hannon GJ, Lai EC. Evolutionary flux of canonical microRNAs and mirtrons in Drosophila. Nat Genet. 2010;42:6–9. doi: 10.1038/ng0110-6. author reply 9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, Zhao Y, Zamore PD, Hannon GJ, Marra Ma, Weng Z, Perrimon N, Lai EC. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21:203–15. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedler JK, Hu W, Tae H, Tu Z. Identification of early zygotic genes in the yellow fever mosquito Aedes aegypti and discovery of a motif involved in early zygotic genome activation. PLoS One. 2012;7:e33933. doi: 10.1371/journal.pone.0033933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedler JK, Tu Z. Evolutionary analysis of the kinesin light chain genes in the yellow fever mosquito Aedes aegypti: gene duplication as a source for novel early zygotic genes. BMC Evol Biol. 2010;10:206. doi: 10.1186/1471-2148-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biryukova I, Ye T, Levashina E. Transcriptome-wide analysis of microRNA expression in the malaria mosquito Anopheles gambiae. BMC Genomics. 2014;15:557. doi: 10.1186/1471-2164-15-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant B, Macdonald W, Raikhel AS. microRNA miR-275 is indispensable for blood digestion and egg development in the mosquito Aedes aegypti. Proc Natl Acad Sci U S A. 2010;107:22391–8. doi: 10.1073/pnas.1016230107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18:501–6. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–15. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ. microRNAs, the cell’s Nepenthe: clearing the past during the maternal-to-zygotic transition and cellular reprogramming. Curr Opin Genet Dev. 2010;20:369–75. doi: 10.1016/j.gde.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg E, Nemlich Y, Markel G. MicroRNAs in Cancer: Lessons from Melanoma. Curr Pharm Des. 2014 doi: 10.2174/1381612820666140128210105. [DOI] [PubMed] [Google Scholar]
- Gu J, Hu W, Wu J, Zheng P, Chen M, James Aa, Chen X, Tu Z. miRNA genes of an invasive vector mosquito, Aedes albopictus. PLoS One. 2013;8:e67638. doi: 10.1371/journal.pone.0067638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra-Assunção JA, Enright AJ. MapMi: automated mapping of microRNA loci. BMC Bioinformatics. 2010;11:133. doi: 10.1186/1471-2105-11-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M, Luo X, Murohara T, Yang B, Dobrev D, Nattel S. MicroRNA Regulation and Cardiac Calcium Signaling: Role in Cardiac Disease and Therapeutic Potential. Circ Res. 2014;114:689–705. doi: 10.1161/CIRCRESAHA.114.301798. [DOI] [PubMed] [Google Scholar]
- Hoeppner MP, White S, Jeffares DC, Poole AM. Evolutionarily stable association of intronic snoRNAs and microRNAs with their host genes. Genome Biol Evol. 2009;1:420–8. doi: 10.1093/gbe/evp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Rana V, Shrinet J, Sharma A, Tridibes A, Sunil S, Bhatnagar RK. Blood Feeding and Plasmodium Infection Alters the miRNome of Anopheles stephensi. PLoS One. 2014;9:e98402. doi: 10.1371/journal.pone.0098402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Peery A, Hall A, Sharma A, Chen XG, Waterhouse RM, Komissarov A, Riehl MM, Shouche Y, Sharakhova MV, Lawson D, Pakpour N, Arensburger P, Davidson VLM, Eiglmeier K, Emrich S, George P, Kennedy RC, Mane SP, Maslen G, Oringanje C, Qi Y, Settlage R, Tojo M, Tubio JMC, Unger MF, Wang B, Vernick KD, Ribeiro JMC, James AA, Michel K, Riehle MA, Luckhart S, Sharakhov IV, Tu Z. Genome analysis of a major urban malaria vector mosquito, Anopheles stephensi. Genome Biol. 2014;15:459. doi: 10.1186/s13059-014-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhn J, James aa. oskar gene expression in the vector mosquitoes, Anopheles gambiae and Aedes aegypti. Insect Mol Biol. 2006;15:363–72. doi: 10.1111/j.1365-2583.2006.00655.x. [DOI] [PubMed] [Google Scholar]
- Kozomara a, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2013:1–6. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski J, Grushko OG, Besansky NJ. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol Phylogenet Evol. 2006;39:417–23. doi: 10.1016/j.ympev.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Kye MJ, Gonçalves I, do CG. The role of miRNA in motor neuron disease. Front Cell Neurosci. 2014;8:15. doi: 10.3389/fncel.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, Gaul U. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Levine MT, Jones CD, Kern AD, Lindfors Ha, Begun DJ. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci U S A. 2006;103:9935–9. doi: 10.1073/pnas.0509809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Mead Ea, Liang S, Tu Z. Direct sequencing and expression analysis of a large number of miRNAs in Aedes aegypti and a multi-species survey of novel mosquito miRNAs. BMC Genomics. 2009;10:581. doi: 10.1186/1471-2164-10-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Lehane S, He X, Lehane M, Hertz-Fowler C, Berriman M, Pickett Ja, Field LM, Zhou JJ. Characterisations of odorant-binding proteins in the tsetse fly Glossina morsitans morsitans. Cell Mol Life Sci. 2010;67:919–29. doi: 10.1007/s00018-009-0221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu CI. The birth and death of microRNA genes in Drosophila. Nat Genet. 2008;40:351–5. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- Maher C, Stein L, Ware D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006;16:510–9. doi: 10.1101/gr.4680506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A. Sex-biased expression of microRNAs in Drosophila melanogaster. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A, Hui JHL, Ronshaugen M, Griffiths-Jones S. Functional shifts in insect microRNA evolution. Genome Biol Evol. 2010;2:686–96. doi: 10.1093/gbe/evq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A, Ninova M, Ronshaugen M, Griffiths-Jones S. Clusters of microRNAs emerge by new hairpins in existing transcripts. Nucleic Acids Res. 2013;41:7745–52. doi: 10.1093/nar/gkt534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead EA, Tu Z. Cloning, characterization, and expression of microRNAs from the Asian malaria mosquito, Anopheles stephensi. BMC Genomics. 2008;9:244. doi: 10.1186/1471-2164-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes ND, Freitas AT, Vasconcelos AT, Sagot MF. Combination of measures distinguishes pre-miRNAs from other stem-loops in the genome of the newly sequenced Anopheles darlingi. BMC Genomics. 2010;11:529. doi: 10.1186/1471-2164-11-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnerat AT, Machado MP, Vale BS, Soares MJ, Lima JBP, Lenzi HL, Valle D. Anopheles albitarsis embryogenesis: morphological identification of major events. Mem Inst Oswaldo Cruz. 2002;97:589–96. doi: 10.1590/s0074-02762002000400026. [DOI] [PubMed] [Google Scholar]
- Mor E, Shomron N. Species-specific microRNA regulation influences phenotypic variability: perspectives on species-specific microRNA regulation. Bioessays. 2013;35:881–8. doi: 10.1002/bies.201200157. [DOI] [PubMed] [Google Scholar]
- Morin RD, Connor MDO, Griffith M, Kuchenbauer F, Delaney A, Prabhu A, Zhao Y, Mcdonald H, Zeng T, Hirst M, Eaves CJ, Marra MA. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. 2008. pp. 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CFM, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O’leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JMC, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–23. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninova M, Ronshaugen M, Griffiths-Jones S. Fast-evolving microRNAs are highly expressed in the early embryo of Drosophila virilis. RNA. 2014;20:360–72. doi: 10.1261/rna.041657.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olena AF, Patton JG. Genomic organization of microRNAs. J Cell Physiol. 2010;222:540–5. doi: 10.1002/jcp.21993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros JC. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007 http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- Piriyapongsa J, Mariño-Ramírez L, Jordan IK. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007;176:1323–37. doi: 10.1534/genetics.107.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–64. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrinet J, Jain S, Jain J, Bhatnagar RK, Sunil S. Next Generation Sequencing Reveals Regulation of Distinct Aedes microRNAs during Chikungunya Virus Development. PLoS Negl Trop Dis. 2014;8:e2616. doi: 10.1371/journal.pntd.0002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Vanlandingham DL, Scholle F, Higgs S, Cullen BR. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genomics. 2010;11:119. doi: 10.1186/1471-2164-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling EA, Peterson KJ. microRNAs and metazoan phylogeny: big trees from little genes. In: Telford MJLD, editor. Animal Evolution—genomes, Trees …. Oxford Univ Press; Oxford: 2009. pp. 157–210. [Google Scholar]
- Tarver JE, Sperling Ea, Nailor A, Heimberg AM, Robinson JM, King BL, Pisani D, Donoghue PCJ, Peterson KJ. miRNAs: Small Genes with Big Potential in Metazoan Phylogenetics. Mol Biol Evol. 2013:1–14. doi: 10.1093/molbev/mst133. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vital W, Rezende GL, Abreu L, Moraes J, Lemos FJa, Vaz IDS, Logullo C. Germ band retraction as a landmark in glucose metabolism during Aedes aegypti embryogenesis. BMC Dev Biol. 2010;10:25. doi: 10.1186/1471-213X-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter F, Edaye S, Hüttenhofer A, Brunel C. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res. 2007;35:6953–62. doi: 10.1093/nar/gkm686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Ren Q, Li C, Wang Y, Sang M, Zhang Y, Li B. Characterization and comparative profiling of MicroRNAs in a sexual dimorphism insect, Eupolyphaga saneness Walker. PLoS One. 2013;8:e59016. doi: 10.1371/journal.pone.0059016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Sharp Pa. Divergent transcription: a driving force for new gene origination? Cell. 2013;155:990–6. doi: 10.1016/j.cell.2013.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Sun X, Liu H, Xie J. MicroRNA genes derived from repetitive elements and expanded by segmental duplication events in mammalian genomes. PLoS One. 2011;6:e17666. doi: 10.1371/journal.pone.0017666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Hussein M, O’Neill SL, Safari S. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proc Natl Acad Sci U S A. 2013;110:10276–81. doi: 10.1073/pnas.1303603110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Flowchart of miRNA prediction steps for Aedes aegypti and Anopheles stephensi. The inputs for each step were shown by arrows. The numbers of miRNAs that have been predicted before and after the first and second of round manual inspections were separated by “/” in parentheses.
Figure S2. Examples showing smRNA reads aligning to aae-miR-2941-1/2941-2 pre-miRNAs. Red color indicates unique miRNA* or isomiRs that can be used to indicate that both copies of the aae-miR-2941 are expressed.
Figure S3. RT-PCR showing the expression profiles of selected novel miRNAs in Aedes aegypti (A) and Anopheles stephensi (B). A). aae-miR-new8 and aae-miR-new 12 showed enriched expression in embryos; the conserved miRNA aae-miR-13 was used as a positive control. B) ast-miR-new24 and ast-miR-new7 showed enriched expression in males; ast-miR-new15 was present in all stages; ast-miR-13 was used as a positive control.
Figure S4. Examples of matchRNA output showing different patterns of smRNA alignment. The name right after the smRNA sequence (eg., Ste-E, Ste-P) indicates the sample origin of the aligned reads. The number after “x” is read count. 100% indicates the reads match the pre-miRNA sequence with 100% identity. A) A typical miRNA pattern with reads aligning to two arms of pre-miRNA hairpin. The boundaries of alignments are clear and there are no reads aligning to other places of pre-miRNA. B) An example where reads align to places other than arms of pre-miRNA. C) An example where the boundaries of alignment are not clear. Candidates as shown in B) and C) were discarded after the first round of manual inspection.
Table S1. Predicted novel and conserved miRNAs in Aedes aegypti
Table S2. Predicted novel and conserved miRNAs in Anopheles stephensi
Table S3. Predicted miRNA candidates in Aedes aegypti and Anopheles stephensi
Table S4. Criteria used in the second round of inspection to identify novel miRNAs from the miRNA candidates in Aedes aegypti and Anopheles stephensi
Table S5. The names of miRNAs that were gained and lost as described in Figure 2