Abstract
Haemophilus ducreyi, the etiologic agent of chancroid, has been previously reported to show genetic variance in several key virulence factors, placing strains of the bacterium into two genetically distinct classes. Recent studies done in yaws-endemic areas of the South Pacific have shown that H. ducreyi is also a major cause of cutaneous limb ulcers (CLU) that are not sexually transmitted. To genetically assess CLU strains relative to the previously described class I, class II phylogenetic hierarchy, we examined nucleotide sequence diversity at 11 H. ducreyi loci, including virulence and housekeeping genes, which encompass approximately 1% of the H. ducreyi genome. Sequences for all 11 loci indicated that strains collected from leg ulcers exhibit DNA sequences homologous to class I strains of H. ducreyi. However, sequences for 3 loci, including a hemoglobin receptor (hgbA), serum resistance protein (dsrA), and a collagen adhesin (ncaA) contained informative amounts of variation. Phylogenetic analyses suggest that these non-sexually transmitted strains of H. ducreyi comprise a sub-clonal population within class I strains of H. ducreyi. Molecular dating suggests that CLU strains are the most recently developed, having diverged approximately 0.355 million years ago, fourteen times more recently than the class I/class II divergence. The CLU strains' divergence falls after the divergence of humans from chimpanzees, making it the first known H. ducreyi divergence event directly influenced by the selective pressures accompanying human hosts.
Introduction
Haemophilus ducreyi is a Gram-negative coccobacillus in the polyphyletic family Pasteurellaceae, and is the etiologic agent of the sexually transmitted infection chancroid, a genital ulcer disease [1]. An obligate human pathogen, H. ducreyi has no known environmental reservoir outside of its host. Like other ulcerative sexually transmitted infections, chancroid facilitates both the transmission and acquisition of HIV-1, making the limitation of chancroidal infection paramount for HIV control efforts [2–5]. Chancroid ulcers are able to facilitate transmission of HIV-1, making chancroid a concern to HIV prevention efforts due to is presence among commercial sex worker populations ([2–5]
H. ducreyi has historically only been known to cause genital ulcer infections as chancroid is transmitted via micro-abrasions developed during sexual intercourse [1]. However, in 1989, an ulcer caused by H. ducreyi was reported on the left foot of a 22-year-old male reported to have returned to Denmark from the Fiji Islands who had no history or sign of primary genital infection [6]. No additional reports of non-genital H. ducreyi infection were reported until 2007, when H. ducreyi was confirmed to be the etiologic agent of cutaneous limb ulcers (CLU) present in three children ranging between 6–9 years of age via phenotypic testing on cultures and by 16S rDNA sequencing [7]. These cases also occurred after travel to the South Pacific, as each child had recently visited Samoa. In 2007, another extra-genital leg ulceration due to H. ducreyi was reported in a 5-year-old Sudanese refugee recently placed in Canada. Symptoms were similar to those seen in previous H. ducreyi leg ulcerations, and the infection was resolved after treatment with oral azithromycin [8]. This case was the first reported outside of the South Pacific, indicating H. ducreyi associated leg ulceration may be more widespread than previously suspected. Chronic limb ulcers caused by H. ducreyi are not limited to children [9]. [10].
Recent studies conducted in the South Pacific Island Countries and Territories (PICT) have shown that H. ducreyi is the etiologic agent of non-sexually transmitted skin ulcers which clinically resemble yaws, an infection by Treponema pallidum subspecies pertenue. A prospective cohort examination conducted in the yaws-endemic area Papua New Guinea revealed H. ducreyi is the cause of 3.2 cases per 100 persons, with H. ducreyi DNA detected in 60% of ulcers and T. pallidum DNA detected in 34% [11]. These data suggest H. ducreyi is a major cause of leg ulcers. While often characterized as fastidious, H. ducreyi is first and foremost a skin pathogen, which has been demonstrated by experimentally established infections in the upper arms of human volunteers [12]. Thus, a non-genital infection would not be entirely unprecedented. Similar findings were also reported in a yaws survey conducted in the Solomon Islands [13].
These new infection patterns prompt a re-examination of the population structure of H. ducreyi. While many Haemophilus species are inherently competent, H. ducreyi is considered non-transformable due to mutations within its competence genes, comA, comB, and comM, which contain an internal stop codon [14]. Thus, rates of recombination and gene transfer are likely lower than other species, generating stable clones among H. ducreyi strains [14,15]. Previous attempts at examining genetic diversity within the species H. ducreyi using PCR and sequencing analysis have shown a large amount of genetic variation between a few key virulence genes within the genome of H. ducreyi, and strains of H. ducreyi fall into the recognized subcategories class I and class II [16–18]. Here we re-examine the relationships among H. ducreyi strains using a Multi Locus Sequence Analysis (MLSA) of 11 genes, including virulence factors and housekeeping genes, to understand the position of strains cultured from cutaneous limb ulcers within the class hierarchy [19].
Methods
Strains and Culture Conditions
H. ducreyi strains were cultured on Columbia Agar supplemented with 5% hemoglobin and 1% IsoVitaleX (Table 1). Strains were revived from frozen stocks and incubated at 35°C with 5% CO2 for 24–48 hours. Strains were sub-cultured no more than once prior to PCR.
Table 1. Strains of H. ducreyi used in this study.
| Strain | Class | Site of Isolation (year) | Reference |
|---|---|---|---|
| NZS1 | CLU 1 | Samoa (2006) | [7] |
| NZS2 | CLU | Samoa (2006) | [7] |
| NZS3 | CLU | Samoa (2006) | [7] |
| NZS4 | CLU | Samoa (2007) | This study |
| SSMC57 | I | Bangladesh (N/A 2 ) | [18] |
| 82–029362 | I | California, USA (1982) | [31] |
| HMC56 | I | Dominican Republic (1995) | [32] |
| HMC60 | I | Florida, USA (1989) | [32] |
| HD188 | I | Kenya (1982) | [31] |
| HMC46 | I | Kenya (1995) | [32] |
| 6644 | I | Massachusetts, USA (1989) | [31] |
| C111 | I | Nairobi, Kenya (N/A) | [18] |
| 85–023233 | I | New York, USA (1985) | [31] |
| HD183 | I | Singapore (1982) | [31] |
| 35000HP | I | Winnipeg, Canada (1972) | [33] |
| DMC111 | II | Bangladesh (N/A) | [18] |
| DMC64 | II | Bangladesh (N/A) | [18] |
| SSMC71 | II | Bangladesh (N/A) | [18] |
| CIP542 | II | Hanoi, Vietnam (1954) | [18] |
| 33921 | II | Nairobi, Kenya (N/A) | [16] |
| HMC112 | II | Unknown (1984) | [18] |
1 H. ducreyi strains isolated from a cutaneous limb ulcer
2 N/A, year of isolation unknown
PCR, Gel Electrophoresis, and Sequencing
Genomic DNA was obtained from each strain using GeneReleaser according to the manufacturer’s specifications (BioVentures, Murfreesboro, TN). Sequences of all loci (16s rDNA, cpxR, wecA, recA, dsrA, ncaA, ompA2, lspA2, fgbA, hgbA, dltA) were obtained from fragments amplified via PCR using the primers listed (Table 2). PCR products were examined for quality and contamination via gel electrophoresis. Gels were stained with SYBR Safe (Life Technologies, Grand Island, NY) and quantified using ImageJ to determine relative concentration of DNA in PCR products prior to sequencing. Monodirectional sequencing was conducted by Eurofins MWG|Operon and Yale University DNA Analysis Facility using the forward primers employed in PCR to amplify loci selected for analysis (Table 2). All sequences for CLU strains were obtained as part of this study. All cpxR sequences were obtained as part of this study, as was the fgbA sequence for strain 82–029362. All other sequences were obtained from GenBank. See S1 Table for a list of accession numbers for all genes used in this study.
Table 2. List of oligonucleotides used in this study.
| Locus | Forward | Reverse | Reference |
|---|---|---|---|
| fgbA | 5′ CCTCAAGTTGAAGAAATGAAACAAACGG 3′ | 5′ CGAATTCGTATTTGGTAATAAATGACCGC 3′ | [34] |
| wecA | 5′ CCGGAATCACCCATAAACAC 3′ | 5′ CCGGAATCACCCATAAACAC 3′ | [17] |
| recA | 5′ CATTATGGCAGCGGATAAAAA 3′ | 5′ TCCTCAAACGCTTCATCAAA 3′ | [17] |
| hgbA | 5′ AGCGATTACTCTCTGTATTTTGGGG 3′ | 5′ GCAGATATTGCTGCATCATCGGAG 3 | [18] |
| lspA2 | 5′ AAGTTTCAGCAAGAGCGGC 3′ | 5′ TATTGGCTGCAAGCTCTG 3′ | [35] |
| ncaA | 5′ GGTTGATTATGTCGAATAATTTG 3′ | 5′ CTAAGCGCGTAAAAATTCGATG 3′ | [18] |
| dsrA | 5′ GACAGCATTCAGTGAATAATGGC 3′ | 5′ AATGAAGTCCGCACCTTTAACGGC 3′ | [18] |
| ompA2 | 5′ GAGGTACCGCGCCACAAGCGGATACTTTTTAT 3′ | 5′ GCTTAAGCGTGGTTTATCTCTTACATTCGCTACA 3′ | [26] |
| dltA | 5′ CGCTTGTACAAGCGGGC 3′ | 5′ CAGCTTACAAAATGATGGGC 3′ | [36] |
| 16s rDNA | 5′ CAAGTCGAACGGTAGCACGAAG 3′ | 5′ TTCTGTGACTAACGTCAATCAATTTTG 3′ | This Study |
| cpxR | 5′ TGAAGATCCAAGAGGCAATG 3′ | 5′ AGGGATTCGTTCAATCGACA 3′ | This study |
Computational Analyses
DNA sequences were edited for base miscalls and trimmed of low-quality bases in Sequencher 5.2 (Genecodes, Ann Arbor, MI). Due to the length of the hgbA locus, internal primers were used to primer walk sequencing as previously described [18]. hgbA sequences were then assembled into consensus sequences representing the entire region of interest. Alignments of each loci were also initially performed in Sequencher 5.2 and all sequences were further trimmed so that each amplicon was of equal length. Alignments were exported in FASTA format, and imported into MEGA 6.0 for phylogenetic analysis (Neighbor-Joining, Minimum Evolution, Maximum Likelihod, and Maximum Parismony) [20]. A bootstrap consensus neighbor-joining tree was generated for loci containing informative variation using the Tamura-3 Parameter statistical method and tested for significance using 1,000 rounds of bootstrap replication. Tests for selection and nucleotide diversity were carried out in MEGA 6.0 as well using Tajima’s Test of Neutrality to calculate Tajima’s D statistic. Nucleotide sequences for dsrA and ncaA were translated to amino acids in MEGA 6.0 and examined for variation. The relative clade divergence date for loci not under positive selection was calculated by dividing the number of single nucleotide polymorphisms (SNPs) by the total number of base pairs then dividing the quotient by the evolutionary rate (1% per million years) of the closest relative possible [21,22].
Results
In this study, a modified MLSA protocol was used to characterize H. ducreyi isolates from non-genital infections (Table 3). The sequences from multiple strains of class I and class II H. ducreyi were previously published for 7 of the loci examined in this study: wecA, recA, fgbA, hgbA, ncaA, dsrA, and lspA2 [17]. To expand this data set, we determined the DNA sequences for all of the above loci from the genomes of four leg ulcer isolates. Four genes previously used to distinguish class I and II (pal, murC, mtrC, sapA) had very limited variation even between class I and II [17], and were therefore excluded from this analysis. In place of these four genes, we examined four additional loci, cpxR, ompA2, dltA, and 16s rDNA, to better distinguish the CLU (cutaneous limb ulcer) strains from class I and class II H. ducreyi. These additional loci also increased the distribution of loci throughout the genome. Thus, the total data set includes 11 unlinked loci with genome wide distribution in class I, class II, and CLU strains (Fig. 1). This number exceeds the standard conventional multilocus sequence analysis that requires the sequencing and comparison of 7 unlinked loci [19], and is sufficient to establish H. ducreyi CLU strains' position among the previously described class hierarchy of strains [17].
Table 3. Cutaneous limb ulcer patient information.
| Strain | Year | Gender | Age (yr) | Clinical Outcome |
|---|---|---|---|---|
| NZS1 | 2006 | F | 11 | ulcer healed following antimicrobial treatment |
| NZS2 | 2006 | F | 5 | ulcer healed following antimicrobial treatment |
| NZS3 | 2006 | F | 6 | ulcer healed following antimicrobial treatment |
| NZS4 | 2007 | F | 15 | ulcer healed following antimicrobial treatment |
Fig 1. Distribution of sequenced loci compared to the 35000HP reference genome.

A graphic depiction of the distribution of loci chosen for MLSA analysis. Sites are aligned to the complete 35000HP reference genome (horizonal bar). All sites chosen for analysis were unlinked, and represent a genome wide distribution of variation. Arrows represent loci chosen for analysis. Numbers represent position in base pairs relative to the reference genome.
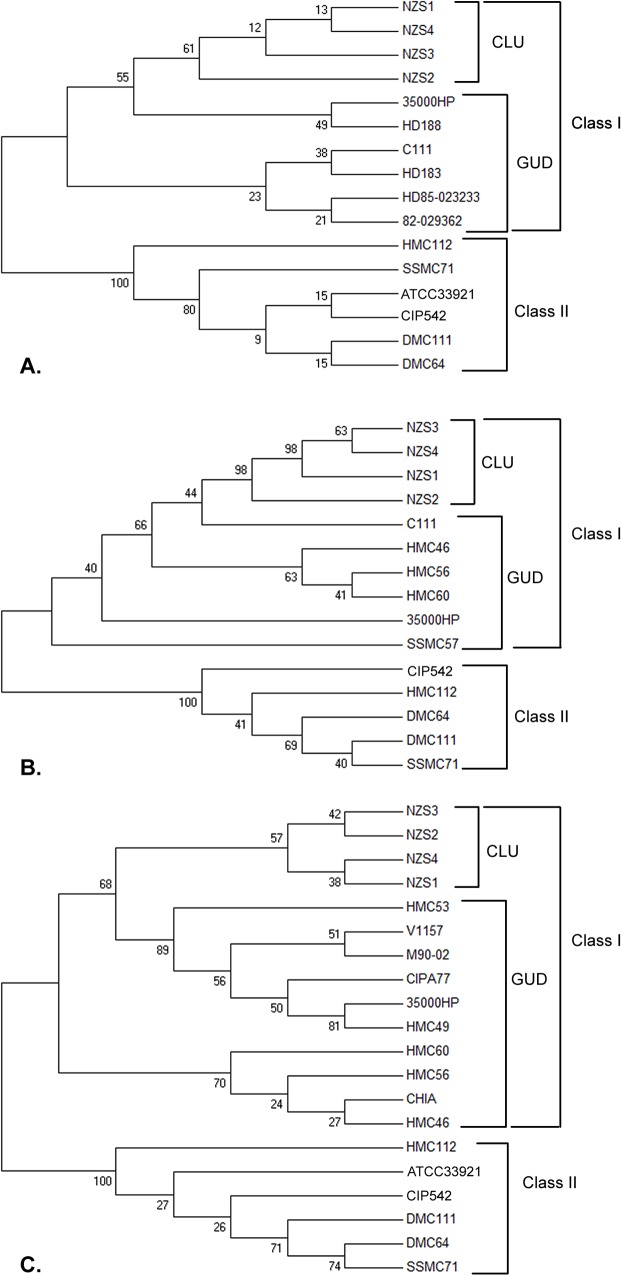
While neighbor-joining trees were used as final phylogenies, other methods of phylogeny reconstruction (Maximum Likelihood, Maximum Parsimony, Minimum Evolution) showed similar topology and positioning of CLU strains among class I H. ducreyi, with minor differences in the bootstrap values and strain positioning within class clades. CLU strains of H. ducreyi form a statistically supported subclade within the class I clade of H. ducreyi. Thus, they are the only series of H. ducreyi strains which form a subclade based on their geographic location (Table 1, Fig. 2). cpxR, ompA2, lspA2, 16s rDNA, dltA, fgbA, recA, and wecA were excluded from phylogenetic reconstruction due to a lack of informative variation needed to produce high fidelity tree branches.
Fig 2. Evolutionary relationships of taxa using the loci (A) ncaA, (B) hgbA, and (C) dsrA.
The evolutionary history was inferred with the Neighbor-Joining method using MEGA6. The bootstrap consensus tree above were inferred from 1,000 replicates. Bootstrap values are located on top of the nodes.
Concatenation of the 11 genes provided a total of 11,398 sites, representing ∼0.67% of the H. ducreyi genome when compared to the size of the published 35000HP genome (NC_002940.2). Variation specific to CLU strains was present within several of the loci surveyed, with 74 sites differentiating CLU strains from class I. Most of the genetic variation was concentrated in 3 of the 11 genes surveyed (ncaA, dsrA, hgbA), all of which code for proteins involved in bacterial virulence in humans. Though variation was present, the majority of nucleotides were conserved between class I and CLU H. ducreyi. wecA, cpxR, fgbA, 16S rDNA, and dltA CLU sequences contained no variability when compared to class I sequences. recA contained 1 polymorphic site, ompA2 had 2 (2.7%), and lspA2 contained 5 variable sites (6.7%) when compared to class I sequences. Of the total variablility specific to the CLU strains, ncaA contained 40 variant sites (54.1%), dsrA contained 18 variant sites (24.3%), and hgbA contained 7 variant sites (9.5%). CLU strains were not monotypic at these loci as differential haplotypes are present within each of the loci, comprised of either one or two CLU strain-specific single nucleotide polymorphisms. When compared to class II strains of H. ducreyi, CLU strain sequences contained no similar SNPs to those characteristic of class II.
With the exception of CLU dsrA (D = 2.773) and hgbA (D = 2.156), which were found to be under positive selection when grouped with only class I strains, all loci studied were under neutral or purifying selection (Table 4). Due to the high amounts of variation between class I and class II H. ducreyi within the dsrA and hgbA loci, only class I sequences were chosen for statistical comparison to CLU H. ducreyi during selection analyses to eliminate bias from class divergence (Table 4). Thus, the Tajima’s D statistic gives a comparative depiction of selection between CLU and class I strains. While genes under positive selection are normally not suitable for MLSA analysis [19], the gene loci under positive selection were included for phylogenetic reconstruction as the CLU sequences contained numerous informative sites while retaining a high level of conservation when compared to class I H. ducreyi. CLU strains contained less nucleotide diversity when compared to class I than when class I is compared to class II H. ducreyi. In between group comparison, CLU and class I strains had an average nucleotide diversity of 0.00220, while the average nucleotide diversity between class I and class II was 0.0479.
Table 4. Nucleotide diversity and selection relative to location and function.
| Locus | Position 1 | Length 2 | Tajima's D 3 | Class I vs. Class II (π) 4 | Class I vs. CLU (π') | Location; Function |
|---|---|---|---|---|---|---|
| 16s rDNA | 10643–12179 | 396 | 0.85056 | 0 | 0.00133 | Cytoplasm; ribosomal subunit |
| cpxR | 1204379–1205101 | 598 | −0.27919 | 0.00447 | 0.00114 | Cytoplasm; response regulator |
| dltA | 586988–597714 | 697 | 1.70281 | 0.00719 | 0 | Outer membrane; Lectin |
| dsrA | 603307–604195 | 1229 | 2.77342 | 0.16910 | 0.01420 | Outer membrane; serum resistance |
| fgbA | 136574–137285 | 648 | 0.29421 | 0.00795 | 0 | Outer membrane; fibrinogen binding |
| hgbA | 1686700–1689521 | 2860 | 2.15658 | 0.01859 | 0.00049 | Outer membrane; hemoglobin receptor |
| lspA2 | 925194–925452 | 461 | −1.40008 | 0.10658 | 0.00166 | Outer membrane; antiphagocytic protein |
| ncaA | 1614393–1614596 | 2138 | −0.78926 | 0.09943 | 0.00140 | Outer membrane; collagen adhesin |
| ompA2 | 41761–42984 | 1000 | 1.30267 | 0 | 0.00040 | Outer membrane; membrane stability |
| recA | 325798–326862 | 975 | −0.81649 | 0.00877 | 0.00055 | Periplasm; recombination promotion |
| wecA | 1551061–1551456 | 396 | 0.33694 | 0.00456 | 0.00091 | Cytoplasm; synththesis of common antigen |
| Weighted Average | 0.0479 | 0.00220 | ||||
1 Position in base pairs relative to the H. ducreyi 35000HP genome (NC_002940.2).
2 PCR amplicon alignment length in base pairs, including indels.
3 Tajima's D statistic of selection; positive values indicate positive selection, and those greater than 2 are significant.
4 Tajima's pi statistic; indicates the relative nucleotide diversity at any given locus.
When comparing the sequence of amino acids in each of the loci used for phylogenetic reconstruction, HgbACLU’s amino acid sequence was completely conserved when compared to class I. However,DsrACLU and NcaACLU contained unique variation within their amino acid sequences (Table 4). NcaA contained 1 amino acid substitution, while DsrACLU contained 13 amino acid shifts within the passenger domain of the protein, which is involved in fibrinogen binding [23]. DsrACLU strains contained a dimer of the NTHNINK motif in the serum resistance domain similar to that of class I strains other than M90-02 and V1157, which contain a trimer, and 35000HP, which contains a monomer (Table 5).
Table 5. Characterization of nonsynonymous amino acid changes in CLU strains relative to 35000HP (I).
| Locus | Location | Change | Significance |
|---|---|---|---|
| ncaA | Not Characterized | Lys47Glu | Charge Inversion |
| dsrA | Passenger Domain | Gly51Asp | Nonpolar to Polar |
| Passenger Domain | Val57Ala | NS 1 | |
| Passenger Domain | Glu59Lys | Charge Inversion | |
| Passenger Domain | Ala72Val | NS | |
| Passenger Domain | Glu76Lys | Charge Inversion | |
| Passenger Domain | Ala92Val | NS | |
| Passenger Domain | Gly94Pro | Amino to Imino | |
| Passenger Domain | Val95Pro | NS | |
| Passenger Domain | Ser96Pro | Amino to Imino | |
| Passenger Domain | Pro99Ser | Imino to Amino | |
| Passenger Domain | Lys136Asn | Positive to Neutral | |
| Passenger Domain | Tyr155His | Hydrophobic to Hydrophilic | |
| Passenger Domain | Asp165Gly | Polar to nonpolar | |
| Serum Resistance Domain | 180–185 NTHNINK monomer to NTHNINK dimer | Unknown |
1 NS, no structural or functional change occurs as a result of this change.
Discussion
In this study, we describe genotyping of cutaneous leg ulcer H. ducreyi strains using an MLSA approach that provides information for the genetic basis of the population structure (Table 2). Although the data set is limited to four CLU strains (most of the prior work done on this syndrome was solely PCR-based, limiting the number of available isolates), it does shed light on the relationship of these strains to genital ulcer disease (GUD) strains of H. ducreyi. The 11 loci around the H. ducreyi chromosome fall into two groups, 16s rDNA, wecA, recA, ompA2, lspA2, fgbA, dltA, and cpxR, which contain little variation from class I, and ncaA, dsrA, and hgbA, which contain information that differentiates CLU strains from class I H. ducreyi (Table 4 and Fig. 2).
With the exception of the dsrA and hgbA loci, all other class I and CLU loci included in the study were either under neutral or purifying selection typically associated with loci in MLSA analysis (Table 3) [19]. The positive selection identified in dsrA and hgbA is likely due to their association with virulence in humans [24,25]. HgbA is necessary due to its role as a hemoglobin receptor, aiding H. ducreyi in the acquisition of iron which is required for survival. DsrA is necessary for full virulence in humans due to its roles in serum resistance and fibrinogen binding, helping the bacterium to evade the innate immune response and adhere to the body during pathogenesis. The various amino acid changes within DsrA’s passenger domain responsible for fibrinogen binding will likely influence its ability to bind to fibrinogen, warranting further investigation (Table 5) [23,26].
H. ducreyi has long been recognized as the etiologic agent of the sexually transmitted infection chancroid, and more recently the causitive agent of chronic, cutaneous limb ulceration afflicting children and adults. H. ducreyi has been previously described to form two classes based on DNA sequencing and proteomic analyses, yet despite this evolutionary divergence, the classes are associated with the same clinical syndrome. The CLU strains mark the first H. ducreyi strains isolated whose infection patterns differ from those of GUD class I and class II strains, though the nature of the bacterium as a skin pathogen makes this change plausible.
The presence of unique and informative nucleotide diversity within CLU strains, which form a differentiable subclade from the class I GUD clade, suggests that these isolates may be evolving differently than others (Fig. 2). Using an evolutionary rate of 1% change per million years [22,27], CLU strains diverged from class I approximately 0.355 million years before present, making them the most recently diverged H. ducreyi strains. Comparatively, class I strains diverged from class II ∼ 5 million years ago, which predates the separation of human and chimpanzee lineages [17,28]. This calculation disproves the notion that the cutaneous strains are older clones of H. ducreyi [29]. Instead this may be due to specific niche or environmental adaptation.
We do not yet know why the CLU strains are closely related phylogenetically to class I strains. There are distinct genotypic and phenotypic differences between class I and II strains [16–18]. In the lab, class I isolates grow faster and the colonies are larger than class II isolates. Class II isolates are underrepresented in strain collections, most likely because they are more difficult to culture. It is possible that class II strains may also cause cutaenous infections but we have not isolated any strains yet due to the difficulty in growing class II strains in vitro.
Previously, H. ducreyi was thought to be exclusively transmitted through micro abrasions that occur during sexual intercourse. However, human challenges for mutant parent comparisons have been conducted for over fifteen years in which volunteers were inoculated on the upper arm [12]. Patient inoculations are delivered via allergy testing lancets as 106 H. ducreyi cells did not infect intact skin in inoculation experiments [30]. Even when infected during human challenge, no cases of secondary transmission occurred in 2,123 subject-days of infection [12]. Thus, the question of why CLU strains are able to transmit at rates of 3.2 per 100 cutaneous ulcer cases presents itself [11]. Whole genome sequencing of multiple class I, class II, and CLU strains would likely provide more detail on the genetic mechanism involved in the CLU strains’ unique proliferation, which is likely due to the modification or addition of virulence factors.
Supporting Information
(DOCX)
Acknowledgments
The authors thank Stephanie Blystone and Emanoel DeJesus for their technical contribution to this research, Lin Sutley for preparation of figures, and Stanley Spinola for thoughtful critique of this manuscript. We would like to offer special thanks to Kristen Webb for technical and bioinformatics assistance, as well as thoughtful critique of the manuscript.
Data Availability
Data will be available on GenBank. Accession numbers will be provided in a supplemental table.
Funding Statement
This study was funded by Allegheny College. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Albritton WL (1989) Biology of Haemophilus ducreyi . Microbiological Reviews 53: 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Steen R, Chersich M, De vals SJ (2012) Periodic presumptive treatment of curable sexually transmitted infections among sex workers: Recent experience with implementation. Current Opinion in Infectious Diseases 25: 100–106. 10.1097/QCO.0b013e32834e9ad1 [DOI] [PubMed] [Google Scholar]
- 3. Fleming DT, Wasserheit JN (1999) From epidemiological synergy to public health policy and practice: The contribution of other sexually transmitted diseases to sexual transmission of HIV infection. Sexually Transmitted Infections 75: 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Galvin SR, Cohen MS (2004) The role of sexually transmitted diseases in HIV transmission. Nature Reviews Microbiology 2: 33–42. [DOI] [PubMed] [Google Scholar]
- 5. Wasserheit JN. Epidemiological synergy. interrelationships between human immunodeficiency virus infection and other sexually transmitted diseases. Sexually Transmitted Diseases 19: 61–77. [PubMed] [Google Scholar]
- 6. Marckmann P, HØjbjerg T, von Eyben FE, Christensen I (1989) Imported pedal chancroid: Case report. Genitourinary Medicine 65: 126–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ussher JE, Wilson E, Campanella S, Taylor SL, Roberts SA (2007) Haemophilus ducreyi causing chronic skin ulceration in children visiting samoa. Clinical Infectious Diseases 44: 85. [DOI] [PubMed] [Google Scholar]
- 8. Humphrey SH, Romney M, Au S (2007) Haemophilus ducreyi leg ulceration in a 5-year old boy. Journal of the American Academy of Dermatology 56: AB121. [Google Scholar]
- 9. McBride WJH, Hannah RCS, Le Cornec GM, Bletchly C (2008) Cutaneous chancroid in a visitor from Vanuatu. Australasian Journal of Dermatology 49: 98–99. 10.1111/j.1440-0960.2008.00439.x [DOI] [PubMed] [Google Scholar]
- 10. Peel TN, Bhatti D, De Boer JC, Stratov I, Spelman DW (2010) Chronic cutaneous ulcers secondary to Haemophilus ducreyi infection. The Medical Journal of Australia 192: 348–50. [DOI] [PubMed] [Google Scholar]
- 11. Mitjà O, Lukeheart SA, Pokowas G, Moses P, Kapa A, Godornes C, et al. (2014) Haemophilus ducreyi as a cause of skin ulcers in children from a yaws-endemic area of Papua New Guinea: A prospective cohort study. Lancet Global Health 2: 235–241. [DOI] [PubMed] [Google Scholar]
- 12. Janowicz DM, Ofner S, Katz BP, Spinola SM (2009) Experimental infection of human volunteers with Haemophilus ducreyi: Fifteen years of clinical data and experience. Journal of Infectious Diseases 199: 1671–9. 10.1086/598966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marks M, Chi KH, Vahi V, Pillay A, Sokana O, Pavluck A, et al. (2014) Haemophilus ducreyi associated with skin ulcers among children, Solomon Islands. Emerging Infectious Diseases 20: 1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Redfield RJ, Findlay WA, Bossé J, Kroll JS, Cameron ADS, Nash JH (2006) Evolution of competence and DNA uptake specificity in the Pasteurellaceae. BMC Evolutionary Biology 6: 82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gioia J, Qin X, Jiang H, Clinkenbeard K, Lo R, Liu Y, et al. (2006) The genome sequence of Mannheimia haemolytica A1: Insights into virulence, natural competence, and Pasteurellaceae phylogeny. Journal of Bacteriology 188: 7257–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Post DMB, Gibson BW (2007) Proposed second class of Haemophilus ducreyi strains show altered protein and lipooligosaccharide profiles. Proteomics 7: 3131–42. [DOI] [PubMed] [Google Scholar]
- 17. Ricotta EE, Wang N, Cutler R, Lawrence JG, Humphreys TL (2011) Rapid divergence of two classes of Haemophilus ducreyi . Journal of Bacteriology 193: 2941–7. 10.1128/JB.01400-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. White CD, Leduc I, Olsen B, Jeter C, Harris C, Elkins C (2005) Haemophilus ducreyi outer membrane determinants, including DsrA, define two clonal populations. Infection and Immunity 73: 2387–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spratt BG (1999) Multilocus sequence typing: Molecular typing of bacterial pathogens in an era of rapid DNA sequencing and the internet. Current Opinion in Microbiology 2: 312–6. [DOI] [PubMed] [Google Scholar]
- 20. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Obbard DJ, Maclennan J, Kim K, Rambaut A, O'Grady PM, Jiggins FM (2012) Estimating divergence dates and substitution rates in the Drosophila phylogeny. Molecular Biology and Evolution 29: 3459–3473. 10.1093/molbev/mss150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moran NA, Munson MA, Baumann P, Ishikawa H (1993) A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proceedings of the Royal Society B 253: 167–171. [Google Scholar]
- 23. Fusco WG, Elkins C, Leduc I (2013) Trimeric autotransporter DsrA is a major mediator of fibrinogen binding in Haemophilus ducreyi . Infection and Immunity 81: 4443–4452. 10.1128/IAI.00743-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Al-Tawfiq JA, Fortney KR, Katz BP, Hood AF, Elkins C, Spinola SM (2000) An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. Journal of Infectious Diseases 181: 1049–54. [DOI] [PubMed] [Google Scholar]
- 25. Bong CT, Throm RE, Fortney KR, Katz BP, Hood AF, Elkins C, et al. (2001) DsrA-deficient mutant of Haemophilus ducreyi is impaired in its ability to infect human volunteers. Infection and Immunity 69: 1488–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leduc I, White CD, Nepluev I, Throm RE, Spinola SM, Elkins C (2008) Outer membrane protein DsrA is the major fibronectin-binding determinant of Haemophilus ducreyi . Infection and Immunity 76: 1608–16. 10.1128/IAI.00994-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ochman H, Wilson AC (1987) Evolution in bacteria: Evidence for a universal substitution rate in cellular genomes. Journal of Molecular Evolution 26: 74–86. [DOI] [PubMed] [Google Scholar]
- 28. Hobolth A, Christensen OF, Mailund T, Schierup MH (2007) Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden markov model. PLoS Genetics 3: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spinola SM (2014) Haemophilus ducreyi as a cause of skin ulcers. The Lancet Global Health 2: e387 10.1016/S2214-109X(14)70250-5 [DOI] [PubMed] [Google Scholar]
- 30. Spinola SM, Wild LM, Apicella MA, Gaspari AA, Campagnari AA (1994) Experimental human infection with Haemophilus ducreyi . Journal of Infectious Diseases 169: 1146–50. [DOI] [PubMed] [Google Scholar]
- 31. Spinola SM, Griffiths GE, Bogdan J, Menegus MA (1992) Characterization of an 18,000-molecular-weight outer membrane protein of Haemophilus ducreyi that contains a conserved surface-exposed epitope. Infection and Immunity 60: 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Haydock AK, Martin DH, Morse SA, Cammarata C, Mertz KJ, Totten PA (1999) Molecular characterization of Haemophilus ducreyi strains from Jackson, Mississippi, and New Orleans, Louisiana. J Infect Dis 179: 1423–32. [DOI] [PubMed] [Google Scholar]
- 33. Al-Tawfiq JA, Thornton AC, Katz BP, Fortney KR, Todd KD, Hood AF, et al. (1998) Standardization of the experimental model of Haemophilus ducreyi infection in human subjects. J Infect Dis 178: 1684–7. [DOI] [PubMed] [Google Scholar]
- 34. Bauer ME, Townsend CA, Doster RS, Fortney KR, Zwickl BW, Katz BP, et al. (2009) A fibrinogen-binding lipoprotein contributes to the virulence of Haemophilus ducreyi in humans. Journal of Infectious Diseases 199: 684–92. 10.1086/596656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ward CK, Lumbley SR, Latimer JL, Cope LD, Hansen EJ (1998) Haemophilus ducreyi secretes a filamentous hemagglutinin-like protein. J Bacteriol 180: 6013–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leduc I, Richards P, Davis C, Schilling B, Elkins C (2004) A novel lectin, DltA, is required for expression of a full serum resistance phenotype in Haemophilus ducreyi . Infection and Immunity 72: 3418–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
Data will be available on GenBank. Accession numbers will be provided in a supplemental table.