Abstract
Key message
5,511 genic small-size PAVs in sorghum were identified and examined, including the pattern and the function enrichment of PAV genes. 325 PAV markers were developed to construct a genetic map.
Abstract
Presence/absence variants (PAVs) correlate closely to the phenotypic variation, by impacting plant genome sizes and the adaption to the environment. To shed more light on their genome-wide patterns, functions and the possibility of using them as molecular markers, we generated next generation genome sequencing data for four sorghum inbred lines and used associated bioinformatic pipelines to identify small-size PAVs (40–10 kb). Five thousand five hundreds and eleven genic PAVs (40–10 kb) were identified and found to affect 3,238 genes. These PAVs were mainly distributed on the sub-telomeric regions, but the highest proportions occurred in the vicinity of the centromeric regions. One of the prominent features of the PAVs is the high occurrence of long terminal repeats retrotransposons and DNA transposons. PAVs caused various alterations to gene structure, primarily including the coding sequence variants, intron variants, transcript ablation, and initiator codon changes. The genes affected by PAVs were significantly enriched in those involved in stress responses and protein modification. We used 325 PAVs polymorphic between two sorghum inbred lines Ji2731 and E-Tian, together with 49 SSR markers, and constructed a genetic map, which consisted of 10 linkage groups corresponding to the 10 chromosomes of sorghum and spanned 1,430.3 cM in length covering 97 % of the physical genome. The resources reported here should be useful for genetic study and breeding of sorghum and related species.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-015-2458-4) contains supplementary material, which is available to authorized users.
Introduction
Single nucleotide polymorphisms (SNPs) and small insertion/deletion polymorphisms (IDPs) have long been thought to be the only means through which most of genetic variations could arise (Feuk et al. 2006). Recently, the central role of structural variation has been gradually emerging through the examination of more sequenced genomes of animals and plants obtained from the next generation sequencing platforms (Tuzun et al. 2005; Kidd et al. 2008; Conrad et al. 2010). Structural variation refers to genomic alterations such as insertions, deletions, duplications, inversions, and translocations covering at least 50 base pairs (bp) (Feuk et al. 2006; Mills et al. 2011). Presence and absence variants (PAVs) are an important type of structural variation present in one genome but entirely missing in the other (Springer et al. 2009), and play an important role in shaping genomes and contribute to phenotypic diversity (Marroni et al. 2014).
PAVs have been widely found in the human genomes (Mills et al. 2011; McKernan et al. 2009; Hastings et al. 2009; Kidd et al. 2008; Korbel et al. 2007; Redon et al. 2006) and have been implicated to cause diseases in humans through the positional effect and the alteration of the gene dosage (Sebat et al. 2007; Zhang et al. 2009). They also contribute to the observed phenotypic variation (Conrad et al. 2010), and determine the fitness with potential evolutionary implications (Stefansson et al. 2005). Compared to the human genome studies, PAVs are less investigated in plants, but are certainly prevalent. For example, in maize (Zea mays), it has been reported that 20 % of genome segments (about 10,000 genes or gene fragments) were not shared between inbred lines B73 and Mo17 (Morgante et al. 2005), while an array of comparative genome hybridization (aCGH) showed that more than 1,000 PAVs affected at least 180 single copy genes (Springer et al. 2009). In rice (Oryza sativa), it has been reported that 2.2 and 3.3 % of indica and japonica genes were absent in the corresponding subspecies, respectively (Yu et al. 2005), and 5.2 % of the genes were found with presence and absence polymorphisms between japonica Nipponbare and indica 93-11 (Ding et al. 2007). A comparison of 18 fully sequenced Arabidopsis (Arabidopsis thaliana) genomes showed that on average 775 genes per accession have more than 50 % regions with deletions or polymorphism relative to the reference accession Col-0 (Gan et al. 2011). Furthermore, a comparison of 80 Arabidopsis genomes revealed that 8.9 % of the total genes in A. thaliana showed PAVs averaging 444 absent genes per accession (Tan et al. 2012). In other plants, such as soybean (Glycine max) and melon (Cucumis melo), recent studies also found that a large number of PAVs were distributed in their genomes (Lam et al. 2010; McHale et al. 2012; Gonzalez et al. 2013). Even since the first description of presence and absence polymorphisms in the RPM1 gene in Arabidopsis (Grant et al. 1998), an increasing number of studies indicated that a dominant feature of the PAV genes is the enrichment in genes involved in the stress responses, especially the disease resistance (Shen et al. 2006; Ding et al. 2007; Ziolkowski et al. 2009; Lam et al. 2010; McHale et al. 2012; Tan et al. 2012; Gonzalez et al. 2013). This feature has also been confirmed to be present in the mammalian (Iafrate et al. 2004; Yalcin et al. 2011), Drosophila (Kern and Begun 2008), and bacteria (Arrach et al. 2008). However, since a large number of fully sequenced individual genomes are the prerequisite for such studies, there is still a lack of a clear estimate of the PAV gene numbers, proportions, and variation patterns.
Sorghum (Sorghum bicolour) is the fifth important cereal crop worldwide after wheat, rice, maize, and barley (FAO 2004). It adapts well in the arid and semi-arid areas due to its ability to tolerate drought stress (Dalal et al. 2012). To deflate tradeoff between food and biofuel production, sweet sorghum, a variant of grain sorghum, has been deemed an ideal dedicated biofuel crop due to its high stem sugar and juice accumulation. Sorghum is an interesting system to study changes in genome structure for a number of reasons. It has a relatively small genome (about 730 Mb) and does not seem to have undergone whole-genome duplications as in other closely related crops such as maize, sugarcane, miscanthus, and switchgrass (Paterson et al. 2009), making it a more tractable system to establish gene–phenotype associations. Furthermore, as a predominantly selfing plant, sorghum accessions representing elite inbred lines, land races (caudatum, durra, bicolor, guinea, and kafir) of S. bicolour ssp. bicolour and ssp. verticilliflorum (arundinaceum, aethiopicum, and verticilliflorum) have greater linkage disequilibrium (LD) and lower sequence variation than in maize as evaluated with sequences of RFLP markers (Hamblin et al. 2004). This notion is further supported by assays using SNPs and IDPs markers (Bekele et al. 2013; Zheng et al. 2011; Zou et al. 2012; Mace et al. 2013). However, different from the situation in the exotic lines/landraces, the genetic diversity among the publically available inbreds tends to be low (Menz et al. 2004). Sorghum has a very high outcrossing tendency (Barnaud et al. 2008), which could potentially increase the genetic variation. Taken together, these characteristics of sorghum genomes offer a unique niche to study structural variation and its impact on important phenotypic traits.
Molecular markers have been a vital tool in both basic and applied research for fingerprinting genotypes, identifying genetic diversity, defining variety identity, assisting breeding selection and phylogenetic analysis (Vos et al. 1995; McCouch et al. 1997; Kolliker et al. 2001). Since early 1990s, a number of DNA-based molecular markers such as restriction fragment length polymorphism (RFLP ) (Hulbert et al. 1990; Whitkus et al. 1992; Xu et al. 1994; Dufour et al. 1997; Kebede et al. 2001; Bowers et al. 2003; Subudhi and Nguyen 2000), random amplified polymorphism (RAPD) (Dahlberg et al. 2002), amplified fragment length polymorphism (AFLP) (Boivin et al. 1999), simple sequence repeat (SSR) (Menz et al. 2002; Wu and Huang 2007), and diversity arrays technology (DArT) (Bouchet et al. 2012; Mace et al. 2008) have been used for constructing genetic maps and assessing intraspecific diversity in sorghum. Recently, with the advance in sequencing and resequencing sorghum genomes, sequence-based molecular markers such as SNPs have been increasingly exploited for genetic mapping (Bekele et al. 2013; Zou et al. 2012). In contrast, PAV markers attract less attention. PAV markers with moderate sizes have the advantages of simple operation and less demanding for small laboratories (Wu et al. 2013; Wang et al. 2014). In particular, within-gene or genic PAVs are potentially functional markers which are a good “translator” from genomic study into improved crop varieties (Andersen and Lübberstedt 2003).
Previously, we analyzed PAVs with size fragments larger than 30 kb in sorghum and showed that large-size PAVs are widespread in sorghum inbred lines of different origins (Zhang et al. 2014). This prompted us to explore further in detail the small-size (40–10 kb) PAVs. Here we reported our study focusing on the genic small-size PAVs, including the distribution over the genome, the function, and the pathway enrichment of genes affected. We used the available genome resequencing data and identified 5,511 genic small-size PAVs. We showed that these PAVs were preferentially distributed on different parts of chromosome, and the affected 3,238 genes were predominantly involved in stress response and protein modification. As a proof of concept, 325 PAV markers from 10 chromosomes were used to construct a genetic map together with 49 SSR markers, which showed the advantages of genome coverage, integration of physical and genetic maps, and a better functionality over maps constructed with other molecular markers used in sorghum.
Materials and methods
Plant materials
Four S. bicolour inbred lines, including two grain sorghum (Ji2731, BTx623) and two sweet sorghum (E-Tian, Keller), were used for small-size PAVs (40–10 kb) discovery and validation in this study. Among them, the genomes of Ji2731, E-Tian, and Keller were resequenced (Zheng et al. 2011), and the reference genome of BTx623 was previously published (Paterson et al. 2009). These four lines exhibit significant phenotypic variation in plant height, grain yield, photoperiods, stem sugar and juice accumulation, and salt stress tolerance. An F2 population consisting of 209 individuals derived from a cross between Ji2731 and E-Tian was developed in Gongzhuling, Jilin province from May to October 2010 and used for genetic linkage group analysis. A 15-row plot of 5 m row length was planted. The inter-row space was 70 cm, and three seeds per hole were sown at 4-cm depth and 20-cm intervals. Full irrigation and timely weeding were carried out as required during the whole growing season, and 225 kg/ha urea was applied followed by irrigation in the week before sowing.
Sequence data sets
The whole-genome sequences of BTx623 were retrieved from the version of S. bicolour v1.4 in the Phytozome 9.0 databases (www.phytozome.org) and used as the reference. The sequence data of three sorghum lines (Ji2731, E-Tian, and Keller) previously resequenced to approximately 12× coverage by Illumina 100-bp paired-end sequencing (Zheng et al. 2011) were acquired at GigaScience (Zheng et al. 2011; www.gigasciencejournal.com). For the detection of PAVs, short insert-size paired-end reads of three sorghum lines genome were first aligned to the reference genome of BTx623 and their assemblies by SOAPaligner, respectively (Li et al. 2009). The ratios of aligned single-end reads to paired-end reads (S/P ratios) were acquired from the alignment results achieved by SOAPcoverage (http://soap.genomics.org.cn). The overall S/P ratio of identified PAVs was evaluated by calculating the number of mapped paired-end reads with the expected orientation and insert size and the unexpected orientation and insert size. Then by calculating the P value using Fisher’s exact test, we tested the significant difference between the S/P ratio of PAVs and the S/P ratio of the whole genome. The PAVs were validated when they meet the conditions of (1) P value <0.05 and (2) their depths were consistent with the type of PAV. In this study, the PAVs were supported by at least six paired-end reads. A total of 5,511 small-size PAVs with explicit physical positions and fragment sizes were obtained from the SVs dataset and used for investigation (Table S1.1). The data of transposon elements (TEs) of sorghum were downloaded from Helmholtz-Muenchen plantDB (ftp://ftpmips.helmholtz-muenchen.de/plants/sorghum/) and used for matching PAV sequences.
DNA preparation and experimental validation of PAVs
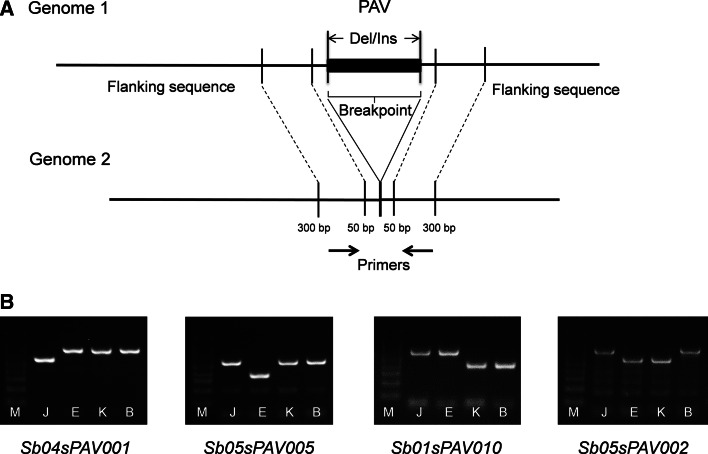
In total, 1,779 PAVs used for experimental validation of polymorphisms between three sorghum lines and BTx623 were randomly selected from the 10 chromosomes of sorghum (Table S1.2). DNA was isolated from young leaves of field-grown plants following a CTAB extraction method (Doyle 1987). Experimental validation of PAVs was executed by polymerase chain reaction (PCR) and agarose gel electrophoresis. The primer pairs used for PCR were designed based on the reference genome sequences of 50–300 bp of up- and down-stream of insertion/deletion breakpoints by the software of Primer 6.0 (http://www.premierbiosoft.com/) (Fig. 1a). A total of 10 μL mixture was used for PCR, containing 1 μL genomic DNA (80–120 ng/μL), 5 μL MasterMix (Biomed, Beijing), 1 μL 10× primer, and 3 μL ultrapure water. The PCR program was set at 94 °C for 5 min, then 34 cycles of 94 °C for 30 s, 55–62 °C for 30 s, and 72 °C for 30 s to 2 min, and followed by a final extension of 10 min at 72 °C. The PCR products were separated on 2–5 % agarose gels depending on the sizes of the fragments and visualized and recorded under UV light.
Fig. 1.
A diagram to show the designing of primers (a) and representative gel images (b) for the verification of PAVs. Four PAVs between three sorghum lines (Ji2731, E-Tian, and Keller) and BTx623 were presented as examples (Sb04sPAV001, Sb05sPAV005, Sb01sPAV010, Sb05sPAV002). A 100-bp DNA ladder marker was on the left of the gel and used for size measurement of PCR products
Gene ontology enrichment analysis
Two different analyses, including gene ontology (GO) enrichment and Pfam, were carried out on the PAV-affected genes. GO identities (IDs ) of PAV genes and corresponding GO term annotations were obtained from Ensemble Biomart (Smedley et al. 2009). The enrichment of GO terms was assessed using hypergeometric distribution by WEGO (Ye et al. 2006) (http://wego.genomics.org.cn/cgi-bin/wego/index.pl) and agriGO (Du et al. 2010) (http://bioinfo.cau.edu.cn/agriGO/). The detailed information of GO enrichment of PAV genes was presented in Table S1.7. Pfam terms were acquired from Pfam v27.0 (Punta et al. 2012) (http://pfam.sanger.ac.uk/).
Genetic linkage map construction
In total, 325 PAVs which showed polymorphisms between Ji2731 and E-Tian in validation experiment and 49 SSR markers obtained from the articles published previously (Bhattramakki et al. 2000; Kong et al. 2000; Yu et al. 2010) were selected for developing molecular makers. An F2 population of 209 individuals derived from a cross between Ji2731 and E-Tian was genotyped and used for genetic linkage map construction. The software Joinmap® 4.0 (Kyazma, Wageningen, The Netherlands) was applied to construct genetic linkage map by regression mapping algorithm. Because PAV and SSR markers are commonly co-dominant markers, they were pooled together for map construction using the same genotype codes (a/c, b/d, h). In the end, a total of 325 PAV markers and 49 SSR markers (Tables S2.1, S2.2) were assigned to 10 linkage groups using a minimal logarithm of the odds (LOD) threshold value of 3.0 as the criterion, corresponding to the 10 sorghum chromosomes. The Kosambi (1943) mapping function was used to calculate the genetic distance. The graphical representation of the map was drawn by using GGT 2.0 software based on the order and genetic distances of markers (van Berloo 2008). The segregation distortion was estimated using Pearson’s Chi-squared test.
Results
Identification of small-size PAVs in sorghum
Resequencing of the genomes of three sorghum lines uncovered a large number of PAVs (Zheng et al. 2011). We previously showed that large-size (>30 kb) PAVs influenced the genome size and varied substantially in sorghum inbred lines (Zhang et al. 2014). We wondered whether the small-size PAVs between 40 and 10 kb within genes are more likely to influence gene function and phenotypic variation. Sequence analysis identified a total of 5,511 genic small-size PAVs affecting 3,238 genes and 9.69 Mb gene sequences (Table S1.1). Among the PAVs identified, 1,635 were commonly shared by the three lines, 777 were shared between Ji2731 and E-Tian, 481 between Ji2713 and Keller, and 620 between E-Tian and Keller (Figure S1). Furthermore, 915, 589, and 494 PAVs were specific to Ji2731, E-Tian, and Keller, respectively.
To test the authenticity of the PAVs obtained from Next Generation Sequencing, we selected 1,779 PAV events with sizes in the range of 100–2,000 bp polymorphic between the reference line BTx623 and the three resequenced sorghum lines for experimental verification (Table S1.2). As shown in Table 1, 313 had no or non-specific amplifications due to primer sequence errors from either the reference genome (BTx623) or the targeted genomes (Ji2731, E-Tian, and Keller). In the end, we had 1,466 clean single PCR products (simple insertion or deletion events), of which 1,142 were consistent between NGS data and PCR results, while 105 were detected in NGS but not in PCRs and 219 were not detected in NGS but detected in PCRs. As such, the overall validation rate is 77.9 %, false positive 7.1 %, and false negative 14.8 %, respectively.
Table 1.
Summary of experimental validation of NGS PAVs in three sorghum lines
| Group | PAV category | Ji2731 | E-Tian | Keller | Sub-total |
|---|---|---|---|---|---|
| PAVs from sequencing data | PAVs | 526 | 434 | 17 | 978 |
| No PAVs | 325 | 417 | 60 | 801 | |
| PAVs from PCR results | PAVs | ||||
| Confirmed PAVs | 396 | 305 | 15 | 716 | |
| False positives | 45 | 58 | 2 | 105 | |
| No PAVs | |||||
| Confirmed no PAVs | 156 | 219 | 51 | 426 | |
| False negatives | 108 | 102 | 9 | 219 | |
| No or non-specific products | 146 | 167 | 0 | 313 | |
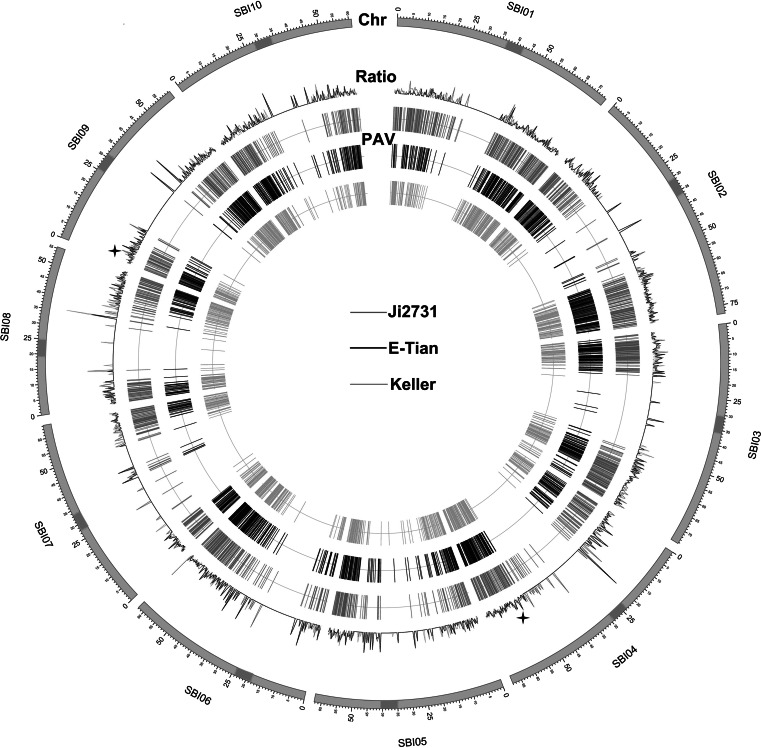
Figure 2 shows the differential distribution of 5,511 PAVs on the 10 chromosomes. We examined the occurrence of the PAVs in relation to the gene density along each chromosome and found that Chromosome 5 had the highest occurrence with one PAV per 15.67 genes, whereas Chromosome 7 had the lowest with one PAV per 22.18 genes (P value <0.05 by Fisher’s exact test) (Fig. 2; Table S1.3). Although the number of PAVs varied within different regions of individual chromosome, the highest proportions of PAVs often occurred around centromeres (Fig. 2; Table S1.4). A 300-kb bin size was used to examine the distribution of PAVs on a finer scale and revealed two incidences of enrichment on Chromosome 4 in Ji2731 (SBI04: 53100001–53400000) and Chromosome 9 in E-Tian (SBI09: 3900001–4200000) (P value <0.05 by Fisher’s exact test) containing 11 and 12 PAVs, respectively (Table S1.4).
Fig. 2.
Genome-wide distribution of 5,511 genic small-size PAVs (40–10 kb) discovered from three sorghum lines. The ratio ring shows the ratio between the number of genes with PAVs and the number of total genes in the 300-kb bin of the sorghum genome. The yellow bands denote the centromeric regions. The star symbols stand for the regions with significant enrichment of PAVs by Fisher’s exact test (P value <0.05). The ten chromosomes are drawn by a scale of Mb (color figure online)
For the distribution of the fragment sizes of 5511 PAVs, we found that the number of PAVs peaked at the 500–999 bp bin with median value of 670 bp (Figure S2). The smallest group of PAVs fell into the 40–99 bp bin. The insertions were much shorter ranging from 43 to 347 bp when compared to deletions which ranged from 259 to 9,991 bp in length. In most of all next generation sequencing data generated using the Illumina platform, it is routine rather than exception that only short insertions could be detected. Such marked asymmetry might be caused by the use of short reads (<500 bp) and by the detection method/algorithm of PAVs.
Sequence features and functional categories of PAV genes
To gain insight into the sequence features of small-size PAVs, we analyzed the signature of TEs of PAV sequences. As shown in Table 2, out of the 5,511 PAVs, 3,742 (67.9 %) contained TEs, influencing about 4.5 Mb genomic sequences. In total, 7,532 TEs were found falling into three major categories: retrotransposons (5,280), DNA transposons (2,227) and unclassified (25). Long terminal repeats (LTR) accounted for the largest proportion (98.3 %) of the retrotransposons, while the DNA transposons contained 73.3 % unclassified elements and 26.7 % of the terminal inverted repeats, respectively. Table 2 also shows that limited full-length LTRs were found, while in the DNA transposons, most of them were full-length elements. Furthermore, most of the full-length/complete TEs were found as single events in a PAV, only limited PAVs contained 2, 3, 4, or 5 TEs (Table S1.5). This is consistent with our PCR results showing that most of the PAVs are simple insertion and deletion events.
Table 2.
Categories of transposon elements in PAVs
| Category | Feature | Number | PAVs involved | Genes affected | Sequences (bp) |
|---|---|---|---|---|---|
| Retransposons | 5280 | 2823 | 1745 | 3502807 | |
| LTR | Full length | 233 | 213 | 128 | 1149923 |
| Partial | 4957 | 2634 | 1644 | 2317591 | |
| LINE | Partial | 24 | 20 | 13 | 14303 |
| Unclassified | Partial | 66 | 60 | 39 | 20990 |
| DNA transposons | 2227 | 1767 | 1121 | 1001317 | |
| TIR | Full length | 370 | 354 | 232 | 561978 |
| Partial | 225 | 128 | 79 | 98989 | |
| Unclassified | Full length | 1308 | 1185 | 768 | 276012 |
| Partial | 324 | 286 | 201 | 64338 | |
| Unclassified | Partial | 25 | 14 | 9 | 5805 |
| Total | 7532 | 3742 | 2270 | 4509929 |
LTR long terminal repeats, LINE long interspersed nuclear elements, TIR terminal inverted repeats
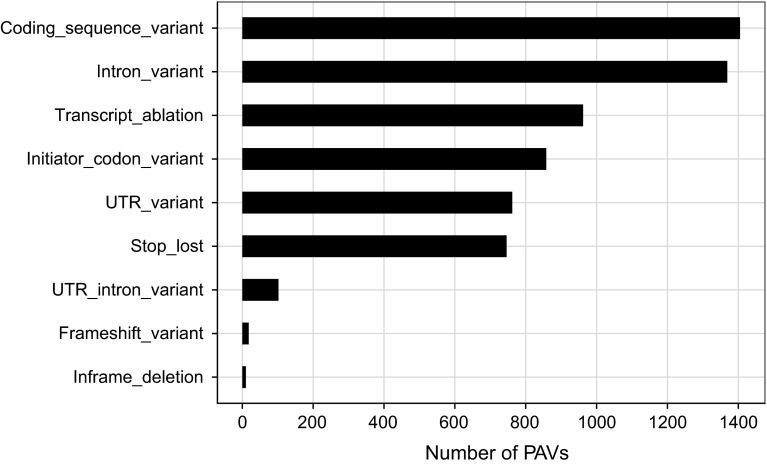
We analyzed the impact of small-size PAVs on the gene structure. By using Variant Effect Predictor at the Ensemble website (http://plants.ensembl.org/tools.html), nine different alterations in gene structure caused by PAVs were found (Fig. 3). Among them, the coding sequence variants, transcript ablation, initiator codon variants, untranslated region (UTR ) variants, stop-lost and frameshift variants occupied 75.9 % of the total variants, which were proposed to have large effects on the gene function or expression. The remaining variants (intron variants, UTR-intron variants, and inframe deletion) occupied 24.1 %. Therefore, it can be seen that small-size PAVs most likely change gene function or expression by altering gene structure (Table 3).
Fig. 3.
Distributions of PAVs in different variants of gene structure. The x axis represents the frequency of PAVs
Table 3.
Summary of the genetic map constructed with PAV and SSR markers in sorghum
| SBI01 | SBI02 | SBI03 | SBI04 | SBI05 | SBI06 | SBI07 | SBI08 | SBI09 | SBI10 | Total | Average | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of markers | 49 | 42 | 33 | 29 | 34 | 36 | 36 | 37 | 38 | 40 | 374 | 37.4 |
| PAV markers | 41 | 33 | 29 | 26 | 29 | 33 | 32 | 32 | 34 | 36 | 325 | 32.5 |
| SSR markers | 8 | 9 | 4 | 3 | 5 | 3 | 4 | 5 | 4 | 4 | 49 | 4.9 |
| Average genetic distance between markers (cM) | 3.87 | 4.01 | 4.78 | 4.58 | 3.7 | 3.74 | 3.65 | 3.33 | 3.4 | 3.21 | 3.83 | |
| Genetic distance (cM) | 193.6 | 168.6 | 157.7 | 137.8 | 125.8 | 134.7 | 131.5 | 123.3 | 129.1 | 128.2 | 1430.3 | 143.0 |
| Genetic distances (cM) in Mace et al. (2008) | 188.1 | 135.6 | 83 | 133.9 | 130.4 | 157.1 | 120.5 | 184.5 | 149.3 | 149.2 | 1431.6 | 143.2 |
| Genetic distances (cM) in Bekele et al. (2013) | 78 | 114.1 | 87.8 | 113.6 | 128.1 | 112.6 | 94.3 | 109.9 | 111.4 | 135.2 | 1085 | 108.5 |
| Physical distance (Mb) | 0.86–72.76 | 0.32–77.31 | 1.85–67.82 | 0.42–66.35 | 0.74–61.17 | 0.39–62.19 | 0.13–63.62 | 0.38–54.74 | 0.02–58.87 | 0.12–60.73 | 640.4 | 64.0 |
| Physical coverage (%) | 97 | 99 | 89 | 97 | 97 | 99 | 99 | 98 | 99 | 99 | 97 |
Results from two recent Sorghum bicolour genetic maps are compared. LG stands for linkage group
To further understand functional features of PAV genes, we examined GO annotations of the 3,238 PAV genes. In total, 44 unique GO terms (FDR < 0.05) were enriched by WEGO (http://wego.genomics.org.cn/cgi-bin/wego/index.pl) and agriGO (http://bioinfo.cau.edu.cn/agriGO/) (Table S1.7), including 9 cellular components, 16 molecular functions, and 19 biological processes (Figure S3). Furthermore, 105 PAV genes (8.0 %) were involved in cell death, similar to those genes containing large-size PAVs (Zhang et al. 2014). PAV genes related to the reproductive cellular processes and regulation of protein modification, albeit only being 1.8 and 2.8 %, were significantly overrepresented in comparison with the overall frequencies of the corresponding genes (Du et al. 2010), being 0.3 and 0.7 % (χ 2 = 92.7, df = 1; χ 2 = 74.5, df = 1).
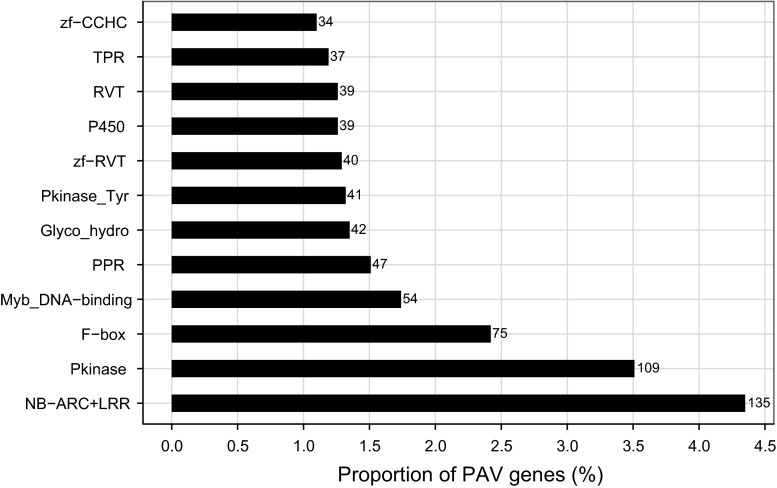
We analyzed protein families and functional domains of PAV genes by Pfam database v27.0 (Punta et al. 2012). In total, 3,105 (95.9 %) PAV genes were classified into 2,529 Pfam categories (Table S1.8). Although most of PAV genes tended to be fragmented across these categories, as shown in Fig. 4, the disease resistance genes with NB-ARC domains and LRR domains were significantly (χ 2 = 73.5, df = 1) overrepresented, accounting for 4.3 % of total PAV genes. Moreover, these resistance genes were predicted to be predominantly involved in the biological process of programmed cell death, receptor signaling, and phosphorylation (FDR < 0.05) (Table S1.9).
Fig. 4.
Distributions of PAV genes in the protein families (Pfam). The x axis represents the proportion of PAV genes in total PAV genes with annotated Pfam. The y axis shows the categories of Pfam. The numbers after bars indicate the number of PAV genes
Construction of genetic map by PAV markers
To develop PAVs into functional markers for genetics and breeding of sorghum, we selected 360 PAVs with identifiable polymorphism between Ji2731 and E-Tian from the experimental validation results. To avoid the overlap in the genetic position, in the end, 325 PAV markers were used for the construction of a genetic map (Table S2.1). An F2 mapping population with 209 individuals was obtained by crossing Ji2731 and E-Tian and screened for the construction of genetic map. For constructing linkers between PAVs markers and other molecular marker systems, 192 SSR markers originated from previous genetic maps of sorghum (Bhattramakki et al. 2000; Kong et al. 2000; Yu et al. 2010) were examined and 49 (25.5 %) of them were polymorphic between Ji2731 and E-Tian (Table S2.2).
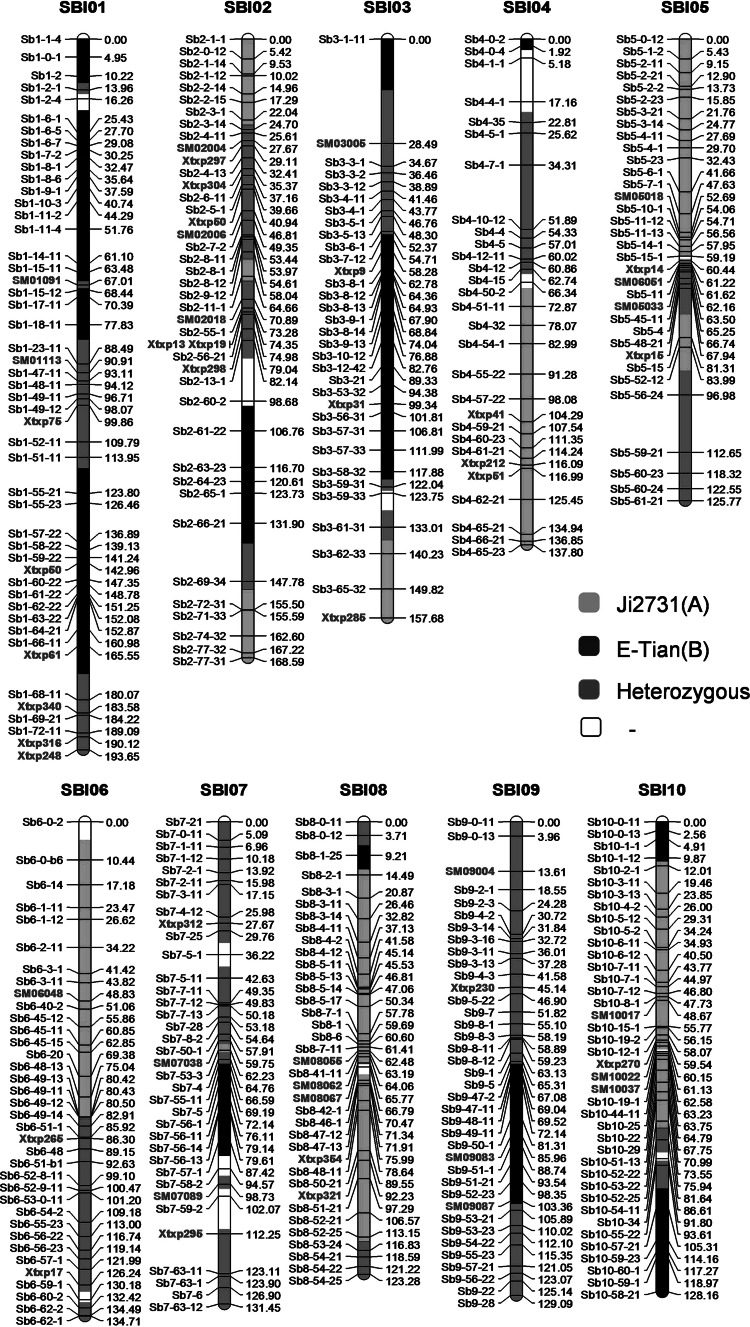
As shown in Fig. 5, a total of 374 markers including 325 PAV markers and 49 SSR markers were assigned to 10 linkage groups, corresponding to the 10 chromosomes of sorghum. This genetic map spanned 1,430.2 cM, with an average marker distance of 3.83 cM and an average PAV marker distance of 4.40 cM. SSR markers interspersed among the PAV markers in each linkage group. According to the physical locations of PAV markers, this map covered 640.44 Mb, which is approximately 97 % of the sorghum genome released from Phytozome v9.1 (http://www.phytozome.org/) with an average physical distance of 1.76 Mb per marker. Nonetheless, on this genetic map, a total of five gaps over 15 cM were encountered on Chromosomes 2 (82.14–98.68 cM; 131.90–147.78 cM), 3 (0.00–28.49 cM), 4 (34.31–51.89 cM), and 5 (96.98–112.65 cM), respectively. We checked the colinearity between the physical and genetic orders of markers. Although the linkage analysis was executed by regression mapping, which was recognized to reduce the conservation of the physical and genetic marker orders compared to the maximum likelihood mapping (MLM) (Cheema and Dicks 2009), the physical and genetic orders of markers on this genetic map were mostly consistent, with little discrepancy by several PAV markers, including one each on Chromosomes 1, 2, and 9, three each on Chromosomes 4 and 8 and four each on Chromosomes 5 and 10 (Fig. 6).
Fig. 5.
An integrated genetic linkage map of Sorghum bicolour. The vertical bars represent the chromosomes of Sorghum bicolour. The codes on the left are the PAV marker loci and the SSR marker loci which are labeled with red color, while the corresponding accumulative genetic distances (cM) are on the right. The discrete segments of the vertical chromosomal bars are color-coded according to the allele colors in the figure legends. Heterozygous stands for alleles heterozygous for parents (Ji2731 and E-Tian) alleles. Ji2731(A) is female allele, while E-Tian(B) is male allele. – is missing value (color figure online)
Fig. 6.
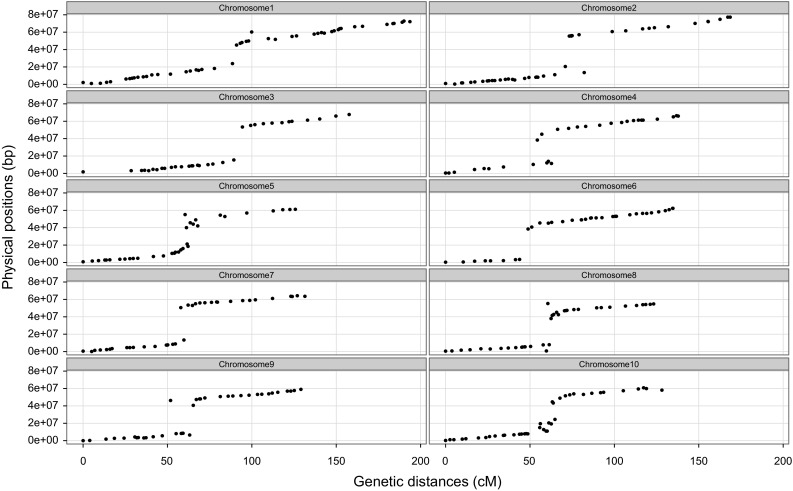
Colinearity between the physical position and the genetic distance of PAV and SSR markers. The x axis shows the accumulative genetic distance (cM) in the genetic linkage map. The y axis represents the physical positions (bp) obtained by aligning with the reference genome of BTx623
We used Chi-squared test to examine the segregation pattern of all the 374 markers. A total of 112 (104 PAVs and 8 SSR) markers were significantly distorted from the expected Mendelian segregation ratio (P value <0.05) (Figure S4; Table S2.3). A detailed exam showed that the majority of the markers with distortion resided on Chromosomes 1 (65.3 %), 2 (38.1 %), 7 (41.7 %), 8 (40.5 %), and 9 (31.6 %), respectively, and tend to form clusters (Figure S4).
Discussion
Intraspecies structural variations in genes have been proposed to play an important role in the adaptation of particular populations to variation in environmental conditions (Feuk et al. 2006). Here we identified 5,511 small-size PAVs affecting 3,238 genes representing 9.5 % of the total genes in sorghum. This proportion is similar to that of the genes with presence and absence polymorphism in Arabidopsis (Tan et al. 2012). It seems that small-size PAVs influence gene sequences greatly in plants.
Small-size PAVs distribution and sequence features
The distribution of PAVs on the chromosomes has been investigated in Arabidopsis and been shown that the number of large InDels (>100 bp) along chromosomes increased gradually from telomere toward centromere (Ziolkowski et al. 2009). Conversely, PAVs were mainly accumulated at the distal ends of chromosomes and few on the pericentromeres (Tan et al. 2012). In sorghum, we found that genic PAVs more often resided in sub-telomeric regions and few were around the centromeres (Fig. 2), which may be related to differences in the gene lengths and recombination frequencies in these two regions. In sorghum, high frequencies of recombination were observed on both the distal ends of chromosomes (Mace and Jordan 2011), and high levels of linkage disequilibrium (LD) have been showed in the heterochromatic regions surrounding the centromeres with a recombination suppression rate of 33 % (Kim et al. 2005). Interestingly, the highest ratios of PAV genes to the total genes were found near the centromeres (Fig. 2), which have also been described in Arabidopsis (Tan et al. 2012). Despite centromere has the conserved function as the site for kinetochore formation and sister chromatid join, the DNA sequences associated with the centromeres are highly variable containing many satellite repeats and transposons (Lee et al. 2005; Murphy et al. 2005), which might cause widespread gene conversion (Shi et al. 2010) and variation in the tandem repetitive sequences in the centromere (Miller et al. 1998; Zwick et al. 2000). It is anticipated that such dynamic changes in the DNA sequences may generate high frequency of insertion or deletion.
One of the prominent sequence features of the small-size PAVs is the enrichment of transposable elements (TEs) and repeat sequences. Since its first discovery in maize (Mcclintock 1948), transposable elements have been shown to be prevalent in plant genomes, particularly revealed through the recent whole-genome sequencing of a number of species (Tenaillon et al. 2010). Since TEs can cause duplication, deletion, transposition of nearby non-TE genes or ectopic recombination by a variety of mechanisms, it has been recognized that transposons have a great impact on the genome structure and gene function in nearly all organisms (Kidwell and Lisch 1997; Bennetzen 2000; Wang et al. 2013). In this study, 67.9 % of PAVs contained TEs, affecting 4.5 Mb sequences out of the total 9.69 Mb PAV sequences (Table 2; Table S1.5). Moreover, the proportions of retroelement and DNA transposon varied. The overall ratio of PAV sequences between them was 3.5, which is much lower than that in the whole genome of sorghum (7.3) (Paterson et al. 2009). Within the full-length TEs, 1,678 DNA transposons resided in 1,504 PAVs, whereas only 233 retrotransposons resided in 213 PAVs (Table 2). It seems that DNA transposons might participate in the generation of the small-size PAVs actively. And these features were also found in the large-size PAVs of sorghum in our previous study (Zhang et al. 2014). It is known that DNA transposons transfer sequences by a cut and paste mechanism, whereas LTRs by a copy and paste mechanism. In this study, we found a lot more deletions than insertions, this could be partially due to experimental procedures and the algorithms and the bioinformatic pipeline used, but could also be related to the high occurrence of DNA transposons in the PAVs examined. Overall, our results imply that TEs might play a key role in the formation of PAVs in sorghum.
Functional features of PAV genes
More attention was given to the functions of genes with structural variation due to their tight relations with phenotypic variation and development of plants. Previous research on PAVs of plants showed that the most predominant feature of the function of the PAV genes was stress response, particularly disease resistance (Ziolkowski et al. 2009; Tan et al. 2012; Gonzalez et al. 2013; Swanson-Wagner et al. 2010; McHale et al. 2012; Bush et al. 2014). Similarly, we showed that the genes for nucleotide binding and protein modification by ubiquitination were enriched (Figure S2), and the NB-LRR genes occupied the largest portion (Fig. 4). This feature was also found within the SNP, small InDel, and large-size PAVs in sorghum (Zheng et al. 2011; Zhang et al. 2014). Such significant enrichments in gene functions and annotated domains have been proposed to reflect the adaptive role of large polymorphic deletions (Bush et al. 2014). However, the proportion of NB-LRR genes with PAVs was not excessively large compared to previous findings in plants (McHale et al. 2012; Tan et al. 2012).
The feature of the PAV genetic map and segregation distortion
A genetic map, composed of PAV markers and SSR markers, was constructed using an F2 population, which was derived from a cross of two resequenced sorghum lines (Ji2731 and E-Tian). This map spanned 1,430.3 cM with average markers distance of 3.83 cM and had average 97 % physical coverage of genome. Compared to the recently published sorghum genetic maps using SNPs and DArT markers (Bekele et al. 2013; Mace et al. 2008), our current map has a larger or approximate genetic distance and physical coverage (Table 2). Moreover, this map with the PAVs markers derived from genes with function annotation might benefit future QTL analysis to quickly establish gene–trait association. The physical positions of markers corresponded to their genetic map orders. Nevertheless, a few disordered markers were distributed on the chromosomes except chromosomes 3, 6, and 7. This phenomenon was also presented in genetic map of sorghum and other plants (Bekele et al. 2013; Ganal et al. 2011; Felcher et al. 2012; Sim et al. 2012; Zhang et al. 2012). It is likely to be caused by the different algorithms for construction of genetic map or the partially inversion of chromosome regions (Felcher et al. 2012; Bekele et al. 2013). Because the recombination rates are suppressed around centromeric regions, it was made that although there is no markers on these regions, the genetic orders were continuous (Fig. 6).
Strongly distorted segregation was detected during the constructing of our current genetic map. 29.9 % of all the markers across the 10 chromosomes showed allele frequencies skewed from their Mendelian expectations (Table S2.3). We found that relatively high proportions occurred prominently on Chromosomes 1, 2, 7, 8, and 9 with the range of 31.6–65.3 %. Some skewed markers were clustered on the long arms of Chromosomes 1 (0–93 cM) and 7 (49–87 cM), respectively (Figure S3). This phenomenon has been reported previously in sorghum and was referred to as the segregation distortion region (SDR) (Murray et al. 2008; Menz et al. 2002; Mace et al. 2009). A recent report confirmed Chromosomes 1 and 7 to be gathered with skewed markers (Felderhoff et al. 2012). Yet, another study further showed that Chromosomes SBI-04 and SBI-08 had more than 50 % of chromosomal regions with segregation distortion (Mace et al. 2009). Previous proposals indicated that these distortions were likely caused by the ablation of gametes or zygotes by a lethal factor (Qi et al. 2004; van Os et al. 2006; Menz et al. 2002) and further study is required to explore the molecular genetics mechanisms.
Author contribution statement
Hai-Chun Jing conceived and designed the experiments. Xin Shen, Zhiquan Liu, and Anne Mocoeur constructed the genetic map. Xin Shen and Yan Xia analyzed the data. Xin Shen and Hai-Chun Jing wrote the first and the final draft.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We would like to thank Xiaoyuan Wu for his assistance in data processing and graphical drawing by circos (http://circos.ca/). We are grateful to Ms. Yu Jiang and Dr. Fengxue Jin for field management of four sorghum lines and their F2 populations in Jilin. The gratitude is also given to Dr. Elikana Anami for the article revision. Other members of Hai-Chun Jing’s lab are acknowledged for offering valuable suggestions throughout this project. This work is partially supported by Grants to Hai-Chun Jing from CNSF (31271797) and MOST (2013BAD22B01).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
X. Shen and Z. Liu have contributed equally to this work.
References
- Andersen JR, Lübberstedt T. Functional markers in plants. Trends Plant Sci. 2003;8(11):554–560. doi: 10.1016/j.tplants.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Arrach N, Porwollik S, Cheng P, Cho A, Long F, Choi SH, McClelland M. Salmonella serovar identification using PCR-based detection of gene presence and absence. J Clin Microbiol. 2008;46(8):2581–2589. doi: 10.1128/JCM.02147-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnaud A, Trigueros G, Mckey D, Joly HI. High outcrossing rates in fields with mixed sorghum landraces: how are landraces maintained? Heredity. 2008;101(5):445–452. doi: 10.1038/hdy.2008.77. [DOI] [PubMed] [Google Scholar]
- Bekele WA, Wieckhorst S, Friedt W, Snowdon RJ. High-throughput genomics in sorghum: from whole-genome resequencing to a SNP screening array. Plant Biotechnol J. 2013;11(9):1112–1125. doi: 10.1111/pbi.12106. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. Transposable element contributions to plant gene and genome evolution. Plant Mol Biol. 2000;42(1):251–269. doi: 10.1023/A:1006344508454. [DOI] [PubMed] [Google Scholar]
- Bhattramakki D, Dong JM, Chhabra AK, Hart GE. An integrated SSR and RFLP linkage map of Sorghum bicolour (L.) Moench. Genome/National Research Council Canada = Genome/Conseil national de recherches Canada. 2000;43(6):988–1002. doi: 10.1139/gen-43-6-988. [DOI] [PubMed] [Google Scholar]
- Boivin K, Deu M, Rami JF, Trouche G, Hamon P. Towards a saturated sorghum map using RFLP and AFLP markers. Theor Appl Genet. 1999;98(2):320–328. doi: 10.1007/s001220051076. [DOI] [Google Scholar]
- Bouchet S, Pot D, Deu M, Rami JF, Billot C, Perrier X, Rivallan R, Gardes L, Xia L, Wenzl P, Kilian A, Glaszmann JC (2012) Genetic structure, linkage disequilibrium and signature of selection in sorghum: lessons from physically anchored DArT markers. PloS One 7(3). doi:10.1371/journal.pone.0033470 [DOI] [PMC free article] [PubMed]
- Bowers JE, Abbey C, Anderson S, Chang C, Draye X, Hoppe AH, Jessup R, Lemke C, Lennington J, Li ZK, Lin YR, Liu SC, Luo LJ, Marler BS, Ming RG, Mitchell SE, Qiang D, Reischmann K, Schulze SR, Skinner DN, Wang YW, Kresovich S, Schertz KF, Paterson AH. A high-density genetic recombination map of sequence-tagged sites for sorghum, as a framework for comparative structural and evolutionary genomics of tropical grains and grasses. Genetics. 2003;165(1):367–386. doi: 10.1093/genetics/165.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush SJ, Castillo-Morales A, Tovar-Corona JM, Chen L, Kover PX, Urrutia AO. Presence–absence variation in A. thaliana is primarily associated with genomic signatures consistent with relaxed selective constraints. Mol Biol Evol. 2014;31(1):59–69. doi: 10.1093/molbev/mst166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheema J, Dicks J. Computational approaches and software tools for genetic linkage map estimation in plants. Brief Bioinform. 2009;10(6):595–608. doi: 10.1093/bib/bbp045. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang YJ, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, MacArthur DG, MacDonald JR, Onyiah I, Pang AWC, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME, Control WTC. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464(7289):704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg JA, Zhang X, Hart GE, Mullet JE. Comparative assessment of variation among sorghum germplasm accessions using seed morphology and RAPD measurements. Crop Sci. 2002;42(1):291–296. doi: 10.2135/cropsci2002.0291. [DOI] [PubMed] [Google Scholar]
- Dalal M, Mayandi K, Chinnusamy V (2012) Sorghum: improvement of abiotic stress tolerance. In: Improving crop resistance to abiotic stress. Wiley-VCH Verlag GmbH & Co. KGaA, pp 923–950. doi:10.1002/9783527632930.ch36
- Ding J, Araki H, Wang Q, Zhang P, Yang S, Chen JQ, Tian D (2007) Highly asymmetric rice genomes. BMC Genomics 8. doi:10.1186/1471-2164-8-154 [DOI] [PMC free article] [PubMed]
- Doyle JJ. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang ZH, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour P, Deu M, Grivet L, D'Hont A, Paulet F, Bouet A, Lanaud C, Glaszmann JC, Hamon P. Construction of a composite sorghum genome map and comparison with sugarcane, a related complex polyploid. Theor Appl Genet. 1997;94(3–4):409–418. doi: 10.1007/s001220050430. [DOI] [Google Scholar]
- FAO (2004) Food and Agriculture Organization of the United Nations, Statistics Division. http://www.fao.org/es/ess/index_en.asp
- Felcher KJ, Coombs JJ, Massa AN, Hansey CN, Hamilton JP, Veilleux RE, Buell CR, Douches DS (2012) Integration of two diploid potato linkage maps with the potato genome sequence. PloS One 7(4). doi:10.1371/journal.pone.0036347 [DOI] [PMC free article] [PubMed]
- Felderhoff TJ, Murray SC, Klein PE, Sharma A, Hamblin MT, Kresovich S, Vermerris W, Rooney WL. QTLs for energy-related traits in a sweet x grain sorghum [Sorghum bicolor (L.) Moench] mapping population. Crop Sci. 2012;52(5):2040–2049. doi: 10.2135/cropsci2011.11.0618. [DOI] [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7(2):85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Gan XC, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, Kahles A, Bohnert R, Jean G, Derwent P, Kersey P, Belfield EJ, Harberd NP, Kemen E, Toomajian C, Kover PX, Clark RM, Ratsch G, Mott R. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477(7365):419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganal MW, Durstewitz G, Polley A, Berard A, Buckler ES, Charcosset A, Clarke JD, Graner EM, Hansen M, Joets J, Le Paslier MC, McMullen MD, Montalent P, Rose M, Schon CC, Sun Q, Walter H, Martin OC, Falque M (2011) A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PloS One 6(12). doi:10.1371/journal.pone.0028334 [DOI] [PMC free article] [PubMed]
- Gonzalez VM, Aventin N, Centeno E, Puigdomenech P (2013) High presence/absence gene variability in defense-related gene clusters of Cucumis melo. BMC Genomics 14. doi:10.1186/1471-2164-14-782 [DOI] [PMC free article] [PubMed]
- Grant MR, McDowell JM, Sharpe AG, Zabala MDT, Lydiate DJ, Dangl JL. Independent deletions of a pathogen-resistance gene in Brassica and Arabidopsis. Proc Natl Acad Sci USA. 1998;95(26):15843–15848. doi: 10.1073/pnas.95.26.15843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin MT, Mitchell SE, White GM, Gallego W, Kukatla R, Wing RA, Paterson AH, Kresovich S. Comparative population genetics of the panicoid grasses: sequence polymorphism, linkage disequilibrium and selection in a diverse sample of Sorghum bicolour. Genetics. 2004;167(1):471–483. doi: 10.1534/genetics.167.1.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10(8):551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert SH, Richter TE, Axtell JD, Bennetzen JL. Genetic mapping and characterization of sorghum and related crops by means of maize DNA probes. Proc Natl Acad Sci USA. 1990;87(11):4251–4255. doi: 10.1073/pnas.87.11.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk T, Van Puymbroeck L, Rivera MN, Listewnik ML, Ying QP, Scherer SW, Lee C. Detection of large-scale variation in the human genome. J Mol Diagn. 2004;6(4):411. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Kebede H, Subudhi PK, Rosenow DT, Nguyen HT. Quantitative trait loci influencing drought tolerance in grain sorghum (Sorghum bicolour L. Moench) Theor Appl Genet. 2001;103(2–3):266–276. doi: 10.1007/s001220100541. [DOI] [Google Scholar]
- Kern AD, Begun DJ. Current deletion and gene presence/absence polymorphism: telomere dynamics dominate evolution at the tip of 3L in Drosophila melanogaster and D-simulans. Genetics. 2008;179(2):1021–1027. doi: 10.1534/genetics.107.078345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, Haugen E, Zerr T, Yamada NA, Tsang P, Newman TL, Tuzun E, Cheng Z, Ebling HM, Tusneem N, David R, Gillett W, Phelps KA, Weaver M, Saranga D, Brand A, Tao W, Gustafson E, McKernan K, Chen L, Malig M, Smith JD, Korn JM, McCarroll SA, Altshuler DA, Peiffer DA, Dorschner M, Stamatoyannopoulos J, Schwartz D, Nickerson DA, Mullikin JC, Wilson RK, Bruhn L, Olson MV, Kaul R, Smith DR, Eichler EE. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453(7191):56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Lisch D. Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci USA. 1997;94(15):7704–7711. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Islam-Faridi MN, Klein PE, Stelly DM, Price HJ, Klein RR, Mullet JE. Comprehensive molecular cytogenetic analysis of sorghum genome architecture: distribution of euchromatin, heterochromatin, genes and recombination in comparison to rice. Genetics. 2005;171(4):1963–1976. doi: 10.1534/genetics.105.048215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolliker R, Jones ES, Drayton MC, Dupal MP, Forster JW. Development and characterisation of simple sequence repeat (SSR) markers for white clover (Trifolium repens L.) Theor Appl Genet. 2001;102(2–3):416–424. [Google Scholar]
- Kong L, Dong J, Hart GE. Characteristics, linkage-map positions, and allelic differentiation of Sorghum bicolour (L.) Moench DNA simple-sequence repeats (SSRs) Theor Appl Genet. 2000;101(3):438–448. doi: 10.1007/s001220051501. [DOI] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, Taillon BE, Chen ZT, Tanzer A, Saunders ACE, Chi JX, Yang FT, Carter NP, Hurles ME, Weissman SM, Harkins TT, Gerstein MB, Egholm M, Snyder M. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318(5849):420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1943;12:172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x. [DOI] [Google Scholar]
- Lam HM, Xu X, Liu X, Chen WB, Yang GH, Wong FL, Li MW, He WM, Qin N, Wang B, Li J, Jian M, Wang JA, Shao GH, Wang J, Sun SSM, Zhang GY. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 2010;42(12):1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- Lee HR, Zhang WL, Langdon T, Jin WW, Yan HH, Cheng ZK, Jiang JM. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci USA. 2005;102(33):11793–11798. doi: 10.1073/pnas.0503863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RQ, Yu C, Li YR, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Mace ES, Jordan DR. Integrating sorghum whole genome sequence information with a compendium of sorghum QTL studies reveals uneven distribution of QTL and of gene-rich regions with significant implications for crop improvement. Theor Appl Genet. 2011;123(1):169–191. doi: 10.1007/s00122-011-1575-y. [DOI] [PubMed] [Google Scholar]
- Mace ES, Xia L, Jordan DR, Halloran K, Parh DK, Huttner E, Wenzl P, Kilian A (2008) DArT markers: diversity analyses and mapping in Sorghum bicolour. BMC Genomics 9. doi:10.1186/1471-2164-9-26 [DOI] [PMC free article] [PubMed]
- Mace ES, Rami JF, Bouchet S, Klein PE, Klein RR, Kilian A, Wenzl P, Xia L, Halloran K, Jordan DR (2009) A consensus genetic map of sorghum that integrates multiple component maps and high-throughput Diversity Array Technology (DArT) markers. BMC Plant Biol 9. doi:10.1186/1471-2229-9-13 [DOI] [PMC free article] [PubMed]
- Mace ES, Tai S, Gilding EK, Li Y, Prentis PJ, Bian L, Campbell BC, Hu W, Innes DJ, Han X, Cruickshank A, Dai C, Frere C, Zhang H, Hunt CH, Wang X, Shatte T, Wang M, Su Z, Li J, Lin X, Godwin ID, Jordan DR, Wang J. Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat Commun. 2013;4:2320. doi: 10.1038/ncomms3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroni F, Pinosio S, Morgante M. Structural variation and genome complexity: is dispensable really dispensable? Curr Opin Plant Biol. 2014;18:31–36. doi: 10.1016/j.pbi.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Mcclintock B. Mutable loci in maize. Carnegie Inst Wash. 1948;47:155–169. [Google Scholar]
- McCouch SR, Chen XL, Panaud O, Temnykh S, Xu YB, Cho YG, Huang N, Ishii T, Blair M. Microsatellite marker development, mapping and applications in rice genetics and breeding. Plant Mol Biol. 1997;35(1–2):89–99. doi: 10.1023/A:1005711431474. [DOI] [PubMed] [Google Scholar]
- McHale LK, Haun WJ, Xu WW, Bhaskar PB, Anderson JE, Hyten DL, Gerhardt DJ, Jeddeloh JA, Stupar RM. Structural variants in the soybean genome localize to clusters of biotic stress-response genes. Plant Physiol. 2012;159(4):1295–1308. doi: 10.1104/pp.112.194605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu YT, Tsung EF, Clouser CR, Duncan C, Ichikawa JK, Lee CC, Zhang Z, Ranade SS, Dimalanta ET, Hyland FC, Sokolsky TD, Zhang L, Sheridan A, Fu HN, Hendrickson CL, Li B, Kotler L, Stuart JR, Malek JA, Manning JM, Antipova AA, Perez DS, Moore MP, Hayashibara KC, Lyons MR, Beaudoin RE, Coleman BE, Laptewicz MW, Sannicandro AE, Rhodes MD, Gottimukkala RK, Yang S, Bafna V, Bashir A, MacBride A, Alkan C, Kidd JM, Eichler EE, Reese MG, De la Vega FM, Blanchard AP. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19(9):1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz MA, Klein RR, Mullet JE, Obert JA, Unruh NC, Klein PE. A high-density genetic map of Sorghum bicolour (L.) Moench based on 2926 AFLP (R), RFLP and SSR markers. Plant Mol Biol. 2002;48(5):483–499. doi: 10.1023/A:1014831302392. [DOI] [PubMed] [Google Scholar]
- Menz MA, Klein RR, Unruh NC, Rooney WL, Klein PE, Mullet JE. Genetic diversity of public inbreds of sorghum determined by mapped AFLP and SSR markers. Crop Sci. 2004;44:1236–1244. doi: 10.2135/cropsci2004.1236. [DOI] [Google Scholar]
- Miller JT, Jackson SA, Nasuda S, Gill BS, Wing RA, Jiang J. Cloning and characterization of a centromere-specific repetitive DNA element from Sorghum bicolour. Theor Appl Genet. 1998;96(6–7):832–839. doi: 10.1007/s001220050809. [DOI] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, Chinwalla A, Conrad DF, Fu YT, Grubert F, Hajirasouliha I, Hormozdiari F, Iakoucheva LM, Iqbal Z, Kang SL, Kidd JM, Konkel MK, Korn J, Khurana E, Kural D, Lam HYK, Leng J, Li RQ, Li YR, Lin CY, Luo RB, Mu XJ, Nemesh J, Peckham HE, Rausch T, Scally A, Shi XH, Stromberg MP, Stutz AM, Urban AE, Walker JA, Wu JT, Zhang YJ, Zhang ZDD, Batzer MA, Ding L, Marth GT, McVean G, Sebat J, Snyder M, Wang J, Ye K, Eichler EE, Gerstein MB, Hurles ME, Lee C, McCarroll SA, Korbel JO, Project G Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470(7332):59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet. 2005;37(9):997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Larkin DM, Everts-van der Wind A, Bourque G, Tesler G, Auvil L, Beever JE, Chowdhary BP, Galibert F, Gatzke L, Hitte C, Meyers SN, Milan D, Ostrander EA, Pape G, Parker HG, Raudsepp T, Rogatcheva MB, Schook LB, Skow LC, Welge M, Womack JE, O’Brien SJ, Pevzner PA, Lewin HA. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309(5734):613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Murray SC, Sharma A, Rooney WL, Klein PE, Mullet JE, Mitchell SE, Kresovich S. Genetic improvement of sorghum as a biofuel feedstock: I. QTL for stem sugar and grain nonstructural carbohydrates. Crop Sci. 2008;48(6):2165–2179. doi: 10.2135/cropsci2008.01.0016. [DOI] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboob ur R, Ware D, Westhoff P, Mayer KF, Messing J, Rokhsar DS. The Sorghum bicolour genome and the diversification of grasses. Nature. 2009;457(7229):551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer ELL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40(D1):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Pittaway TS, Lindup S, Liu H, Waterman E, Padi FK, Hash CT, Zhu J, Gale MD, Devos KM. An integrated genetic map and a new set of simple sequence repeat markers for pearl millet, Pennisetum glaucum. Theor Appl Genet. 2004;109(7):1485–1493. doi: 10.1007/s00122-004-1765-y. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen WW, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang FT, Zhang JJ, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JD, Araki H, Chen LL, Chen JQ, Tian DC. Unique evolutionary mechanism in R-genes under the presence/absence polymorphism in Arabidopsis thaliana. Genetics. 2006;172(2):1243–1250. doi: 10.1534/genetics.105.047290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JH, Wolf SE, Burke JM, Presting GG, Ross-Ibarra J, Dawe RK (2010) Widespread gene conversion in centromere cores. Plos Biol 8(3). doi:10.1371/journal.pbio.1000327 [DOI] [PMC free article] [PubMed]
- Sim SC, Durstewitz G, Plieske J, Wieseke R, Ganal MW, Van Deynze A, Hamilton JP, Buell CR, Causse M, Wijeratne S, Francis DM (2012) Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PloS One 7(7). doi:10.1371/journal.pone.0040563 [DOI] [PMC free article] [PubMed]
- Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A (2009) BioMart—biological queries made easy. BMC Genomics 10. doi:10.1186/1471-2164-10-22 [DOI] [PMC free article] [PubMed]
- Springer NM, Ying K, Fu Y, Ji TM, Yeh CT, Jia Y, Wu W, Richmond T, Kitzman J, Rosenbaum H, Iniguez AL, Barbazuk WB, Jeddeloh JA, Nettleton D, Schnable PS (2009) Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. Plos Genet 5(11). doi:10.1371/journal.pgen.1000734 [DOI] [PMC free article] [PubMed]
- Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, Baker A, Jonasdottir A, Ingason A, Gudnadottir VG, Desnica N, Hicks A, Gylfason A, Gudbjartsson DF, Jonsdottir GM, Sainz J, Agnarsson K, Birgisdottir B, Ghosh S, Olafsdottir A, Cazier JB, Kristjansson K, Frigge ML, Thorgeirsson TE, Gulcher JR, Kong A, Stefansson K. A common inversion under selection in Europeans. Nat Genet. 2005;37(2):129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- Subudhi PK, Nguyen HT. Linkage group alignment of sorghum RFLP maps using a RIL mapping population. Genome/National Research Council Canada = Genome/Conseil national de recherches Canada. 2000;43(2):240–249. doi: 10.1139/g99-112. [DOI] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Eichten SR, Kumari S, Tiffin P, Stein JC, Ware D, Springer NM. Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 2010;20(12):1689–1699. doi: 10.1101/gr.109165.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SJ, Zhong Y, Hou H, Yang SH, Tian DC (2012) Variation of presence/absence genes among Arabidopsis populations. BMC Evol Biol 12. doi:10.1186/1471-2148-12-86 [DOI] [PMC free article] [PubMed]
- Tenaillon MI, Hollister JD, Gaut BS. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 2010;15(8):471–478. doi: 10.1016/j.tplants.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, Olson MV, Eichler EE. Fine-scale structural variation of the human genome. Nat Genet. 2005;37(7):727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- Van Berloo R. GGT 2.0: versatile software for visualization and analysis of genetic data. J Hered. 2008;99(2):232–236. doi: 10.1093/jhered/esm109. [DOI] [PubMed] [Google Scholar]
- Van Os H, Andrzejewski S, Bakker E, Barrena I, Bryan GJ, Caromel B, Ghareeb B, Isidore E, de Jong W, van Koert P, Lefebvre V, Milbourne D, Ritter E, van der Voort JNAMR, Rousselle-Bourgeois F, van Vliet J, Waugh R, Visser RGF, Bakker J, van Eck HJ. Construction of a 10,000-marker ultradense genetic recombination map of potato: providing a framework for accelerated gene isolation and a genomewide physical map. Genetics. 2006;173(2):1075–1087. doi: 10.1534/genetics.106.055871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. Aflp—a new technique for DNA-fingerprinting. Nucleic Acids Res. 1995;23(21):4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Weigel D, Smith LM (2013) Transposon variants and their effects on gene expression in Arabidopsis. Plos Genet 9(2). doi:10.1371/journal.pgen.1003255 [DOI] [PMC free article] [PubMed]
- Wang Y, Lu J, Chen S, Shu L, Palmer RG, Xing G, Li Y, Yang S, Yu D, Zhao T. Exploration of presence/absence variation and corresponding polymorphic markers in soybean genome. J Integr Plant Biol. 2014;56:1009–1019. doi: 10.1111/jipb.12208. [DOI] [PubMed] [Google Scholar]
- Whitkus R, Doebley J, Lee M. Comparative genome mapping of sorghum and maize. Genetics. 1992;132(4):1119–1130. doi: 10.1093/genetics/132.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YQ, Huang YH. An SSR genetic map of Sorghum bicolour (L.) Moench and its comparison to a published genetic map. Genome/National Research Council Canada = Genome/Conseil national de recherches Canada. 2007;50(1):84–89. doi: 10.1139/g06-133. [DOI] [PubMed] [Google Scholar]
- Wu DH, Wu HP, Wang CS, Tseng HY, Hwu KK. Genome-wide InDel marker system for application in rice breeding and mapping studies. Euphytica. 2013;192(1):131–143. doi: 10.1007/s10681-013-0925-z. [DOI] [Google Scholar]
- Xu GW, Magill CW, Schertz KF, Hart GE. A RFLP linkage map of Sorghumbicolour (L) Moench. Theor Appl Genet. 1994;89(2–3):139–145. doi: 10.1007/BF00225133. [DOI] [PubMed] [Google Scholar]
- Yalcin B, Wong K, Agam A, Goodson M, Keane TM, Gan XC, Nellaker C, Goodstadt L, Nicod J, Bhomra A, Hernandez-Pliego P, Whitley H, Cleak J, Dutton R, Janowitz D, Mott R, Adams DJ, Flint J. Sequence-based characterization of structural variation in the mouse genome. Nature. 2011;477(7364):326–329. doi: 10.1038/nature10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng HK, Zhang Y, Chen J, Zhang ZJ, Wang J, Li ST, Li RQ, Bolund L, Wang J. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li SG, Li H, Zhou J, Ni PX, Dong W, Hu SN, Zeng CQ, Zhang JG, Zhang Y, Li RQ, Xu ZY, Li ST, Li XR, Zheng HK, Cong LJ, Lin L, Yin JN, Geng JN, Li GY, Shi JP, Liu J, Lv H, Li J, Wang J, Deng YJ, Ran LH, Shi XL, Wang XY, Wu QF, Li CF, Ren XY, Wang JQ, Wang XL, Li DW, Liu DY, Zhang XW, Ji ZD, Zhao WM, Sun YQ, Zhang ZP, Bao JY, Han YJ, Dong LL, Ji J, Chen P, Wu SM, Liu JS, Xiao Y, Bu DB, Tan JL, Yang L, Ye C, Zhang JF, Xu JY, Zhou Y, Yu YP, Zhang B, Zhuang SL, Wei HB, Liu B, Lei M, Yu H, Li YZ, Xu H, Wei SL, He XM, Fang LJ, Zhang ZJ, Zhang YZ, Huang XG, Su ZX, Tong W, Li JH, Tong ZZ, Li SL, Ye J, Wang LS, Fang L, Lei TT, Chen C, Chen H, Xu Z, Li HH, Huang HY, Zhang F, Xu HY, Li N, Zhao CF, Li ST, Dong LJ, Huang YQ, Li L, Xi Y, Qi QH, Li WJ, Zhang B, Hu W, Zhang YL, Tian XJ, Jiao YZ, Liang XH, Jin JA, Gao L, Zheng WM, Hao BL, Liu SQ, Wang W, Yuan LP, Cao ML, McDermott J, Samudrala R, Wang J, Wong GKS, Yang HM. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3(2):266–281. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C-Z, Zhai G-W, Zou G-H, Y-z TAO, Wang H. Assessment of genetic diversity among 41 sorghum varieties using SSR markers. Jiangsu J Agric Sci. 2010;2:007. [Google Scholar]
- Zhang F, Gu WL, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WW, Pan JS, He HL, Zhang C, Li Z, Zhao JL, Yuan XJ, Zhu LH, Huang SW, Cai R. Construction of a high density integrated genetic map for cucumber (Cucumis sativus L.) Theor Appl Genet. 2012;124(2):249–259. doi: 10.1007/s00122-011-1701-x. [DOI] [PubMed] [Google Scholar]
- Zhang LM, Luo H, Liu ZQ, Zhao Y, Luo JC, Hao DY, Jing HC. Genome-wide patterns of large-size presence/absence variants in sorghum. J Integr Plant Biol. 2014;56(1):24–37. doi: 10.1111/jipb.12121. [DOI] [PubMed] [Google Scholar]
- Zheng LY, Guo XS, He B, Sun LJ, Peng Y, Dong SS, Liu TF, Jiang SY, Ramachandran S, Liu CM, Jing HC (2011) Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolour). Genome Biol 12(11). doi:10.1186/Gb-2011-12-11-R114 [DOI] [PMC free article] [PubMed]
- Ziolkowski PA, Koczyk G, Galganski L, Sadowski J. Genome sequence comparison of Col and Ler lines reveals the dynamic nature of Arabidopsis chromosomes. Nucleic Acids Res. 2009;37(10):3189–3201. doi: 10.1093/nar/gkp183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou GH, Zhai GW, Feng Q, Yan S, Wang A, Zhao Q, Shao JF, Zhang ZP, Zou JQ, Han B, Tao YZ. Identification of QTLs for eight agronomically important traits using an ultra-high-density map based on SNPs generated from high-throughput sequencing in sorghum under contrasting photoperiods. J Exp Bot. 2012;63(15):5451–5462. doi: 10.1093/jxb/ers205. [DOI] [PubMed] [Google Scholar]
- Zwick MS, Islam-Faridi MN, Zhang HB, Hodnett GL, Gomez MI, Kim JS, Price HJ, Stelly DM. Distribution and sequence analysis of the centromere-associated repetitive element CEN38 of Sorghum bicolour (Poaceae) Am J Bot. 2000;87(12):1757–1764. doi: 10.2307/2656825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.