Abstract
HMGB1 is an alarmin that can stimulate the innate immune system alone or in a complex with other inflammatory mediators. Given the recent interest in HMGB1 with respect to the pathogenesis of eosinophil-associated disorders, including asthmatic inflammation and chronic rhinosinusitis, we have explored the role of this mediator and in promoting eosinophil activation. HMGB1 receptors RAGE and TLR4 but not TLR2 were detected on freshly isolated human eosinophils from healthy donors. Physiologic and relevant pathophysiologic levels of biologically-active HMGB1 had no effect on survival of human eosinophils alone or in combination with pro-survival cytokines IL-5, IL-3, or GM-CSF, and increasing concentrations of HMGB1 had no impact on surface expression of RAGE, TLR2 or TLR4. Similarly, HMGB1 did not elicit chemotaxis of human eosinophils alone and had no effect in combination with the eosinophil chemotactic agent, eotaxin-2 (CCL24). However, surface expression of TLR2 and TLR4 increased in response to cell stress, notably on eosinophils that remain viable after 48 hours without IL-5. As such, HMGB1 signaling on eosinophils may be substantially more detailed, and may involve complex immunostimulatory pathways other than or in addition to those evaluated here.
Introduction
High mobility group box 1 protein (HMGB1) is an evolutionarily ancient protein that was originally characterized as a chromatin stabilizing nuclear DNA-binding protein. Wang and colleagues [1] were the first to identify an extracellular role for HMGB1, specifically its participation in cellular activation and pro-inflammatory responses (reviewed in [2–10]). Containing 215 amino acids comprising three distinct protein domains, HMGB1 is expressed ubiquitously, is released from dead and dying cells, and serves as an alarmin or damage-associated molecular pattern (DAMP) molecule, stimulating the innate immune system by itself or via immunostimulatory complexes with endotoxin, nucleic acids, or proinflammatory cytokines and chemokines [3,11]. Additionally, activated immune cells (macrophages, monocytes, dendritic cells and natural killer cells) and endothelial cells secrete HMGB1 in response to pro-inflammatory stimuli [2].
HMGB1 signals through multiple surface receptors; TLR2, TLR4, and RAGE, receptor for advanced glycation end product, are the best characterized [2,12] but HMGB1 can also signal through human CD24 / Siglec-10 [13]. A series of recent studies has revealed a role for HMGB1 in sensing and responding to exogenous and endogenous nucleic acids (double-stranded RNA, single-stranded RNA, CpG-containing oligodeoxynucleotides) and amplifying the responses of these ligands to pattern recognition receptors TLR3, TLR7, and TLR9 (reviewed in [9]). Interestingly, while HMGB1 gene-deleted mice die in infancy [14], mice with conditional ablation of HMGB1 in myeloid cells develop normally, although they are more sensitive to endotoxin shock compared with control mice [15].
There is substantial interest in HMGB1 signaling and inflammation associated acute and chronic disease, notably in diseases associated with eosinophilic inflammation [7,16]. Elevated levels of HMGB1 have been detected in sputum, plasma and nasal lavage of eosinophilic asthmatics as compared to normal controls, with levels of HMGB1 correlating with both sputum levels of IL-5, IL-13, and eosinophil counts [17–19]. Additionally, there is a negative correlation between HMGB1 levels and pulmonary function [20]. Similarly, HMGB1 has been implicated in the pathogenesis of chronic rhinosinusitis, an asthma co-morbidity characterized by eosinophils in nasal polyps and in mucous drainage [21]. Expression of HMGB1 was detected in paranasal sinus mucosae of individuals with this condition [22,23], with levels correlating directly with those of serum IL-5 and blood eosinophil counts [24].
HMGB1 expression is ubiquitous and serum levels in normal individuals are on the order of <5–30 ng/ml but can rise 3-fold or more under conditions associated with eosinophil activation and recruitment (Table 1). Lotfi et al. [25] demonstrated that human eosinophils are mobilized and activated in response to supraphysiologic concentrations of HMGB1 (104 ng/ml) and proposed a role for this DAMP in the induction of eosinophilic inflammation. At these supraphysiologic concentrations, which are similar to those described by Ueno et al. [26] in lung epithelial lining cells responding to acute lung injury in response to sepsis, HMGB1 promoted survival, chemotaxis and degranulation of eosinophils. We were interested in expanding the findings of Lotfi et al. [25] and in exploring the effects of HMGB1 on human eosinophils at concentrations similar to those identified in acute and chronic states directly associated with eosinophilic disease. Additionally, we wanted to explore the role of HMGB1 in combination with eosinophil active cytokines as it is known that HMGB1 can form immunostimulatory complexes with other soluble mediators. Here, we examine the expression of HMGB1 receptors on human eosinophils isolated from peripheral blood and explore the impact of HMGB1 in promoting eosinophil chemotaxis and survival.
Table 1. HMGB1 concentrations associated with inflammatory and eosinophil-related diseases.
| Disease | Reference | Body fluid | Conditions evaluated | [HMGB1] ng/mL |
|---|---|---|---|---|
| Vasculitis | [18] | Serum | Controls | 30 ± 16 |
| Urticarial vasculitis | 51 ± 32 | |||
| Allergic vasculitis | 53 ± 37 | |||
| Henoch-Schonlein purpura | 57 ± 41 | |||
| [27] | Serum | Controls | < 5 | |
| Wegener's granulomatosis | 12 ± 9 | |||
| Dermatitis | [28] | Serum | Controls | 28 ± 18 |
| Psoriasis | 78 ± 66 | |||
| Atopic dermatitis | 35 ± 20 | |||
| Allergy/Asthma | [29] | Nasal lavage | Controls | 9 ± 4 |
| Allergic rhinitis | 97 ± 19 | |||
| [20] | Plasma | Controls | 3.7 | |
| Eosinophilic asthma | 4.7 | |||
| [20] | Sputum | Controls | 0.4 | |
| Eosinophilic asthma | 3.8 | |||
| Autoimmune | [30] | Serum | Controls | 13 ± 10 |
| Lupus nephritis | 108 ± 48 | |||
| [31] | Serum | Controls | 18 | |
| Rheumatoid arthritis | 71 |
Materials and Methods
Isolation of eosinophils from normal donors
One hundred milliliter (100 mL) samples of peripheral blood from normal donors were obtained on the Laboratory of Allergic Diseases normal donor protocol (NIAID Protocol Number: NIH 09-I-0049). This protocol is designed to assure adequate and complete informed consent, counseling, and protection of the study subjects according the NIAID Institutional Review Board, Office of Human Subjects Research (OHSR), Office for Human Research Protections (OHRP) and other applicable Federal regulatory standards. As part of this protocol, written consent is obtained from each normal volunteer allowing their sample to be used in this study. Eosinophils were isolated from the blood according to the protocol of Percopo et al. [32]. Essentially granulocytes were separated on a 67% Percoll gradient (GE #17-0891-02) and eosinophils were isolated by negative selection using the Miltenyi eosinophil isolation kit as per manufacturer’s directions. The eosinophils were either used immediately or cultured for 48 hours in the presence or absence of the anti-apoptotic cytokine, IL-5 (100 pg/mL) with or without HMGB1 as indicated. The monocytic cell fraction from the Percoll gradient was collected and used to determine the titer of the anti-human TLR2 (CD282) and anti-human TLR4 (CD284) antibodies. Additionally, the granulocytes, prior to isolation of eosinophils, were fixed in 4% paraformaldehyde (EMS, Hatfield, PA) and used to determine the titer of the anti-human RAGE antibody.
Flow cytometry
Non-specific antibody binding was blocked by incubating human eosinophils with FcR blocking reagent (Miltenyi). Antibody directed against the indicated HMGB1 receptor—RAGE, TLR2 or TLR4 is shown in Table 2. After incubation with antibodies for 30 minutes at 4°C, the cells were washed with 3 ml of 0.1% BSA/PBS and then fixed in 4% paraformaldehyde. The samples were stored at 4°C in the dark until analyzed. A minimum of 100,000 events was collected on an LSRII flow cytometer (BD Biosciences) and data was analyzed in FlowJo (Tree Star, Inc). Gating was performed by comparison to isotype control for each antibody. Isotype controls (fluorescence minus one) were subtracted from each data point.
Table 2. Antibodies used to examine surface expression of HMGB1 receptors on human eosinophils.
| Antigen | CloneVendor | Concentration(106 cells) | Notes |
|---|---|---|---|
| TLR2/CD282 | TL2.1 Ebioscience | 0.5 μg / 100 μL | Titered on monocytes |
| TLR4/CD284 | HTA125 Ebioscience | 0.5 μg / 100 μL | Titered on monocytes |
| RAGE | 176907 R&D | 0.5 μg / 100 μL | Used on 4% paraformaldehyde fixed cells; Secondary rat anti-mouse IgG1—APC |
HMGB1 activity assay
This assay was modified from that described by Gero et al. [33] RAW264.7 (ATCC, TIB71) were plated at 100,000 cells per well in a 96-well plate in 200 μL of media containing RPMI1640 (Life Technologies), 10% FBS (Lonza), 100 IU/mL penicillin, and 10 μg/mL streptomycin (Life Technologies) and grown overnight at 37°C in a cell culture incubator. The culture media was removed and 100 μL Optimem (Life Technologies) containing 0–100 ng/mL HMGB1 (R&D Systems) was added. Lipopolysaccharide (LPS; Sigma) was used at 50 ng/mL as a positive control. After 7 hours, the plate was centrifuged at 300 x g for 5 minutes at room temperature to remove the cells and the cell free supernatant was stored at −80°C. The samples were diluted 1:10 in Optimem prior to determining TNFα concentrations by ELISA (R&D) according the manufacturer’s recommendations. We determined that recombinant HMGB1 used in our studies was biologically active, with 100 ng/mL eliciting 177 pg/mL TNF-alpha, ∼3-fold over background levels at t = 7 hrs.
Viability assay
Eosinophils were suspended in media with the indicated cytokines (HMBG1, IL-3, IL-5 and GM-CSF; R&D) and were incubated for in a 37°C in a humidified CO2 incubator for 48 hours. The cells were washed one time in 3 mL PBS (Life Technologies) and stained with Violet live-dead (Life Technologies) for 30 minutes in the dark. The samples were washed with 3 mL 0.1% BSA/PBS and fixed with 200 μL 4% paraformaldehyde at 4°C. Prior to running the samples on a LSRII flow cytometer, they were washed once to remove the paraformaldehyde and suspended in 100 μl 0.1% BSA/PBS. Total cell number per sample and percent viability was obtained.
Chemotaxis Assay
The chemotaxis assay was performed as previously described [34]. Briefly, eosinophils were suspended at 106/mL in chemotaxis buffer and 100 μL was placed in the top well of a 5-μm 96-well Transwell plate (Corning). The 100 μL of the chemoattractant, diluted in chemotaxis buffer, was placed in the bottom well. The plate was incubated for 3 hours at 37°C in a humidified CO2 incubator. Cells that migrate to the lower well through the membrane are collected and enumerated by collecting events on LSR II at high flow rate for 60 seconds. Data is reported as cell number or chemotactic index (CI = cells migrating in response to chemoattractant / cells migrating in response to vehicle). Recombinant human eotaxin (rhE2, R&D) was used as a positive control for eosinophil chemotaxis.
Results and Discussion
Expression of HMGB1 receptors on eosinophils
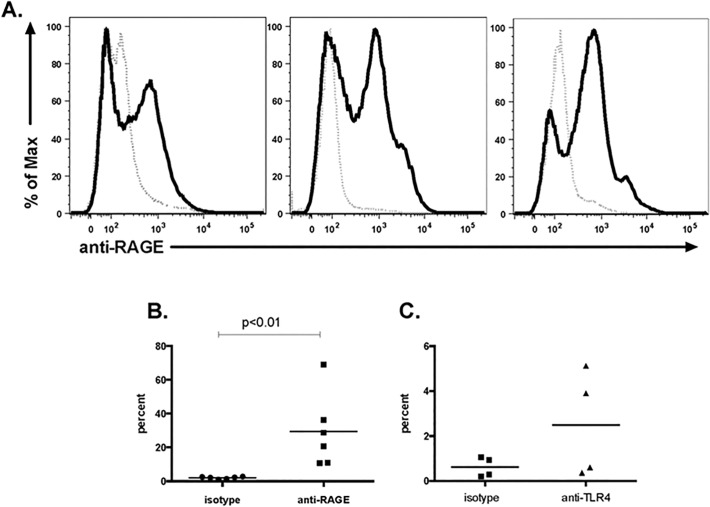
Human eosinophils were isolated from the peripheral blood of normal donors and surface expression of RAGE, TLR2 and TLR4 was examined. RAGE expression was variable among normal donors and on average 30% of the eosinophils isolated from normal human donors express RAGE (range 11–69%, n = 6 donors, Fig. 1A and 1B ). The expression of pattern recognition receptors on eosinophils has recently been reviewed [4]. Our results are consistent with those of others who have shown documented expression of RAGE on human eosinophils [25,35]; however, this is the first study documenting the heterogeneous nature of RAGE expression on eosinophils isolated from multiple normal donors.
Fig 1. Expression of HMGB1 receptors on eosinophils.
Human eosinophils were isolated from peripheral blood of normal donors. Surface expression of RAGE and TLR4 was evaluated by flow cytometry. (A) RAGE expression (heavy solid line) in comparison to isotype control (dotted line) in three individual donors. (B) RAGE expression in from 6 normal donors shown in comparison to isotype control. (C) TLR4 expression in from 4 normal donors shown in comparison to isotype control.
TLR4 was detected on a small population (0.3–5%) of eosinophils from normal donors (Fig. 1C). Our findings are consistent with those of Wong et al. [36] who detected TLR4 by both Western blot and flow cytometry. In contrast, we did not detect TLR2 on freshly isolated normal human eosinophils. Our findings are consistent with those of Nagase et al. [37] and Sabroe et al. [38] who were unable to detect surface expression of TLR2 by flow cytometry. Interestingly Wong et al. [36] reported detection of TLR2 on and in human eosinophils with intracellular expression being higher then surface expression. Likewise Wong et al. [36] reported that peptidoglycan (PGN), a TLR2 ligand, signaled through this receptor; in contrast, even though TLR4 was present, its ligand LPS did not elicit a response. Similarly, Sabroe et al. [26] failed to elicit a response from eosinophils with LPS stimulation.
Eosinophils were isolated from peripheral blood of normal donors and cultured for 48 hrs either in the presence or absence of IL-5; living cells (87.9 ± 2.0% of total in response to 100 pg/mL IL-5 culture and 42.1 ± 5.9% in the absence of IL-5) were evaluated for the expression of RAGE, TLR4, and TLR2. At the same time, both cultures were treated with increasing concentrations of HMGB1 (from 0 to 100 ng/mL). As shown, HMGB1 had no independent effect on the cell surface expression of RAGE, TLR4 or TLR2. In contrast, IL-5 had a clear and differential impact on receptor expression. While IL-5 had little to no effect on the surface expression of RAGE on live eosinophils (Fig. 2A), this was not the case for surface expression of TLR4 (Fig. 2B) or TLR2 (Fig. 2C). Detection was minimal (TLR2) to non-existent (TLR4) after 48 hrs in culture with 100 pg / mL IL-5. In contrast, immunoreactive TLR2 and TLR4 was detected on ∼20% and ∼10% of the live cells, respectively, after 48 hrs in cultures devoid of IL-5. This finding is intriguing and requires further study.
Fig 2. HMGB1 has no impact on cell surface expression of RAGE, TLR2 or TLR4.
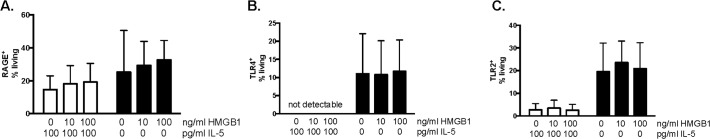
Eosinophils isolated from peripheral blood of normal donors incubated in the presence (open bars) or absence (closed bars) of IL-5 with 0, 10 or 100 ng/ml HMGB1for 48 hours and then evaluated for the expression of (A) RAGE, (B) TLR4, and (C) TLR2. Data represent mean ± SEM of percent population of live eosinophils that are positive for indicated receptor minus staining with isotype control; n = 3 independent donors.
Recombinant human HMGB1 does not enhance viability or chemotaxis of isolated human eosinophils
As noted in the Introduction, very high concentrations (104 ng/mL), were utilized in an earlier publication documenting HMGB1-mediated eosinophil activation [25]. This concentration exceeds physiologic serum levels (typically measured at 0–30 ng/mL), and likewise exceeds levels measured in serum, plasma and tissue fluids of individuals with eosinophil-associated disorders (Table 1).
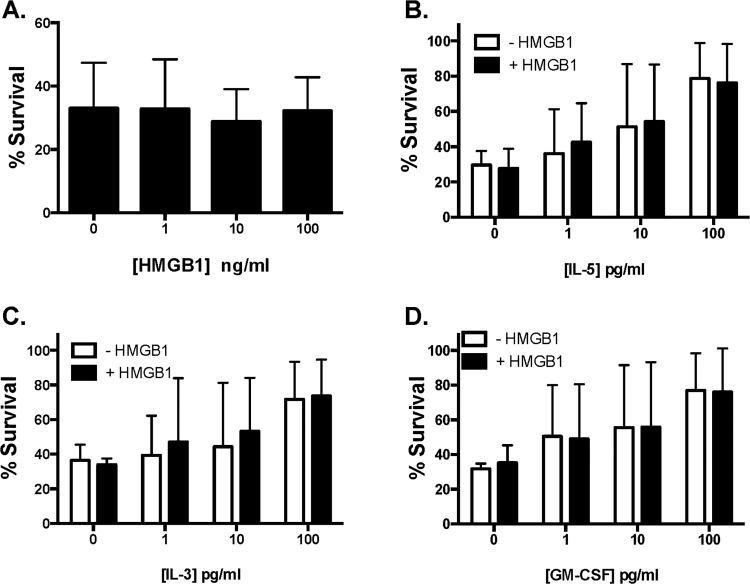
When evaluated alone, in the absence pro-survival cytokines (ie, under conditions of pro-apoptotic stress [39]), HMGB1 had no impact on survival of human eosinophils (Fig. 3A). Similarly, HMGB1 at 10 ng/mL had no impact on eosinophil survival promoted by IL-5, IL-3, or GM-CSF (Fig. 3B-D).
Fig 3. HMGB1 has no impact on eosinophil survival ex vivo.
Eosinophils isolated from peripheral blood of normal donors incubated with (A) HMGB1 alone or 10 ng/mL HMGB1 in combination with increasing concentrations of pro-survival cytokines (B) IL-5, (C) IL-13 or (D) GM-CSF for 48 hours and then evaluated with Violet live-dead for the assessment of viability.
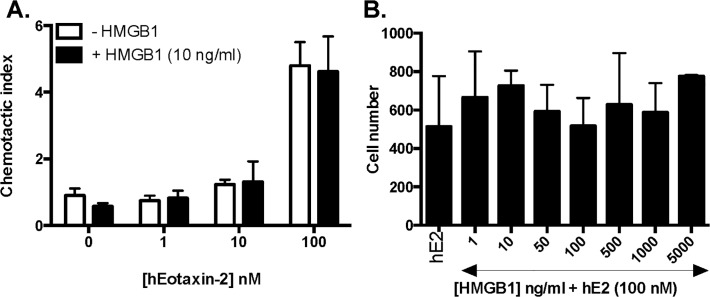
Likewise, as shown in Fig. 4A, HMGB1 at 10 ng/ml elicited no chemotaxis of human eosinophils and had no impact on chemotaxis elicited by the eosinophil chemotactic agent, eotaxin-2 (CCL24). Eosinophils incubated for 30 minutes with HMGB1 prior to introduction of an eotaxin-2 gradient migrated to the same extent as did the eosinophils pre-incubated with buffer alone. Furthermore, varying concentrations of HMGB1, including supraphysiologic concentrations (1–5000 ng/ml) had no impact on chemotaxis elicited by 100 nM eotaxin (Fig. 4B). Interestingly, this is in direct contrast to the inhibitory effect of HMGB1 observed when measuring neutrophil chemotaxis mediated via RAGE both at baseline at moderate concentrations (50–100 ng/mL) and in response to these concentrations in the presence of gradients of neutrophil-specific chemoattractants fMLP- and IL-8 [40].
Fig 4. HMGB1 has no impact on eotaxin-2-mediated chemotaxis.
Chemotaxis of peripheral blood eosinophils from normal donors evaluated in response to (A) increasing concentrations of human eotaxin-2 in the presence or absence of 10 ng/mL HMGB1 or (B) 100 nM human eotaxin-2 (hE2) in the presence of increasing concentrations of HMGB1.
Summary
At physiologic and pathophysiologic concentrations related to eosinophil-associated disease, recombinant HMGB1 has no impact on survival of freshly isolated human eosinophils, or on the expression of its receptors, RAGE, TLR2 or TLR4, either alone or in conjunction with IL-3, IL-5, or GM-CSF, in experiments performed ex vivo, although a greater fraction of viable cells express TLR2 and TLR4 in cultures undergoing pro-apoptotic stress. Additionally, HMGB1 has no apparent impact on eotaxin 2-mediated chemotaxis, in direct contrast to what has been reported for HMGB1 and human neutrophils [40]. HMGB1 also did not have an impact on cytokine-mediated survival over a wide range of concentrations when used in combination with known eosinophil-active cytokines. From our findings, we must consider the possibility that HMGB1 may be generating complexes in vivo with other cytokines or with nucleic acid ligands that modulate chemotaxis and prolong survival and/or that may have an impact on eosinophil biology via outcomes other than those explored here.
Data Availability
Data are contained within the paper.
Funding Statement
This work is supported by the NIAID Division of Intramural Research #AI000941 to HFR. Funding supported the design, execution and interpretation of the results. Source of funding did not play a role in the performance, decision to publish or preparation of the manuscript.
References
- 1. Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285: 248–251. [DOI] [PubMed] [Google Scholar]
- 2. Kang R, Chen R, Zhang Q, Hou W, Wu S, et al. HMGB1 in health and disease. Mol Aspects Med. 2014;40: 1–116. 10.1016/j.mam.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Keyel PA. How is inflammation initiated? Individual influences of IL-1, IL-18 and HMGB1. Cytokine. 2014;69: 136–145. 10.1016/j.cyto.2014.03.007 [DOI] [PubMed] [Google Scholar]
- 4. Kvarnhammar AM, Cardell LO. Pattern-recognition receptors in human eosinophils. Immunology. 2012;136: 11–20. 10.1111/j.1365-2567.2012.03556.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Musumeci D, Roviello GN, Montesarchio D. An overview on HMGB1 inhibitors as potential therapeutic agents in HMGB1-related pathologies. Pharmacol Ther. 2014;141: 347–357. 10.1016/j.pharmthera.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 6. Rosenberg HF, Dyer KD, Foster PS. Eosinophils: changing perspectives in health and disease. Nat Rev Immunol. 2013;13: 9–22. 10.1038/nri3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sukkar MB, Ullah MA, Gan WJ, Wark PA, Chung KF, et al. RAGE: a new frontier in chronic airways disease. Br J Pharmacol. 2012;167: 1161–1176. 10.1111/j.1476-5381.2012.01984.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tsung A, Tohme S, Billiar TR. High mobility group box-1 in sterile inflammation. J Intern Med. 2014;276: 425–443. 10.1111/joim.12276 [DOI] [PubMed] [Google Scholar]
- 9. Yanai H, Taniguchi T. Nucleic acid sensing and beyond: virtues and vices of HMGB1. J Intern Med. 2014;276: 444–453. 10.1111/joim.12285 [DOI] [PubMed] [Google Scholar]
- 10. Yang H, Antoine DJ, Andersson U, Tracey KJ. The many faces of HMGB1: molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J Leukoc Biol. 2013;93: 865–873. 10.1189/jlb.1212662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harris HE, Andersson U, Pisetsky DS. HMGB1: a multifunctional alarmin driving autoimmune and inflammatory disease. Nat Rev Rheumatol. 2012;8: 195–202. 10.1038/nrrheum.2011.222 [DOI] [PubMed] [Google Scholar]
- 12. Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5: 331–342. [DOI] [PubMed] [Google Scholar]
- 13. Chen GY, Tang J, Zheng P, Liu Y. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science. 2009;323: 1722–1725. 10.1126/science.1168988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Calogero S, Grassi F, Aguzzi A, Voigtlander T, Ferrier P, et al. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999;22: 276–280. [DOI] [PubMed] [Google Scholar]
- 15. Yanai H, Matsuda A, An J, Koshiba R, Nishio J, et al. Conditional ablation of HMGB1 in mice reveals its protective function against endotoxemia and bacterial infection. Proc Natl Acad Sci U S A. 2013;110: 20699–20704. 10.1073/pnas.1320808110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lotze MT, Zeh HJ, Rubartelli A, Sparvero LJ, Amoscato AA, et al. The grateful dead: damage-associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunol Rev. 2007;220: 60–81. [DOI] [PubMed] [Google Scholar]
- 17. Bellussi LM, Iosif C, Sarafoleanu C, Jianu E, Duda R, et al. Are HMGB1 protein expression and secretion markers of upper airways inflammatory diseases? J Biol Regul Homeost Agents. 2013;27: 791–804. [PubMed] [Google Scholar]
- 18. Chen T, Guo ZP, Wang WJ, Qin S, Cao N, et al. Increased serum HMGB1 levels in patients with Henoch-Schonlein purpura. Exp Dermatol. 2014;23: 419–423. 10.1111/exd.12422 [DOI] [PubMed] [Google Scholar]
- 19. Shim EJ, Chun E, Lee HS, Bang BR, Kim TW, et al. The role of high-mobility group box-1 (HMGB1) in the pathogenesis of asthma. Clin Exp Allergy. 2012;42: 958–965. 10.1111/j.1365-2222.2012.03998.x [DOI] [PubMed] [Google Scholar]
- 20. Hou C, Zhao H, Liu L, Li W, Zhou X, et al. High mobility group protein B1 (HMGB1) in Asthma: comparison of patients with chronic obstructive pulmonary disease and healthy controls. Mol Med. 2011;17: 807–815. 10.2119/molmed.2010.00173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schleimer RP, Kato A, Kern R. Eosinophils and Chronic Rhinosinusitis In: Lee JJ, Rosenberg HF, editors. Eosinophils in Health and Disease. San Diego: Academic Press; 2013. pp. 508–519. [Google Scholar]
- 22. Hong SM, Cho JS, Um JY, Shin JM, Park IH, et al. Increased expression of high-mobility group protein B1 in chronic rhinosinusitis. Am J Rhinol Allergy. 2013;27: 278–282. 10.2500/ajra.2013.27.3909 [DOI] [PubMed] [Google Scholar]
- 23. Paris G, Pozharskaya T, Asempa T, Lane AP. Damage-associated molecular patterns stimulate interleukin-33 expression in nasal polyp epithelial cells. Int Forum Allergy Rhinol. 2014;4: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen D, Mao M, Bellussi LM, Passali D, Chen L. Increase of high mobility group box chromosomal protein 1 in eosinophilic chronic rhinosinusitis with nasal polyps. Int Forum Allergy Rhinol. 2014;4: 453–462. 10.1002/alr.21294 [DOI] [PubMed] [Google Scholar]
- 25. Lotfi R, Herzog GI, DeMarco RA, Beer-Stolz D, Lee JJ, et al. Eosinophils oxidize damage-associated molecular pattern molecules derived from stressed cells. J Immunol. 2009;183: 5023–5031. 10.4049/jimmunol.0900504 [DOI] [PubMed] [Google Scholar]
- 26. Ueno H, Matsuda T, Hashimoto S, Amaya F, Kitamura Y, et al. Contributions of high mobility group box protein in experimental and clinical acute lung injury. Am J Respir Crit Care Med. 2004;170: 1310–1316. [DOI] [PubMed] [Google Scholar]
- 27. Wibisono D, Csernok E, Lamprecht P, Holle JU, Gross WL, et al. Serum HMGB1 levels are increased in active Wegener's granulomatosis and differentiate between active forms of ANCA-associated vasculitis. Ann Rheum Dis. 2010;69: 1888–1889. 10.1136/ard.2009.119172 [DOI] [PubMed] [Google Scholar]
- 28. Chen T, Guo ZP, Li L, Wang L, Jia RZ, et al. Increased HMGB1 serum levels and altered HMGB1 expression in patients with psoriasis vulgaris. Arch Dermatol Res. 2013;305: 263–267. 10.1007/s00403-013-1330-0 [DOI] [PubMed] [Google Scholar]
- 29. Salpietro C, Cuppari C, Grasso L, Tosca MA, Miraglia Del Giudice M, et al. Nasal high-mobility group box-1 protein in children with allergic rhinitis. Int Arch Allergy Immunol. 2013;161: 116–121. 10.1159/000345246 [DOI] [PubMed] [Google Scholar]
- 30. Zickert A, Palmblad K, Sundelin B, Chavan S, Tracey KJ, et al. Renal expression and serum levels of high mobility group box 1 protein in lupus nephritis. Arthritis Res Ther. 2012;14: R36 10.1186/ar3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goldstein RS, Bruchfeld A, Yang L, Qureshi AR, Gallowitsch-Puerta M, et al. Cholinergic anti-inflammatory pathway activity and High Mobility Group Box-1 (HMGB1) serum levels in patients with rheumatoid arthritis. Mol Med. 2007;13: 210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Percopo CM, Dyer KD, Killoran KE, Rosenberg HF. Isolation of human eosinophils: microbead method has no impact on IL-5 sustained viability. Exp Dermatol. 2010;19: 467–469. 10.1111/j.1600-0625.2009.00974.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gero D, Szoleczky P, Modis K, Pribis JP, Al-Abed Y, et al. Identification of pharmacological modulators of HMGB1-induced inflammatory response by cell-based screening. PLoS One. 2013;8: e65994 10.1371/journal.pone.0065994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dyer KD, Garcia-Crespo KE, Percopo CM, Sturm EM, Rosenberg HF. Protocols for identifying, enumerating, and assessing mouse eosinophils. Methods Mol Biol. 2013;1032: 59–77. 10.1007/978-1-62703-496-8_5 [DOI] [PubMed] [Google Scholar]
- 35. Curran CS, Bertics PJ. Human eosinophils express RAGE, produce RAGE ligands, exhibit PKC-delta phosphorylation and enhanced viability in response to the RAGE ligand, S100B. Int Immunol. 2011;23: 713–728. 10.1093/intimm/dxr083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wong CK, Cheung PF, Ip WK, Lam CW. Intracellular signaling mechanisms regulating toll-like receptor-mediated activation of eosinophils. Am J Respir Cell Mol Biol. 2007;37: 85–96. [DOI] [PubMed] [Google Scholar]
- 37. Nagase H, Okugawa S, Ota Y, Yamaguchi M, Tomizawa H, et al. Expression and function of Toll-like receptors in eosinophils: activation by Toll-like receptor 7 ligand. J Immunol. 2003;171: 3977–3982. [DOI] [PubMed] [Google Scholar]
- 38. Sabroe I, Jones EC, Usher LR, Whyte MK, Dower SK. Toll-like receptor (TLR)2 and TLR4 in human peripheral blood granulocytes: a critical role for monocytes in leukocyte lipopolysaccharide responses. J Immunol. 2002;168: 4701–4710. [DOI] [PubMed] [Google Scholar]
- 39. Ilmarinen P, Moilanen E, Kankaanranta H. Regulation of spontaneous eosinophil apoptosis-a neglected area of importance. J Cell Death. 2014;7: 1–9. 10.4137/JCD.S13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Berthelot F, Fattoum L, Casulli S, Gozlan J, Marechal V, et al. The effect of HMGB1, a damage-associated molecular pattern molecule, on polymorphonuclear neutrophil migration depends on its concentration. J Innate Immun. 2012;4: 41–58. 10.1159/000328798 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are contained within the paper.