Abstract
The Salicaceae family comprises a large number of high-biomass species with remarkable genetic variability and adaptation to ecological niches. Salix caprea survives in heavy metal contaminated areas, translocates and accumulates Zn/Cd in leaves. To reveal potential selective effects of long-term heavy metal contaminations on the genetic structure and Zn/Cd accumulation capacity, 170 S. caprea isolates of four metal-contaminated and three non-contaminated middle European sites were analysed with microsatellite markers using Wright’s F statistics. The differentiation of populations North of the Alps are more pronounced compared to the Southern ones. By grouping the isolates based on their contamination status, a weak but significant differentiation was calculated between Northern metallicolous and non-metallicolous populations. To quantify if the contamination and genetic status of the populations correlate with Zn/Cd tolerance and the accumulation capacity, the S. caprea isolates were exposed to elevated Cd/Zn concentrations in perlite-based cultures. Consistent with the genetic data nested anova analyses for the physiological traits find a significant difference in the Cd accumulation capacity between the Northern and Southern populations. Our data suggest that natural populations are a profitable source to uncover genetic mechanisms of heavy metal accumulation and biomass production, traits that are essential for improving phytoextraction strategies.
Keywords: cadmium, cross-genera transferability, genetic diversity, microsatellite, Salix caprea, soil contamination, zinc
INTRODUCTION
Phytoextraction is an emerging technology targeting on heavy metal removal from soils by accumulation in the harvestable part of plants (McGrath & Zhao 2003). Fast-growing metal-accumulating trees were considered as potential candidates for phytoextraction trials (e.g. Wieshammer et al. 2007). Members of the Salicaceae family comprise a large number of high-biomass species and hybrids with remarkable genetic variability (Pulford & Dickinson 2005). In previous field screenings, Salix caprea was identified as Zn/Cd-accumulating species (Lepp & Madejón 2007; Unterbrunner et al. 2007). Further pot experiments have shown that several isolates of S. caprea are able to accumulate large amounts of Zn (up to 2210 mg kg−1) and Cd (up to 340 mg kg−1) in their leaves (Dos Santos Utmazian & Wenzel 2007; Dos Santos Utmazian et al. 2007a; Wieshammer et al. 2007). In contrast with other tested Salix species, the growth of S. caprea isolates was less affected by high metal concentrations in soils (Dos Santos Utmazian & Wenzel 2007). However, it was unclear if the origin and the genetic makeup of S. caprea isolates correlate with Zn/Cd accumulation.
The only genetic analysis of S. caprea to date presented a high level of variation within European populations, revealing a trend that the number of chloroplastic haplotypes in the North is higher than in the South (Palmé, Semerikov & Lascoux 2003). Nuclear microsatellites, also known as simple sequence repeats (SSRs), are widely used because of their co-dominance and high variability for genetic mapping studies and population genetic analyses (Zhang & Hewitt 2003; Ellegren 2004; Schlötterer 2004; Kuroda et al. 2006). Although the number of SSR repeat motifs is highly variable, the flanking regions are conserved between related species and used to design primers for cross-species/-genera amplification (Karhu, Dieterich & Savolainen 2000; Arnold et al. 2002; Clauss, Cobban & Mitchell-Olds 2002; Tuskan et al. 2004). For several species of the Salicaceae family, SSR makers have been developed (Dayanandan, Rajora & Bawa 1998; Rahman, Dayanandan & Rajora 2000; van der Schoot et al. 2000; Lian et al. 2001; Smulders et al. 2001; Barker et al. 2003; Stamati et al. 2003; Tuskan et al. 2004). In particular, 4200 Populus SSRs are available from the International Populus Genome Consortium. Since the sequencing of the P. trichocarpa genome has been published, the chromosomal positions of the SSRs are known (Tuskan et al. 2006). In addition, Populus shares a high degree of macrosynteny (shared order of genes on chromosomes between species, families and even genera) with the genus Salix (Hanley, Mallott & Karp 2006). Roughly 100 SSRs have been tested for cross-species/-genera amplifications in Salix species with a success rate of 30–50% (Tuskan et al. 2004; http://www.ornl.gov/sci/ipgc/ssr_resource.htm). However, neither S. caprea was included in this survey, nor the amplification products confirmed by either sequencing or evaluated for their degree of polymorphisms.
Most of the population genetic studies that address heavy metal tolerance and accumulation have focused on herbaceous plants such as Thlaspi caerulescens (Meerts & van Isacker 1997; Basic et al. 2006), T. pindicum and T. alpinum (Taylor & Macnair 2006) and Arabidopsis halleri (Van Rossum et al. 2004; Pauwels et al. 2005, 2006, 2008). Heavy metal tolerance in A. halleri is largely a constitutive property and non-metallicolous populations also have a high frequency of enhanced tolerance (Bert et al. 2000; Pauwels et al. 2006). Isolates of the facultative metallophyte T. caerulescense from normal soil had a lower tolerance but were able to accumulate more Zn (Meerts & Van Isacker 1997; Escarré et al. 2000; Assunção et al. 2003; Frérot et al. 2003) and Basic et al. (2006) demonstrated that T. caerulescense populations with contrasting Cd accumulation capacities can be distinguished based on amplified fragment-length polymorphism and chloroplast markers.
Quantitative trait loci (QTL) analyses with T. caerulescens revealed that only few loci explain most of the variance for Zn and Cd concentrations (Assunção et al. 2006; Deniau et al. 2006). A comparable analysis involving a cross between A. halleri and the non-tolerant A. lyrata relative identified only three major QTLs responsible for Zn tolerance. One of the loci mapped to an already known gene involved in heavy metal uptake and this heavy metal transporting ATPase 4 (HMA4) is known to be higher expressed in A. halleri compared to Arabidopsis. lyrata sp. petraea (Courbot et al. 2007; Willems et al. 2007). Hanikenne et al. (2008) recently showed that the Zn hyperaccumulation and tolerance to Cd and Zn depend on HMA4 and that its enhanced expression is due to a combination of modified cis-regulatory sequences and copy number expansion.
To reveal potential effects of centuries of heavy metal contaminations on the structure and accumulation capacity in S. caprea, the objectives of this study were:
to evaluate the Zn/Cd accumulation potential of S. caprea in standarized conditions;
to investigate the differences in Zn/Cd accumulation in S. caprea isolates from contaminated and non-contaminated sites
to screen for the most effective isolates that exhibit a high Zn/Cd phytoextraction efficiency (i.e. heavy metal concentration × leaf biomass).
to establish microsatellite markers and elucidate the genetic structure of central European S. caprea populations from historic contaminated and non-contaminated sites.
to evaluate the genetic differentiation between populations in the concept of local adaptation.
MATERIALS AND METHODS
Selection of S. caprea isolates
In this study, we used seven S. caprea populations from three countries in Central Europe. These were: in Austria [one metal contaminated (Arnoldstein – A) and two non-contaminated sites (Forchtenstein – F, Völkermarkt – V)], in the Czech Republic {two contaminated [Příbram – PR, Kutná Hora – KH] and one non-contaminated site [Prague (P)]} and in Slovenia [one contaminated (Mežica – M) site] (Table 1). The contaminated sites were exposed to atmospheric heavy metal deposition for centuries because of the activity of local metal smelters. In the territories of the Czech Republic, lead ores were mined and processed between the Middle Ages and the year 1994 (Czech Republic Ministry of Environment 2005). The deposition of heavy metal in the area of Kutná Hora and Příbram has been well-documented (Grmela & Rapantova 2005). A similar situation exists in Mežica, where Zn and Pb mining was operating for more than 300 years until 1989 (Prestor, Strucl & Pungartnik 2003; Druzina 2006). In Arnoldstein, Pb and Zn smelting started at the end of the 15th century and was active until 1991. Information on the sites, their soil characteristics and the number of isolates is presented in Table 1 (Supporting Information Table S1, Fig. S3).
Table 1.
Site characteristics of the populations selected for the sampling of S. caprea isolates. Total Zn and Cd was determined in aqua regia extracts, whereas labile Zn and Cd were measured in 1 M NH4NO3 (1:2.5; m/v) extracts.All heavy metal concentrations are given in mg kg−1 (range)
| Population | Kutna Hora (K) Czech Republic North | Pribram (PR) Czech Republic North | Arnoldstein (A) Austria South | Mezica (M) Slovenia South | Prague (P) Czech Republic North | Forchtenstein (F) Austria South | Völkermarkt (V) Austria South |
|---|---|---|---|---|---|---|---|
| Contamination | Yes | Yes | Yes | Yes | No | No | No |
| Coordinates (lat/long) | 49° 58′ / 15° 17′ | 49° 42′ / 14° 58′ | 46° 33′ / 13° 41′ | 46° 28′ / 14° 52′ | 50° 10′ / 14° 19′ | 47° 42′ / 16° 21′ | 46° 36′ / 14° 41′ |
| Altitude (m) | 200–230 | 440–500 | 570 | 500 | 250–270 | 400–500 | 480 |
| Soil (WRB) | Cambisols | Fluvisols | Cambisols and Leptosols | Cambisols and Leptosols | Fluvisols | Cambisols | Cambisols |
| pH | 5.12–8.46 | 4.88–7.35 | 6.23–7.97 | 6.64–7.57 | 5.83–7.62 | 4.30–7.47 | 6.50–6.80 |
| Total Cd | 1.50–84.0 | 4.70–38.9 | 3.91–31.2 | 1.93–59.4 | 0.19–0.41 | 0.37–0.79 | 0.45–0.58 |
| Labile Cd | 0.005–1.21 | 0.11–4.69 | 0.03–5.06 | 0.07–0.63 | 0.005–0.0016 | 0.003–0.0052 | 0.001–0.10 |
| Total Zn | 105–8603 | 202–4390 | 871–4465 | 404–9530 | 59.9–87.4 | 80.9–519 | 58.4–63.5 |
| Labile Zn | 0.88–83.0 | 1.20–221 | 7.57–117 | 0.70–29.4 | 0.12–0.85 | 0.15–76.5 | 0.58–1.40 |
| No. specimen* | 21 (23) | 17 (20) | 21 (38) | 21 (27) | 20 (21) | 13 (20) | 19 (21) |
Number of isolates genotyped are shown in parenthesis.
Green cuttings of 20–25 individuals were collected and represent isolates. The group of isolates from each site constitutes a population. For the individual isolates, a soil sample (0–20 cm depth) below the canopy of the tree was included. In case of small, i.e. few meters, distance between the individual trees a composite soil sample was obtained and analysed (Supporting Information Table S1).
Propagation and perlite-based soil-less cultures
Green cuttings with four internodes were used for vegetative propagation in late June/early July 2004. The cuttings were rooted in quartz sand for 6 weeks in a shadowed greenhouse chamber (14/20 °C day/night temperature; 16 h light period;>90% air moisture).The final number of isolates per population that could be successfully propagated is presented in Table 1. Up to 10 rooted cuttings with similar properties, i.e. root length and diameter, were selected for each isolate and transplanted into 1 L plastic pots containing perlite. Climatic conditions were as described earlier, but with reduced air moisture (80%) and without shadowing. A Cd-free nutrient solution, containing normal Zn concentration, was provided for a period of 8 weeks.This nutrient solution contained (in μM) 1000 Ca(NO3)2, 500 Mg(SO4)2, 50 KH2PO4, 100 KCl, 5 H3BO3, 0.2 H24Mo7N6O24, 10 MnSO4, 2.5 CuSO4, 0.25 NiSO4, 2.5 ZnSO4 and 50 Fe (III)-EDDHA (ethylenediamine-di(o-hydroxyphenylacetic acid) (Shen, Zhao & McGrath 1997). Solution pH was maintained at around 6.0 with 1 mM MES as potassium salt. Each pot was surface-watered with 300 mL of this nutrient solution twice a week, following the removal of excess previously-added solution from the saucer in which the pot stood.After 8 weeks, three to five clones of each isolate with similar shoot length were selected and the height of the shoots was reduced to uniform length. Subsequently, the plants were provided twice a week with a modified solution containing elevated concentrations of Zn (5 mg L−1) and Cd (0.5 mg L−1) for a further 8 weeks.
Analysis of plant and soil material
At harvest, plant material was divided into roots, shoots and leaves. Shoot length and number of leaves was quantified (Fig. S1). Leaf samples were washed with distilled water before drying at 80 °C.After recording the DW, leaves were ground in a metal-free mill (IKA® – Werke, MF 10) and digested in a mixture of HNO3 (puriss. p. a., Sigma-Aldrich, Vienna, Austria) and HClO4 (puriss. p. a., Sigma-Aldrich) (4:1, v/v) using an automated heating block (Digester DK 42/26, Velp Scientifica, Milano, Italy). Soil samples were air-dried and passed through a 2 mm stainless-steel mesh. The sieved fraction was analysed for pH and total (aqua regia extract; Österreichisches Normungsinstitut 1999) and labile (1 M NH4NO3 extract, 1:2.5 m/v; DIN-Deutsches Institut für Normung 1995) Zn and Cd concentrations (Supporting Information Table S1). Measurement of Zn and Cd concentration was performed by flame atomic absorption spectrometry (AAS, Perkin Elmer 2100, Waltham, MA, USA). For quality assurance, replicate samples, blanks and standardized reference materials (Eurosoil 7; internal plant reference material) were included in all analyses.
DNA preparation
Genomic DNA was isolated from roughly 150 mg frozen leaves of 170 isolates after a modified protocol of Rogers & Bendich (1988). In brief, leaves were ground in liquid nitrogen in 15 mL tubes. 5 mL of 2X CTAB (2% (w/v) hexadecyltrimethylammonium bromide, Sigma-Aldrich), 100 mM Tris/HCl pH 8.0, 20 mM ethylenediaminetetraacetic acid (EDTA), 1.4 M NaCl, 1% (w/v) polyvinylpyrrolidone (PVP40T, Sigma-Aldrich) was added, vortexed and incubated at 65 °C for 20 min. Equal amount of chloroform was added and vortexed for 2 min. Following centrifugation at 4250 g for 10 min at 4 °C, the supernatant was transferred to a new tube and the DNA was precipitated by addition of 0.8 volumes isopropanol (Carl-Roth, Karlsruhe, Germany). The pellet was washed with 70% ethanol and dissolved in 100 μL TE (10 mM Tris-HCl pH 8.5, 1 mM EDTA) with 3 μL RNase (10 mg/mL, DNase free, Roth). The integrity and the quantity of the genomic DNA were evaluated on 0.8% (w/v) agarose gels containing 0.5 mg/mL of ethidium bromide.
PCR amplification, electrophoresis and sequencing
PCR amplifications were carried out in a total volume of 20 μL containing 20 ng genomic DNA, 200 μM of each dNTP, 5 pmol of forward and reverse primers (Supporting Information Table S2), 1 U TAQ polymerase and 1X PCR buffer (10 mM Tris-HCl pH 8.5, 50 mM KCl, 2 mM MgCl2, 0.15% Triton X-100). PCR was conducted in a Master Cycler-Gradient PCR machine (Eppendorf, Germany) with an initial cycle of 94 °C/3 min and 35 cycles of 15 s/50 °C, 30 s/72 °C, 15 s/94 °C. Size and amount of the PCR products were evaluated on 2% (w/v) agarose gels containing 0.5 mg/mL of ethidium bromide.
SSR amplicons were directly sequenced with the DYEnamic ET Terminator Cycle Sequencing Kit (Amersham Pharmacia Biosciences, Pittsburgh, PA, USA) and according to the ABI Prism® BigDye® Terminator Cycle Sequencing Ready Reaction Kit on the ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Carlsbad, CA, USA). Sequences were analysed by BLAST algorithm (Altschul et al.1997) at NCBI (http://www.ncbi.nlm.nih.gov) and JGI (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html). Sequence alignments were done using the MegAlign software of the DNASTAR program package (DNASTAR Inc.,Madison,WI,USA).To evaluate the degree of polymorphism in the S. caprea populations, SSR amplicons from 18 isolates (three from six populations) were separated on 5% non-denaturing polyacrylamide gels,as described previously (Hauser et al. 1998).
Genotyping
One of each primer pair was labelled with 5-HEX (5-hexachlorofluorescin), 6-FAM (6-carboxylfluorescin) or 5-NED (5-benzofluorotrichlorocarboxylfluorescin). PCR amplicons were diluted in distilled water with a ratio of 1:2 to 1:10 (depending on the amplicon concentration). For multiplexing, PCR amplicons with two different labels and fragment sizes were mixed 1:1. 1 μL and suspended in 10.3 μL consisting of 10 μL Hi-Di™ formamide (Applied Biosystems) and 0.3 μL of the size standard Genescan 400HD-ROX (Applied Biosystems). After denaturation at 94 °C for 3 min. and immediate chilling on ice, the products were analysed with the ABI Prism 3100 Genetic Analyzer (Applied Biosystems). The chromatograms were analysed using ABI Prism® Genotyper® 3.7 NT Software (Applied Biosystems).
Statistical analyses
A nested analysis of variance (general linear model) was performed to detect significant differences between populations. The factors were ‘contamination’ (which was defined ‘yes’ for contaminated sites and ‘no’ for non-contaminated) and ‘population within contamination’ (i.e. the factor ‘population’ was treated as a nested, that means second hierarchy level below the factor ‘contamination’).A level of significance of α = 0.05 was used to determine significant effects. Pearson correlations were calculated and visualized with Microsoft Excel 2002.The significance of the correlation r was evaluated with the Pearson’s table.
To assess the differences between the individual populations, a post hoc comparison of means was performed using the Scheffé test (P < 0.05). Correlation analysis (Pearson r) was performed between leaf biomass and foliar Zn and Cd concentration. All statistical analyses were performed using Statistica 6.0 for Windows (StatSoft Inc. 2001).
Allele number and frequencies for each marker, population and categorical group, expected heterozygosity (Hexp), the global and pairwise F statistics were calculated with the program SPAGeDi (Spatial Pattern Analysis of Genetic Diversity), version 1.2 after Weir & Cockerham (1984).This program operates with genotype data of any ploidy level (Hardy & Vekemans 2002). The Weir & Cockerham algorithm for the F statistics is based on a nested anova, where populations are weighted according to their sample size. Bonferroni correction was applied to correct for multiple-comparisons (Sokal & Rohlf 1995). Observed heterozygosity (Hobs) was computed using the MSA program (Dieringer & Schlötterer 2003). Significance (P values) of non-random distributions of specific alleles was calculated with a permutation test of 1000 replications in Microsoft Office® Excel and a Fisher’s exact test.
We assessed evidences of selection (i.e. adaptation in contaminated populations) using the lnRH test and calculated the average gene diversity Hexp of each locus for the contaminated (μc) and uncontaminated populations (μu) and the formula Θ = (1/(1 − Hexp))2 − 1 as described by Kauer, Dieringer & Schlötterer (2003). The Θ values were normalized and Bonferroni corrected to get significance values (Supporting Information Table S3).
RESULTS
For all isolates tested in the perlite-based soil-less culture (n = 132), the average (mean ± standard deviation) foliar concentrations were 812 ± 238 mg Zn kg−1 and 115 ± 56.0 mg Cd kg−1. Mean leaf biomass was 2.91 ± 1.55 g plant−1.
Influence of isolate origin
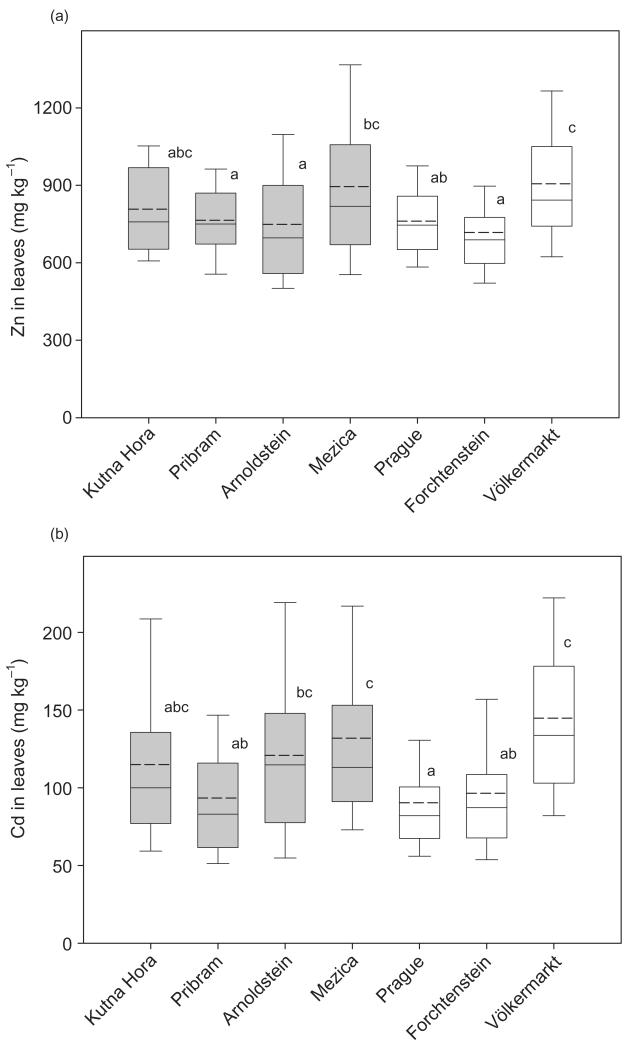
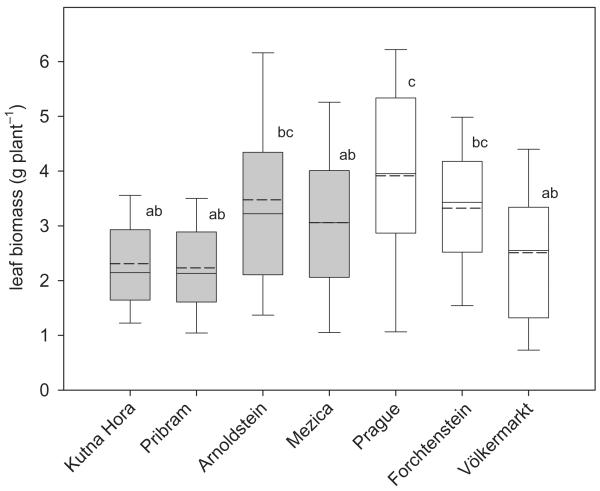
The factor ‘contamination’ had no significant influence on the Cd and Zn concentration in leaves. In contrast, leaf DW of isolates from contaminated sites was significantly lower than for isolates from non-contaminated sites (Table 2).The factor ‘population within contamination’ had a significant influence on all the investigated variables (Table 2). In spite of this, the differences of foliar Zn and Cd concentration (Fig. 1), as well as of leaf biomass (Fig. 2), shoot length and leaf number (Supporting Information Fig. S1) between the individual populations were rather small and only a few significant differences were detected. Comparing the results of the perlite-based soil-less culture with the soil characteristics of the isolate’s origin, we found a positive and significant trend. However, this correlation was only positive up to a contamination level of 20 mg kg−1 total Cd. Isolates from soils above this total Cd concentration did not increase their accumulation capacities in perlite-based soil-less cultures (Supporting Information Fig. S5).
Table 2.
Main effects of the factors ‘contamination’ and ‘population within contamination’ determined by a nested analysis of variance (general linear model, type III decomposition). F values were treated as fixed. F values were calculated by dividing the mean square (MQ) of either factor by the mean square of the error.A significant effect is indicated by a P value below 0.05
| Variable | Factor | SS* | df | MQ | F | P |
|---|---|---|---|---|---|---|
| Biomass (g plant−1) | Constant | 4 768 | 1 | 4 768 | 2 335 | 0.00 |
| Contamination | 37.8 | 1 | 37.7 | 18.4 | 0.00 | |
| Population within contamination | 177 | 5 | 35.3 | 17.3 | 0.00 | |
| Error | 1 170 | 573 | 2.02 | – | – | |
| Cd (mg kg−1) | Constant | 6 912 645 | 1 | 6 912 645 | 2 451 | 0.00 |
| Contamination | 4 163 | 1 | 4 163 | 1.48 | 0.22 | |
| Population within contamination | 194 709 | 5 | 38 942 | 13.8 | 0.00 | |
| Error | 1 616 241 | 573 | 2 821 | – | – | |
| Zn (mg kg−1) | Constant | 348 160 948 | 1 | 348 160 948 | 6 566 | 0.00 |
| Contamination | 7 073 | 1 | 7 073 | 0.13 | 0.72 | |
| Population within contamination | 240 9978 | 5 | 481 996 | 9.09 | 0.00 | |
| Error | 30 384 698 | 573 | 53 027 | – | – |
Sum of squares.
Figure 1.
Foliar Zn (a) and Cd (b) concentration in S. caprea isolates derived from seven different populations. Boxes represent the median (vertical solid line), the arithmetic mean (vertical dashed line), and 25–75% percentile.Whiskers represent the 90th and 10th percentile. Significant differences were determined by a post hoc comparison of means (Scheffé test after nested anova; P < 0.05) and are indicated by different letters.
Figure 2.
Total amount of leaf biomass per plant for S. caprea isolates derived from seven different populations. Boxes represent the median (vertical solid line), the arithmetic mean (vertical dashed line), and 25–75% percentile.Whiskers represent the 90th and 10th percentile. Significant differences were determined by a post hoc comparison of means (Scheffé test after nested anova; P < 0.05) and are indicated by different letters.
Comparison of individual isolates and correlations
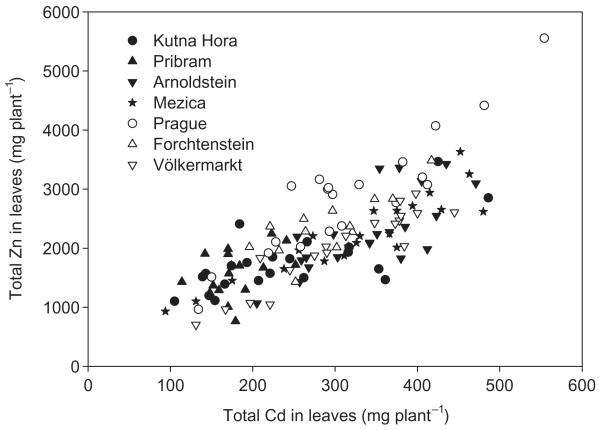
Figure 3 shows metal contents (metal concentration × leaf biomass) in leaves of each isolate, which ranged from 704 to 5560 μg Zn plant−1 and from 94.1 to 554 μg Cd plant−1, respectively. The highest total Cd and Zn contents were found for an isolate from the non-contaminated population Prague. Both the foliar concentration of Zn and Cd (r = 0.79) and the total content of Zn and Cd in leaves (r = 0.79) correlated significantly. The leaf biomass had a higher influence (r = 0.85 and 0.58, respectively) on the total foliar Zn and Cd contents than the corresponding Zn and Cd concentrations (r = 0.10 and 0.22, respectively).
Figure 3.
Correlation between total Zn and total Cd in leaves of the individual isolates (n = 132; r = 0.79; P < 0.001).
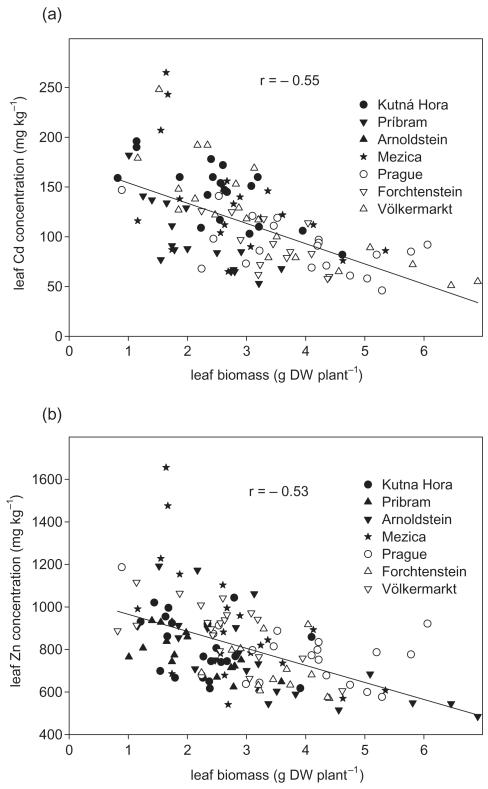
Correlation between growth and Zn/Cd concentration
A negative correlation was found between foliar Zn and Cd concentration in leaves and the total leaf biomass (Fig. 4), indicating a decrease of shoot growth at increasing foliar Zn and Cd concentration. This observation could be due to Zn and/or Cd toxicity or might be caused by a ‘dilution effect’. Further potential signs of toxicity, i.e. chlorotic and necrotic spots on leaves, were detected only for a few specimens and were not correlated with foliar Zn and Cd concentrations (data not shown).
Figure 4.
Relationship between the leaf biomass and the foliar Cd (A) and Zn (B) concentration.
Comparing the results of the perlite-based soil-less culture with the soil characteristics of the isolate’s origin, we found that no significant dependency existed between biomass production and the Zn and Cd levels in the isolates derived from contaminated sites (Supporting Information Fig. S4). However, the biomass negatively correlates with the much lower total Cd levels of uncontaminated soils. Thus the isolates of uncontaminated sites are more sensitive to elevated Cd and Zn exposures while isolates from contaminated sites seem to be more resistant to higher doses of Cd and Zn under soil-less condition and might have been selected to withstand higher Cd and Zn concentrations.
Transferability of P. trichocarpa, P. nigra, S. lanata and S. burjatica SSR markers is low
To characterize the genetic structure of the S. caprea populations, a total of 83 Populus ssp. and 10 Salix ssp. SSRs loci were tested for cross-amplification (Table 3). Whereas around 70% of the Populus and all Salix SSR could be amplified, only half of these amplicons had roughly the expected fragment size and contained SSR repeats (Table 3).
Table 3.
Summary of cross-genera and -species amplification of SSR markers in S. caprea
| P. trichocarpa | P. nigra | S. burjatica | S. lanata | |
|---|---|---|---|---|
| SSRs tested | 62 | 21 | 3 | 7 |
| Amplified | 45 | 14 | 3 | 6 |
| Unexpected amplicon size | 15 | 3 | – | 1 |
| Sequenced | 23 | 11 | 3 | 4 |
| SSR present | 12 | 7 | 3 | 4 |
| Wrong amplicon | 7 | – | – | – |
| No repeat | 5 | 3 | – | – |
| Different repeat | 4 | – | – | – |
| Polymorphic | 3 | 4 | 3 | 1 |
| Success rate (%) | 4.8 | 19.1 | 100 | 14.3 |
Sequencing of the PCR-amplified loci revealed a high degree of conservation in the flanking regions of the SSRs between genera and species. Interestingly, five of P. trichocarpa and three of the P. nigra loci lost completely the repeat region in S. caprea, although the rest of the sequence was conserved. Four loci had altered repeat motives: While in P. trichocarpa, the repeat motif of ORPM_62 was [AT]4.[ATTTT]3, it exists as [AT]2… [T]8 in S. caprea. ORPM_86 changed from [CTT]5 to [CTT]2[CAT]8, ORPM_137 from [AT]7 to [GT]4 and ORPM_312 from [CCT]6 to [CTT]5 in S. caprea. Six SSR amplicons had no homology to any Populus SSR locus (in Table 3 indicated as wrong amplicons, Supporting Information Table S2). Only 11 SSR loci were polymorphic enough to be used for the genetic structure analyses of S. caprea. Assuming a high degree of synteny between P. trichocarpa and S. caprea, these loci are located on nine different chromosomes (Supporting Information Table S2). The alleles of these 11 loci differed mainly in unit size, although some alleles did not fit the standard unit series similar to previous reports for P. tremuloides (Cole 2005).
Beside the overall low transferability for Populus (below 10%) and Salix SSRs (40%), the 11 SSR markers could also be amplified in S. cinerea, S. fragilis, S. viminalis and S. purpurea.
Allelic variation of S. caprea is high and, for some populations, specific
All 11 SSRs were highly polymorphic in 170 isolates. In total, 202 alleles were identified and their number per locus varied from 5 to 30 (Table 4).The level of heterozygosity of each locus was between Hexp 0.34 and 0.89 (Table 4). Three highly polymorphic SSR markers (WMPS_21, SB_38, gSIMCT024) were sufficient to unambiguously distinguish the genotypes among individuals. Interestingly, none of the isolates shared the same genotype. In contrast to some Salix species that spread clonally or have a mixed reproduction system, our genetic results are in accordance with the fact that no asexual propagation occurred in the sampled S. caprea populations. On average, 19 and 29% of the isolates had more than two alleles or only one allele of a particular SSR, respectively (Table 4). Of the 1870 loci evaluated, 15 and 2.8% had three and four alleles, respectively.Therefore, the genetic diversity and population differentiation was calculated with a program that accepts data sets containing individuals with multialleles and different ploidy levels (Hardy & Vekemans 2002).
Table 4.
Allelic variations, heterozygosity, FST and P-values and frequencies of multi- and single allele loci
| Locus | No. of Alleles | H obs | H exp | % multiallele loci | % single allele loci | F ST | P value |
|---|---|---|---|---|---|---|---|
| ORPM_62 | 14 | 0.49 | 0.74 | 20.00 | 50.63 | 0.035 | 0.0005 |
| ORPM_312 | 18 | 0.81 | 0.82 | 12.80 | 19.02 | 0.012 | 0.012 |
| ORPM_446 | 25 | 0.56 | 0.81 | 13.94 | 44.24 | 0.009 | 0.174 |
| WMPS_12 | 5 | 0.34 | 0.33 | 0.61 | 66.06 | 0.052 | 0.000 |
| WMPS_14 | 21 | 0.85 | 0.84 | 39.52 | 14.97 | 0.007 | 0.062 |
| WMPS_19 | 30 | 0.84 | 0.88 | 33.13 | 16.27 | 0.007 | 0.032 |
| WMPS_21 | 12 | 0.79 | 0.76 | 31.36 | 21.30 | 0.014 | 0.003 |
| SB_24 | 15 | 0.89 | 0.77 | 10.65 | 11.24 | 0.005 | 0.196 |
| SB_38 | 30 | 0.82 | 0.91 | 11.15 | 17.47 | 0.016 | 0.000 |
| SB_199 | 16 | 0.61 | 0.76 | 23.64 | 38.18 | 0.011 | 0.023 |
| gSIMCT024 | 18 | 0.85 | 0.80 | 9.41 | 14.71 | 0.009 | 0.040 |
| All loci | 0.0143 | 0.000 |
The occurrence of multiallele loci may be the results of local genome amplifications. To determine if the frequency of multiallele loci is significantly different in contaminated versus uncontaminated populations, we used the Student’s t-test (Table 5). While no significance was detected if all loci were compared, contaminated populations had significantly more multiples alleles at loci ORPM_312 and SB_24, indicating that genome amplification may have occurred. Assuming a high degree of synteny between Populus and Salix, these two loci are not on the same chromosome (Supporting Information Table S2). Another marker on the same chromosome, SB_24, does not show this correlation indicating that partial genome duplication may have occurred at specific loci and chromosomal regions.
Table 5.
Frequency of multiallele (ma) loci in S. caprea isolates of contaminated (A, M, KH, Pr) and uncontaminated (F, P,V) populations. n indicates the number of individuals. A P value < 0.05 indicates that significantly more multiples alleles are present at these loci
| n | ORPM_62 | ORPM_312 | ORPM_446 | WMPS_12 | WMPS_14 | WMPS_19 | WMPS_21 | SB_24 | SB_38 | SB_199 | gSIMCT024 | Mean | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 38 | 22.86 | 16.22 | 21.05 | 0.00 | 27.03 | 43.24 | 36.84 | 13.51 | 13.16 | 15.79 | 5.26 | 19.5 |
| M | 27 | 8.70 | 23.08 | 3.70 | 3.70 | 51.85 | 11.54 | 25.93 | 18.52 | 7.69 | 37.04 | 3.70 | 17.8 |
| KH | 23 | 13.04 | 9.05 | 4.35 | 0.00 | 17.39 | 47.83 | 27.27 | 8.70 | 13.04 | 18.18 | 21.74 | 17.3 |
| Pr | 20 | 15.79 | 15.00 | 0.00 | 0.00 | 60.00 | 60.00 | 45.00 | 15.00 | 21.05 | 10.53 | 5.00 | 22.5 |
| F | 20 | 31.58 | 5.00 | 10.00 | 0.00 | 60.00 | 15.79 | 25.00 | 5.00 | 10.00 | 16.67 | 5.00 | 16.7 |
| P | 21 | 33.33 | 4.76 | 10.00 | 0.00 | 52.38 | 38.10 | 52.38 | 0.00 | 5.00 | 28.57 | 14.29 | 21.7 |
| V | 21 | 10.00 | 5.56 | 0.00 | 0.00 | 15.79 | 14.29 | 4.76 | 9.52 | 9.52 | 33.33 | 14.29 | 10.6 |
| Mean | 20.00 | 12.80 | 13.94 | 0.61 | 39.52 | 33.13 | 31.26 | 10.65 | 11.15 | 23.64 | 9.41 | ||
| P value | 0.23 | 0.0016 | 0.93 | 0.44 | 0.83 | 0.25 | 0.64 | 0.041 | 0.17 | 0.50 | 0.71 | 0.38 |
Furthermore, at two loci, specific alleles have been detected for the Northern (ORPM_446/246, WMPS_19/174) and one (gSIMCT024/114) for the contaminated populations (Table 6). Using Fisher’s exact and a permutation test, the probability that these three alleles occurred by chance is below the significance level (Table 6).
Table 6.
Frequencies and significance (P value) of population-specific alleles
| Populations | ORPM_446/246 | WMPS_19/174 | gSIMCT024/114 |
|---|---|---|---|
| A | 0.092 | 0.023 | 0.013 |
| M | 0.038 | 0.077 | 0.019 |
| F | 0.075 | 0.079 | 0.000 |
| V | 0.139 | 0.024 | 0.000 |
| KH | 0.000 | 0.000 | 0.058 |
| PR | 0.000 | 0.000 | 0.077 |
| P | 0.000 | 0.000 | 0.000 |
| P values | 0.001 | 0.010 | 0.0 |
Genetic differentiation of S. caprea isolates is significant for the contamination status, the geographic origin and for isolates with a high leaf biomass production
Differentiation within a random mating population is correlated with a loss of genetic variation and a decrease in heterozygosity between subpopulations. To measure the degree of differentiation between the geographically distinct S. caprea populations Wright’s F statistics was used. Isolates from Prague (P) showed the highest average pairwise FST values, and thus this population is significantly different from all others. Because of the Prague populations, the differentiation is more pronounced North of the Alps (KH, PR, P) compared to the populations South of the Alps (A, M, V) (Table 7). The population of Forchtenstein (F) although situated North-East of the Alps is genetically closer to the Southern populations (Table 1).
Table 7.
Pairwise FST and P values among S. caprea populations. FST values in bold and italic are significant Bonferroni corrected and not significant, respectively
|
F
ST
|
|||||||
|---|---|---|---|---|---|---|---|
| A | M | V | F | KH | PR | P | |
| P | |||||||
| A | 0.0118 | 0.0041 | 0.0076 | 0.0108 | 0.0108 | 0.0176 | |
| M | 0.0025** | 0.0155 | 0.0035 | 0.0090 | 0.0117 | 0.0303 | |
| V | 0.3599 | 0.0121* | 0.0136 | 0.0171 | 0.0272 | 0.0256 | |
| F | 0.0579 | 0.5586 | 0.0299* | 0.0079 | 0.0123 | 0.0184 | |
| KH | 0.0085* | 0.0365* | 0.0039* | 0.1053 | 0.0123 | 0.0309 | |
| PR | 0.0059* | 0.0119* | 0.0000** | 0.0145* | 0.0063* | 0.0235 | |
| P | 0.0000** | 0.0000** | 0.0000** | 0.0005** | 0.0000** | 0.0000** | |
P < 0.05 with Bonferroni correction;
P < 0.05 without Bonferroni correction.
By grouping the populations based on the contamination status, a weak but significant differentiation was calculated between metallicolous and non-metallicolous isolates (Table 8). This differentiation is mainly due to the markers ORPM_62, WMPS_12, WMPS_21 and SB_38.
Table 8.
Summary of the FST calculations for individual and all loci
| Category | F ST | P-value | Bonferroni |
|---|---|---|---|
| Metallicolous versus non-metallicolous | |||
| ALL LOCI | 0.0053 | 0.0003 | * |
| ORPM_62 | 0.0346 | 0.0009 | * |
| WMPS_12 | 0.0517 | 0.0007 | * |
| WMPS_21 | 0.0141 | 0.0035 | * |
| SB_38 | 0.0160 | 0.0003 | * |
| North-South | |||
| ALL LOCI | 0.005 | 0.0005 | * |
| ORPM_446 | 0.0145 | 0.0095 | |
| WMPS_21 | 0.0112 | 0.0033 | * |
| North: metallicolous versus non-metallicolous | |||
| ALL LOCI | 0.0232 | 0.0000 | * |
| WMPS_14 | 0.0235 | 0.0043 | * |
| Dry weight* | |||
| ALL LOCI | 0.0124 | 0.0219 | |
| gSIMCT024 | 0.0617 | 0.0007 | * |
Isolates with leaf dry weights higher than 5 mg/kg were compared to the rest.
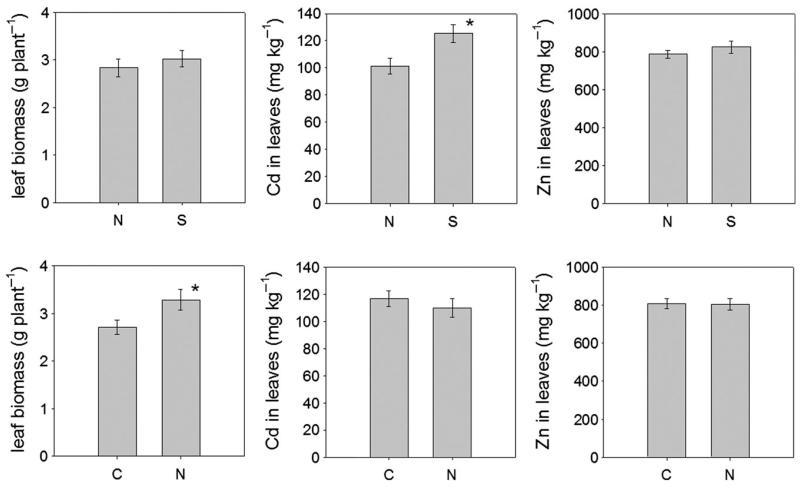
Separated FST values for individual loci were also calculated by grouping Southern and Northern populations. The differentiation was mainly due to the marker WMPS_12. Within the Northern populations, differentiation was found between the metal contaminated (KH, PR) and non-contaminated (P) populations. However, the differentiation between Southern metallicolous (A, M) and nonmetallicolous (V) populations was not significant, although it is significant between Arnoldstein (A) and Mežica (M), which are separated by the Karawanken mountains, but not between Arnoldstein (A) and Völkermarkt (V), which are in the same valley. These genetic data are consistent with the geographic differentiation of the nested anova analyses for the physiological traits that find a significant difference in the Cd accumulation capacity between the Northern and Southern populations (Fig. 5).
Figure 5.
Comparison of leaf biomass and Zn/Cd concentration of Salix caprea between isolates from Northern (N) and Southern (S) populations as well as between specimen from contaminated (C) and non-contaminated (N) sites. Error bars represent the standard error of the mean.A significant difference (P < 0.05) is indicated by an asterisk.
Next, we grouped S. caprea individuals according to their physiological characteristics such as biomass (e.g. below and above 5 mg kg−1 leaf dry weight). For this trait the marker gSIMCT024 had a significant FST value.
This first genetic analysis of S. caprea with nuclear marker shows that there is a weak but significant geographic differentiation of populations North and South the Alps. Moreover, S. caprea populations of contaminated areas are genetically distinct. Apart from significant FST values for individual SSR markers, our data suggest that local genome amplifications might occur during adaptation to heavy metal contaminated environments.
DISCUSSION
We are aware of the fact that some significant limitations may be associated with soil-less experiments (e.g. regarding the extrapolation of results to soil conditions). Some problems associated with hydroponic and soil-lees studies have been previously discussed by Stoltz & Greger (2002), Watson, Pulford & Riddel-Black (2003) and Dos Santos Utmazian et al. (2007b). However, we have chosen this approach since Watson et al. (2003) also claim that soil-less hydroponic screening tests comprise a method for differentiation between isolates under standardized conditions. In order to overcome problems that were found for S. caprea when growing in nutrient solution (Dos Santos Utmazian et al. (2007b), we have decided to use a perlite-based soil-less culture.
The first pot experiments with S. caprea isolates from contaminated sites revealed a large Cd and Zn accumulation capacity of this species (Dos Santos Utmazian & Wenzel 2007; Dos Santos Utmazian et al. 2007a). Interestingly, similar concentrations were found in a specimen from a tree nursery (Wieshammer et al. 2007). Therefore, the question arose if the Cd and Zn accumulation capacity is a constitutive or an adaptive property of S. caprea. The comparison of seven populations from four contaminated and three non-contaminated sites suggests that it is likely a constitutive property, because isolates obtained from non-contaminated sites had similar Zn and Cd concentrations in their leaves compared with those from contaminated sites (Fig. 1). Landberg & Greger (1996) and Vysloužilová et al. (2006) have reported similar observations for other Salix species.
The Cd and Zn translocation factor was investigated for four selected isolates: two with the lowest and two with the highest foliar Cd and Zn concentration. For the two low accumulator isolates, the ratio was 0.6, but for the two high-accumulator isolates, the ratio was 1 and 1.8, respectively. This demonstrates the high capacity of some S. caprea isolates to translocate Cd from roots to shoots. Even higher shoot:root ratios were reported for a S. caprea isolate grown in contaminated soil (up to 2.3; Dos Santos Utmazian & Wenzel 2007). Regarding the uptake of Cd, the correlation analysis between foliar Cd and Zn or Ca concentrations suggest that Cd is taken up more likely by the carrier system for Zn (r = 0.79) than for Ca (r = 0.51). Previous reports suggest that Cd may be taken up by both Zn and Ca carriers (Welch & Norvell 1999; Zhao et al. 2002; Antosiewicz & Hennig 2004; Pittman et al. 2004; Korenkov et al. 2007, 2009; Parameswaran et al. 2007; Moreno et al. 2008; Mei et al. 2009; Morel et al. 2009; Oomen et al. 2009; Küpper & Kochian 2010).
Based on the anova analysis with isolates grouped according to the contamination status, no significant difference was found for the foliar Zn and Cd concentration (Fig. 5). Nevertheless, signs of adaptation were revealed through the significant positive correlation between the Cd concentration of the soils and the isolates capability to accumulate Cd in leaves in soil-less perlite conditions. However, this correlation was only significant for total Cd concentrations of up to 20 mg kg−1 soil. Isolates from soils above this level did not exhibit a further enhancement of their Cd accumulation capabilities (Fig. S5). Leaf biomass was, on average, larger for the isolates from non-contaminated sites.These findings suggest that: (1) most S. caprea isolates are good extractors and accumulators of Zn and Cd; and (2) the contamination of the site of isolate origin has a significant influence on foliar biomass production and Cd but not Zn accumulation. The constitutive property of metal accumulation has been previously reported for the hyperaccumulators A. halleri (Bert et al. 2000, 2002; Macnair 2002) and T. caerulescens (Meerts & Van Isacker 1997; Escarré et al. 2000). Our data suggest a similar trait for S. caprea.
The significant influence of the factor ‘population’ on biomass production and foliar Cd and Zn concentrations suggests that the isolates from each site represent a distinct population. This geographical difference was supported by genetic analyses, which show that the Southern populations are distinct from the Northern ones (Table 8). However, the differences between the populations are less pronounced (sometimes even not significantly different) than expected. The average foliar Cd concentration in the population with the lowest mean value (Prague) is only 37% lower compared to the population with the highest mean value (Völkermarkt). For Zn, the difference between Mežica (highest) and Forchtenstein (lowest) is even less (22%).
Among the populations from non-contaminated sites, plants from Völkermarkt are characterized by higher Cd concentrations than those from Prague or Forchtenstein. This may be explained by the rather short geographical distance between Völkermarkt and contaminated sites in the vicinity (including Mežica and Arnoldstein). The hypothesis that populations from Völkermarkt and Mežica or Arnoldstein are genetically closely related was supported by our genetic analysis that could not detect a significant differentiation between Völkermarkt and Arnoldstein and Völkermarkt and Mežica (Table 7).
Signs of differentiation and adaptation are best detectable by genetic analyses with molecular marker such as the co-dominant nuclear SSRs. Since Tuskan et al. (2004) published that 30–50% of the Populus SSRs can be amplified in different Salix species, we used the rich source of mapped SSRs of the fully sequenced Populus genome (Tuskan et al. 2006). Although 73% of the Populus SSRs could be amplified in S. caprea, only 10% were polymorphic and could be used for the genetic analysis of our populations. This low rate of transferability of Populus SSRs for Salix ssp. implies that Populus SSRs are not recommendable for whole genome association studies in Salix.
To date, few data are available on the population structures of S. caprea (Palmé et al. 2003) and other Salix species (Reisch, Schurm & Poschlod 2007). Our genetic analysis of seven middle European populations found a very low level of differentiation with FST values between 0.0118 and 0.052 (Tables 7 and 8) and agrees with data of maternally inherited chloroplast markers (PCR-RFLPs) that revealed a low genetic differentiation (GST = 0.09) between S. caprea populations (Palmé et al. 2003). Palmé et al. (2003) also reported that the genetic diversity was higher in the Northern populations, a finding that is consistent to our significant differentiation of the populations North and South the Alps. Our data are also comparable with an SSR analysis on P. tremuloides populations, where the FST values varied between 0.006 and 0.045 (Cole 2005).
The frequency of multialleles differed between the populations and loci. At two loci, it was significantly higher in population of contaminated areas, independent of the geographic origin. This result may come from an ancient introgression event with a polyploid Salix species and subsequent loss of most of the introduced chromosomes. On the other hand, the presence of multialleles might point to local genome amplifications, a phenomenon that has been identified in all the recently sequenced plant genomes including that of P. trichocarpa (Tuskan et al. 2006). DNA quantification with flow cytometry showed that S. caprea had the highest intraspecific variation of DNA content among Salix species (Thibault 1998). Correspondingly, our flow cytometric DNA quantification of all S. caprea isolates found a similar variation (data not shown). Although further Southern blot or sequence analyses are needed, the data are indicative that S. caprea may use gene number expansion in the adaptation process to specific environments in a similar manner as it has been described for A. halleri by Hanikenne et al. (2008).
To test if other loci have signs of selection, the lnRH test was used that compares significant reductions in variability of each locus in each population in relation to all loci (Supporting Information Table S3). Effects of demography, such as bottlenecks, are removed by the lnRH test because these would affect all loci at the same degree. According to this analysis only for the WMPS_14 locus a sign of selection was revealed that lost the significance after Bonferroni. However, the lnRH test was designed for genome scans analysing a large number of loci.With the moderate number of loci used in this study, the chances to detect a genomic region subjected to selection are small. It is also unclear how the partial deviation from a strict diploid genome affects the FST – heterozygosity correlation and the lnRH value. A simulation test is, however, beyond the scope of this paper. Other signs of selection are the significant correlations between specific loci and physiological traits (Table 8), and the detection of three alleles that are specific for the geographic localization and the contamination status of the populations (Table 6).
To our knowledge, this is the first study on the population structure of S. caprea based on nuclear markers that associates phenotypic with genotypic variability. The weak indications of differentiation between populations and among isolates of specific physiological characteristics suggest that selection might have shaped the populations. To increase the resolution of the genetic analysis, a larger number of polymorphic sequences are needed. Our results exemplify that deeper molecular analyses of isolates with contrasting performance could be an effective measure for understanding the molecular basis of Zn/Cd tolerance, accumulation as well as biomass production all traits that are essential for improving the phytoextraction efficiency.
Supplementary Material
Figure S1. Shoot length (A) and leaf number (B) of S. caprea isolates. Boxes represent the median (vertical solid line), the arithmetic mean (vertical dashed line), and 25–75% percentile. Whiskers represent the 90th and 10th percentile. Significant differences were determined by a post hoc comparison of means (Scheffé test after nested anova; P < 0.05) and are indicated by different letters.
Figure S2. Geographic map indicating the location of the S. caprea populations. South the Alps in Slovenia the contaminated population in Mežica – M, and in Austria the contaminated population Arnoldstein – (A) and the non-contaminated sites, Völkermarkt – V and Forchtenstein – F. North the Alps in the Czech Republic the two contaminated populations Příbram – PR and Kutná Hora – KH and the non-contaminated site near Prague (P).
Figure S3. The graphs show that the amount of labile Zn and Cd depends with a significance level for Pearson’s correlation of P < 0.01 on the total Zn and Cd concentration in the soil. In fact, 40% of the variation of labile Zn and 36% for Cd are explained by the total Zn and Cd concentration, respectively. Other factors influencing the labile heavy metal fractions are pH, content of clay, carbonate and organic matter.
Figure S4. Pearson’s correlations between the level of contamination where the isolate originated and the biomass production in perlite cultures exposed to elevated levels of Cd and Zn.A and B are the graphs for the uncontaminated, and C and D for the contaminated sites. The only significant negative correlation between Cd concentration and biomass production was seen at uncontaminated sites (A). Isolates from the contaminated sites did not show such a correlation indicating that they might have been selected to withstand higher Cd and Zn concentrations. Note that two soil samples from the uncontaminated sites had very high Zn concentrations probably because of a nearby rusty fence that leached into the soil (B).
Figure S5. Pearson’s correlations between the level of soil contamination and Cd concentration in leaves after the exposure of the isolates in perlite to Cd and Zn.Although a significant trend was found between soil contamination and accumulation capacity in perlite cultures for soil contamination below 20 mg kg−1 (A, B), above this soil Cd contamination level, the trend diminished.
Table S1. Summary of the soil characteristics for each isolate and their growth and accumulation behaviour in perlite-based soil-less cultures exposed to elevated levels of Cd (0.5 mg L−1) and Zn (5 mg L−1).
Table S2. Overview of the SSR marker characteristics.
Table S3. Heterozygosities, lnRH and normalized lnRH values of the 11 loci and 7 populations. Loci with Norm. Θ-values of larger +1.95 and smaller −1.95 have a significant reduced variability and thus might have been under selection and contribute to adaptation in the contamination area.
ACKNOWLEDGMENTS
We gratefully acknowledge Christian Schlötterer for his valuable advices and critical reading of the manuscript. We are thankful to Claire Arnold and Nada Hamza for sharing some of the S. burjatica and S. lanata primers, Daniel Dieringer for his help with the sequence analyser and some statistical analyses and Agata Mansfeld for her kind introduction into the polyacrylamide gel electrophoresis, Elisabeth Netherer for her help with the heavy metal quantifications and Joseph Glössl for his general support. We thank Ewald Brauner,Alex Dellantonio,Xiaojiang Gao, Evi Oburger, Gorana Todorovic and Siegfried Sowboda for their help in greenhouse and laboratory work.This study was supported by the Vienna Fund for Science and Technology (WWTF LS-149) and the FWF (project L433_B17).
Footnotes
SUPPORTING INFORMATION Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
REFERENCES
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antosiewicz DM, Hennig J. Overexpression of LCT1 in tobacco enhances the protective action of calcium against cadmium toxicity. Environmental Pollution. 2004;129:237–245. doi: 10.1016/j.envpol.2003.10.025. [DOI] [PubMed] [Google Scholar]
- Arnold C, Rossetto M, McNally J, Henry RJ. The application of SSRs characterized for grape (Vitis vinifera) to conservation studies in Vitaceae. American Journal of Botany. 2002;89:22–28. doi: 10.3732/ajb.89.1.22. [DOI] [PubMed] [Google Scholar]
- Assunção AGL, Bookum WM, Nelissen HJM, Vooijs R, Schat H, Ernst WHO. Differential metal-specific tolerance and accumulation patterns among Thlaspi caerulescens populations originated from different soil types. New Phytologist. 2003;159:411–419. doi: 10.1046/j.1469-8137.2003.00819.x. [DOI] [PubMed] [Google Scholar]
- Assunção AGL, Pieper B, Vromans J, Lindhout P, Aarts MG, Schat H. Construction of a genetic linkage map of Thlaspi caerulescens and quantitative trait loci analysis of zinc accumulation. New Phytologist. 2006;170:21–32. doi: 10.1111/j.1469-8137.2005.01631.x. [DOI] [PubMed] [Google Scholar]
- Barker JHA, Pahlich A, Trybush S, Edwards KJ, Karp A. Microsatellite markers for diverse Salix species. Molecular Ecology Notes. 2003;3:4–6. [Google Scholar]
- Basic N, Salamin N, Keller C, Galland N, Besnard G. Genetic differentiation of Thlaspi caerulescens natural populations in relation to their cadmium hyperaccumulation capacity. Biochemical Systematics and Ecology. 2006;34:667–677. [Google Scholar]
- Bert V, Macnair MR, de Laguérie P, Saumitou-Laprade P, Petit D. Zinc tolerance and accumulation in metallicolous and non-metallicolous populations of Arabidopsis halleri (Brassicaceae) New Phytologist. 2000;146:225–233. doi: 10.1046/j.1469-8137.2000.00634.x. [DOI] [PubMed] [Google Scholar]
- Bert V, Bonnin I, Saumitou-Laprade P, de Laguérie P, Petit D. Do Arabidopsis halleri from non-metallicolous populations accumulate zinc and cadmium more effectively than those from metallicolous populations? New Phytologist. 2002;155:47–57. doi: 10.1046/j.1469-8137.2002.00432.x. [DOI] [PubMed] [Google Scholar]
- Clauss MJ, Cobban H, Mitchell-Olds T. Cross-species microsatellite markers for elucidating population genetic structure in Arabidopsis and Arabis (Brassicaeae) Molecular Ecology. 2002;11:591–601. doi: 10.1046/j.0962-1083.2002.01465.x. [DOI] [PubMed] [Google Scholar]
- Cole CT. Allelic and population variation of microsatellite loci in aspen (Populus tremuloides) New Phytologist. 2005;167:155–164. doi: 10.1111/j.1469-8137.2005.01423.x. [DOI] [PubMed] [Google Scholar]
- Courbot M, Willems G, Motte P, Arvidsson S, Roosens N, Saumitou-Laprade P, Verbruggen N. A major quantitative trait locus for cadmium tolerance in Arabidopsis halleri colocalizes with HMA4, a gene encoding a heavy metal ATPase. Plant Physiology. 2007;144:1052–1065. doi: 10.1104/pp.106.095133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech Republic Ministry of Environment Report on the environment in the Czech Republic. Ministerstvo životního prostředí – Referenční informační středisko, Praha 10, Czech Republic. 2005.
- Dayanandan S, Rajora OP, Bawa KS. Isolation and characterization of microsatellites in trembling aspen (Populus tremuloides) Theoretical and Applied Genetics. 1998;96:950–956. [Google Scholar]
- Deniau AX, Pieper B, Ten Bookum WM, Lindhout P, Aarts MG, Schat H. QTL analysis of cadmium and zinc accumulation in the heavy metal hyperaccumulator Thlaspi caerulescens. Theoretical and Applied Genetics. 2006;113:907–920. doi: 10.1007/s00122-006-0350-y. [DOI] [PubMed] [Google Scholar]
- Dieringer G, Schlötterer C. Microsatellite analyzer (MSA): a platform independent analysis tool for large microsatellite data sets. Molecular Ecology Notes. 2003;3:167–169. [Google Scholar]
- DIN-Deutsches Institut für Normung . Soil Quality Extraction of Trace Elements with Ammonium Nitrate Solution, DIN 19730. Beuth Verlag; Berlin, Germany: 1995. [Google Scholar]
- Dos Santos Utmazian MN, Wenzel WW. Cadmium and zinc accumulation in willow and poplar species grown on polluted soils. Journal of Plant Nutrition and Soil Science. 2007;70:265–272. [Google Scholar]
- Dos Santos Utmazian MN, Schweiger P, Sommer P, Gorfer M, Strauss J, Wenzel WW. Influence of Cadophora finlandica and other microbial treatments on cadmium and zinc uptake in willows grown on polluted soil. Plant Soil and Environment. 2007a;53:158–166. [Google Scholar]
- Dos Santos Utmazian MN, Wieshammer G, Vega R, Wenzel WW. Hydroponic screening for metal resistance and accumulation of cadmium and zinc in twenty clones of willows and poplars. Environmental Pollution. 2007b;148:155–165. doi: 10.1016/j.envpol.2006.10.045. [DOI] [PubMed] [Google Scholar]
- Druzina B. The state of contaminated sites issues in Slovenia; NATO CCMS Pilot Study Meeting; Athens, Greece. 2006. [Google Scholar]
- Ellegren H. Microsatellites, simple sequences with complex evolution. Nature Reviews Genetics. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- Escarré J, Lefèbvre C, Gruber W, Leblanc M, Lepart J, Rivière Y, Delay B. Zinc and cadmium hyperaccumulation by Thlaspi caerulescens from metalliferous and nonmetalliferous sites in the Mediterranean area, implications for phytoremediation. New Phytologist. 2000;145:429–437. doi: 10.1046/j.1469-8137.2000.00599.x. [DOI] [PubMed] [Google Scholar]
- Frérot H, Petit C, Lefèbvre C, Gruber W, Collin C, Escarré J. Zinc and cadmium accumulation in control crosses between metallicolous and non-metallicolous populations of (Brassicaceae) New Phytologist. 2003;157:643–648. doi: 10.1046/j.1469-8137.2003.00701.x. [DOI] [PubMed] [Google Scholar]
- Grmela A, Rapantova N. Wolkersdorfer C, Bowell R, editors. Mine water issues in the Czech Republic. Contemporary Reviews of mine water studies in Europe, part 3. Mine Water and the Environment. 2005;24:58–76. [Google Scholar]
- Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, Motte P, Kroymann J, Weigel D, Krämer U. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature. 2008;453:391–395. doi: 10.1038/nature06877. [DOI] [PubMed] [Google Scholar]
- Hanley SJ, Mallott MD, Karp A. Alignment of a Salix linkage map to the Populus genomic sequence reveals macrosynteny between willow and poplar genomes. Tree Genetics & Genomes. 2006;3:35–48. [Google Scholar]
- Hardy OJ, Vekemans X. SPAGeDI: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2002;2:618–620. [Google Scholar]
- Hauser MT, Adhami F, Dorner M, Fuchs E, Glössl J. Generation of co-dominant PCR-based markers by duplex analysis on high resolution gels. The Plant Journal. 1998;16:117–125. doi: 10.1046/j.1365-313x.1998.00271.x. [DOI] [PubMed] [Google Scholar]
- Karhu A, Dieterich JH, Savolainen O. Rapid expansion of microsatellite sequences in Pines. Molecular Biology and Evolution. 2000;17:259–265. doi: 10.1093/oxfordjournals.molbev.a026305. [DOI] [PubMed] [Google Scholar]
- Kauer MO, Dieringer D, Schlötterer C. A microsatellite variability screen for positive selection associated with the ‘out of Africa’ habitat expansion of Drosophila melanogaster. Genetics. 2003;165:1137–1148. doi: 10.1093/genetics/165.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenkov V, Park S, Cheng NH, Sreevidya C, Lachmansingh J, Morris J, Hirschi K, Wagner GJ. Enhanced Cd2+ -selective root-tonoplast-transport in tobaccos expressing Arabidopsis cation exchangers. Planta. 2007;225:403–411. doi: 10.1007/s00425-006-0352-7. [DOI] [PubMed] [Google Scholar]
- Korenkov V, King B, Hirschi K, Wagner GJ. Root-selective expression of AtCAX4 and AtCAX2 results in reduced lamina cadmium in field-grown Nicotiana tabacum L. Plant Biotechnology Journal. 2009;7:219–226. doi: 10.1111/j.1467-7652.2008.00390.x. [DOI] [PubMed] [Google Scholar]
- Küpper H, Kochian LV. Transcriptional regulation of metal transport genes and mineral nutrition during acclimatization to cadmium and zinc in the Cd/Zn hyperaccumulator, Thlaspi caerulescens (Ganges population) New Phytologist. 2010;185:114–129. doi: 10.1111/j.1469-8137.2009.03051.x. [DOI] [PubMed] [Google Scholar]
- Kuroda Y, Kaga A, Tomooka N, Vaughan DA. Population genetic structure of Japanese wild soybean (Glycine soja) based on microsatellite variation. Molecular Ecology. 2006;15:959–974. doi: 10.1111/j.1365-294X.2006.02854.x. [DOI] [PubMed] [Google Scholar]
- Landberg T, Greger M. Differences in uptake and tolerance to heavy metals in Salix from unpolluted and polluted areas. Applied Geochemistry. 1996;11:175–180. [Google Scholar]
- Lepp NW, Madejón P. Cadmium and zinc in vegetation and litter of a voluntary woodland that has developed on contaminated sediment-derived soil. Journal of Environmental Quality. 2007;36:1123–1131. doi: 10.2134/jeq2006.0218. [DOI] [PubMed] [Google Scholar]
- Lian C, Nara K, Nakaya H, Zhou Z, Wu B, Miyashita N, Hogetsu T. Development of microsatellite markers in polyploid Salix reinii. Molecular Ecology Notes. 2001;1:160–161. [Google Scholar]
- McGrath SP, Zhao FJ. Phytoextraction of metals and metalloids from contaminated soils. Current Opinion in Biotechnology. 2003;14:1–6. doi: 10.1016/s0958-1669(03)00060-0. [DOI] [PubMed] [Google Scholar]
- Macnair MR. Within and between population genetic variation for zinc accumulation in Arabidopsis halleri. New Phytologist. 2002;155:59–66. doi: 10.1046/j.1469-8137.2002.00445.x. [DOI] [PubMed] [Google Scholar]
- Meerts P, Van Isacker N. Heavy metal tolerance and accumulation in metallicolous and non-metallicolous populations of Thlaspi caerulescens from continental Europe. Plant Ecology. 1997;133:221–231. [Google Scholar]
- Mei H, Cheng NH, Zhao J, Park S, Escareno RA, Pittman JK, Hirschi KD. Root development under metal stress in Arabidopsis thaliana requires the H+/cation antiporter CAX4. New Phytologist. 2009;183:95–105. doi: 10.1111/j.1469-8137.2009.02831.x. [DOI] [PubMed] [Google Scholar]
- Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, Richaud P. AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiology. 2009;149:894–904. doi: 10.1104/pp.108.130294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno I, Norambuena L, Maturana D, Toro M, Vergara C, Orellana A, Zurita-Silva A, Ordenes VR. AtHMA1 is a thapsigargin-sensitive Ca2+/heavy metal pump. The Journal of Biological Chemistry. 2008;283:9633–9641. doi: 10.1074/jbc.M800736200. [DOI] [PubMed] [Google Scholar]
- Oomen RJ, Wu J, Lelièvre F, Blanchet S, Richaud P, Barbier-Brygoo H, Aarts MG, Thomine S. Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens. New Phytologist. 2009;181:637–650. doi: 10.1111/j.1469-8137.2008.02694.x. [DOI] [PubMed] [Google Scholar]
- Österreichisches Normungsinstitut . Chemische Bodenuntersuchungen. Säureextrakt zur Bestimmung von Nähr- und Schadelementen: ÖNORM L 1085. Österreichisches Normungsinstitut; Vienna, Austria: 1999. [Google Scholar]
- Palmé AE, Semerikov V, Lascoux M. Absence of geographical structure of chloroplast DNA variation in sallow. Salix caprea L. Heredity. 2003;91:465–474. doi: 10.1038/sj.hdy.6800307. [DOI] [PubMed] [Google Scholar]
- Parameswaran A, Leitenmaier B, Yang M, Kroneck PM, Welte W, Lutz G, Papoyan A, Kochian LV, Küpper H. A native Zn/Cd pumping P(1B) ATPase from natural overexpression in a hyperaccumulator plant. Biochemical and Biophysical Research Communications. 2007;363:51–56. doi: 10.1016/j.bbrc.2007.08.105. [DOI] [PubMed] [Google Scholar]
- Pauwels M, Saumitou-Laprade P, Holl AC, Petit D, Bonnin I. Multiple origin of metallicolous populations of the pseudometallophyte Arabidopsis halleri (Brassicaceae) in central Europe: the cpDNA testimony. Molecular Ecology. 2005;14:4403–4414. doi: 10.1111/j.1365-294X.2005.02739.x. [DOI] [PubMed] [Google Scholar]
- Pauwels M, Frérot H, Bonnin I, Saumitou-Laprade P. A broad-scale analysis of population differentiation for Zn tolerance in an emerging model species for tolerance study, Arabidopsis halleri (Brassicaceae) Journal of Evolutionary Biology. 2006;19:1838–1850. doi: 10.1111/j.1420-9101.2006.01178.x. [DOI] [PubMed] [Google Scholar]
- Pauwels M, Willems G, Roosens N, Frérot H, Saumitou-Laprade P. Merging methods in molecular and ecological genetics to study the adaptation of plants to anthropogenic metal-polluted sites: implications for phytoremediation. Molecular Ecology. 2008;17:108–119. doi: 10.1111/j.1365-294X.2007.03486.x. [DOI] [PubMed] [Google Scholar]
- Pittman JK, Shigaki T, Marshall JL, Morris JL, Cheng NH, Hirschi KD. Functional and regulatory analysis of the Arabidopsis thaliana CAX2 cation transporter. Plant Molecular Biology. 2004;56:959–971. doi: 10.1007/s11103-004-6446-3. [DOI] [PubMed] [Google Scholar]
- Prestor J, Strucl S, Pungartnik M. Mežica lead and zinc mine closure impact on hydrogeological conditions in upper Meža valley. Materials and Geoenvironment. 2003;50:313–316. [Google Scholar]
- Pulford ID, Dickinson NM. Phytoremediation technologies using trees. In: Prasad MNV, Sajwan KS, Naidu R, editors. Trace Elements in the Environment. Taylor and Francis; Boca Raton, USA: 2005. pp. 383–403. [Google Scholar]
- Rahman MH, Dayanandan S, Rajora OP. Microsatellite DNA markers in Populus tremuloides. Genome. 2000;43:293–297. [PubMed] [Google Scholar]
- Reisch C, Schurm S, Poschlod P. Spatial genetic structure and clonal diversity in an alpine population of Salix herbacea (Salicaceae) Annals of Botany. 2007;99:647–651. doi: 10.1093/aob/mcl290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of DNA from plant tissues. Plant Molecular Biology Manual. 1988;A6:1–10. doi: 10.1007/BF00020088. [DOI] [PubMed] [Google Scholar]
- Schlötterer C. The evolution of molecular markers – just a matter of fashion? Nature Reviews Genetics. 2004;5:63–69. doi: 10.1038/nrg1249. [DOI] [PubMed] [Google Scholar]
- van der Schoot J, Pospíšková M, Vosman B, Smulders MJM. Development and characterization of microsatellite markers in black poplar (Populus nigra L.) Theoretical and Applied Genetics. 2000;101:317–322. [Google Scholar]
- Shen ZG, Zhao FJ, McGrath SP. Uptake and transport of zinc in the hyperaccumulator Thlaspi caerulescens and the non-hyperaccumulator Thlaspi ochroleucum. Plant Cell & Environment. 1997;20:898–906. [Google Scholar]
- Smulders MJM, van der Shoot J, Arens P, Vosman B. Trinucleotide repeat microsatellite markers for black poplar (Populus nigra L.) Molecular Ecology Notes. 2001;1:188–190. [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. Freeman WH; New York: 1995. [Google Scholar]
- Stamati K, Blackie S, Brown JWS, Russell J. A set of polymorphic SSR loci for subarctic willow (Salix lanata, S. lapponum and S. herbacea) Molecular Ecology Notes. 2003;3:280–282. [Google Scholar]
- StatSoft Inc. STATISTICA for Windows. StatSoft, Inc.; Tulsa, OK, USA: 2001. [Google Scholar]
- Stoltz E, Greger M. Accumulation properties of As, Cd, Cu, Pb and Zn by four wetland plant species growing on submerged mine tailings. Environmental and Experimental Botany. 2002;47:271–280. [Google Scholar]
- Taylor SI, Macnair MR. Within and between population variation of zinc and nickel accumulation in two species of Thlaspi (Brassicaceae) New Phytologist. 2006;169:505–514. doi: 10.1111/j.1469-8137.2005.01625.x. [DOI] [PubMed] [Google Scholar]
- Thibault J. Nuclear DNA amount in pure species and hybrid willows (Salix): a flow cytometric investigation. Canadian Journal of Botany. 1998;76:157–165. [Google Scholar]
- Tuskan GA, Gunter LE, Yang ZK, Yin T, Sewell M, DiFazio P. Characterization of microsatellites revealed by genomic sequencing of Populus trichocarpa. Canadian Journal of Forest Research. 2004;34:85–93. [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, et al. The genome of black Cottonwood Populus trichocarpa (Torr.&Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Unterbrunner R, Puschenreiter M, Sommer P, Wieshammer G, Tlustoš P, Zupan M, Wenzel WW. Heavy metal accumulation in trees growing on contaminated sites in Central Europe. Environmental Pollution. 2007;148:107–114. doi: 10.1016/j.envpol.2006.10.035. [DOI] [PubMed] [Google Scholar]
- Van Rossum FV, Bonnin I, Fénart S, Pauwels M, Petit D, Saumitou-Laprade P. Spatial genetic structure within a metallicolous population of Arabidopsis halleri, a clonal, self-and heavy-metal-tolerant species. Molecular Ecology. 2004;13:2959–2967. doi: 10.1111/j.1365-294X.2004.02314.x. [DOI] [PubMed] [Google Scholar]
- Vysloužilová M, Puschenreiter M, Wieshammer G, Wenzel WW. Rhizosphere characteristics, heavy metal accumulation and growth performance of two willow (Salix x rubens) isolates. Plant, Soil Environment. 2006;52:353–361. [Google Scholar]
- Watson C, Pulford ID, Riddel-Black D. Screening of willow species for resistance to heavy metals: comparison of performance in a hydroponics system and field trials. International Journal of Phytoremediation. 2003;5:351–365. doi: 10.1080/15226510309359042. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Welch RM, Norvell WA. Mechanisms of cadmium uptake, transpopulation and deposition in plants. In: McLaughlin MJ, Sing BR, editors. Cadmium in Soils and Plants. Kluwer Academic Publishers; Dordrecht, the Netherlands: 1999. pp. 125–150. [Google Scholar]
- Wieshammer G, Unterbrunner R, Bañares García T, Zivkovic MF, Puschenreiter M, Wenzel WW. Phytoextraction of Cd and Zn from agricultural soils by Salix ssp. and intercrop-ping of Salix caprea and Arabidopsis halleri. Plant and Soil. 2007;298:255–264. [Google Scholar]
- Willems G, Dräger DB, Courbot M, Godé C, Verbruggen N, Saumitou-Laprade P. The genetic basis of zinc tolerance in the metallophyte Arabidopsis halleri ssp. halleri (Brassicaceae): an analysis of quantitative trait loci. Genetics. 2007;176:659–674. doi: 10.1534/genetics.106.064485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Hewitt GM. Nuclear DNA analyses in genetic studies of populations: practice, problems and prospects. Molecular Ecology. 2003;12:563–584. doi: 10.1046/j.1365-294x.2003.01773.x. [DOI] [PubMed] [Google Scholar]
- Zhao FJ, Hamon RE, Lombi E, McLaughlin MJ, McGrath SP. Characteristics of cadmium uptake in two contrasting ecotypes of the hyperaccumulator Thlaspi caerulescens. Journal of Experimental Botany. 2002;53:535–543. doi: 10.1093/jexbot/53.368.535. Received 23 March, 2010; accepted for publication 14 April, 2010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Shoot length (A) and leaf number (B) of S. caprea isolates. Boxes represent the median (vertical solid line), the arithmetic mean (vertical dashed line), and 25–75% percentile. Whiskers represent the 90th and 10th percentile. Significant differences were determined by a post hoc comparison of means (Scheffé test after nested anova; P < 0.05) and are indicated by different letters.
Figure S2. Geographic map indicating the location of the S. caprea populations. South the Alps in Slovenia the contaminated population in Mežica – M, and in Austria the contaminated population Arnoldstein – (A) and the non-contaminated sites, Völkermarkt – V and Forchtenstein – F. North the Alps in the Czech Republic the two contaminated populations Příbram – PR and Kutná Hora – KH and the non-contaminated site near Prague (P).
Figure S3. The graphs show that the amount of labile Zn and Cd depends with a significance level for Pearson’s correlation of P < 0.01 on the total Zn and Cd concentration in the soil. In fact, 40% of the variation of labile Zn and 36% for Cd are explained by the total Zn and Cd concentration, respectively. Other factors influencing the labile heavy metal fractions are pH, content of clay, carbonate and organic matter.
Figure S4. Pearson’s correlations between the level of contamination where the isolate originated and the biomass production in perlite cultures exposed to elevated levels of Cd and Zn.A and B are the graphs for the uncontaminated, and C and D for the contaminated sites. The only significant negative correlation between Cd concentration and biomass production was seen at uncontaminated sites (A). Isolates from the contaminated sites did not show such a correlation indicating that they might have been selected to withstand higher Cd and Zn concentrations. Note that two soil samples from the uncontaminated sites had very high Zn concentrations probably because of a nearby rusty fence that leached into the soil (B).
Figure S5. Pearson’s correlations between the level of soil contamination and Cd concentration in leaves after the exposure of the isolates in perlite to Cd and Zn.Although a significant trend was found between soil contamination and accumulation capacity in perlite cultures for soil contamination below 20 mg kg−1 (A, B), above this soil Cd contamination level, the trend diminished.
Table S1. Summary of the soil characteristics for each isolate and their growth and accumulation behaviour in perlite-based soil-less cultures exposed to elevated levels of Cd (0.5 mg L−1) and Zn (5 mg L−1).
Table S2. Overview of the SSR marker characteristics.
Table S3. Heterozygosities, lnRH and normalized lnRH values of the 11 loci and 7 populations. Loci with Norm. Θ-values of larger +1.95 and smaller −1.95 have a significant reduced variability and thus might have been under selection and contribute to adaptation in the contamination area.