Abstract
APOBEC3 (apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3) proteins are cellular DNA deaminases that restrict a broad spectrum of lentiviruses. This process is counteracted by Vif (viral infectivity factor) of lentiviruses, which binds APOBEC3s and promotes their degradation. CBF-β (core binding factor subunit β) is an essential co-factor for the function of human immunodeficiency virus type 1 Vif to degrade human APOBEC3s. However, the requirement for CBF-β in Vif-mediated degradation of other mammalian APOBEC3 proteins is less clear. Here, we determined the sequence of feline CBFB and performed phylogenetic analyses. These analyses revealed that mammalian CBFB is under purifying selection. Moreover, we demonstrated that CBF-β is dispensable for feline immunodeficiency virus Vif-mediated degradation of APOBEC3s of its host. These findings suggested that primate lentiviruses have adapted to use CBF-β, an evolutionary stable protein, to counteract APOBEC3 proteins of their hosts after diverging from other lentiviruses.
Several human APOBEC3 (apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3) proteins, notably APOBEC3D, APOBEC3F, APOBEC3G and APOBEC3H, have the capacity to restrict human immunodeficiency virus type 1 (HIV-1) replication (Albin & Harris, 2010; Desimmie et al., 2014; Feng et al., 2014; Kitamura et al., 2011; Refsland & Harris, 2013). These restriction factors are incorporated into the progeny virions and enzymically convert viral cDNA cytosines to uracils during reverse transcription, which can debilitate viral function (Albin & Harris, 2010; Desimmie et al., 2014; Feng et al., 2014; Kitamura et al., 2011; Refsland & Harris, 2013). Although original studies focused on human APOBEC3G (Harris et al., 2003; Mariani et al., 2003; Sheehy et al., 2002; Zhang et al., 2003), subsequent work revealed that all placental mammals have APOBEC3 enzymes (Münk et al., 2012), albeit different numbers, and that these enzymes have the potential to attenuate the infectivity of a broad spectrum of viruses, including simian immunodeficiency virus (SIV) (Mariani et al., 2003), feline immunodeficiency virus (FIV) (Münk et al., 2008; Zielonka et al., 2010), bovine immunodeficiency virus (BIV) (LaRue et al., 2010) and small ruminant lentiviruses (SRLVs; e.g. Maedi-Visna virus and caprine arthritis encephalitis virus) (LaRue et al., 2010). However, to counteract APOBEC3-mediated restriction, these lentiviruses encode a protein called Vif (viral infectivity factor). Vif recruits a cellular E3 ubiquitin ligase complex including cullin 5 (CUL5) and elongin B/C (ELOB/C), and degrades host APOBEC3 proteins through a ubiquitin/proteasome-dependent pathway (Albin & Harris, 2010; Desimmie et al., 2014; Feng et al., 2014; Kitamura et al., 2011; Refsland & Harris, 2013).
Several studies have demonstrated that CBF-β (core binding factor subunit β) is an essential co-factor for HIV-1 and SIV Vif proteins to form the ubiquitin ligase complex that degrades human and rhesus monkey APOBEC3 enzymes (Hultquist et al., 2012a; Jäger et al., 2012; Zhang et al., 2012). CBF-β normally functions as a transcription factor for haematopoiesis, T-cell differentiation and bone development through binding with Runt-related transcription factors 1, 2 and 3 (RUNX1, 2 and 3) (Adya et al., 2000; de Bruijn & Speck, 2004; Ito, 2008). Structural studies have provided additional compelling evidence that CBF-β is an integral part of the HIV-1 Vif ubiquitin ligase complex (Guo et al., 2014; Kim et al., 2013). In particular, the surface area of the CBF-β/HIV-1 Vif heterodimeric interface is nearly 5000 Å2, which is larger than that in the CBF-β/RUNX complex (Guo et al., 2014).
Despite these advances, it is not clear whether CBF-β is required for non-primate lentiviral Vif proteins to degrade the APOBEC3 proteins of their hosts. For instance, although FIV Vif also degrades feline APOBEC3 proteins through a ubiquitin/proteasome-dependent pathway (Wang et al., 2011), a role for CBF-β in this complex has yet to be investigated. Moreover, the feline CBF-β gene (CBFB) has yet to be characterized. Here, we sequenced feline CBFB and performed molecular phylogenetic analyses to show it is under purifying selection. We further demonstrated that CBF-β was dispensable for FIV Vif function in degrading feline APOBEC3 proteins.
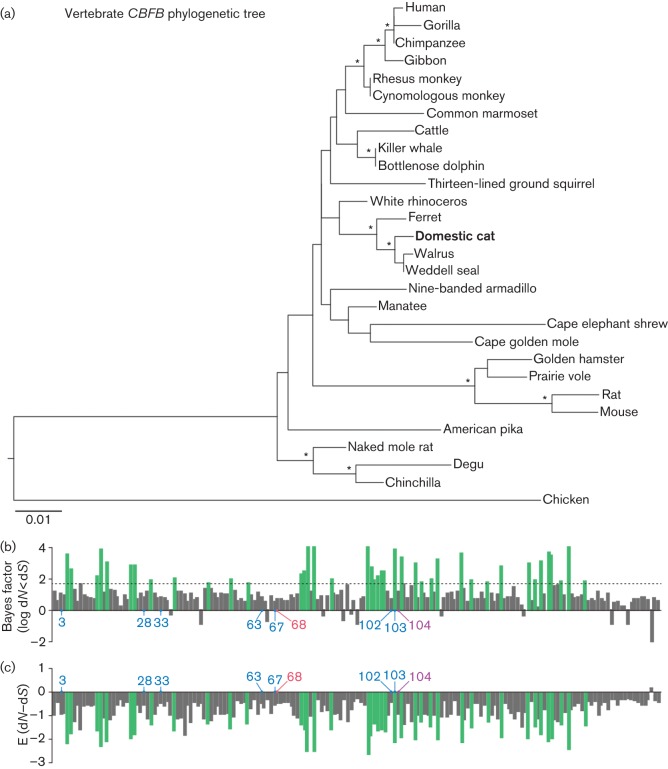
To investigate the conservation of CBFB in mammals, including domestic cats, in depth, we set out to determine the sequence of feline CBFB. The open reading frame (ORF) of domestic cat (Felis catus) CBFB was amplified by PCR using a cDNA library of the MYA-1 cell line (Shimojima et al., 2004) as the template and the following primers: fCBFb-F2, 5′-ATGCCGCGCGTCGTGCCCG-3′ and fCBFb-R1, 5′-TTAACGAAGTTTGAGGTCATCACCAC-3′. PCR was performed by using PrimeStar GXL DNA polymerase (TaKaRa) according to the manufacturer’s protocol. The obtained DNA fragment was cloned into pCR-Blunt II-TOPO plasmid by using a Zero Blunt TOPO PCR cloning kit (Life Technologies) according to the manufacturer’s protocol. The sequence of domestic cat CBFB was determined by a DNA sequencing service (Fasmac) and the data were analysed with Sequencher v5.1 software (Gene Codes). The domestic cat CBFB ORF was aligned with 27 mammalian and one avian CBFB sequences using mafft implemented in the guidance server (Penn et al., 2010). The sequences used in this study are listed in Table 1 and the resulting alignment was verified manually at the amino acid level. Then the phylogenetic tree of the 29 CBFB genes was reconstructed using the maximum-likelihood method with PhyML (Guindon et al., 2010). As shown in Fig. 1(a), domestic cat CBFB formed a cluster with those of ferret, Weddell seal and walrus, which is supported by a relatively high bootstrap value (91 %).
Table 1. GenBank accession numbers of the CBFB genes used in this study.
The common name of each primate is identical to that in Fig. 1(a).
| Common name | Scientific name | GenBank accession no. |
| Human | Homo sapiens | AK290462 |
| Gorilla | Gorilla gorilla | XM_004057790 |
| Chimpanzee | Pan troglodytes | GABD01004603 |
| Gibbon | Nomascus leucogenys | XM_003262871 |
| Rhesus monkey | Macaca mulatta | JU336882 |
| Cynomolgus monkey | Macaca fascicularis | XM_005592190 |
| Common marmoset | Callithrix jacchus | XM_002761058 |
| Cattle | Bos taurus | NM_001191435 |
| Killer whale | Orcinus orca | XM_004280877 |
| Bottlenose dolphin | Tursiops truncatus | XM_004311020 |
| Thirteen-lined ground squirrel | Ictidomys tridecemlineatus | XM_005318290 |
| White rhinoceros | Ceratotherium simum | XM_004431701 |
| Ferret | Mustela putorius furo | XM_004744336 |
| Domestic cat | Felis catus | LC003231 (this study) |
| Walrus | Odobenus rosmarus | XM_004393406 |
| Weddell seal | Leptonychotes weddellii | XM_006741505 |
| Nine-banded armadillo | Dasypus novemcinctus | XM_004460890 |
| Manatee | Trichechus manatus | XM_004371510 |
| Cape elephant shrew | Elephantulus edwardii | XM_006878708 |
| Cape golden mole | Chrysochloris asiatica | XM_006863598 |
| Golden hamster | Mesocricetus auratus | XM_005076189 |
| Prairie vole | Microtus ochrogaster | XM_005345497 |
| Rat | Rattus norvegicus | BC081946 |
| Mouse | Mus musculus | D14571 |
| American pika | Ochotona princeps | XM_004583922 |
| Naked mole rat | Heterocephalus glaber | XM_004843071 |
| Degu | Octodon degus | XM_004625892 |
| Chinchilla | Chinchilla lanigera | XM_005403704 |
| Chicken | Gallus gallus | AF472513 |
Fig. 1.
Molecular phylogenetic analyses of CBFB. (a) Phylogenetic tree of 29 CBFB genes reconstructed using the maximum-likelihood method. Chicken CBFB was used as an outgroup. Nodes with >70 % bootstrap values are indicated with asterisks. (b, c) Negative selection in 29 CBFB genes inferred by the REL method in HyPhy. The Bayes factor for dN<dS (negative selection) (b) and the E (dN–dS) value (c) are shown. RUNX1-binding sites (aa 3, 28, 33, 63, 67, 102 and 103; blue) (Bravo et al., 2001; Tahirov et al., 2001; Yan et al., 2004), Vif-binding site (aa 68; red) (Hultquist et al., 2012b), and the site binding to both RUNX1 and Vif (aa 104; purple) are indicated. The green bars represent the 41 negatively selected sites identified with Bayes factor >50. The dotted line in (b) indicates the Bayes factor threshold of 50 specified for the REL analysis.
We then conducted the analysis to estimate selective pressure acting on CBFB genes. The random effects likelihood (REL) method available in the HyPhy package was employed (Pond & Frost, 2005). As shown in Fig. 1(b), 41 sites were found to be under negative selection that had a Bayes factor >50 for dN<dS. In addition, the values of E (dN–dS) at all the codons except for codon 180 in CBFB were <0 (Fig. 1c). RUNX1-binding sites (Bravo et al., 2001; Tahirov et al., 2001; Yan et al., 2004) and Vif-binding sites (Du et al., 2013) were highly conserved in CBF-β (Fig. 1c). Moreover, although the identity between human CBFB and domestic cat CBFB is 96.5 % at the nucleotide level, the amino acid sequences were 100 % identical (data not shown). Taken together, these results indicated that mammalian CBFB is under strong purifying selection and stably maintained in mammals.
It is known that domestic cat (F. catus) expresses five kinds of APOBEC3 proteins, APOBEC3Z2a, APOBEC3Z2b, APOBEC3Z2c, APOBEC3Z3 and APOBEC3Z2Z3 (LaRue et al., 2010; Münk et al., 2008; Stern et al., 2010; Zielonka et al., 2010), and that feline APOBECZ3 and APOBEC3Z2Z3 have the ability to restrict FIV infectivity (Zielonka & Münk, 2011). To determine whether CBF-β was required for FIV Vif as a co-factor to degrade feline APOBEC3s, we constructed a FLAG-tagged FIV Vif expression plasmid as follows: the codon-optimized ORF of FIV strain Petaluma with a FLAG-tag at the C terminus (the sequence is available upon request) was obtained from GeneArt Gene Synthesis service (Life Technologies). The obtained DNA was digested with BglII and SalI, and then was inserted into the BamHI/SalI sites of pDON-AI plasmid (TaKaRa). Expression plasmids for haemagglutinin (HA)-tagged feline APOBEC3Z2b (AY971954), feline APOBEC3Z3 (EU011792), feline APOBEC3Z2Z3 (HM100128) (Münk et al., 2008) and human CBF-β (Jäger et al., 2012) were also used. We co-transfected each feline APOBEC3-HA expression plasmid and FIV Vif-FLAG expression plasmid with or without human CBF-β expression plasmid into HEK293T-shCBFB cells, a HEK293T cell line stably expressing small hairpin RNA targeting the 5′ UTR of endogenous human CBFB (Jäger et al., 2012), using Lipofectamine 2000 according to the manufacturer’s procedure (Life Technologies). Cells were harvested at 48 h post-transfection, and extracts were analysed by SDS-PAGE and Western blotting as described previously (Kobayashi et al., 2014a, b) with the following antibodies: anti-HA mAb (3F10; Roche), anti-FLAG polyclonal antibody (OctA; Santa Cruz), anti-CBF-β mAb (sc-56751; Santa Cruz) and anti-α-tubulin (TUBA) mAb (DM1A; Sigma). As described in Fig. 2(a), the levels of feline APOBECZ2b, APOBEC3Z3 and APOBEC3Z2Z3 were decreased by FIV Vif regardless of the presence or absence of CBF-β. These results showed that FIV Vif did not require CBF-β for feline APOBEC3 degradation.
Fig. 2.
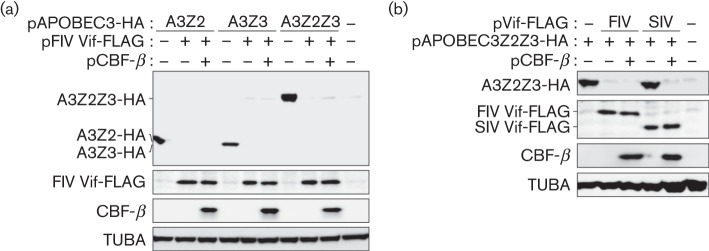
Functional analyses of the requirement for CBF-β. (a) Feline APOBEC3-HA (150 ng) and FIV Vif-FLAG expression plasmids (500 ng) were co-transfected with or without human CBF-β expression plasmid (400 ng) into HEK293T-shCBFB cells, and the transfected cell lysates were analysed by SDS-PAGE and Western blotting. As negative controls, pDON-AI and pcDNA3.1 plasmids were used in place of FIV Vif-FLAG and CBF-β expression plasmids, respectively. A3Z2, feline APOBEC3Z2b; A3Z3, feline APOBEC3Z3; A3Z2Z3, feline APOBEC3Z2Z3. (b) Feline APOBEC3Z2Z3-HA expression plasmid (150 ng) and either FIV Vif-FLAG or SIV Vif-FLAG expression plasmid (500 ng) were co-transfected with or without human CBF-β expression plasmid (400 ng) into HEK293T-shCBFB cells, and the transfected cells were analysed by SDS-PAGE and Western blotting. As negative controls, pDON-AI and pcDNA3.1 plasmids were used in place of Vif-FLAG and CBF-β expression plasmids, respectively. FIV, FIV Vif-FLAG; SIV, SIVmac Vif-FLAG.
Interestingly, it has been reported that the Vif protein of SIVmac can degrade feline APOBEC3Z2Z3 (Stern et al., 2010). This raises the possibility that feline APOBEC3Z2Z3 degradation may be governed directly by the APOBEC3–Vif interaction (i.e. SIVmac Vif may be able to degrade feline APOBEC3Z2Z3 without CBF-β). To address this, we constructed a FLAG-tagged SIVmac239 Vif expression plasmid as follows: PCR was performed by using PfuUltra High-Fidelity DNA Polymerase (Agilent Technologies) with pSIVmac239 (M33262) (kindly provided by Dr Tomoyuki Miura) as the template and the following primers: forward, 5′-TTTTTTTTGGATCCGCCACCATGGAGGAGGAAAAGAGG-3′ and reverse, 5′-TTTTTTTTTTGTCGACTCACTTATCGTCGTCATCCTTGTAATCTGCCAGTATTCCCAAGAC-3′. The obtained SIVmac239 Vif-FLAG ORF fragment was digested with BamHI and SalI, and then was inserted into the BamHI/SalI site of pDON-AI plasmid. Plasmid integrity was confirmed by DNA sequencing, as described above. We co-transfected the feline APOBEC3Z2Z3-HA expression plasmid and either a FIV Vif-FLAG or a SIVmac Vif-FLAG expression plasmid with or without a human CBF-β expression plasmid into HEK293T-shCBFB cells (Jäger et al., 2012). As shown in Fig. 2(b), we found that CBF-β was essential for SIVmac Vif to degrade feline APOBEC3Z2Z3. These results showed that CBF-β was essential for the Vif protein of SIVmac, a primate lentivirus, to degrade APOBEC3, regardless of the origin of the APOBEC3 protein.
Thus, we have shown that CBFB is highly conserved in mammals, including the hosts of lentiviruses such as human, gorilla, chimpanzee, rhesus monkey, cattle, and domestic cat (Fig. 1). We have also demonstrated that FIV Vif does not use CBF-β for feline APOBEC3 degradation (Fig. 2). These data corroborate recent reports suggesting that not only FIV Vif but also BIV and SRLV Vif proteins do not require CBF-β to degrade APOBEC3 proteins of their hosts (Ai et al., 2014; Zhang et al., 2014). These observations indicate that only primate lentiviruses require CBF-β for APOBEC3 degradation and further suggest that non-primate lentiviruses have either maintained an ancestral CBF-β-independent mechanism or have evolved to use another co-factor in the same way to degrade APOBEC3 proteins of their ancestral hosts. Interestingly, it has been recently reported that HIV-1 Vif can utilize CBF-β proteins from flies (Drosophila melanogaster) and worms (Saccoglossus kowalevskii) as the co-factor for human APOBEC3 degradation (Han et al., 2014). This further suggests that the usability of CBF-β is not dependent on the hosts and that primate lentiviral Vif has adapted to use CBF-β as an essential co-factor during its evolution.
In comparison with BIV and SRLVs, the feline lentivirus FIV is phylogenetically closer to primate lentiviruses (Gifford, 2012). Therefore, our findings strongly suggest that the requirement of CBF-β for primate lentiviral Vif arose after the divergence with the FIV lineage. As CBF-β is under purifying selection in mammals (Fig. 1), it would be difficult for CBF-β to change in order to evade primate lentiviral Vif-mediated hijacking. This may be advantageous for the virus because CBF-β is an evolutionarily stable protein.
FIV Vif is very different from HIV/SIV Vif. For instance, FIV Vif has ~60 extra amino acids compared with HIV/SIV Vif, and the identity and the similarity of FIV Vif (strain Petaluma) to HIV-1 Vif (strain NL4-3) are only 16.6 and 64.7 %, respectively. These characteristics imply that FIV Vif is structurally dissimilar to HIV/SIV Vif. Additionally, Ai et al. (2014) have shown that the conserved CBF-β interaction sequences in SIV/HIV Vif are not present in FIV Vif. Furthermore, Wang et al. (2011) have reported that the FIV Vif has neither a CUL5 box nor HCCH zinc-binding motif, despite a functional requirement for CUL5 in APOBEC3 degradation. Due to these extensive differences with primate lentiviral Vif proteins, it is not clear whether FIV Vif has adapted to use another cellular protein(s) or whether it is capable of functioning without a CBF-β-like co-factor. More work will be needed to distinguish between these possibilities.
Acknowledgements
We would like to thank Ms. Naoko Misawa and Mr Yuichi Kimura (Laboratory of Viral Pathogenesis, Institute for Virus Research, Kyoto University) for experimental support, Dr Tomoyuki Miura (Laboratory of Primate model, Institute for Virus Research, Kyoto University) for providing pSIVmac239, Dr Terumasa Ikeda (University of Minnesota) for helpful suggestions and Ms Kotubu Misawa for dedicated support. This study was supported in-part by grants from the following: the Aihara Innovative Mathematical Modelling Project, the Japan Society for the Promotion of Science through the ‘Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST)’, initiated by the Council for Science and Technology Policy of Japan (to K. S.); CREST, Japan Science and Technology Agency (to K. S.); Ministry of Health, Labor and Welfare of Japan (Health Labor Sciences Research Grant 26361601) (to K. S.); Takeda Science Foundation (to K. S.); Sumitomo Foundation Research Grant (to K. S.); Senshin Medical Research Foundation (to K. S.); Ichiro Kanehara Foundation (to K. S.); Kanae Foundation for the Promotion of Medical Science (to K. S.); Ansmann Foundation for AIDS Research (to C. M.); National Institutes of Health (R01 AI064046 and P01 GM091743) (to R. H.); Japan Society for the Promotion of Science (Grants-in-Aid for Scientific Research B24390112 and S22220007) (to Y. K.); Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant-in-Aid for Scientific Research on Innovative Areas 24115008) (to Y. K.); and Research on HIV/AIDS from Ministry of Health, Labor and Welfare of Japan (to Y. K.).
References
- Adya N., Castilla L. H., Liu P. P. (2000). Function of CBFβ/Bro proteins. Semin Cell Dev Biol 11, 361–368. 10.1006/scdb.2000.0189 [DOI] [PubMed] [Google Scholar]
- Ai Y., Zhu D., Wang C., Su C., Ma J., Ma J., Wang X. (2014). Core-binding factor subunit beta is not required for non-primate lentiviral Vif-mediated APOBEC3 degradation. J Virol 88, 12112–12122. 10.1128/JVI.01924-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin J. S., Harris R. S. (2010). Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev Mol Med 12, e4. 10.1017/S1462399409001343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J., Li Z., Speck N. A., Warren A. J. (2001). The leukemia-associated AML1 (Runx1)–CBF β complex functions as a DNA-induced molecular clamp. Nat Struct Biol 8, 371–378. 10.1038/86264 [DOI] [PubMed] [Google Scholar]
- de Bruijn M. F., Speck N. A. (2004). Core-binding factors in hematopoiesis and immune function. Oncogene 23, 4238–4248. 10.1038/sj.onc.1207763 [DOI] [PubMed] [Google Scholar]
- Desimmie B. A., Delviks-Frankenberrry K. A., Burdick R. C., Qi D., Izumi T., Pathak V. K. (2014). Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol 426, 1220–1245. 10.1016/j.jmb.2013.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Zhao K., Rui Y., Li P., Zhou X., Zhang W., Yu X. F. (2013). Differential requirements for HIV-1 Vif-mediated APOBEC3G degradation and RUNX1-mediated transcription by core binding factor beta. J Virol 87, 1906–1911. 10.1128/JVI.02199-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Baig T. T., Love R. P., Chelico L. (2014). Suppression of APOBEC3-mediated restriction of HIV-1 by Vif. Front Microbiol 5, 450. 10.3389/fmicb.2014.00450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford R. J. (2012). Viral evolution in deep time: lentiviruses and mammals. Trends Genet 28, 89–100. 10.1016/j.tig.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59, 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Guo Y., Dong L., Qiu X., Wang Y., Zhang B., Liu H., Yu Y., Zang Y., Yang M., Huang Z. (2014). Structural basis for hijacking CBF-β and CUL5 E3 ligase complex by HIV-1 Vif. Nature 505, 229–233. 10.1038/nature12884 [DOI] [PubMed] [Google Scholar]
- Han X., Liang W., Hua D., Zhou X., Du J., Evans S. L., Gao Q., Wang H., Viqueira R. & other authors (2014). Evolutionarily conserved requirement for core binding factor beta in the assembly of the human immunodeficiency virus/simian immunodeficiency virus Vif-cullin 5-RING E3 ubiquitin ligase. J Virol 88, 3320–3328. 10.1128/JVI.03833-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. S., Bishop K. N., Sheehy A. M., Craig H. M., Petersen-Mahrt S. K., Watt I. N., Neuberger M. S., Malim M. H. (2003). DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809. 10.1016/S0092-8674(03)00423-9 [DOI] [PubMed] [Google Scholar]
- Hultquist J. F., Binka M., LaRue R. S., Simon V., Harris R. S. (2012a). Vif proteins of human and simian immunodeficiency viruses require cellular CBFβ to degrade APOBEC3 restriction factors. J Virol 86, 2874–2877. 10.1128/JVI.06950-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist J. F., McDougle R. M., Anderson B. D., Harris R. S. (2012b). HIV type 1 viral infectivity factor and the RUNX transcription factors interact with core binding factor β on genetically distinct surfaces. AIDS Res Hum Retroviruses 28, 1543–1551. 10.1089/aid.2012.0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y. (2008). RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res 99, 33–76. 10.1016/S0065-230X(07)99002-8 [DOI] [PubMed] [Google Scholar]
- Jäger S., Kim D. Y., Hultquist J. F., Shindo K., LaRue R. S., Kwon E., Li M., Anderson B. D., Yen L. & other authors (2012). Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature 481, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Y., Kwon E., Hartley P. D., Crosby D. C., Mann S., Krogan N. J., Gross J. D. (2013). CBFβ stabilizes HIV Vif to counteract APOBEC3 at the expense of RUNX1 target gene expression. Mol Cell 49, 632–644. 10.1016/j.molcel.2012.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura S., Ode H., Iwatani Y. (2011). Structural features of antiviral APOBEC3 proteins are linked to their functional activities. Front Microbiol 2, 258. 10.3389/fmicb.2011.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Koizumi Y., Takeuchi J. S., Misawa N., Kimura Y., Morita S., Aihara K., Koyanagi Y., Iwami S., Sato K. (2014a). Quantification of deaminase activity-dependent and -independent restriction of HIV-1 replication mediated by APOBEC3F and APOBEC3G through experimental-mathematical investigation. J Virol 88, 5881–5887. 10.1128/JVI.00062-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Takeuchi J. S., Ren F., Matsuda K., Sato K., Kimura Y., Misawa N., Yoshikawa R., Nakano Y. & other authors (2014b). Characterization of red-capped mangabey tetherin: implication for the co-evolution of primates and their lentiviruses. Sci Rep 4, 5529. 10.1038/srep05529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue R. S., Lengyel J., Jónsson S. R., Andrésdóttir V., Harris R. S. (2010). Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross-species activity. J Virol 84, 8193–8201. 10.1128/JVI.00685-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani R., Chen D., Schröfelbauer B., Navarro F., König R., Bollman B., Münk C., Nymark-McMahon H., Landau N. R. (2003). Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114, 21–31. 10.1016/S0092-8674(03)00515-4 [DOI] [PubMed] [Google Scholar]
- Münk C., Beck T., Zielonka J., Hotz-Wagenblatt A., Chareza S., Battenberg M., Thielebein J., Cichutek K., Bravo I. G. & other authors (2008). Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol 9, R48. 10.1186/gb-2008-9-3-r48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münk C., Willemsen A., Bravo I. G. (2012). An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol Biol 12, 71. 10.1186/1471-2148-12-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn O., Privman E., Ashkenazy H., Landan G., Graur D., Pupko T. (2010). guidance: a web server for assessing alignment confidence scores. Nucleic Acids Res 38 (Web Server issue), W23–W28. 10.1093/nar/gkq443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond S. L., Frost S. D. (2005). Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533. 10.1093/bioinformatics/bti320 [DOI] [PubMed] [Google Scholar]
- Refsland E. W., Harris R. S. (2013). The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol 371, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002). Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650. 10.1038/nature00939 [DOI] [PubMed] [Google Scholar]
- Shimojima M., Miyazawa T., Ikeda Y., McMonagle E. L., Haining H., Akashi H., Takeuchi Y., Hosie M. J., Willett B. J. (2004). Use of CD134 as a primary receptor by the feline immunodeficiency virus. Science 303, 1192–1195. 10.1126/science.1092124 [DOI] [PubMed] [Google Scholar]
- Stern M. A., Hu C., Saenz D. T., Fadel H. J., Sims O., Peretz M., Poeschla E. M. (2010). Productive replication of Vif-chimeric HIV-1 in feline cells. J Virol 84, 7378–7395. 10.1128/JVI.00584-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov T. H., Inoue-Bungo T., Morii H., Fujikawa A., Sasaki M., Kimura K., Shiina M., Sato K., Kumasaka T. & other authors (2001). Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell 104, 755–767. 10.1016/S0092-8674(01)00271-9 [DOI] [PubMed] [Google Scholar]
- Wang J., Zhang W., Lv M., Zuo T., Kong W., Yu X. (2011). Identification of a Cullin5-ElonginB-ElonginC E3 complex in degradation of feline immunodeficiency virus Vif-mediated feline APOBEC3 proteins. J Virol 85, 12482–12491. 10.1128/JVI.05218-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Liu Y., Lukasik S. M., Speck N. A., Bushweller J. H. (2004). CBFbeta allosterically regulates the Runx1 Runt domain via a dynamic conformational equilibrium. Nat Struct Mol Biol 11, 901–906. 10.1038/nsmb819 [DOI] [PubMed] [Google Scholar]
- Zhang H., Yang B., Pomerantz R. J., Zhang C., Arunachalam S. C., Gao L. (2003). The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424, 94–98. 10.1038/nature01707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du J., Evans S. L., Yu Y., Yu X. F. (2012). T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481, 376–379. [DOI] [PubMed] [Google Scholar]
- Zhang W., Wang H., Li Z., Liu X., Liu G., Harris R. S., Yu X. F. (2014). Cellular requirements for bovine immunodeficiency virus Vif-mediated inactivation of bovine APOBEC3 proteins. J Virol 88, 12528–12540. 10.1128/JVI.02072-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielonka J., Münk C. (2011). Cellular restriction factors of feline immunodeficiency virus. Viruses 3, 1986–2005. 10.3390/v3101986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielonka J., Marino D., Hofmann H., Yuhki N., Löchelt M., Münk C. (2010). Vif of feline immunodeficiency virus from domestic cats protects against APOBEC3 restriction factors from many felids. J Virol 84, 7312–7324. 10.1128/JVI.00209-10 [DOI] [PMC free article] [PubMed] [Google Scholar]