Abstract
Light stimulates carotenoid biosynthesis in the ascomycete fungus Fusarium fujikuroi through transcriptional activation of the structural genes of the pathway carRA, carB, and cart, but the molecular basis of this photoresponse is unknown. The F. fujikuroi genome contains genes for different predicted photoreceptors, including the WC protein WcoA, the DASH cryptochrome CryD and the Vivid-like flavoprotein VvdA. We formerly found that null mutants of wcoA, cryD or vvdA exhibit carotenoid photoinduction under continuous illumination. Here we show that the wild type exhibits a biphasic response in light induction kinetics experiments, with a rapid increase in carotenoid content in the first hours, a transient arrest and a subsequent slower increase. The mutants of the three photoreceptors show different kinetic responses: the wcoA mutants are defective in the rapid response, the cryD mutants are affected in the slower response, while the fast and slow responses were respectively enhanced and attenuated in the vvdA mutants. Transcriptional analyses of the car genes revealed a strong reduction of dark and light-induced transcript levels in the wcoA mutants, while minor or no reductions were found in the cryD mutants. Formerly, we found no change on carRA and carB photoinduction in vvdA mutants. Taken together, our data suggest a cooperative participation of WcoA and CryD in early and late stages of photoinduction of carotenoid biosynthesis in F. fujikuroi, and a possible modulation of WcoA activity by VvdA. An unexpected transcriptional induction by red light of vvdA, cryD and carRA genes suggest the participation of an additional red light-absorbing photoreceptor.
Introduction
Filamentous fungi are a widespread group of lower eukaryotes able to grow in a large diversity of natural habitats, where they use light as a key environmental signal to modulate physiological and developmental processes. Some fungi stand out for their ability to produce a wide range of secondary metabolites [1]. Species of the genus Fusarium, an ubiquitous group of phytopathogenic fungi able to produce an extensive array of compounds, belong to this class of fungi [2]. A representative example is Fusarium fujikuroi, well known for its capacity to produce gibberellins (GAs), growth-promoting plant hormones with biotechnological applications [3]. This fungus produces many other compounds, including pigments that provide characteristic colors to their mycelia [4]. When grown in the light, F. fujikuroi colonies acquire a characteristic orange pigmentation due to the synthesis of carotenoids (see wild type in Fig. 1), with the xanthophyll neurosporaxanthin as major component [5]. The biosynthetic carotenoid pathway of this fungus has been investigated in detail, and all the enzymatic genes have been identified and characterized [6–9].
Fig 1. Colonies of the wild type and representative ΔwcoA (SF226), ΔcryD (SF236) and ΔvvdA (SF258) mutants.
The strains were grown for 7 days at 30°C in the dark or under continuous illumination on DGasn medium. The brownish color of the ΔwcoA mutants in the dark and the reddish color of the ΔcryD mutants in the light are due to the production of secondary metabolite pigments unrelated to carotenoids, such as bikaverins and other polyketides.
The regulation of carotenoid biosynthesis has been object of special attention in several fungi, including F. fujikuroi [10]. Light was found to induce the synthesis of carotenoids in different Fusarium species [11]. This photoinduction was first investigated in Fusarium aquaeductuum [12,13], and later described in F. fujikuroi [14], F. verticillioides [15] and F. oxysporum [16]. A second regulatory signal is nitrogen availability, which plays a central role in the control of secondary metabolism in this fungus [17–19], including a negative effect on the production of carotenoids [20]. The only regulatory gene identified affecting carotenoid biosynthesis in Fusarium is carS, whose mutation leads to transcriptional up-regulation of the structural car genes [20,21], and accumulation of a high amount of carotenoids under different culture conditions [20,22]. carS encodes a protein of the ring finger family [16,23], whose molecular mechanism of action remains to be established.
Photoinduction of carotenogenesis has been investigated in detail in Neurospora crassa, where it is mediated by the heterodimeric White Collar (WC) complex [24]. The WC complex is activated by light through the WC-1 flavin-binding LOV/PAS domain and binds upstream promoter sequences of light-regulated genes to induce their transcription [25]. Similar WC complexes control diverse photoresponses in other fungi [26], including photoinduction of β-carotene production in the mucorales P. blakesleeanus and M. circinelloides. Against the predictions, the mutational loss of the WC-1-like protein WcoA of F. fujikuroi [27] or Wc1 of F. oxysporum [28] did not impede the photoinduction of the carotenoid pathway in these species, indicating the participation of other photoreceptors. A detailed action spectrum for photocarotenogenesis in Fusarium aquaeductuum was consistent with the participation of a flavin-based photoreceptor [12]. Besides the WcoA/Wc1 protein, the Fusarium genomes contain genes for three other presumptive flavin photoreceptors with counterparts in N. crassa, a small VIVID-like protein, a photolyase, and a DASH-cryptochrome [11]. Additionally, the F. fujikuroi genome includes a gene coding for a cryptochrome/photolyase, cry1 (B. Tudzysnki, unpublished), with no ortholog in N. crassa.
Cryptochromes are blue light/UV-A photoreceptors possibly evolved from photolyases, able to bind two chromophores, flavin adenine dinucleotide (FAD) and pterin 5,10-methenyltetrahydrofolate (MTHF) [29,30]. Most cryptochromes have C-terminal extensions that are absent in photolyases, which play regulatory roles related to light control of growth, development, cell signaling or circadian rhythm in different taxonomic groups [30]. DASH-cryptochromes (abbreviated hereafter as cry-DASHs), a subgroup in this family, differ from cryptochromes in their ability to repair DNA lesions in single-stranded DNA [31] or loop-structures of duplex DNA [32]. A recent functional analysis of the F. fujikuroi cry-DASH gene cryD showed that it is strongly induced by light in the wild type but not in ΔwcoA disruption mutants [33]. Targeted ΔcryD mutants exhibited light-dependent alterations in the production of some secondary metabolites, as bikaverin and gibberellins, but they were not apparently affected in the synthesis of carotenoids.
Vivid-like photoreceptors are small flavin binding proteins represented by VVD (VIVID) of N. crassa and ENV1 (ENVOY) of Hypocrea jecorina. In N. crassa, VVD participates in photoadaptation of light-regulated genes [34,35], a function achieved through the modulation of the activity of the WC complex [36]. As a consequence, targeted Δvvd mutants exhibit a deeper pigmentation and accumulate more carotenoids under light than the wild type [37,38]. ENV1 plays a more relevant role in H. jecorina, as indicates the drastic effect of the env1 mutation on growth and morphology under light [39]. Additionally, ENV1 was found to participate in the regulation by light of cellulose degradation [39], sexual cycle [40], and photoadaptation of some light-induced genes [41]. Our recent investigation of the vvd gene of F. fujikuroi, vvdA, revealed significant differences in its function with VVD and ENV1: as found for cryD, vvdA expression is strongly induced by light in a WcoA-dependent manner, but the ΔvvdA mutants produced less carotenoids than the control strains and exhibited light-dependent alterations in conidiation and mycelia development [42]. However, expression analyses found no alterations in photoinduction or photoadaptation of the car genes.
Here we report on the role of the WcoA, CryD and VvdA photoreceptors in the regulation by light of carotenoid biosynthesis in F. fujikuroi. We found that this light response has two separate components in this fungus, a fast response in the first hours of illumination and a slower response upon more prolonged light exposure. The kinetics of the photoresponse in mutants for these putative photosensors indicates that WcoA is the major photoreceptor in F. fujikuroi carotenogenesis, and that CryD mediates a slower and less sensitive photoresponse, which explains the photoinduction exhibited by the ΔwcoA mutants under continuous illumination. The kinetics of the response is also altered by the loss of VvdA, which seems to play negative and positive effects on WcoA and CryD functions, respectively. Our results represent the first report on the participation of a cryptochrome in the regulation of carotenoid biosynthesis in any microorganism, and point to a complex organization of the light-detection machinery that controls carotenoid biosynthesis in Fusarium.
Results
Effect of wcoA, cryD, and vvdA mutations on regulation of carotenogenesis under constant illumination
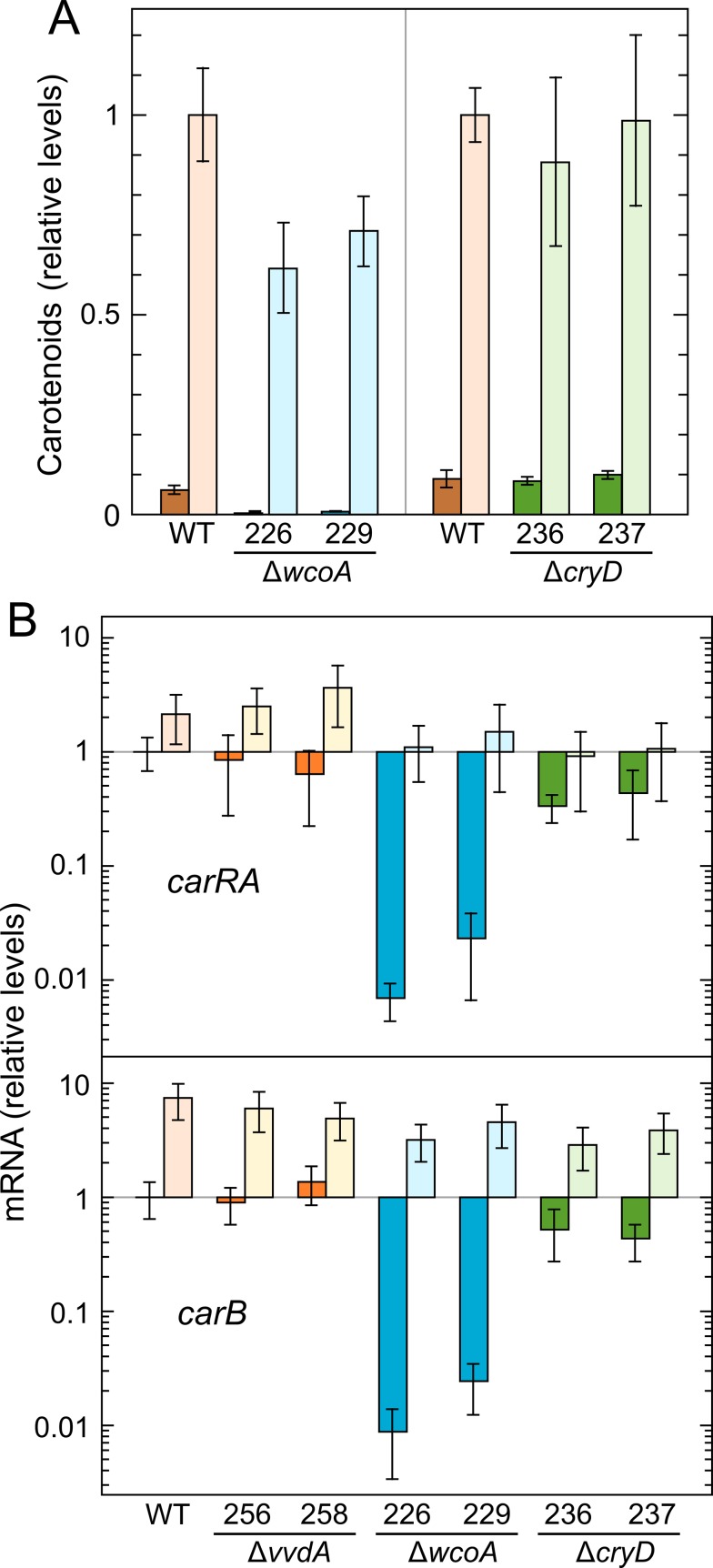
The mutants of the genes for the predicted photoreceptors WcoA, CryD and VvdA exhibit differences in pigmentation and morphology (7-day old cultures shown in Fig. 1). Our former studies on these mutants included carotenoid analyses that showed the persistence of photoinduction in all the tested strains. Thus, ΔvvdA mutants accumulated in the light about 55–60% of the carotenoids produced by the wild type, while the synthesis in the dark was unaffected [42]. On the other hand, the ΔwcoA and ΔcryD mutants accumulated similar amounts of carotenoids in the light compared to the wild type [27,33]. However, in the case of the ΔwcoA mutants the experiment was done under different culture conditions and carotenoid levels in the dark were not determined. For more reliable comparison, we performed new carotenoid analyses of the ΔwcoA and ΔcryD mutants under the same conditions, 7-day incubations in the dark or under constant illumination (Fig. 2A). As recently found for the ΔvvdA strains [42], the ΔwcoA colonies produced less carotenoids than those of the wild type, albeit they maintained a clear photoinduction. Interestingly, the amounts of carotenoids in the dark in the ΔwcoA mutants were remarkably low compared to those of the wild type, suggesting a regulatory role for WcoA in the dark. However, in agreement with former observations [33], the total amount of carotenoids in the ΔcryD mutants remained very similar to those of the wild type, either in the dark or in the light.
Fig 2. Effect of the mutation of the photoreceptor genes cryD, wcoA and vvdA on carotenogenesis in the dark versus constant illumination.
A: Total concentration of carotenoids in the wild type and in parallel cultures of the ΔwcoA and ΔcryD mutants grown for 7 days at 30°C on DGasn medium in the dark or under continuous illumination. Data are the means and standard deviations of four determinations from two biological replicates, taking as 1 the value of the wild type under light. Data for ΔvvdA under the same culture conditions were recently published [42]. B: Real-time RT-PCR analyses of the genes carRA and carB in RNA samples of the wild-type and the indicated strains grown for 7 days at 30°C on DGasn medium in the dark or under continuous illumination. Relative expression for each gene was referred to the value in the wild type grown in the dark. Data are the means and standard deviations of six determinations from two biological replicates. Here, and in following figures, the wild type and the mutants for each gene are represented by different colors (brown, wild type; reddish-orange, ΔvvdA; blue, ΔwcoA; green, ΔcryD). Lighter versions of the colors are used for illuminated samples. Statistical analysis for differences between data of wild type and mutants are displayed in S1 Table.
To correlate these data with the expression of the genes of the carotenoid pathway, we determined the carRA and carB mRNA levels under the same culture conditions (Fig. 2B). The results showed a significant down-regulation of carRA and carB transcripts in the dark in the ΔwcoA mutants, consistent with their lower carotenoid content in vivo. A significant carB photoinduction, ranging from 3- to 7-fold compared to wild-type dark levels, was observed in all the strains tested. In the case of carRA, the increase was hardly detectable in the wild-type and ΔvvdA strains (2 to 3-fold), and inexistent in the ΔwcoA and ΔcryD mutants. However, because of their lower mRNA levels in the dark, carRA photoinduction was also manifest in these strains, most especially in the ΔwcoA mutants. In summary these experiments indicate the occurrence of light responses of carRA and carB expression in the mutants for the three photoreceptors, with differences in their dark and light levels.
Effect of ΔwcoA, ΔcryD, and ΔvvdA mutations on kinetics of carotenoid accumulation upon illumination of dark-grown mycelia
The lower amounts of carotenoids in the light in the ΔwcoA and ΔvvdA mutants and the differences observed in carRA and carB mRNA levels in the ΔwcoA and ΔcryD mutants led us to investigate in more detail the responses of these strains to light. The wild type and the mutants for each presumed photoreceptor were incubated in parallel for three days in the dark, and illuminated afterwards up to 48 hours. Time-course of light-induced carotenoid accumulation was determined for each strain (Fig. 3A). Since the colonies grow further during illumination, the analysis was restricted to the central part of the colonies, i.e., the mycelium formed during the three days in the dark (see Materials and Methods). The results showed a biphasic response for the wild-type strain, with a rapid increase in the carotenoid content in the first 6 hours (first stage) followed by an arrest of the synthesis (6–12 h), and its resumption after a longer light exposure (12–48 h, second stage). The mutants for the photoreceptor genes showed different alterations of this pattern. Thus, the ΔcryD mutants responded to light as fast as the wild type in the first stage and paused the synthesis at the same time, but they exhibited a slower accumulation of carotenoids in the second stage. On the other hand, the response of the ΔvvdA mutants was faster in the first stage and slower in the second stage, but not as slow as in the ΔcryD mutants. In contrast with the former strains, the ΔwcoA mutants exhibited a much slower monophasic response, resulting after 48 h illumination in about 1/3 of the carotenoids accumulated by the wild type. In congruence with the data displayed in Fig. 2B the carotenoid content of the ΔwcoA mutants was much lower than those of the other strains at the start of illumination; so, paradoxically, the induction by light was proportionally higher in the ΔwcoA strains.
Fig 3. Light-induced carotenogenesis in wild type and mutants of the photoreceptor genes cryD, wcoA and vvdA.
A: Kinetics of carotenoid accumulation after illumination of the wild type, ΔwcoA mutants SF226 and SF229, ΔcryD mutants SF236 and SF237, and ΔvvdA mutants SF256 and SF258. The strains were incubated for three days in the dark on DGasn agar and exposed to white light for the time indicated in abscissae. Each point is the mean and standard deviation of four determinations from two biological replicates. In this and in other figures, overlapping data were separated for better visualization. B: Real-time RT-PCR analyses of the genes carRA (pale bars) and carB (dark bars) in RNA samples of the same strains under the same culture conditions after 48 h illumination. Relative expression for each gene was referred to the value in the wild type grown in the dark. Data are the means and standard deviations of six determinations from two biological replicates.
To gain more insights on the alterations of carotenoid photoinduction in the mutants, we determined the carRA and carB mRNA levels in the mycelia of the experiments described above after 48h illumination and the data were referred to the value of the wild-type strain in the dark (Fig. 3B). The mutants for the three photoreceptor genes exhibited a significant photoinduction of the transcript levels. However, the amounts were about 4-fold lower in the ΔcryD and ΔvvdA mutants and about 10-fold lower in the ΔwcoA mutants compared to those of the wild type. These data are consistent with the lower carotenoid accumulation exhibited by the different mutants in the later stage of the photoinduction kinetics.
Since cryD is transcriptionally photoinduced by WcoA [33], we checked the effect of expressing cryD in the ΔwcoA mutant in a WcoA-independent manner (S1 Fig.). For this purpose, the ΔwcoA mutant SF226 was transformed with a plasmid containing a tagged cryD ORF under control of the gpdA promoter of A. nidulans. After molecular analysis of several transformants, one strain was identified with the expected plasmid sequences (S1A Fig.) and exhibiting a strong cryD expression, with ca. 50-fold higher transcript levels in the dark and similar levels after one hour of illumination compared to the ΔwcoA control (S1B Fig.). The cryD-overexpressing strain exhibited a similar monophasic time-course of light-induced carotenoid accumulation as the control ΔwcoA mutant, but it accumulated about 70% more carotenoids (S1C Fig.), confirming the participation of CryD in photocarotenogenesis.
The results described above are consistent with a cooperative participation of the WcoA and CryD photoreceptors in the photoinduction of carotenogenesis in F. fujikuroi. To confirm this hypothesis, experiments were carried out to obtain a double ΔwcoA ΔcryD mutant. Two plasmids were constructed to obtain the targeted disruption of wcoA by homologous recombination in a ΔcryD background. In one of them the wcoA coding sequence was replaced by the nitrate reductase gene niaD, and in the other it was replaced by a geneticin resistance cassette. For niaD selection, a niaD –mutant was obtained from the ΔcryD mutant SF237 by chlorate resistance. After three transformation attempts of the niaD - SF237 derivative, none of the transformants obtained either through protoplasts (39 niaD + colonies tested from two independent transformations) or through a biolistc approach (7 niaD + colonies tested from one transformation), exhibited the expected ΔwcoA phenotypic pattern, i.e., purple pigmentation in the dark and reduced mycelial hydrophobicity, and PCR tests confirmed the presence of an intact wcoA gene. Five additional protoplast-mediated transformation experiments achieved upon geneticin resistance selection, in this case using SF236, led to the isolation of 16 transformants. All of them resulted from ectopic plasmid integrations, and no wcoA mutants were obtained. As an alternative approach, mutagenesis experiments were done with SF236 and SF237 and a total of 15 mutants with a ΔwcoA-like pigmentation were analyzed. The wcoA alleles from four mutants, those exhibiting a phenotype more similar to that of the wcoA mutant SF226, were cloned and sequenced, but no mutations were found in their respective wcoA coding sequences. Taken together, these results suggest a lack of viability for the double ΔwcoA ΔcryD mutant.
Effect of light intensity on photoinduction of carotenogenesis in ΔwcoA, ΔcryD, and ΔvvdA mutants
The predicted photoreceptors under investigation may differ in their sensitivities to light. To obtain more information on the alterations resulting from the loss of each photoreceptor we used neutral filters to reduce the light intensity in the photoinduction kinetics experiments. To distinguish the effects on the fast and slow stages of the photoresponse (Fig. 3A), we determined the carotenoid content in the mycelia in the dark and after 6 h or 48 h illumination under 0.07, 0.7 and 7 W m-2 (Fig. 4 and S2 Fig.). The results confirmed the different responses of the mutants after the first and second kinetics stages. After 6 h illumination, the ΔcryD mutants contained similar amounts of carotenoids than those found in the wild type while the ΔwcoA mutants produced lower amounts. On the other hand, after 48 h the ΔcryD mutants produced less carotenoids than the wild type, but their levels were similar to those of the ΔwcoA mutants. Also, the carotenoid content of the ΔvvdA strains was higher than those of the wild type after 6 h, and lower after 48h, indicating that these mutants accumulate less carotenoids than the wild type upon prolonged incubation, as found in the cultures under constant illumination [42].
Fig 4. Effect of light intensity on photoinduction of carotenoid accumulation in the wild type and ΔwcoA mutants SF226 and SF229, ΔcryD mutants SF236 and SF237, and ΔvvdA mutants SF256 and SF258.
The strains were incubated for three days in the dark in DGasn medium (left bars, darker colors) and exposed to 0.07 W m-2, 0.7 W m-2 or 7 W m-2 (brighter colors from left to right) of white light for 6 hours (above) and 48 h (below). Data show means and standard deviations of four determinations from two biological replicates. Statistical analysis for differences between data of wild type and mutants are displayed in S1 Table.
The results were consistent with differences in light sensitivity between different photoreceptors. No significant photoinduction was found in the ΔwcoA mutants under 0.07 and 0.7 W m-2, indicating that CryD is able to respond only to high light intensities. However, the ΔcryD mutants showed a significant photoinduction even at the lower light intensity, consistent with a high sensitivity to light of the WcoA photoreceptor. Interestingly, after 6 h illumination the response to low or very low light intensity was enhanced in the ΔvvdA mutants compared to the wild type. Thus, these mutants accumulated more carotenoids after this time under 0.7 W m-2 than the wild type under 7 W m-2.
To test if the function of VvdA is performed on transcription of the other photoreceptor genes, we determined the effect of the ΔvvdA mutation on cryD and wcoA mRNA levels. For more complete information, in this case the analysis was done with three independent ΔvvdA mutants (Fig. 5). The results confirmed the strong photoinduction of cryD mRNA, as formerly reported [33]. However, the cryD mRNA levels were visibly enhanced in the ΔvvdA strains compared to the wild type. As already found for carRA and carB [33], the ΔvvdA mutants exhibited a similar photoadaptation of cryD mRNA levels compared to wild type. On the other hand, a minor photoinduction was found for wcoA in the same samples. This induction, which was disregarded in former analysis [27], was basically unaffected in the vvdA mutants.
Fig 5. Effect of the vvdA mutation on expression of the photoreceptor genes cryD and wcoA.
Real-time RT-PCR analyses of genes cryD and wcoA in total RNA samples from the wild type (brown squares) and the ΔvvdA mutants SF256 (red circles), SF257 (red rhombs) and SF258 (red triangles) grown for three days in DGasn medium in the dark or after 15 min, 30 min, 1 h, 2 h, or 4 h exposure to 7 W m-2 white light. Relative levels are referred to the value of the wild type in the dark. Data show means and standard deviations for nine measurements from three biological replicates. Statistical analysis for differences between data of wild type and mutants are displayed in S1 Table.
Effect of the mutation of the wcoA and cryD genes on expression of photoregulated genes
Recently, it was shown that the mutation of the vvdA gene does not affect the photoinduction pattern of the structural genes carRA and carB [42]. For better understanding of the phenotypic effects of the ΔwcoA and ΔcryD mutations on the photoinduction kinetics of carotenoid accumulation, we investigated the effect of light on carRA, carB and carT mRNA levels in these mutants in comparison to the wild type. Formerly, it was reported that the carB gene was down-regulated in wcoA mutants, while the result was less clear for carRA [27]. We have analyzed in more detail two ΔwcoA mutants, extending the analysis up to 4 h of illumination (Fig. 6). As expected, the RT-PCR data for the wild type showed a strong photoinduction of carRA and carB transcript levels, exceeding one hundred fold the levels in the dark after one hour, and decaying in the following hours. A more modest induction was exhibited by carT, which did not reach a tenfold increase, returning afterwards to background levels. In agreement with the lower carotenoid content of the ΔwcoA mutants in the dark (Figs. 2 and 3), the car mRNA levels were much lower in the dark in these strains than in the wild type. The photoinduction was also very low, with carRA, carB and carT transcript levels still much lower in the wcoA mutants after illumination than in the wild type in the dark. These results fit the slow accumulation of carotenoids by the ΔwcoA mutants in the kinetics experiments (Fig. 3A), which could be explained by a slow up-regulating activity of CryD.
Fig 6. Effect of the wcoA mutation on expression of genes of the carotenoid pathway.
Real-time RT-PCR analyses of genes carB, carRA and carT in total RNA samples from the wild type (brown squares) and the ΔwcoA mutants SF226 (blue circles) and SF229 (blue triangles) grown for three days in DGasn medium in the dark or after 15 min, 30 min, 1 h, 2 h, or 4 h exposure to 7 W m-2 white light. Relative levels are referred to the value of the wild type in the dark. Data show means and standard deviations for nine measurements from three biological replicates.
In contrast to the ΔwcoA mutants, the expression of the genes carRA and carB was strongly photoinduced in the ΔcryD mutants. However, there was a decrease of at least five-fold in the photoinduction levels of carRA (Fig. 7), but this effect was less manifest on carB and hardly detectable for carT, that only reached a 4-fold induction in the wild type in this set of experiments. As found for carB, the ΔcryD mutation produced minor or no effects in the photoinduction of other light-regulated genes, such as carO, carX and phr1, encoding a rhodopsin protein [21], a retinal-producing carotenoid oxygenase [8], and a CPD-photolyase [43], respectively. On the other hand, no photoinduction was found for carRA and carB in the ΔwcoA mutant overexpressing cryD in the dark (S1C Fig.). Taken together, the results indicate that CryD does not play an important role in the transcriptional regulation of light-induced genes in F. fujikuroi.
Fig 7. Effect of the cryD mutation on expression of genes of the carotenoid pathway and other light-regulated genes of F. fujikuroi.
Real-time RT-PCR analyses of genes carB, carRA, carT, carO, carX and phr1 in total RNA samples from the wild type (brown squares) and the ΔcryD mutants SF236 (green circles) and SF237 (green triangles) grown for three days in DGasn medium in the dark or after 15 min, 30 min, 1 h, 2 h, or 4 h exposure to 7 W m-2 white light. Relative levels are referred to the value of the wild type in the dark. Data show means and standard deviations for nine measurements from three biological replicates. Statistical analysis for differences between data of wild type and mutants are displayed in S1 Table. For better comparison, mean values for mRNA levels of genes carB, carRA, carO and carX in wild type and mutants at the point of maximal expression (one hour of illumination) are indicated in the figure.
Effect of blue and red light on expression of carRA and photoreceptor genes
The ΔcryD mutants exhibit light-dependent alterations in morphology and pigmentation. However, these alterations were not observed under red light [33]. CryD, WcoA and VvdA are predicted blue-light photoreceptors. To check the dependence of F. fujikuroi photocarotenogenesis on light wavelength, we compared the effect of blue, red and white lights on transcript levels of the structural gene carRA, and on those of the photoreceptor genes cryD, wcoA and vvdA (Fig. 8). As expected blue light was as efficient as white light to induce the expression of the gene carRA; however, red light produced a significant photoinduction, with transcript levels reaching about 10% of the induction produced by white or blue light. A similar pattern was observed for the gene vvdA, with blue light giving the higher efficiency. Surprisingly, red light was particularly efficient to induce cryD mRNA levels, being as efficient as white light during the first 30 min of induction. In contrast, the minor induction of wcoA was less apparent either with blue or red light. Congruently with these results, the mycelia incubated for one week under red light contained about 54 μg g-1 dry weight compared to 7 μg g-1 dry weight in the dark or 197 μg g-1 dry weight under white light. Taken together, the data suggest the participation of a red-light photoreceptor in the transcriptional regulation of the genes for carotenogenesis and in those for the VvdA and CryD photoreceptors.
Fig 8. Effect of light wavelength on photoinduced expression of the photoreceptor genes cryD, wcoA and vvdA, and the carotenogenic gene carRA.
Real-time RT-PCR analyses of genes cryD, wcoA, vvdA and carRA in total RNA samples from the wild type grown for three days in DGasn medium in the dark or after 15 min, 30 min, 1 h, 2 h, or 4 h exposure to white light (black symbols), blue light (blue symbols) or red light (red symbols). Relative levels are referred to the value of the wild type in the dark. Data show means and standard deviations for six measurements from two biological replicates. The inset shows the transmittance spectra of the uncolored, blue and red filters used for illumination in this experiment (adapted from [33]).
Discussion
Induction of carotenogenesis by light is a well-known photoresponse in some filamentous fungi. Former data indicated that the regulatory mechanism for this photoinduction in F. fujikuroi differs from that of other fungal models, such as N. crassa, P. blakesleeanus and M. circinelloides, where photocarotenogenesis is mediated by a White Collar complex (reviewed by [44]). The persistence of photoinduction of carotenogenesis in the ΔwcoA mutants of F. fujikuroi [27] led us to investigate other predicted blue-light photoreceptors, as the cry-DASH CryD and the small flavoprotein VvdA. The loss of WcoA has a drastic effect on the ability of the car genes to respond to light, a result that contrast with the persistence of photocarotenogenesis in these mutants under constant illumination. Moreover, the down-regulatory effect of the ΔwcoA mutation is also noticeable in the dark, fitting former observations on light-independent functions for this protein [27]. The impact of the ΔwcoA mutation on carRA and carB expression was more drastic than in our former analyses [27]. This difference may be attributed to the higher resolution of the methodology used in this work, that led also to detect higher photoinduction levels in the wild type.
The drastic transcriptional effect of the wcoA mutation clearly differs from the milder effect of the cryD mutation, that produced an appreciable reduction in the mRNA levels of the gene carRA, but it did not prevent its photoinduction. This gene plays a key role in carotenoid biosynthesis, since it encodes the enzyme required for the synthesis of phytoene, the uncolored precursor of all carotenoids. However, the mutants lacking a functional CryD basically conserved the induction pattern of other light-regulated genes, including carB, and accumulated similar amounts of carotenoids than the wild type upon continuous illumination. The persistence of light-induced carotenogenesis in the ΔcryD and ΔwcoA mutants indicates that neither WcoA nor CryD act as single photoreceptors for this response.
For a better understanding of the photoinduction mechanism, we investigated the kinetics of carotenoid accumulation resulting from the illumination of young dark-grown colonies from the wild type and mutants of three different photoreceptors. The ΔwcoA and ΔcryD mutants exhibited alterations in the production of bikaverin [27,33], a red pigment that could interfere with the absorption of blue light by the putative photoreceptors, but the young colonies used in our experiments contain low concentrations of these pigments. These experimental conditions led us to identify two different induction stages: a rapid response achieved during the first 6 hours, and a subsequent slower response holding at least for 36 h. Former experiments with submerged cultures of F. fujikuroi showed simple induction kinetics, with a significant increase of the carotenoid content after 3 h illumination and the reach of maximum levels after 24 h [14]. Similar induction curves were described in submerged conditions for F. aquaeductuum [12] and N. crassa [45], although a significant photoinduction was detected in the latter after one-hour light exposure. A more detailed analysis revealed however a two-step kinetics response in N. crassa. Under similar light intensities to those used in our experiment (8 W m-2), a first induction step occurred in this fungus during the first minute of illumination [46], a result consistent with a rapid activity stimulation of an enzymatic set already available in the dark. A simple photoinduction curve of carotene accumulation was also observed in P. blakesleeanus, but fluence-response curves achieved with different wavelengths revealed a two-step response, suggesting the participation of two photosystems of different sensitivity thresholds [47]. The single mutants of the WC complex MadA/MadB still exhibit photoinduction, but the double mutants become blind [48], indicating that this WC complex is responsible for photocarotenogenesis in this fungus.
The alteration in the kinetics curves of carotenoid accumulation in mutants devoid of functional WcoA, CryD or VvdA proteins provide valuable clues on the participation of these predicted photoreceptors in carotenoid photoinduction in F. fujikuroi. The results are consistent with the participation of WcoA in a highly sensitive photoinduction mechanism, fast but transitory, while CryD is involved in a less sensitive but more sustained light-mediated stimulation. The first mechanism could be involved in the rapid response to sudden changes of light conditions and its subsequent adaption, as indicates the transience of the transcriptional photoinduction of the structural car genes. On the other hand, the latter mechanism could be involved in the response to persistent day-light illumination conditions. Both mechanisms seem to operate simultaneously under long standing illumination, as indicates the low but detectable increase in carB mRNA levels of the car genes in 7-day-old colonies of either the wild type or the ΔwcoA, ΔcryD or ΔvvdA mutants incubated under constant light. The low mRNA levels under these conditions suggest the operation of an efficient long-standing adaptation mechanism, in which VvdA apparently does not participate.
The phenotypic effect of the cryD mutation on carotenoid accumulation in F. fujikuroi is the first report on the participation of a cryptochrome on light regulation of carotenogenesis in any microorganism. The conservation of high photoinduction levels for the car genes in the ΔcryD mutants suggests that this regulation is achieved mostly through a non-transcriptional mechanism. The ability of cry-DASHs to bind nucleic acids [30], and the presence in the CryD C-terminal extension (CryD sequence: GenBank HE650104) of several RGG motifs (M. Castrillo et al., manuscript in preparation), formerly associated to RNA binding [49], is suggestive of a possible role for CryD promoting the stability or the translation of mRNAs for the car genes. Thus, a lower stability could explain the minor reductions observed in photoinduced carRA or carB mRNA levels in the ΔcryD mutants in Fig. 7. A similar mechanism could explain the lack of correlation between bikaverin biosynthesis and mRNA levels for the structural genes of the bikaverin pathway in the ΔcryD mutants grown in the light [33]. In contrast, the regulation of carotenogenesis by light in N. crassa appears to be under exclusive control of the WC complex, and no effect on this photoresponse was reported for the mutant of the cryD ortholog cry-1 [50]. Another cry-DASH protein, Cry1, was investigated in Sclerotinia sclerotium. In this fungus, cry1 transcripts are found at higher levels in light-exposed stages of apothecia development [51], but the cry1 mutation has only a minor effect in this developmental process. Interestingly, S. sclerotium produces β-carotene and its production is induced by light coupled to sclerotia formation [52]. However, the consequences of the cry1 mutation on light-induced β-carotene production in this fungus have not been reported.
The results support the participation of VvdA in the regulation of photocarotenogenesis in F. fujikuroi. As already mentioned, the vvdA mutants accumulate less carotenoids than the wild type under constant illumination [42]. As the wild type, the ΔvvdA mutants exhibit a biphasic photoresponse; however, the response is enhanced in its first stage and it is attenuated in the second stage. Moreover, the ΔvvdA mutants become more sensitive than the wild type under low light intensity. This result would be consistent with the function of VvdA as a negative modulator of WcoA activity, a regulatory scenario that reminds the down-regulation of WC-1 by VVD in N. crassa [36]. This is apparently contradicted by the lack of effect of the ΔvvdA mutation on the photoinduction of carRA and carB levels [42]. However, the latter experiments were achieved with submerged cultures while the carotenoid kinetics experiments were achieved on surface colonies. Differences in VvdA levels in the dark between both culture conditions might explain this discrepancy.
The slower response of the ΔvvdA mutants in the second stage of the induction kinetics could be explained by the interaction of VvdA with the WC complex or by a positive regulatory effect on CryD activity. In support of the first hypothesis, upon illumination VVD inactivates the WC complex in N. crassa forming a stable VVD-WCC heterodimer, which at the same time protects the light-induced WC complex from degradation [42]. It is very likely that a similar interaction occurs in F. fujikuroi, allowing the persistence of WC proteins that could eventually get free from VvdA to allow a slower photocarotenogenesis. Therefore, the light-activated WC complex would not be restrained by VvdA in the ΔvvdA mutant, leading to an enhanced carotenoid photoinduction in an early stage, but it could be more rapidly degraded afterwards, explaining the lower synthesis in a later stage. Taken together, the phenotypic effects of the vvdA mutation suggest that the participation of VvdA in carotenoid photoinduction is mainly achieved interfering with WcoA function, albeit a possible role on CryD function is not discarded.
In conclusion, the data provided in this work suggest a global model for carotenoid photoinduction in F. fujikuroi (Fig. 9). In this model, WcoA modulates the synthesis of carotenoids in the dark allowing a basal transcription rate for the structural genes, represented here by carRA and carB. A sudden light stimulus produces a rapid response in which WcoA is activated and promotes the expression of the car genes, together with those of cryD and vvdA. The activation of the car genes results in a rapid carotenoid accumulation. These WcoA functions are probably achieved as a WC complex with the WC-2-like partner WcoB, not represented in the scheme. However, this scenario changes with the persistence of the light stimulus in the following hours, during which light-activated VvdA restrains WcoA activity and light-activated CryD up-regulates carB and carRA, seemingly through a non-transcriptional mechanism rather than at transcription level. The coordinated participation of both photoreceptors explains the slow induction of carotenoids after illumination in the absence of WcoA and the slower response after prolonged illumination in the absence of CryD.
Fig 9. A hypothetic model for regulation of carotenogenesis by light in F. fujikuroi.
Circles correspond to proteins and squares to genes. The sizes of the circles represent low or high protein amounts in the corresponding conditions. In the absence of information on PhyA regulation, the same size is used for this protein. Non-activated proteins are indicated in pale colors with black letters and light-activated proteins are indicated in dark colors with white letters. The thickness of the arrows roughly represents predicted intensity of the regulatory effects, while positive and negative signs (arrows and truncated lines) represent inducing or repressing effects. Effect of VvdA could be achieved by protein/protein interactions, while CryD would up-regulate the car genes post-transcriptionally. The final effect on carotenoid biosynthesis is indicated below.
Upon sustained light exposure, VvdA keeps WcoA partially inactivated. However, WcoA would still mediate a certain transcriptional photoinduction in later stages, since carotenoid production is not totally prevented in ΔcryD mutants during long-term illumination. VvdA could play a role under these conditions preventing the inactivated WC complexes from degradation, thus contributing to a more persistent but attenuated photoinduction. Alternatively, VvdA could have a positive influence on CryD activity by an unknown molecular mechanism. Finally, the data suggest the participation of the only phytochrome found in the Fusarium proteomes [11], whose F. fujikuroi ortholog is called here PhyA (GenBank CCT69959). The activation of carRA transcripts by red light requires a functional WcoA, as indicates the drastic effect of the ΔwcoA mutation under white light, which includes red wavelengths. Tentatively, PhyA could act as an accessory red-light absorbing pigment that would transfer the signal to the putative WcoA/WcoB complex.
The regulatory system in F. fujikuroi reminds that of N. crassa in that a WC protein, susceptible to be activated by light and modulated by a VVD protein, is the only specific transcriptional activator that controls the expression of the structural genes of the carotenoid pathway. However, F. fujikuroi exhibits a more sophisticated regulation with some distinctive features: WcoA mediates a basal transcription level in the dark, allowing a detectable carotenoid biosynthesis under these conditions, and other photoreceptors add to WcoA to stimulate the synthesis in response to light, as CryD and probably also PhyA. This phytochrome could account for a supplementary response to red light, but its participation is just speculative and remains to be experimentally demonstrated in future works.
Materials and Methods
Strains and culture conditions
Fusarium fujikuroi FKMC1995 (Kansas State University Collection, Manhattan, KS, USA) was used as the wild-type strain. F. fujikuroi mutants for the wcoA, cryD and vvdA genes were previously described: wcoA mutants (SF226 and SF229), referred as ΔwcoA, were generated by interruption of the wcoA coding region with a hygR resistance cassette [27]. ΔcryD mutants (SF236 and SF237) and ΔvvdA mutants (SF256, SF257 and SF258) were obtained by replacement of the 5’ coding sequence [33] or the whole coding sequence [42], respectively, with the same hygR cassette. A strain constitutively expressing cryD was generated in this work and described in a later section.
DG minimal medium, that contains 3 g l-1 of NaNO3 as nitrogen source, has been previously described [53]. Unless otherwise stated, the strains were grown in DGasn medium, with the same composition of DG minimal medium except that the 3 g l-1 of NaNO3 were replaced by 3 g l-1 asparagine. Agar cultures were inoculated with sterile toothpicks in seven symmetrical positions on the agar surface, and liquid cultures were inoculated with 106 fresh conidia. To obtain conidia, the strains were incubated on solid medium for 7 days at 22°C under indirect illumination. Conidia were collected by washing the agar surface with water; afterwards they were separated from mycelial debris by filtration, washed with water by centrifugation and counted in a hemocytometer (Bürker chamber, Blau Brand, Germany).
Illumination experiments were achieved under four fluorescent tubes Philips TL-D 18W/840, placed at a distance of ca. 60 cm. For carotenoid analysis the strains were grown at 30°C in darkness or under 7 W m-2 white light for the times indicated. For carotenoid kinetics assays the strains were grown on solid medium for 3 days in complete darkness at 30°C and exposed afterwards for 3, 6, 12, 24 or 48 h to 7 W m-2 white light, or for 6 or 48 h to 7 W m-2, 0.7 W m-2 or 0.07 W m-2 white light. Reduction of light intensities was achieved with neutral grey plastic filters, whose transmittance was determined spectrophotometrically. Since the colonies keep expanding after light onset, mycelial samples were restricted to the dark-grown colony areas, avoiding colony borders grown under continuous illumination. For this purpose, the border of the colonies after the 3-day dark incubation was marked on the back of the Petri dishes and mycelia out of the delimited area, grown during light exposure, was removed with a blade.
For light induction analyses of gene expression, the strains were grown in 15 Ø cm Petri dishes containing 80 ml of liquid medium. The dishes were incubated for 3 days at 30°C in darkness and illuminated afterwards with 7 W m-2 white light for 15, 30, 60, 120 and 240 minutes. When indicated, the light was filtered through transparent, blue or red cellophane sheets in appropriately sealed boxes, resulting in 6 W m-2, 1.5 W m-2, and 1.7 W m-2 respectively [33]. Expression analyses were also performed with mycelia from 7-day-old colonies grown either in the dark or under continuous illumination, or from colonies grown in the dark and illuminated with 7 W m-2 white light for 48 h.
RT-PCR expression analyses
For expression assays in liquid media, the mycelial layer growing on the bottom of the plate was removed with a rod, immediately frozen in liquid nitrogen and stored at -80°C. For 7-day incubation experiments on solid medium, samples were collected from the surface of the colony with a sterile blade and immediately frozen in liquid nitrogen and stored at -80°C.
RNA was extracted with the RNeasy Plant Mini Kit (Qiagen, Chatsworth, CA, USA) and concentration was estimated with a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). To avoid possible DNA contaminations, 2.5 μg RNA samples were treated with 10 u of DNase I (Affymetrix USB products, Santa Clara, CA, USA) for 15 min at 25°C, followed by 10 minutes of inactivation at 65°C. DNAse I-treated RNA samples were converted to cDNA with the Transcriptor First Strand cDNA synthesis Kit (Roche, Mannheim, Germany) and final concentrations were set to 25 ng μl-1. Quantitative expression analyses were performed on cDNA samples in a Lightcycler 480 equipment (Roche). For amplification and detection, LightCycler 480 SYBR Green I Master (Roche) was used following manufacturer reaction protocol. The genes analyzed in these studies and the corresponding forward and reverse primers used in the RT-PCR amplifications were carB (5’-TCGGTGTCGAGTACCGTCTCT-3’ and 5’-TGCCTTGCCGGTTGCTT-3’), carRA (5’-CAGAAGCTGTTCCCGAAGACA-3’ and 5’-TGCGATGCCCATTTCTTGA-3’), carT (5’-CGGCACCAACACCAGACA-3’ and 5’-TGGACTAGGAATGGCAAGGACTT-3’), carX (5’-GCCGCCCATGAGGATACA-3’ and 5’-TCAGCTTCAACACCGTCGAA-3’), carO (5’-TGGGCAACGCAGTGACAT-3’ and 5’-TGCGCAGACAGCCCAGTA-3’), wcoA (5’-TGAGATTGTCGGCCAGAATTG-3’ and 5’-GAGCCCGCTTCGACTTTG-3’), cryD (5’-CGGGACTACATGCGATTGTG-3’ and 5’- CTTGAAAAGACGTGAGCCAAACT-3’), and vvdA (5’-GCACCACCAGGGCATGA-3’ and 5’-GCGGTGTGAAGCGACCTT-3’). β-tubulin gene (5’-CCGGTGCTGGAAACAACTG-3’ and 5’-CGAGGACCTGGTCGACAAGT-3’) was used as an internal control to normalize the amount of cDNA in each reaction.
Search of double ΔcryD ΔwcoA mutants
Different strategies were followed two obtain double mutants of the photoreceptor genes cryD and wcoA. Plasmid pDMWC was obtained from pALEX7 [27] by replacing the hygromycin resistance cassette by the F. fujikuroi niaD gene (GenBank X90699) as a selectable marker. A spontaneous niaD - mutant was obtained from the ΔcryD mutant SF237 by chlorate resistance as described [54]. Protoplasts of the niaD - ΔcryD strain were transformed with 30 μg of SpeI-linearized pDMWC and transformants were selected on DG medium, which contains nitrate as nitrogen source. Biolistic transformation was achieved with a Biolistic PDS-1000/He Particle Delivery System (Biorad, Hercules, CA, USA). 15 μg of plasmid pDMWC were shot on 106 conidia on DG plates. For selection with an alternative resistance marker, the hygR cassette from pALEX7 [27] and pDcry [33] were replaced by the nptII cassette (neomycin phosphotransferase II) from plasmid pNTP2 [33] to yield plasmids pDwc2 and pGcry7, respectively. The occurrence of double recombination events was expected to generate ΔwcoA mutants with pDwc2 and ΔcryD mutants with pGcry7. To generate double ΔcryD ΔwcoA mutants, five transformations were performed incubating protoplasts of the ΔcryD mutant SF236 with pDwc2, and two transformations were performed with the opposite combination, incubating protoplasts of the ΔwcoA mutant SF226 with pGcry7.
Mutagenesis experiments of the ΔcryD mutants SF236 and SF237 were achieved either by UV-radiation or by exposure to N-methyl-N’nitro-N-nitrosoguanidine (NG) following standard protocols [53]. Plates with 103 irradiated or NG-treated conidia were incubated in the dark for 3 days and screened for the formation of ΔwcoA-like purple-pigmented colonies in the dark.
Constitutive cryD expression
The cryD coding region was expressed in the ΔwcoA mutant SF226 under control of the constitutive promoter of the glyceraldehyde-3-phospate dehydrogenase gene of A. nidulans (gpdA). For this purpose, plasmid pOEcry (9.6 kb), containing a tagged version of the cryD gene under control of the gpdA promoter and trpC terminator was generated. The coding region of the cryD gene was obtained by PCR on F. fujikuroi cDNA template with primers crycDNA-1F (5’-GGTACCTGGGAATAAGCTCCTCGTCTATC-3’) and crycDNA-1R (5’-GCGGCCGCATGGGGTCCAAGGTGAGGAGG-3’), modified to contain KpnI and NotI restriction sites. These and other PCR reactions were performed with Expand High fidelity polymerase (Roche, Mannheim, Germany). The product was cloned into pGEM-T (Promega, Mannheim, Germany), the resulting plasmid was digested with KpnI and NotI, and the corresponding fragment was cloned in pET51b+ (Novagen, Darmstadt, Germany), a plasmid used for protein targeting with Strep-tag in the N-term and a His10-tag in the C-term. The 2.2 kb sequence of the tagged cryD gene was amplified from the resulting plasmid with primers crytag-3F (5’-GCAGGATCCTAGGTTAATTAGT-3’) and crytag-3R (5’-GAAGGAGATATACTAGTGCAAG-3’), which include a BamHI and a SpeI site, respectively. A reverse PCR was performed on pAN7–1 plasmid [55] to remove the hph coding region of the hygromycin resistance cassette with primers pAN7–1–3F (5’-GACCGCGGGTCCACTTAA-3’), which includes a BamHI site, and pAN7–1–3R (5’-GGGAAATACTAGTTCTTGGATGG-3’), which includes a SpeI site. The 5.6 kb PCR fragment was digested with BamHI and SpeI and ligated with the 2.2 kb cryD BamHI/SpeI fragment, resulting in a 7.8 kb plasmid. pOEcry was obtained introducing in the EcoRI site of this plasmid a geneticin resistance cassette (1.8 kb) obtained by EcoRI digestion of the PCR product obtained from plasmid pNTP2 [33] with primers neo-2F (5’-CTCGTCTACTCCAAGAATTCC-3’) and neo-3R (5’-TCTAGAACTAGTGGATCCCC-3), which include EcoRI sites.
To obtain the cryD-overexpressing strain, 30 μg of pOEcry were used to transform SF226 protoplast according to Proctor et al. [56], and the transformants were selected on a medium supplemented with geneticin (G418 disulfate salt, Sigma) as described [33]. The presence of the geneticin resistance cassette in the transformants was confirmed by PCR with primers neo-3F (5’- GAACAAGATGGATTGCACGC-3’) and neo-4R (5’-CGCTCAGAAGAACTCGTCAA-3’), which amplify the 0.8 kb nptII coding region. The presence of the PgpdA-cryD sequence was verified by PCR with primers gpd-2F (5’-ggctcaaatcaataagaagaacgc-3’) and cry-4R (5’-CATTCAGCTCCGTAGCGC-3’), which yield a 3.9 kb PCR fragment.
Carotenoid analyses
Mycelial samples were separated from the agar cultures with a sterile blade, frozen at -20°C and freeze-dried for 24 h in a VirTis SP Scientific sentry 2.0 equipment (SP Industries, Warminster, PA, USA). The dry samples were weighed and disrupted in a FAST-PREP 24 device (MP biomedicals, Irvine, CA) or in a Precellys 24 homogenizer (Bertin Technologies, Montigny le Bretonneux, France) with 1 ml of acetone as described [57]. Extractions were repeated up to total bleaching of the samples (3–6 extractions depending on carotenoid content) and the extracted solvent was dried in a Concentrator Plus equipment (Eppendorf, Hamburg, Germany). Dry samples were resuspended either in 1 ml or 0.1 ml hexane, depending on carotenoid concentrations, and subjected to spectrophotometrical determinations in the range of 350–650 nm (Shimadzu UV spectrophotometer 1800). Maximal absorbance at 482 nm was used to estimate the total amount of carotenoids, based on an average maximal E (1 mg l-1, 1 cm) of 200, and normalized according to the dried weight and dilution of the sample. When required (i.e., in analyses of ΔcryD or ΔwcoA mutants), bikaverins were removed from the carotenoid fraction as described [27].
Statistical analyses
To check if differences between different data values were significant, a statistical analysis was performed with the GraphPad Prism program (GraphPad Software, Inc. La Jolla, CA, USA). An ANOVA test (ANalysis Of VAriance) to compare several groups at the same time was used. The threshold of significance was set to a p value of 0.05 and three levels of significance were distinguished: p< 0.05, 0.05>p>0.001 and p<0.001. In the case of statistical significance (at least one group has a different average value), a Bonferroni post-correction test was applied to determine to which particular times and strains these differences were due in comparison to the wild type. The results of the statistical analysis are displayed in S1 Table.
Supporting Information
A. Left: Schematic representation of the relevant segment in plasmid pOECry containing the cryD gene under control of the A. nidulans gpdA promoter. The geneticin resistance marker nptII is indicated in yellow. Colored arrowheads indicate the primers used in the analysis of the transformant. Right: Agarose gel electrophoresis of PCR amplification products obtained from DNA samples of the ΔwcoA mutant SF226 and the SF226-derived cryD overexpressing strain (oe) with the primers sets indicated under the picture. M: Markers. Relevant sizes of markers and PCR products are shown in kb. B. Real-time RT-PCR analyses of the genes cryD (left panel), carRA (central panel) and carB (right panel) in RNA samples of the wild type, the ΔwcoA mutant SF226 and the SF226-derived cryD overexpressing strain (oe). For each strain, the left bar (dark color) corresponds to 3-day incubation in DGasn medium in the dark and the right bar (pale color) stands for one hour of illumination. Relative expression for each gene was referred to the value in the wild type grown in the dark. Data are the means and standard deviations of six determinations from two biological replicates. C. Kinetics of carotenoid accumulation after illumination of the ΔwcoA mutant SF226 and the SF226-derived cryD overexpressing strain (oe). The strains were incubated for three days in the dark on DGasn agar and exposed to white light for the time indicated in abscissae. Each point is the mean and standard deviation of four determinations from two biological replicates.
(PDF)
The strains were incubated for three days in the dark in DGasn medium and exposed to 0.07 W m-2 (1%), 0.7 W m-2 (10%) or 7 W m-2 (100%) of white light for 6 hours (above) and 48 h (below). Relative positions of the strains are schematized on the left.
(PDF)
Strains and conditions in Figs. 2, 4, 5 and 7 for which the differences with the equivalent data in the wild-type strain were statistically significant according to the ANOVA test (see Materials and Methods).
(PDF)
Acknowledgments
We are grateful to the Spanish Government for FPU fellowship granted to MC.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by European funds (European Regional Development Fund, ERDF), the Spanish Government (Ministerio de Economía y Competitividad, projects BIO2006-01323 and BIO2009-11131), and Andalusian Government (Consejería de Economía, Innovación, Ciencia y Empleo, projects P07-CVI-02813 and CTS-6638). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. URLs: European Regional Development Fund: http://ec.europa.eu/regional_policy/thefunds/regional/index_en.cfm, Ministerio de Economía y Competitividad: http://www.mineco.gob.es/, Consejería de Economía, Innovación, Ciencia y Empleo: http://www.juntadeandalucia.es/organismos/economiainnovacioncienciayempleo.html.
References
- 1. Hoffmeister D, Keller NP. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep. 2007;24: 393–416. [DOI] [PubMed] [Google Scholar]
- 2. Desjardins AE, Proctor RH. Molecular biology of Fusarium mycotoxins. Int J Food Microbiol. 2007;119: 47–50. [DOI] [PubMed] [Google Scholar]
- 3. Rademacher W. Gibberellins In: Anke T, editor. Fungal biotechnology. New York: Chapman & Hall; 1997. pp. 193–205. [Google Scholar]
- 4. Avalos J, Cerdá-Olmedo E, Reyes F, Barrero AF. Gibberellins and other metabolites of Fusarium fujikuroi and related fungi. Curr Org Chem. 2007;11: 721–737. [Google Scholar]
- 5. Avalos J, Prado-Cabrero A, Estrada AF. Neurosporaxanthin production by Neurospora and Fusarium . Methods Mol Biol. 2012;898: 263–274. 10.1007/978-1-61779-918-1_18 [DOI] [PubMed] [Google Scholar]
- 6. Linnemannstöns P, Prado MM, Fernández-Martín R, Tudzynski B, Avalos J. A carotenoid biosynthesis gene cluster in Fusarium fujikuroi: the genes carB and carRA . Mol Genet Genomics. 2002;267: 593–602. [DOI] [PubMed] [Google Scholar]
- 7. Thewes S, Prado-Cabrero A, Prado MM, Tudzynski B, Avalos J. Characterization of a gene in the car cluster of Fusarium fujikuroi that codes for a protein of the carotenoid oxygenase family. Mol Genet Genomics. 2005;274: 217–228. [DOI] [PubMed] [Google Scholar]
- 8. Prado-Cabrero A, Estrada AF, Al-Babili S, Avalos J. Identification and biochemical characterization of a novel carotenoid oxygenase: elucidation of the cleavage step in the Fusarium carotenoid pathway. Mol Microbiol. 2007;64: 448–460. [DOI] [PubMed] [Google Scholar]
- 9. Díaz-Sánchez V, Estrada AF, Trautmann D, Al-Babili S, Avalos J. The gene carD encodes the aldehyde dehydrogenase responsible for neurosporaxanthin biosynthesis in Fusarium fujikuroi . FEBS J. 2011;278: 3164–3176. 10.1111/j.1742-4658.2011.08242.x [DOI] [PubMed] [Google Scholar]
- 10. Avalos J, Díaz-Sánchez V, García-Martínez J, Castrillo M, Ruger-Herreros M, Limón MC. Carotenoids In: Martín JF, García-Estrada C, Zeilinger S, editors. Biosynthesis and molecular genetics of fungal secondary metabolites. New York: Springer; 2014. pp. 149–185. [Google Scholar]
- 11. Avalos J, Estrada AF. Regulation by light in Fusarium . Fungal Genet Biol. 2010;47: 930–938. 10.1016/j.fgb.2010.05.001 [DOI] [PubMed] [Google Scholar]
- 12. Rau W. Untersuchungen über die lichtabhängige Carotinoidsynthese. I. Das Wirkungsspektrum von Fusarium aquaeductuum . Planta. 1967;72: 14–28. [DOI] [PubMed] [Google Scholar]
- 13. Bindl E, Lang W, Rau W. Untersuchungen über die lichtabhängige Carotinoidsynthese. VI. Zeitlicher Verlauf der Synthese der einzelnen Carotinoide bei Fusarium aquaeductuum unter verschiedenen Induktionsbedingungen. Planta. 1970;94: 156–174. 10.1007/BF00387760 [DOI] [PubMed] [Google Scholar]
- 14. Avalos J, Schrott EL. Photoinduction of carotenoid biosynthesis in Gibberella fujikuroi . FEMS Microbiol Lett. 1990;66: 295–298. [Google Scholar]
- 15. Ádám AL, García-Martínez J, Szücs EP, Avalos J, Hornok L. The MAT1–2–1 mating-type gene upregulates photo-inducible carotenoid biosynthesis in Fusarium verticillioides . FEMS Microbiol lett. 2011;318: 76–83. 10.1111/j.1574-6968.2011.02241.x [DOI] [PubMed] [Google Scholar]
- 16. Rodríguez-Ortiz R, Michielse C, Rep M, Limón MC, Avalos J. Genetic basis of carotenoid overproduction in Fusarium oxysporum . Fungal Genet Biol. 2012;49: 684–696. 10.1016/j.fgb.2012.06.007 [DOI] [PubMed] [Google Scholar]
- 17. Tudzynski B. Gibberellin biosynthesis in fungi: genes, enzymes, evolution, and impact on biotechnology. Appl Microbiol Biotechnol. 2005;66: 597–611. [DOI] [PubMed] [Google Scholar]
- 18. Wiemann P, Willmann A, Straeten M, Kleigrewe K, Beyer M, Humpf HU, et al. Biosynthesis of the red pigment bikaverin in Fusarium fujikuroi: genes, their function and regulation. Mol Microbiol. 2009;72: 931–946. 10.1111/j.1365-2958.2009.06695.x [DOI] [PubMed] [Google Scholar]
- 19. Díaz-Sánchez V, Avalos J, Limon MC. Identification and regulation of fusA, the polyketide synthase gene responsible for fusarin production in Fusarium fujikuroi . Appl Environ Microbiol. 2012;78: 7258–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rodríguez-Ortiz R, Limón MC, Avalos J. Regulation of carotenogenesis and secondary metabolism by nitrogen in wild-type Fusarium fujikuroi and carotenoid-overproducing mutants. Appl Environ Microbiol. 2009;75: 405–413. 10.1128/AEM.01089-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prado MM, Prado-Cabrero A, Fernández-Martín R, Avalos J. A gene of the opsin family in the carotenoid gene cluster of Fusarium fujikuroi . Curr Genet. 2004;46: 47–58. [DOI] [PubMed] [Google Scholar]
- 22. Avalos J, Cerdá-Olmedo E. Carotenoid mutants of Gibberella fujikuroi . Curr Genet. 1987;25: 1837–1841. [Google Scholar]
- 23. Rodríguez-Ortiz R, Limón MC, Avalos J. Functional analysis of the carS gene of Fusarium fujikuroi . Mol Genet Genomics. 2013;288: 157–173. 10.1007/s00438-013-0739-7 [DOI] [PubMed] [Google Scholar]
- 24. Linden H, Ballario P, Macino G. Blue light regulation in Neurospora crassa . Fungal Genet Biol. 1997;22: 141–150. [DOI] [PubMed] [Google Scholar]
- 25. He Q, Liu Y. Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation. Genes Dev. 2005;19: 2888–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Corrochano LM. Fungal photoreceptors: sensory molecules for fungal development and behaviour. Photochem Photobiol Sci. 2007;6: 725–736. [DOI] [PubMed] [Google Scholar]
- 27. Estrada AF, Avalos J. The White Collar protein WcoA of Fusarium fujikuroi is not essential for photocarotenogenesis, but is involved in the regulation of secondary metabolism and conidiation. Fungal Genet Biol. 2008;45: 705–718. 10.1016/j.fgb.2007.12.003 [DOI] [PubMed] [Google Scholar]
- 28. Ruiz-Roldán MC, Garre V, Guarro J, Mariné M, Roncero MI. Role of the white collar 1 photoreceptor in carotenogenesis, UV resistance, hydrophobicity, and virulence of Fusarium oxysporum . Eukaryot Cell. 2008;7: 1227–1230. 10.1128/EC.00072-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103: 2203–2237. [DOI] [PubMed] [Google Scholar]
- 30. Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, et al. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 2011;62: 335–364. 10.1146/annurev-arplant-042110-103759 [DOI] [PubMed] [Google Scholar]
- 31. Selby CP, Sancar A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc Natl Acad Sci USA. 2006;103: 17696–17700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pokorny R, Klar T, Hennecke U, Carell T, Batschauer A, Essen LO. Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc Natl Acad Sci USA. 2008;105: 21023–21027. 10.1073/pnas.0805830106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Castrillo M, García-Martínez J, Avalos J. Light-dependent functions of the Fusarium fujikuroi CryD DASH cryptochrome in development and secondary metabolism. Appl Environ Microbiol. 2013;79: 2777–2788. 10.1128/AEM.03110-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shrode LB, Lewis ZA, White LD, Bell-Pedersen D, Ebbole DJ. vvd is required for light adaptation of conidiation-specific genes of Neurospora crassa, but not circadian conidiation. Fungal Genet Biol. 2001;32: 169–181. [DOI] [PubMed] [Google Scholar]
- 35. Schwerdtfeger C, Linden H. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 2003;22: 4846–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Malzahn E, Ciprianidis S, Káldi K, Schafmeier T, Brunner M. Photoadaptation in Neurospora by competitive interaction of activating and inhibitory LOV domains. Cell. 2010;142: 762–772. 10.1016/j.cell.2010.08.010 [DOI] [PubMed] [Google Scholar]
- 37. Youssar L, Schmidhauser TJ, Avalos J. The Neurospora crassa gene responsible for the cut and ovc phenotypes encodes a protein of the haloacid dehalogenase family. Mol Microbiol. 2005;55: 828–838. [DOI] [PubMed] [Google Scholar]
- 38. Navarro-Sampedro L, Yanofsky C, Corrochano LM. A genetic selection for Neurospora crassa mutants altered in their light regulation of transcription. Genetics. 2008;178: 171–183. 10.1534/genetics.107.079582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmoll M, Franchi L, Kubicek CP. Envoy, a PAS/LOV domain protein of Hypocrea jecorina (Anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light. Eukaryot Cell. 2005;4: 1998–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seibel C, Tisch D, Kubicek CP, Schmoll M. ENVOY is a major determinant in regulation of sexual development in Hypocrea jecorina (Trichoderma reesei). Eukaryot Cell. 2012;11: 885–895. 10.1128/EC.05321-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Castellanos F, Schmoll M, Martínez P, Tisch D, Kubicek CP, Herrera-Estrella A, et al. Crucial factors of the light perception machinery and their impact on growth and cellulase gene transcription in Trichoderma reesei . Fungal Genet Biol. 2010;47: 468–476. 10.1016/j.fgb.2010.02.001 [DOI] [PubMed] [Google Scholar]
- 42. Castrillo M, Avalos J. Light-mediated participation of the VIVID ortholog of Fusarium fujikuroi VvdA in pigmentation and development. Fungal Genet Biol. 2014;71: 9–20. 10.1016/j.fgb.2014.08.004 [DOI] [PubMed] [Google Scholar]
- 43. Alejandre-Durán E, Roldán-Arjona T, Ariza RR, Ruiz-Rubio M. The photolyase gene from the plant pathogen Fusarium oxysporum f. sp. lycopersici is induced by visible light and alpha-tomatine from tomato plant. Fungal Genet Biol. 2003;40: 159–165. [DOI] [PubMed] [Google Scholar]
- 44. Corrochano LM, Avalos J. Light sensing In: Borkovich K, Ebbole D, Momany M, editors. Cellular and Molecular Biology of Filamentous Fungi. Washington: ASM press; 2010. pp. 417–441. [Google Scholar]
- 45. Rau W, Lindemann I, Rau-Hund A. Untersuchungen über die lichtabhängige Carotinoidsynthese. III Die Farbstoffbildung von Neurospora crassa in Submerskultur. Planta. 1968;80: 309–316. [Google Scholar]
- 46. Schrott EL. Fluence response relationship of carotenogenesis in Neurospora crassa . Planta. 1980;150: 174–179. 10.1007/BF00582363 [DOI] [PubMed] [Google Scholar]
- 47. Bejarano ER, Avalos J, Lipson ED, Cerdá-Olmedo E. Photoinduced accumulation of carotene in Phycomyces . Planta. 1991;183: 1–9. 10.1007/BF00197560 [DOI] [PubMed] [Google Scholar]
- 48. López-Díaz I, Cerdá-Olmedo E. Relationship of photocarotenogenesis to other behavioural and regulatory responses in Phycomyces . Planta. 1980;150: 134–139. 10.1007/BF00582356 [DOI] [PubMed] [Google Scholar]
- 49. Godin KS, Varani G. How arginine-rich domains coordinate mRNA maturation events. RNA Biol. 2007;4: 69–75. [DOI] [PubMed] [Google Scholar]
- 50. Froehlich AC, Chen CH, Belden WJ, Madeti C, Roenneberg T, Merrow M, et al. Genetic and molecular characterization of a cryptochrome from the filamentous fungus Neurospora crassa . Eukaryot Cell. 2010;9: 738–750. 10.1128/EC.00380-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Veluchamy S, Rollins JA. A CRY-DASH-type photolyase/cryptochrome from Sclerotinia sclerotiorum mediates minor UV-A-specific effects on development. Fungal Genet Biol. 2008;45: 1265–1276. 10.1016/j.fgb.2008.06.004 [DOI] [PubMed] [Google Scholar]
- 52. Georgiou CD, Tairis N, Polycratis A. Production of β-carotene by Sclerotinia sclerotiorum and its role in sclerotium differentiation. Mycol Res. 2001;105: 1110–1115. [Google Scholar]
- 53. Avalos J, Casadesús J, Cerdá-Olmedo E. Gibberella fujikuroi mutants obtained with UV radiation and N-methyl-N'-nitro-N-nitrosoguanidine. Appl Environ Microbiol. 1985;49: 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Klittich CJR, Leslie JF. Nitrate reduction mutants of Fusarium moniliforme (Gibberella fujikuroi). Genetics. 1988;118: 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CAMJJ. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli . Gene. 1987;56: 117–124. [DOI] [PubMed] [Google Scholar]
- 56. Proctor RH, Hohn TM, McCormick SP. Restoration of wild-type virulence to Tri5 disruption mutants of Gibberella zeae via gene reversion and mutant complementation. Microbiology. 1997;143: 2583–2591. [DOI] [PubMed] [Google Scholar]
- 57. Arrach N, Schmidhauser TJ, Avalos J. Mutants of the carotene cyclase domain of al-2 from Neurospora crassa . Mol Genet Genomics. 2002;266: 914–921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Left: Schematic representation of the relevant segment in plasmid pOECry containing the cryD gene under control of the A. nidulans gpdA promoter. The geneticin resistance marker nptII is indicated in yellow. Colored arrowheads indicate the primers used in the analysis of the transformant. Right: Agarose gel electrophoresis of PCR amplification products obtained from DNA samples of the ΔwcoA mutant SF226 and the SF226-derived cryD overexpressing strain (oe) with the primers sets indicated under the picture. M: Markers. Relevant sizes of markers and PCR products are shown in kb. B. Real-time RT-PCR analyses of the genes cryD (left panel), carRA (central panel) and carB (right panel) in RNA samples of the wild type, the ΔwcoA mutant SF226 and the SF226-derived cryD overexpressing strain (oe). For each strain, the left bar (dark color) corresponds to 3-day incubation in DGasn medium in the dark and the right bar (pale color) stands for one hour of illumination. Relative expression for each gene was referred to the value in the wild type grown in the dark. Data are the means and standard deviations of six determinations from two biological replicates. C. Kinetics of carotenoid accumulation after illumination of the ΔwcoA mutant SF226 and the SF226-derived cryD overexpressing strain (oe). The strains were incubated for three days in the dark on DGasn agar and exposed to white light for the time indicated in abscissae. Each point is the mean and standard deviation of four determinations from two biological replicates.
(PDF)
The strains were incubated for three days in the dark in DGasn medium and exposed to 0.07 W m-2 (1%), 0.7 W m-2 (10%) or 7 W m-2 (100%) of white light for 6 hours (above) and 48 h (below). Relative positions of the strains are schematized on the left.
(PDF)
Strains and conditions in Figs. 2, 4, 5 and 7 for which the differences with the equivalent data in the wild-type strain were statistically significant according to the ANOVA test (see Materials and Methods).
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.