Abstract
The CRISPR/Cas9 system has proven to be an efficient gene-editing tool for genome modification of cells and organisms. However, the applicability and efficiency of this system in pig embryos have not been studied in depth. Here, we aimed to remove porcine OCT4 function as a model case using the CRISPR/Cas9 system. Injection of Cas9 and single-guide RNA (sgRNA) against OCT4 decreased the percentages of OCT4-positive embryos to 37–50% of total embryos, while ~100% of control embryos exhibited clear OCT4 immunostaining. We assessed the mutation status near the guide sequence using polymerase chain reaction (PCR) and DNA sequencing, and a portion of blastocysts (20% in exon 2 and 50% in exon 5) had insertions/deletions near protospacer-adjacent motifs (PAMs). Different target sites had frequent deletions, but different concentrations of sgRNA made no impact. OCT4 mRNA levels dramatically decreased at the 8-cell stage, and they were barely detectable in blastocysts, while mRNA levels of other genes, including NANOG, and CDX2 were not affected. In addition, the combination of two sgRNAs led to large-scale deletion (about 1.8 kb) in the same chromosome. Next, we injected an enhanced green fluorescent protein (eGFP) vector targeting the OCT4 exon with Cas9 and sgRNA to create a knockin. We confirmed eGFP fluorescence in blastocysts in the inner cell mass, and also checked the mutation status using PCR and DNA sequencing. A significant portion of blastocysts had eGFP sequence insertions near PAM sites. The CRISPR/CAS9 system provides a good tool for gene functional studies by deleting target genes in the pig.
Introduction
The introduction of mammalian genome sequences including those of humans [1], mice [2], and domestic animals, including cows [3] and pigs [4], has increased the importance and necessity of functional genomic tools to study the roles of genes in the genome. While functional genetic studies using genome targeting technologies, including knockouts and knockins, have been achieved via homologous recombination using embryonic stem cells (ES cells) [5, 6]; however, the unavailability of ES cells in other animals, especially commercially important domestic animal species like cows and pigs, hinders the advance of functional genomic studies in these animals.
To overcome these limitations of ES cell-mediated genome targeting, genome editing technologies using novel programmable DNA endonucleases, including zinc-finger nuclease (ZFN)[7] and transcription activator-like effector nuclease (TALEN)[8], have emerged recently. Both technologies rely on the DNA recognition domain derived from ZFN or transcription activator-like effector (TALE) to specifically recognize DNA elements longer than 15 bp, and they both utilize the DNA cleavage domain from the restriction enzyme FokI for DNA cleavages [9]. Both techniques have been successfully utilized to generate gene-specific knockout or knockin in various animals, including mice [10, 11], pigs [12], and cows [13].
However, several limitations in both techniques have been reported. For example, both techniques rely on the generation of a pair of DNA-recognition modules. For ZFN, generation and selection of DNA-specific ZF modules are time-consuming processes [14]. For TALEN, the modular nature of TALE eases the difficulties of design and selection of specific DNA-binding modules [15, 16], but the repetitive nature of TALE sequences demands a complicated gene synthesis step. Therefore, the time and cost required for obtaining the proper TALEN gene is a major bottleneck to utilize these technologies to generate knockout animals [17, 18]. Another huge hurdle for both techniques is the efficiency of programmed nuclease cleavage. Because of the low cleavage efficiency of both nucleases, genome modification of livestock animals must be performed via somatic cell nuclear transfer (SCNT) [12, 13], which suffers from low efficiency caused by developmental defects from partially reprogrammed embryos [19]. Recently, genome editing via direct injection of TALEN mRNA in porcine zygotes has been reported [20], but the editing frequency in the embryo is as low as 7%.
Recently, the bacterial clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) system, which is known as the bacterial adaptive immune system that confers resistance against bacteriophages, was demonstrated as an efficient gene-targeting technology with the potential for multiplexed genome editing [21–23]. In addition to mammalian cell lines, the CRISPR/Cas9 system can be used to efficiently generate knockout or knockin organisms via zygotic injections of Cas9 and single guide RNA (sgRNA) in many organisms, including the mouse [24, 25], rat[26], zebrafish [27, 28], nematode [29], frog [30], pig [31], and monkey [32], indicating the versatility and universality of the CRISPR/Cas9 system in genome editing.
In addition to generating knockout/knockin animals, CRISPR/Cas9-mediated gene knockout/knockin can be useful to functionally characterize embryogenesis-related genes[33]. Functional characterization of genes involved in early embryogenesis using loss-of-function approaches has relied on using knockdown or antisense oligonucleotide injections [34, 35] because knockout in early embryogenesis-related genes causes embryonic lethality, and therefore maintenance of the mutant animal is impossible.
Porcine parthenogenetic preimplantation embryos have been utilized as model systems for embryogenesis [36–39]. In the present study, we tested the feasibility of the CRISPR/Cas system in studying embryogenesis-related genes in porcine parthenogenetic embryos. We chose the OCT4/POU5f1 gene, encoding an essential transcription factor for stem cell maintenance and pluripotency [40], for the knockout/knockin study using the CRISPR/Cas9 system in porcine parthenogenetic embryos.
Materials and Methods
Chemicals
Unless otherwise described, all of chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Generation of Cas9 mRNA and sgRNAs
A previously described plasmid (pCAG-T3-hCAS-pA)[41], which contains human codon-optimized Cas9 plasmid cloned into a T3 promoter, was obtained from Addgene (Addgene #41815) and used to generate Cas9 mRNA. Briefly, the plasmid was linearized with digestion of SphI and transcribed using an mMessage Machine T3 Kit (Life Technologies; Foster City, CA, USA). After transcription, template DNA was removed by treatment with Turbo-DNase (Life Technologies; Foster City, CA, USA). Resulting transcripts were purified by phenol-chloroform extraction and isopropanol precipitation and stored at −80°C until used.
Guide sequences for sgRNAs corresponding to exons of OCT4 were selected using the CRISPR Design Tool [42] (http://crisp.mit.edu) and are shown in S1 Table. Forward polymerase chain reaction (PCR) primers containing the T7 promoter, guide sequences, and portions of the Cas9 handle were hybridized with a reverse PCR primer containing the Cas9 handle and Streptococcus pyrogenes terminator sequences, and this was amplified by PCR using Phusion DNA polymerase (Thermo Fisher Scientific; Waltham, MA, USA) T7 promoter primer and the terminator primers are listed in S1 Table. Resulting PCR products (123 bp) were purified with gel extraction and used as templates for in vitro transcription using T7 Mega-shortscript kits (Life Technologies; Foster City, CA, USA) and purified with phenol/chloroform extraction, as done with the Cas9 mRNAs.
Generation of OCT4-green fluorescent protein (GFP) knockin constructs
To generate a C-terminal fusion of GFP in the porcine OCT4 locus via homology-dependent repair (HDR), an OCT4-GFP fusion construct was generated using Gibson Assembly techniques [43]). Briefly, a 2103-bp fragment containing exon 5 of porcine OCT4 (spanning 27,266,974–27,269,103 bp at chromosome 7) was amplified using PCR and cloned into the pCRII Topo vector (Life Technology; Foster City, CA, USA).GFP fragments were amplified by PCR with primers containing junction sequences of OCT4 and eGFP, and OCT4-pCRII Topo vector was amplified by inverse-PCR using primers corresponding to the last codon of the OCT4 coding sequence. The amplified OCT4 vector and eGFP fragments were assembled using Gibson Assembly Master Mix (New England BioLabs; Beverly, MA, USA) and transformed in Escherichia coli DH5α. The resulting construct, which contained the C-terminal eGFP fusion at the end of OCT4 exon 5, was confirmed with DNA sequencing.
In vitro porcine oocyte maturation
Prepubertal porcine ovaries were obtained from a local slaughterhouse(Farm Story dodram B&F, um0sung, chungbuk, Korea). Cumulus-oocyte complexes (COCs) were obtained from follicles that were 3–6-mm in diameter using 18-gauge microneedles. Oocytes with evenly granulated cytoplasm and a compact surrounding cumulus mass were collected and washed three times with TL-HEPES-PVA medium (Tyrode’s lactate–HEPES medium supplemented with 0.01% polyvinyl alcohol). After washing, 70–80 COCs were transferred into 500 mL of IVM medium (TCM-199; Invitrogen, Carlsbad, CA) supplemented with 20 ng/mL epidermal growth factor, 1 g/mL insulin, 75 g/mL kanamycin, 0.91 mM Na pyruvate, 0.57 mM l-cysteine, and 10% (v/v) porcine follicular fluid. After 22 h of culture, the COCs were transferred into IVM medium without hormones and cultured for an additional 22 h at 38.5°C in an atmosphere containing 5% CO2 and 100% humidity.
Parthenogenetic activation and in vitro culture (IVC)
After maturation (44 h), cumulus cells were removed by repeated pipetting in the presence of 1 mg/mL hyaluronidase for 2–3 min. Denuded oocytes were activated by an electric pulse (1.0 kV/cm for 60 ms) in activation medium (280 mM mannitol, 0.01 mM CaCl2, and 0.05 mM MgCl2), followed by 3 h of incubation in PZM3 medium containing 2 mM cytochalasin B. About 70–80 post-activation oocytes were cultured in 4-well dishes containing 500 mL of PZM3 for 168 h. Embryo culture conditions were maintained at 38.5°C in an atmosphere containing 5% CO2 and 100% humidity.
Cas9/sgRNA injections in porcine parthenogenetic zygotes
After 8 h of parthenogenetic activation, zygotes were microinjected with a mixture of Cas9 mRNA (100 ng/μL) and different concentrations of sgRNA (10, 50, or 100 ng/μL) under a Nikon TE2000-U inverted microscope (Nikon Corporation; Tokyo, Japan) using a FemtoJet microinjector (Eppendorf; Hamburg, Germany). For large-scale deletion of OCT4, embryos were microinjected with mixtures of Cas9 mRNA (100 ng/μL) and different concentrations of two sgRNAs (10 ng/μL sgRNA targeting exon 2 and 10 ng/μL or 100 ng/μL sgRNA targeting exon 5). To generate OCT4-eGFP knockin embryos, embryos were microinjected with mixtures of Cas9 mRNA (100 ng/μL), sgRNAs (10 ng/μL), and different concentrations of OCT4-eGFP knockin construct DNA (20, 50, or 100 ng/μL). As a control, Cas9 mRNA (100 ng/μL) without sgRNA was injected. After microinjections, zygotes were cultured in PZM3 medium for 168 h at 38.5°C in an atmosphere containing 5% CO2 and 100% humidity.
Preparation of genomic DNA from blastocysts and PCR amplification
To prepare genomic DNA, blastocysts were washed twice in phosphate-buffered saline (PBS)/PVA, treated with proteinase K, and incubated at 50°C for 3 h. Portions of genomic DNA containing guide sequences for sgRNAs (exon 2 or exon 5 of porcine OCT4) were amplified by PCR using the PCR primers listed in Table 1 and Pfu-x DNA polymerase (Solgent; Daejun, Korea). Resulting PCR products (~300 bp) were purified by gel purification, and PCR products were cloned into the pCR-II-Topo vector (Life Technologies; Foster City, CA, USA) or sequenced directly using the PCR primers used for amplification.
Table 1. Developmental competence and targeting efficiency of CRISPR/Cas9 mediated Porcine OCT4 locus.
Cas9 mRNA and sgRNAs were injected in combinations of different concentrations, and cleavage and blastocyst formation rates of each groups are presented.
| Gene | Cas9/sgRNA ng/μl) | Cleavage/injected zygotes | Blastocyst/injected zygotes | OCT4 targeting fficiency | |
|---|---|---|---|---|---|
| Immunostaning | Sequencing | ||||
| OCT4 (Exon 2) | 100/0 | 206/264(78.03%) | 83/264(31.44%) | 0/15(0%) | 0/10(0%) |
| 100/10 | 333/445(74.83%) | 138/445(31.01%) | 3/8(37.5%) | 2/10(20.0%) | |
| 100/50 | 310/391(79.28%) | 123/391(31.46%) | 7/15(46.6%) | 5/25(20.0%) | |
| OCT4 (Exon 5) | 100/0 | 91/120(75.83%) | 43/120(35.83%) | 0/12(0%) | 0/13(0%) |
| 100/10 | 149/196(76.02%) | 65/196(33.16%) | 13/25(52.0%) | 12/24(50.0%) | |
OCT4 targeting efficiency was measured using the presence of OCT4 immunostaining signal in nucleus or PCR amplification and sequencing of single blastocysts. Statistical significance was tested using chi-square test.
Real-time reverse transcription-PCR
Porcine OCT4, CDX2, or NANOG gene expression levels were analyzed by real-time quantitative (q)PCR using the ΔΔCT method[28]. Total RNA was extracted from 50 oocytes using a Dynabead mRNA DIRECT Kit (Life Technologies; Foster City, CA, USA). First-strand cDNA synthesis was completed using a cDNA Synthesis Kit (Takara; Kyoto, Japan) and oligo(dT) 12–18 primers. The PCR primers used to amplify OCT4, CDX2, and NANOG genes are listed in S1 Table. Real-time PCR was performed with SYBR Green in a final reaction volume of 20 μL (qPCR kit, Finnzymes; Vantaa, Finland). PCR conditions were as follows: 95°C for 3 min, followed by 39 cycles of 95°C for 15 s, 57°C for 15 s, 72°C for 45 s, and a final extension of 72°C for 5 min. Finally, relative gene expression levels were quantified by normalizing to the respective GAPDH mRNA level. Experiments were conducted in triplicate.
Immunofluorescence staining and confocal microscopy
For immunostaining of OCT4 or CDX2, blastocysts were fixed in 4% paraformaldehyde dissolved in PBS and then transferred to a membrane permeabilization solution (0.5% Triton X-100) for 1 h. After 1 h in blocking buffer (PBS containing 1% bovine serum albumin), blastocysts were incubated overnight at 4°C with anti-OCT3/4 antibody (sc-8628, Santa Cruz Biotechnology; Santa Cruz, CA, USA) diluted 1:200, or CDX2 antibody (MU392A-UC, BioGenex Laboratories Inc.; San Ramon, CA, USA) diluted 1:200. The blastocysts were washed three times in PBS containing 0.1% Tween 20 and 0.01% Triton X-100 for 2 min, and then the samples were co-stained with Hoechst 33342 (10 mg/mL in PBS) for 15 min and washed three times in washing buffer. Samples were mounted onto glass slides and examined using a confocal laser-scanning microscope (Zeiss LSM 710 META; Jena, Germany). At least 30 blastocysts were examined per group.
Data analysis
For each treatment, at least three replicates were perfomed. Statistical analyses were conducted using pearson’s chi-square test or an analysis of variance(ANOVA) followed by Tukey’s multiple comparisons of means by R (R Development Core Team, Vienna, Austria). Data are expressed as mean ± standard error of the mean and p < 0.05 was considered significant.
Results
CRISPR/Cas9-mediated targeting of the OCT4 gene in porcine zygotes
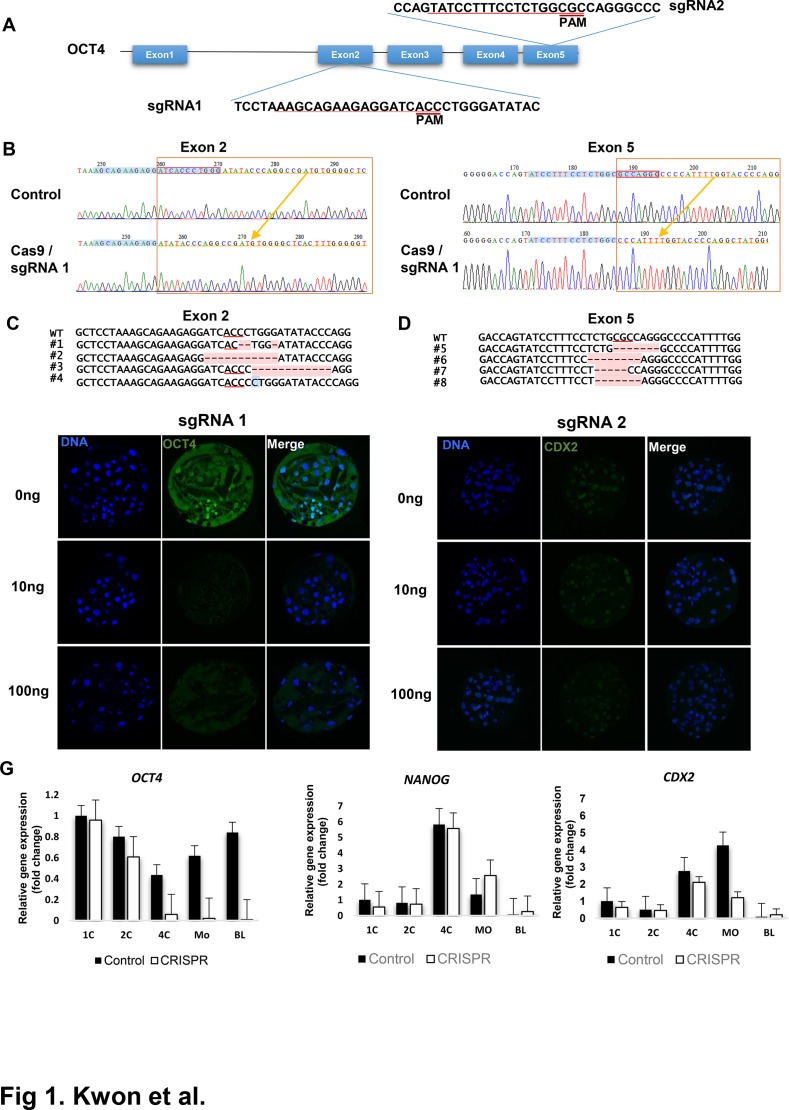
To investigate the usefulness of the CRISPR/Cas9 system in functional characterization of genes involved in mammalian preimplantation development, we chose to use porcine parthenogenetic zygotes and OCT4 as a model system and gene, respectively. In previous studies using mouse embryos, knockout or of OCT4 itself did not impair early blastocyst formation [40, 44] although formation of the inner cell mass and its pluripotency were impaired [25]. To test targeting efficiency of the porcine OCT4 gene, we chose exons 2 and 5 and selected two target regions to introduce indels via non-homologous end repair(NHEJ), as shown in Fig. 1a and Table 2. We injected 100 ng/μL Cas9 and 10, 50, or 100 ng/μL sgRNA for exon 2 and exon 5 of OCT4 into porcine parthenogenetic zygotes, or we injected the Cas9 alone as a control. As shown in Table 1, all groups displayed similar levels of developmental competency. We checked the mutation status using pooled genomic DNA from embryos near the guide sequence by PCR and DNA sequencing. As shown in Table 1, a portion of clones (20% for exon 2 and 50% for exon 5) had an insertion/deletion near the PAM. As shown in Fig. 1b, most clones had 1–12-bp deletions near PAM regions. In some cases, single base insertions were observed (Fig. 1b). To assess OCT4 knockout, we checked OCT4 protein levels in the inner cell mass (ICM) of blastocysts by immunostaining with an anti-OCT4 antibody. As shown in Table 1 and Fig. 1e, while control embryos (Cas9 injection only) had a clear OCT4 signal in ICM regions, injection of Cas9 and sgRNAs decreased OCT4 positivity in embryos to 37–50% of control embryos. These results indicate that injection of Cas9 and sgRNAs designed to target porcine OCT4 successfully targeted the OCT4 locus and depleted OCT4 protein at 30–50% efficiency in embryos.
Fig 1. CRISPR/Cas9 mediated OCT4 targeting in porcine embryos.
(A) Design of sgRNAs for targeting exon 2 or exon 5. Guide sequences correspond to sgRNAs are marked as underlined and the protospacer adjacent motif(PAM) sites for each guide sequences are marked. (B) Sequencing of PCR amplification product confirmed the introduction of indel in exon 2 or exon 5. Locations of guide sequences are marked as blue and bases deleted in Cas9/sgRNAs injected embryos are marked in red box. (C-D) Various deletion/insertions induced by Cas9/sgRNA injections in exon 2 (C) or exon 5(D). Positions of PAM sites are marked as underlined and deleted base was marked as red. Note that insertion can be induced in some case(marked in blue). (E-F) Detections of OCT4 and CDX2 protein using immunostaining in targeted porcine embryos. Embryos were injected with 5–10pl of 100ng/μl of Cas9 mRNA mixed with 0, 10, 100ng/μl of sgRNA1 or sgRNA2, respectively. Location of nucleus was stained with Hoechst 33342 (blue). OCT4(left panel) and CDX2(right panel) are presented as green. (G) mRNA expression levels of OCT4, CDX2, and NANOG measured by qRT-PCR. Expression levels are presented as relative expression levels to those in control embryo at 1 cell stages. Control: Cas9 mRNA injection only; CRISPR: Cas9 mRNA and sgRNA 2 injected. In each developmental stages, 20 embryos were collected for RNA extraction.
Table 2. Developmental competence and targeting efficiency of CRISPR/Cas9 mediated large-scale deletions in Porcine OCT4 locus.
| Gene | Cas9/Exon 2 sgRNA/Exon 5 sgRNA(ng/μl) | Cleavage/injected zygotes | Blastocyst/injected zygotes | Efficiency | |
|---|---|---|---|---|---|
| Immunostaning | Sequencing | ||||
| OCT4 (Exon 2 and Exon5) | 100/0 | 103/140(73.57%) | 37/140(26.43%) | 0/15(0%) | 0/10(0%) |
| 100/10/10 | 213/280(76.07%) | 77/280(27.50%) | 11/23(47.82%) | 6/50(12.0%) | |
sgRNA pair corresponds to exon 2 or exon 5 were coinjected with Cas9 mRNA. Cleavage and blastocyst formation rates of each groups are presented. OCT4 targeting efficiency was measured using the presence of OCT4 immunostaining signal in nucleus or PCR amplification and sequencing of single blastocysts. Statistical significance was tested using chi-square test.
Next, we checked the mRNA levels of OCT4 and other pluripotency related genes, including NANOG, and CDX2 in Cas9/sgRNAs-injected and control embryos. As shown in Fig. 1g, OCT4 mRNA levels in control embryos decreased until the 4-cell stage and increased in the 8-cell and blastocyst stages, presumably by zygotic genomic activation [45]. For embryos injected with Cas9 and sgRNA, maternal OCT4 mRNA was sustained until the 4-cell stage, but mRNA levels dramatically decreased at the morula stage and were barely detectable in the blastocyst stage, while mRNA levels of other genes, including NANOG, and CDX2 were not affected. These results suggest that OCT4 knockout in embryos abolished OCT4 expression in embryonic genome activation, and its effects were specific for OCT4.
Large-scale genomic region deletion in the OCT4 gene using two sgRNA pairs
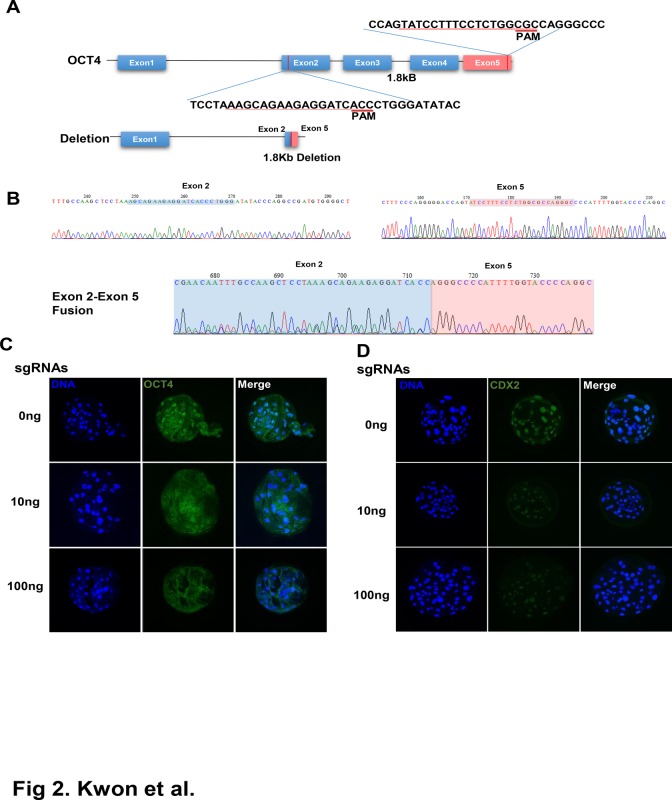
In addition to introduction of small indels or point mutations, CRISPR/Cas9 has been used to introduce large structural variations, including ~10-kb deletions [41] and chromosomal translocations [46, 47]. We tested the induction of large-scale deletions in the porcine genome using two sgRNAs targeting regions located in the same chromosome. As shown in Fig. 2a, the distance between the guide sequences of sgRNA 1 (targeting exon 2) and sgRNA 2 (targeting exon 5) was about 1.8 kb, and using these two sgRNA pairs, we attempted to delete 1.8-kb regions between exon 2 and exon 5 in the porcine OCT4 gene. We injected two sgRNAs (10 ng each) with Cas9 RNA (100 ng/μL) in porcine zygotes and checked the mutation status after sgRNA/Cas9 injection. As shown in Table 2, developmental competency of embryos injected with sgRNAs and Cas9 was not significantly different from that of the control. When genomic DNA from embryos injected with both sgRNAs and Cas9 was screened for large-scale deletions, 12% (6/50) of embryos showed the expected 1.8-kb deletion, as shown in Fig. 2b. In immunostaining results shows that OCT-4 signal in blastocysts was not detected as shown in Fig. 2c. These results showed that large-scale structural variation, including exon deletions or chromosomal rearrangements, could be introduced in the porcine genome.
Fig 2. Targeted deletion of region between exon 2 and exon 5 of porcine OCT4 locus using sgRNA pairs.
(A) Location of sgRNA pair in OCT4 locus and scheme for resulting deletion (B) Sequencing of PCR amplification product confimed the deletion of 1.8kb region between exon 2 and exon 5 of OCT4 locus. Locations of guide sequences in exon 2 and exon 5 are marked as blue(exon 2) or red (exon 5) respectively, and resulting exon 2—exon 5 fusions are shown. (C, D) Deletion of OCT4 and CDX2 protein using immunostaining in targeted porcine embryos. Embryos were injected with 5–10pl of 100ng/μl of Cas9 mRNA mixed with 0, 10, 100ng/μl of sgRNA 1 and sgRNA 2, respectively. Location of nucleus was stained with Hoechst 33342 (blue). OCT4(C) and CDX2 (D) are presented as green.
Generation of OCT4-GFP knockins using homology-dependent repair
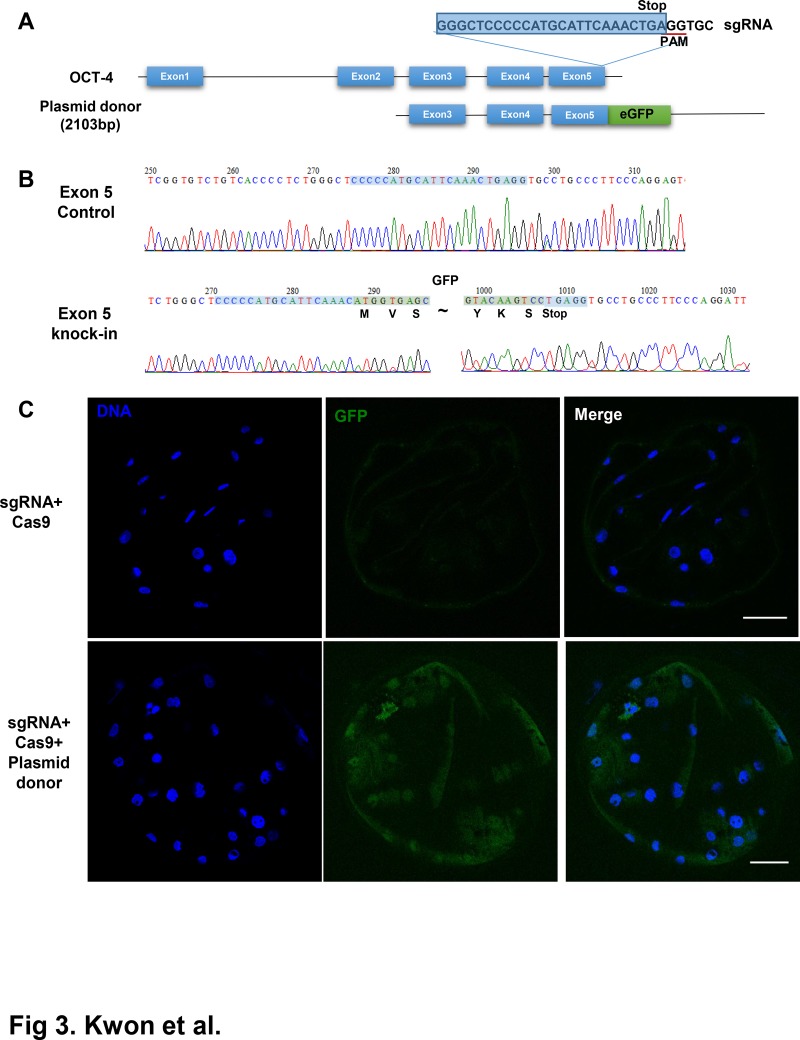
In addition to generating knockouts in the genomic locus, CRISPR/Cas9 systems have been employed to introduce precise genomic DNA knockins, including site-specific mutations, incorporation of loxP sequences, or generation of specific marker gene fusions like those with GFP [24, 25, 48]. We evaluated the possibility that the CRISPR/Cas9 system could be used to introduce exogenous DNA sequences in a targeted manner. We designed an OCT4-GFP targeting vector with a GFP fusion in the 3´-end of the OCT4-coding region, creating an in-frame fusion (Fig. 3a). Then, we injected the targeting vector as well as sgRNA/Cas9 to introduce cleavage near the 3´-end of OCT4. When a high concentration of targeting vector (100 ng/μL) was used, the developmental competency of injected embryos decreased (Table 3). When lower concentrations of targeting vector (20 or 50 ng/μL) were used, the developmental competencies of embryos were similar to those of the control group. We assessed the localization of GFP using a confocal microscope. As shown in Fig. 3c, the GFP signal appeared in Blastocyst-stage embryos and the GFP signal was detected in the ICM, indicating that the OCT4-GFP fusion protein was expressed in the ICM. Next, we assessed the presence of the GFP sequence in embryonic genomic DNA. As shown in Fig. 3b, the eGFP sequence was precisely integrated after the GFP sequence. Knockin efficiency was 2.44% (1/41) (Table 3), which is comparable with that in previous studies using mouse zygotes [25].
Fig 3. Generation of in-frame fusion of eGFP locus in Porcine OCT4 using CRISPR/Cas9 mediated homology dependent repair (HDR).
(A) Scheme for the HDR-mediated integration of eGFP into OCT4 locus. Donor vector for HDR was consisted with the left homology arm (1kb) spanning exon 3—exon 5, eGFP fused after exon 5 as in-frame fusion and the right homology arm (1kb). Guide sequence for sgRNA was designed near stop codon of OCT4 located at exon 5. Location of PAM are underlined and stop codon of OCT4 is marked. Note that eGFP was inserted between stop codon and last codon of OCT4 and PAM of sgRNA was located just after stop codon. Therefore donor vector cannot recognize and digested with Cas9/sgRNA. (B) Confirmation of insertion of eGFP locus in genomic DNA of porcine embryo. PCR amplification spanning exon 5 confirmed the presence of in-frame fusion of eGFP at the end of OCT4 coding region. (C) Expression of eGFP fused Oct4 in HDR mediated eGFP knockin porcine embryos. eGFP expression was detected byConfocal microscopy. Control (Cas9 injected) and Targeted (Cas9/sgRNA/Donor Plasmid) have been compared. Note that localization of eGFP signal in nucleus.
Table 3. Developmental competence and eGFP knockin efficiency in Porcine OCT4 locus.
| Cas9/Exon 2 sgRNA/ donor plasmid (ng/μl) | Cleavage/injected zygotes | Blastocyst/injected zygotes | Efficiency |
|---|---|---|---|
| 100/0 | 121/169 (71.60%) | 37/140(26.43%) | 0/8 (0%) |
| 100/10/20 | 149/203(73.39%) | 53/203(26.11%) | 1/41(2.44%) |
| 100/10/50 | 153/202(75.74%) | 46/202(22.77%) | 2/24(8.33%) |
Different combination of Cas9, sgRNA and donor plasmid were injected as shown. Cleavage and blastocyst formation rates of each groups are presented. OCT4 targeting efficiency was measured by PCR amplification and sequencing of single blastocysts. Statistical significance was tested using chi-square test.
Discussion
Recent advances in genome editing technologies, including ZFN, TALEN, and CRISPR/Cas9, enable precise gene targeting techniques including genetic knockout and knockin in many organisms, including domestic animals like cows [48] or pigs [49, 50].
In addition to the generation of knockout or knockin animals, CRISPR-mediated genome editing technology could be useful for functional characterization of genes involved in development [48], including pre-implantation development of mammals. However, no report has described the use of the CRISPR/Cas9 system for this purpose. To test the possibility of utilizing the CRISPR/Cas9 system in studying mammalian development, we chose porcine parthenogenetic embryos and the OCT4 gene as models, because in vitro fertilization of porcine embryo is technically demanding[51], therefore parthenogenetic porcine embryo would be ideal model systems to test efficiency of sgRNA/Cas9 pairs before generation of knockout animal.
Our results showed that introduction of a single sgRNA/Cas9 pair yielded robust efficiency (20–50%, depending on the guide sequence) in modification. As previously reported [52, 53], cleavage and mutation efficiency were affected by the guide sequences. Specifically, Doench et al. [52] suggested that nucleotides near PAM (NGG) sites, especially at the -1 and +1 base within the NGG, were the most important elements affecting cleavage efficiency, and A/G was a favorable base for cleavage at the -1 position, and G was unfavorable at the +1 position. For sgRNA 1, which targets exon 2 of OCT4, the guide sequence was 5´-AGAGAAGAGGATCACCCTGGGCCC-3´. The NGG, GGG, is underlined. There was a T at the -1 position and an A at the +1 position. For sgRNA 2, which targets exon 5 of OCT4, there was an A at the -1 position and a C at the +1 position. It is noteworthy that sgRNA 2 yielded a much higher mutation frequency (50%) compared to sgRNA 1 (20%), and sgRNA 1 had the unfavorable base T at the -1 position, while sgRNA 2 had an A at that position. We do not know that these difference solely caused the difference in mutation efficiency between the two sgRNAs, but recent results concerning active sgRNA design suggest that careful choices at sgRNA sites are crucial for efficient cleavage, mutation, and achievement of high knockout efficiency, especially for functional studies of genes involved in embryogenesis.
Previous functional studies of genes involved in early embryogenesis have been performed using RNA interference or morpholino antisense oligonucleotides. A drawback of these techniques is that they ablate expression of both maternal and zygotic genes. However, as shown in Fig. 1g, knockout of OCT4 by CRISPR/Cas9 did not affect maternal OCT4 mRNA levels, while OCT4 knockout only ablated embryonic gene expression. These characteristics of the CRISPR/Cas9 system would be useful to study genes involved in embryogenesis, especially those involved in zygotic genome activation.
In addition to simple knockout of a single locus, we showed that more sophisticated genome engineering, including large-scale genome deletion or knockin of eGFP at the OCT4 locus to generate an OCT4-EGFP fusion protein, would be possible with the CRISPR/Cas9 system. Fluorescence marker proteins for visualization of biological molecules and live cell imaging are now essential tools to elucidate protein function, especially those involved in development [54, 55]. However, generation of knockin embryos with fluorescence proteins has been cumbersome and time-consuming, and use of transient fluorescent protein fusion construct expression may lead to artifacts caused from overexpression of the fluorescence protein [33]. Considering these disadvantages, fluorescent tagging of endogenous genes by CRISPR-mediated knockin and imaging of tagged endogenous protein would facilitate imaging of embryogenesis-related genes during embryogenesis. But HDR-mediated integration efficiency of GFP locus in OCT4 gene is far lower than that of NHEJ-mediated knockout, as previously reported in mouse[25]. Enhancement of HDR-mediated knockin efficiency in mammalian would be essential for the generation of more sophisticated transgenesis. Recently reported techniques including NHEJ-mediated transgene integeration[56, 57] in zebrafish may be applicable in mammalian system.
In summary, we established a knockout and knockin system in porcine parthenogenetic embryos using the CRISPR/Cas9 system, and demonstrated that this system could facilitate investigation of genes involved in pre-implantation development or generation of knockin or knockout pigs.
Supporting Information
sgRNAs: OCT4 exon 2 and exon 5 targeting sgRNA primer; PCR sequencing: Target site mutation check primer; qRT-PCR: mRNA expression check primer.
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by a grant from the Next Generation Biogreen 21 Program (PJ011126), RDA, Republic of Korea. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. International Human Genome Sequencing C. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–45. [DOI] [PubMed] [Google Scholar]
- 2. Mouse Genome Sequencing C, Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–62. [DOI] [PubMed] [Google Scholar]
- 3. Bovine Genome S, Analysis C, Elsik CG, Tellam RL, Worley KC, Gibbs RA, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324(5926):522–8. 10.1126/science.1169588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Groenen MA, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491(7424):393–8. 10.1038/nature11622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51(3):503–12. [DOI] [PubMed] [Google Scholar]
- 6. Hacking DF. 'Knock, and it shall be opened': knocking out and knocking in to reveal mechanisms of disease and novel therapies. Early human development. 2008;84(12):821–7. 10.1016/j.earlhumdev.2008.09.011 [DOI] [PubMed] [Google Scholar]
- 7. Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(3):1156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–8. 10.1038/nbt.1755 [DOI] [PubMed] [Google Scholar]
- 9. Boch J. TALEs of genome targeting. Nat Biotechnol. 2011;29(2):135–6. 10.1038/nbt.1767 [DOI] [PubMed] [Google Scholar]
- 10. Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, et al. Targeted genome modification in mice using zinc-finger nucleases. Genetics. 2010;186(2):451–9. 10.1534/genetics.110.117002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, et al. Knockout mice created by TALEN-mediated gene targeting. Nature biotechnology. 2013;31(1):23–4. 10.1038/nbt.2477 [DOI] [PubMed] [Google Scholar]
- 12. Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, et al. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(29):12013–7. 10.1073/pnas.1106422108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, et al. Efficient TALEN-mediated gene knockout in livestock. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(43):17382–7. 10.1073/pnas.1211446109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nature reviews Genetics. 2010;11(9):636–46. 10.1038/nrg2842 [DOI] [PubMed] [Google Scholar]
- 15. Carlson DF, Fahrenkrug SC, Hackett PB. Targeting DNA With Fingers and TALENs. Molecular therapy Nucleic acids. 2012;1:e3 10.1038/mtna.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nature protocols. 2012;7(1):171–92. 10.1038/nprot.2011.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JA. Precise, correlated fluorescence microscopy and electron tomography of lowicryl sections using fluorescent fiducial markers. Methods in cell biology. 2012;111:235–57. 10.1016/B978-0-12-416026-2.00013-3 [DOI] [PubMed] [Google Scholar]
- 18. Schmid-Burgk JL, Schmidt T, Kaiser V, Honing K, Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nature biotechnology. 2013;31(1):76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Narbonne P, Miyamoto K, Gurdon JB. Reprogramming and development in nuclear transfer embryos and in interspecific systems. Current opinion in genetics & development. 2012;22(5):450–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lillico SG, Proudfoot C, Carlson DF, Stverakova D, Neil C, Blain C, et al. Live pigs produced from genome edited zygotes. Scientific reports. 2013;3:2847 10.1038/srep02847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–2. 10.1038/nbt.2507 [DOI] [PubMed] [Google Scholar]
- 24. Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–8. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–9. 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):681–3. 10.1038/nbt.2661 [DOI] [PubMed] [Google Scholar]
- 27. Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(34):13904–9. 10.1073/pnas.1308335110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–9. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cho SW, Lee J, Carroll D, Kim JS, Lee J. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics. 2013;195(3):1177–80. 10.1534/genetics.113.155853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo X, Zhang T, Hu Z, Zhang Y, Shi Z, Wang Q, et al. Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development. 2014;141(3):707–14. 10.1242/dev.099853 [DOI] [PubMed] [Google Scholar]
- 31. Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell research. 2014;24(3):372–5. 10.1038/cr.2014.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156(4):836–43. 10.1016/j.cell.2014.01.027 [DOI] [PubMed] [Google Scholar]
- 33. Harrison MM, Jenkins BV, O'Connor-Giles KM, Wildonger J. A CRISPR view of development. Genes & development. 2014;28(17):1859–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Svoboda P, Stein P, Hayashi H, Schultz RM. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development. 2000;127(19):4147–56. [DOI] [PubMed] [Google Scholar]
- 35. Kanzler B, Haas-Assenbaum A, Haas I, Morawiec L, Huber E, Boehm T. Morpholino oligonucleotide-triggered knockdown reveals a role for maternal E-cadherin during early mouse development. Mechanisms of development. 2003;120(12):1423–32. [DOI] [PubMed] [Google Scholar]
- 36. Brevini TA, Pennarossa G, Attanasio L, Vanelli A, Gasparrini B, Gandolfi F. Culture conditions and signalling networks promoting the establishment of cell lines from parthenogenetic and biparental pig embryos. Stem cell reviews. 2010;6(3):484–95. 10.1007/s12015-010-9153-2 [DOI] [PubMed] [Google Scholar]
- 37. Zheng Z, Zhao MH, Jia JL, Heo YT, Cui XS, Oh JS, et al. Knockdown of maternal homeobox transcription factor SEBOX gene impaired early embryonic development in porcine parthenotes. The Journal of reproduction and development. 2013;59(6):557–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou N, Cao Z, Wu R, Liu X, Tao J, Chen Z, et al. Dynamic changes of histone H3 lysine 27 acetylation in pre-implantational pig embryos derived from somatic cell nuclear transfer. Animal reproduction science. 2014;148(3–4):153–63. 10.1016/j.anireprosci.2014.06.023 [DOI] [PubMed] [Google Scholar]
- 39. Cao Z, Zhou N, Zhang Y, Zhang Y, Wu R, Li Y, et al. Dynamic reprogramming of 5-hydroxymethylcytosine during early porcine embryogenesis. Theriogenology. 2014;81(3):496–508. 10.1016/j.theriogenology.2013.10.025 [DOI] [PubMed] [Google Scholar]
- 40. Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95(3):379–91. [DOI] [PubMed] [Google Scholar]
- 41. Fujii W, Kawasaki K, Sugiura K, Naito K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic acids research. 2013;41(20):e187 10.1093/nar/gkt772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–32. 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods. 2009;6(5):343–5. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 44. Sakurai N, Fujii T, Hashizume T, Sawai K. Effects of downregulating oct-4 transcript by RNA interference on early development of porcine embryos. The Journal of reproduction and development. 2013;59(4):353–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hyttel P, Laurincik J, Rosenkranz C, Rath D, Niemann H, Ochs RL, et al. Nucleolar proteins and ultrastructure in preimplantation porcine embryos developed in vivo. Biology of reproduction. 2000;63(6):1848–56. [DOI] [PubMed] [Google Scholar]
- 46. Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, Rodriguez-Perales S. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nature communications. 2014;5:3964 10.1038/ncomms4964 [DOI] [PubMed] [Google Scholar]
- 47. Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nature communications. 2014;5:3728 10.1038/ncomms4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heo Y, Quan X, Xu Y, Baek S, Choi H, Kim N, et al. CRISPR/Cas9 nuclease-mediated gene knock-in in bovine pluripotent stem cells and embryos. Stem cells and development. 2014. [DOI] [PubMed] [Google Scholar]
- 49. Whitworth KM, Lee K, Benne JA, Beaton BP, Spate LD, Murphy SL, et al. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biology of reproduction. 2014;91(3):78 10.1095/biolreprod.114.121723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou X, Xin J, Fan N, Zou Q, Huang J, Ouyang Z, et al. Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cellular and molecular life sciences: CMLS. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Funahashi H. Polyspermic penetration in porcine IVM-IVF systems. Reproduction, fertility, and development. 2003;15(3):167–77. [DOI] [PubMed] [Google Scholar]
- 52. Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nature biotechnology. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bae S, Kweon J, Kim HS, Kim JS. Microhomology-based choice of Cas9 nuclease target sites. Nature methods. 2014;11(7):705–6. 10.1038/nmeth.3015 [DOI] [PubMed] [Google Scholar]
- 54. Landecker H. Seeing things: from microcinematography to live cell imaging. Nature methods. 2009;6(10):707–09. [DOI] [PubMed] [Google Scholar]
- 55. Schnell U, Dijk F, Sjollema KA, Giepmans BN. Immunolabeling artifacts and the need for live-cell imaging. Nature methods. 2012;9(2):152–8. 10.1038/nmeth.1855 [DOI] [PubMed] [Google Scholar]
- 56. Auer TO, Duroure K, Concordet JP, Del Bene F. CRISPR/Cas9-mediated conversion of eGFP- into Gal4-transgenic lines in zebrafish. Nature protocols. 2014;9(12):2823–40. 10.1038/nprot.2014.187 [DOI] [PubMed] [Google Scholar]
- 57. Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome research. 2014;24(1):142–53. 10.1101/gr.161638.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sgRNAs: OCT4 exon 2 and exon 5 targeting sgRNA primer; PCR sequencing: Target site mutation check primer; qRT-PCR: mRNA expression check primer.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.