Abstract
Background
Mutations in the sulfate transporter gene SLC26A2 (DTDST) cause a continuum of skeletal dysplasia phenotypes that includes achondrogenesis type 1B (ACG1B), atelosteogenesis type 2 (AO2), diastrophic dysplasia (DTD), and recessive multiple epiphyseal dysplasia (rMED). In 1972, de la Chapelle et al reported two siblings with a lethal skeletal dysplasia, which was denoted “neonatal osseous dysplasia” and “de la Chapelle dysplasia” (DLCD). It was suggested that DLCD might be part of the SLC26A2 spectrum of phenotypes, both because of the Finnish origin of the original family and of radiographic similarities to ACG1B and AO2.
Objective
To test the hypothesis whether SLC26A2 mutations are responsible for DLCD.
Methods
We studied the DNA from the original DLCD family and from seven Finnish DTD patients in whom we had identified only one copy of IVS1+2T>C, the common Finnish mutation. A novel SLC26A2 mutation was found in all subjects, inserted by site-directed mutagenesis in a vector harbouring the SLC26A2 cDNA, and expressed in sulfate transport deficient Chinese hamster ovary (CHO) cells to measure sulfate uptake activity.
Results
We identified a hitherto undescribed SLC26A2 mutation, T512K, homozygous in the affected subjects and heterozygous in both parents and in the unaffected sister. T512K was then identified as second pathogenic allele in the seven Finnish DTD subjects. Expression studies confirmed pathogenicity.
Conclusions
DLCD is indeed allelic to the other SLC26A2 disorders. T512K is a second rare “Finnish” mutation that results in DLCD at homozygosity and in DTD when compounded with the milder, common Finnish mutation.
In 1972, de la Chapelle et al described a family with two siblings affected by a distinct and previously unrecognised lethal skeletal dysplasia. The clinical phenotype was characterised by severe micromelia, small thorax, cleft palate, and bilateral clubfoot; radiologically, the main features were short and bowed limb bones, unusually hypoplastic ulna and fibula, and spinal and pelvic underossification.1 In 1986, Whitley et al reported two more patients with what they felt was the same entity, and called this entity “de la Chapelle dysplasia” (DLCD; MIM 256050).2 Autosomal recessive inheritance was considered likely. Whitley et al also reported the histopathological features of cartilage and bone in DLCD, which showed strong similarities with achondrogenesis type 1B (ACG1B; MIM 600972).
In 1987, Sillence et al separated a group of patients who had been considered as having severe diastrophic dysplasia and called them “atelosteogenesis type 2” (AO2; MIM 256050).3 In 1994, Schrander-Stumpel et al described a further case of DLCD and reviewed 10 cases of AO2 pointing to the overlap between these two conditions and to the clinical, radiographic, and histopathological similarities with diastrophic dysplasia (DTD; MIM 222600). The authors hypothesised that DLCD might be a severe form of DTD, with the same genetic and pathophysiological bases.4 This hypothesis could not be proven at that time and the discussion remained open in the subsequent literature.5,6
Following the identification of a sulfation defect in ACG1B7 and of mutations in the sulfate transporter SLC26A2 (also known as DTDST) gene as the cause of DTD,8 it was found that ACG1B, AO2, DTD and recessive multiple epiphyseal dysplasia (rMED; MIM 226900) were allelic disorders comprising a continuum of clinical phenotypes of different severities, correlated to the severity of mutations and their effect on sulfate transport.6 Pathogenic mutations in the SLC26A2 gene proved to reduce the activity of the sulfate transporter and thus sulfation of proteoglycans in cartilage tissue.9–12 Several mutations have been described so far13; five recurrent mutations account for about 2/3 of SLC26A2 pathogenic alleles. Of these, IVS1+2T>C is the most common mutation in the Finnish population (and is therefore called “Finnish mutation”), and the second most frequent in the non-Finnish population. Here we tested the hypothesis that DLCD is indeed part of the SLC26A2 dysplasia spectrum and identified a novel SLC26A2 mutation that seems to be specific to the Finnish population, and causes DLCD when homozygous and DTD when in compound heterozygosity with the common Finnish mutation IVS1+2T>C.
METHODS
Patients and DNA samples
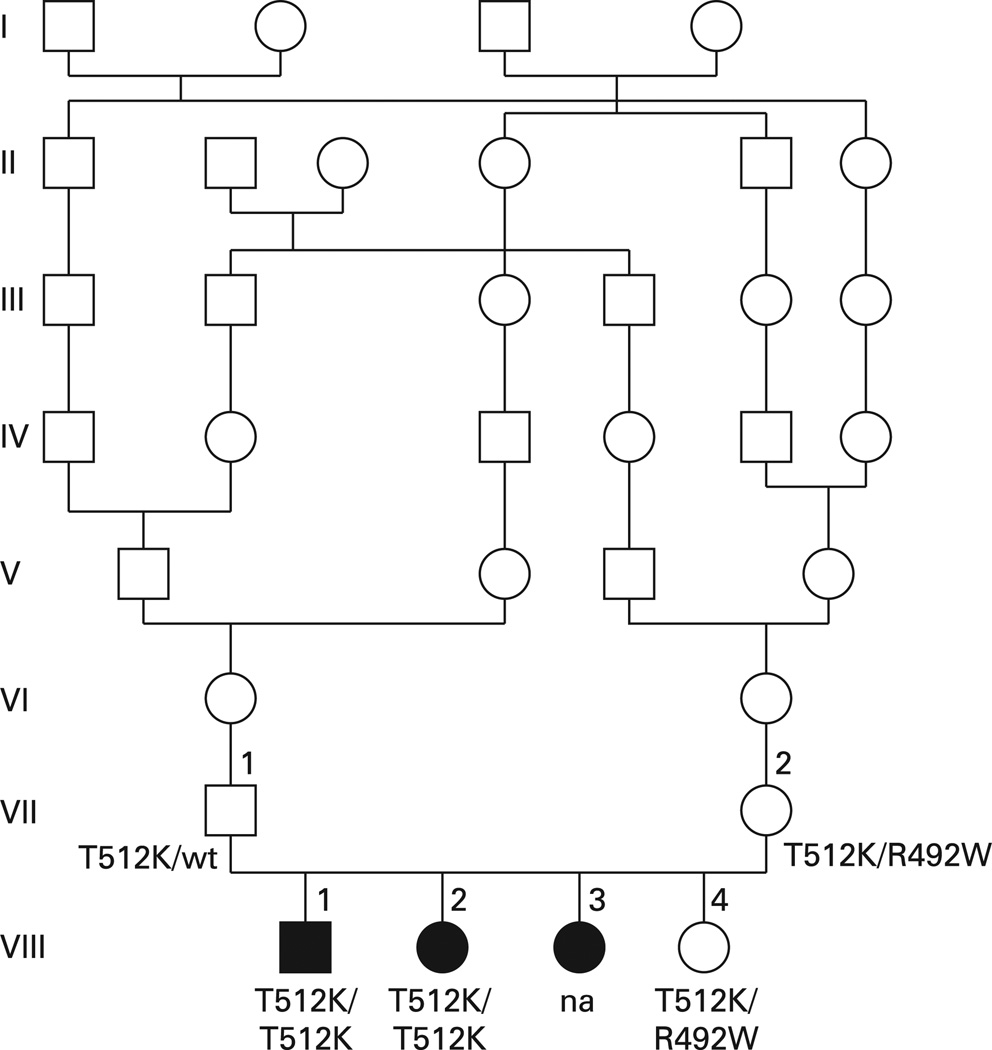
We studied the SLC26A2 gene in the DNA of the original family described by de la Chapelle et al in 1972 (figs 1 and 2). Genomic DNA was obtained from blood from the parents and the unaffected daughter, whereas DNA of two affected siblings was obtained from postmortem paraffin tissue blocks. No material was available from the third affected sibling, whose clinical, radiographic and pathological description is reported by Whitley et al.2 DNA was extracted from blood and paraffin blocks according to standard methods.
Figure 1.
Pedigree of the original family reported by de la Chapelle et al.1 There is pronounced consanguinity in the ancestry. Subject VIII3, whose DNA was not available (na) for this study, presented the same skeletal phenotype as subject VIII1 and VIII2.2
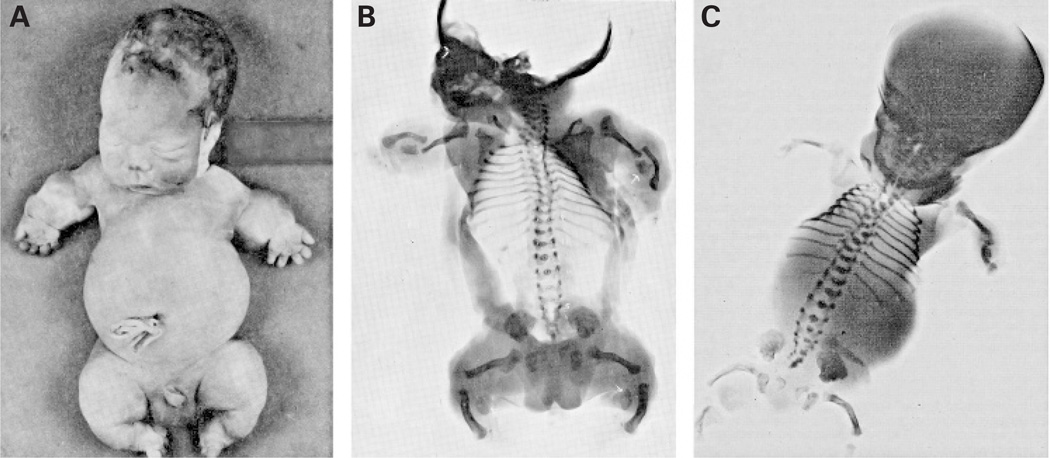
Figure 2.
Phenotypic features of de la Chapelle dysplasia. (A) Photograph of subject VIII1 (fig 1), a male baby born at term with weight 2750 g and length 40 cm, who died shortly after birth. There is severe micromelia, bilateral clubfoot, short fingers and toes, short trunk with protuberant abdomen, and micrognathia. (B) Radiograph of subject VIII1 (fig 1): long bones are shortened and bowed with metaphyseal flaring, with conserved major axis; the humerus is V shaped, and there is pronounced hypoplasia of the ulnae and fibulae which have a triangular shape; the spine is poorly ossified with progressive narrowing of the interpediculate distance in the lower lumbosacral region; the ilia are hypoplastic. (C) Radiograph of subject VIII2 (fig 1), a female baby, born near to term with weight 2450 g and length 37 cm: the radiographic findings are similar to those of subject VIII1, with generalised micromelia, very hypoplastic ulna and apparent absence of the fibula, small ilia and underossified spine. Reproduced from Chapelle et al,1 with permission of the publisher.
We also studied the SLC26A2 gene in seven Finnish patients affected by diastrophic dysplasia in whom only one heterozygous mutation had been found, in the parents of two of them, as well as in 200 unrelated Finnish and 150 unrelated non-Finnish Caucasian controls.
Sequencing of the SLC26A2 gene
The whole coding region of the SLC26A2 gene was amplified in 10 amplicons and analysed by bidirectional direct sequencing, using an ABI 3100-Avant automatic sequencer and the BigDye v1.1 kit (Applied Biosystems, Foster City, California, USA). An additional fragment in the non-coding exon 1, containing the IVS1+2T>C (“Finnish mutation”), was amplified and studied by restriction enzyme digestion and gel electrophoresis, with positive and negative controls. Primers (designed on GenBank sequence NM_000112), polymerase chain reaction (PCR) and restriction enzyme digestion conditions are available upon request.
Paraffin tissue DNA of the two siblings of family 1 was amplified by nested PCR with the following primers: F1 (5′-ATCAACAGGCTGCCATACTCA-3′), R1 (5′-AAACAAACCCCAACAAGTAG-3′), amplicon 270 bp; F2 (5′-CAGCTTTCTGGTGTGGTAACAG-3′), R2 (5′-TTCAGTACTTAGCAGTGCAG-3′), amplicon 225 bp. Amplification was carried out in both steps with an annealing temperature of 52°C and 35 cycles.
PCR cloning
PCR cloning was performed using the TOPO-TA cloning kit (version pCR II-TOPO Invitrogen). The DNAs of mother and daughter (in fig 1: unaffected females in generation VII and VIII, respectively), as well as of one control, were amplified using primers F1 and R1; ligation of the PCR product in the TOPO vector was performed by topoisomerase I. Chemically competent TOP10 E coli cells were transformed with the vector harbouring the PCR product from the three amplified DNAs (mother, daughter, control) and with positive and negative vector controls. Transformed bacteria were incubated overnight on LB-agar and selected by ampicillin resistance. Eighty separated colonies (35 transformed with the mother’s DNA, 35 with the daughter’s DNA, 10 with the control’s DNA) were picked and resuspended in 3 µl of sterile water; 1 µl was used as the template for the PCR test, in order to identify which colonies effectively contained the fragment. Fifty-seven (25 transformed with mother’s PCR product, 25 with daughter’s PCR product, and 7 with PCR product from a control) out of 80 colonies resulted to be transformed with the pCR II vector containing the PCR fragment and were cultured in LB-ampicillin medium. After overnight bacterial growth, plasmidic DNA was extracted from each colony using the NucleoSpin Plasmid DNA purification kit (Macherey-Nagel Inc, Bethlehem, Pennsylvania, USA).
Direct sequencing of the plasmidic DNA of the 57 colonies was performed using a vector specific primer (M13 Forward) on a ABI 3100-Avant (Applied Biosystems).
Expression plasmid construction
The full length human SLC26A2 coding sequence, obtained by retrotranscription of total RNA from control fibroblasts followed by PCR amplification with specific SLC26A2 primers, was cloned into the pcDNA3.1 vector (pCR 3.1, Invitrogen Corp, Carlsbad, California, USA).
The SLC26A2 sequence changes R492W and T512K were introduced with the Quick-Change II XL Site-Directed Mutagenesis kit according to the manufacturer’s instructions (Stratagene, La Jolla, California, USA). Oligonucleotides containing each of both missense mutations, designed for mutagenesis, are shown in table 1. Nucleotide and amino acid numbering is based on the sequence published by Hästbacka et al.8 Plasmid DNA purification was performed by using the EndoFree Plasmid Purification Maxi Kit according to the manufacturer’s protocol (Qiagen, Valencia, California, USA). PCR generated constructs were verified by sequence analysis.
Table 1.
Direct mutagenesis oligonucleotides
| Oligonucleotide | Target sequence 5′–3′* |
|---|---|
| R492W-F | gtgtgatcacaattgtaaatctatggggagcccttc |
| R492W-R | gaagggctccccatagatttacaattgtgatcacac |
| T512K-F | aaatgtggagtattagtagaatggataaagttatctggtttgttactatg |
| T512K-R | catagtaacaaaccagataactttatccattctactaatactccacattt |
Changing bases are in bold.
Cell culture and transfection
Sulfate transport deficient mutants of Chinese hamster ovary (CHO) cells previously described by Esko et al14 were obtained by the American Type Culture Collection (ATCC n. CRL-2245). Cells were maintained in Ham’s nutrient mixture F12 Kaighn’s modification (Ham’s F-12K) supplemented with 10% fetal bovine serum and antibiotics at 37°C and 5% CO2.
CRL-2245 cells were transfected with vector alone, with the plasmid containing the wild-type SLC26A2, the R492W or T512K mutants using Lipofectamine Reagent and Plus Reagent according to the manufacturer’s instructions (Invitrogen). Stable transfection was achieved maintaining transfected cells under selection with G418.
Sulfate uptake in transfected cells
To measure the rate of sulfate uptake, CRL-2245 cells stable transfected with vector alone, wild-type SLC26A2, mutated SLC26A2 for R492W or T512K were seeded in 10 cm2 petri dishes (2×105 cells/dish) and incubated in Ham’s F-12K supplemented with 10% fetal bovine serum and antibiotics for 48 h at 37°C in 5% CO2.
Sulfate uptake was performed as described previously9,15 using low ionic strength buffer (300 mM sucrose, 1 mM MgCl2, 10 mM Tris-Hepes, pH 7.5) containing concentrations of Na2SO4 ranging from 2–250 µM. Before the assay, cells were washed two times with pre-warmed sulfate-free low ionic strength buffer and pre-incubated for 2 min in the same buffer at 37°C. Cells were then incubated for 2 min at 37°C in 0.7 ml of low ionic strength buffer containing different concentrations of Na2SO4 and a constant concentration of carrier-free Na2[35S]O4 (0.1 µM, corresponding to 150 µCi/ml). The uptake medium was removed and the cells were washed four times with ice cold medium containing 100 mM sucrose, 100 mM NaNO3, 1 mM MgCl2, and 10 mM Tris-Hepes, pH 7.5. Finally, cells were lysed in 2% SDS. Lysates were collected, heat denatured, and centrifuged. Supernatants were assayed for radioactivity by scintillation spectroscopy and for protein content (BCA Protein Assay, Pierce Thermo Fisher Scientific, Rockford, Illinois, USA).
RESULTS
Sequencing of the SLC26A2 gene
Both parents and the unaffected daughter of family 1 were heterozygous for the c.1535C>A change (p.T512K) (fig 1); this mutation has not been described before. Both mother and daughter were heterozygous also for a second sequence change, c.1474C>T (p.R492W) (fig 1), previously found in other DTD patients and considered as a putative pathogenic mutation.13
Nested PCR allowed amplification of DNA extracted from paraffin tissue blocks of the two affected siblings originally reported by de la Chapelle et al.1 Direct sequencing of the amplified fragment showed homozygosity for T512K in both subjects, whereas R492W was not found (fig 1). These results were confirmed in three separate amplification products for each subject.
All seven Finnish individuals who had diastrophic dysplasia (DTD) who were heterozygous for IVS1+2T>C (the common Finnish mutation), but in whom a second mutation had not been previously identified, turned out to be compound heterozygous for T512K. Independent segregation of IVS1+2T>C and T512K was confirmed in two patients by analysis of parental DNA.
None of the 200 Finnish controls carried T512K, whereas seven of them were heterozygous for R492W.
PCR cloning
In order to ascertain the cis/trans (coupling/repulsion) phase of the two mutations (T512K and R492W) found at the heterozygote state in mother and daughter (fig 1), we performed PCR cloning of the amplification products of both individuals. The results showed that 18 colonies (nine from the mother’s DNA and nine from the daughter’s) out of 50 colonies harboured T512K, and the other 32 (16 from the mother’s DNA and 16 from the daughter’s) harboured R492W, thus confirming the trans phase of the mutations—that is, the two mutations are located on two different chromosomes in both the unaffected mother and the unaffected daughter. All colonies transformed with control DNA harboured the wt sequence of the amplified fragment.
Sulfate uptake activity of T512K and R492W mutants in CHO cells
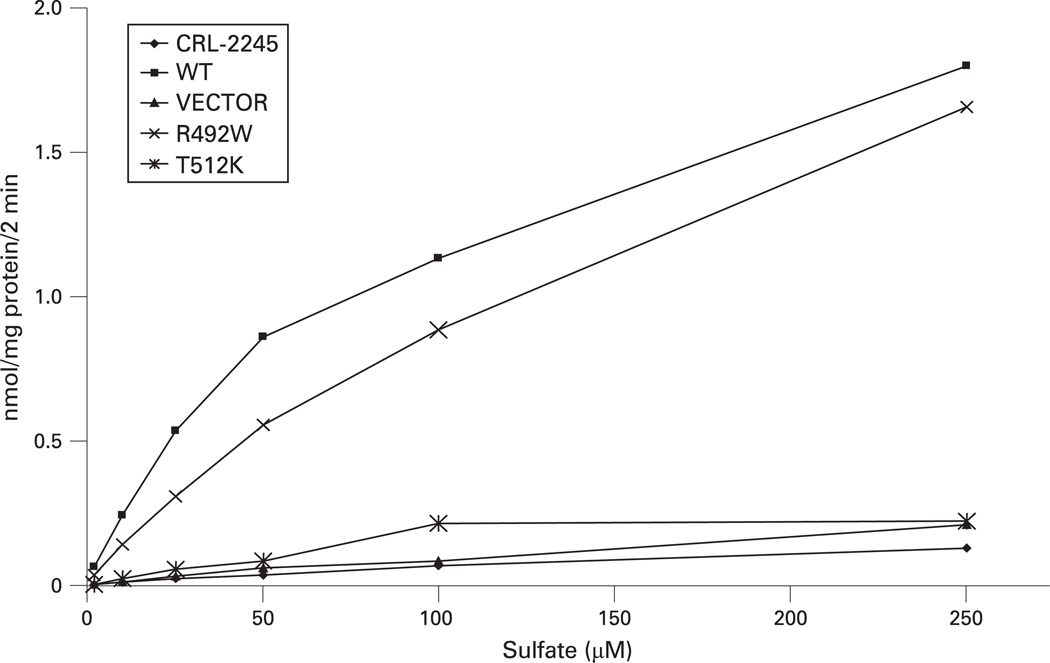
In order to confirm the pathogenic role of T512K and the polymorphic role of R492W, both mutants were expressed in sulfate transport deficient CHO cells. Sulfate uptake in cells transfected with R492W mutant and with wt SCL26A2 was very similar, thus confirming that R492W is not pathogenic. On the contrary, cells expressing T512K mutant showed no sulfate uptake, as seen also in the negative controls (cells transfected with the vector alone and non-transfected cells) (fig 3). These results confirm pathogenicity of T512K and are compatible with the severe phenotype of DLCD.
Figure 3.
Sulfate uptake in sulfate transport deficient Chinese hamster ovary (CHO) cells transfected with the two SLC26A2 mutants (R492W and T512K), with wt SLC26A2 (WT), with vector alone (VECTOR), and in non-transfected cells (CRL-2245). The activity of T512K sulfate transporter overlaps with that of non-transfected cells or transfected with the vector alone, whereas R492W shows normal sulfate uptake activity. These results confirm that T512K is pathogenic and R492W is a polymorphic variant.
DISCUSSION
The concept of “chondrodysplasia families” comprising distinct phenotypes with different degrees of severity and caused by mutations in a single gene was proposed by Spranger in 1988.16 The subsequent identification of rich mutational heterogeneity resulting in bone dysplasia families from the genes encoding for collagen II,17 FGFR3,18 COMP,19 and more recently filamin B20 validated this concept. In such bone dysplasia families the phenotypic variability of the single disorders slightly overlap with each other, leading to a continuum of clinical severity.
It is now clear that structural mutations which completely abolish the sulfate transporter activity on both alleles of the SLC26A2 (DTDST) gene cause the severe lethal phenotypes of ACG1B and AO2, whereas compound heterozygosity of null alleles with missense mutations or other types of mutations preserving a residual transporter activity cause milder phenotypes compatible with life (DTD).6 Mild mutations located out of the transmembrane domains and bearing a significant residual activity of the protein, either in homozygosity or in compound heterozygosity with a similar mild allele, indeed cause the very mild phenotype of rMED, which is associated with normal stature in a significant proportion of patients.21 Intermediate phenotypes exist between lethal AO2 and severe DTD, or between mild DTD and rMED, whose genotypes are more difficult to interpret.
Several features of DLCD overlap with AO2: shortened and curved limb bones with metaphyseal flaring, V shaped distal humerus, bowed radius and tibia, hypoplasia of one or more long bones, underossification of the spine with coronal clefts, hypoplastic ilia with flat acetabulum, cleft palate, micrognathia, and lethality in the perinatal period (fig 2). The shortening of the limb bones is less severe than in ACG1B, with a major axis still recognisable; the underossification of the ilia is also less severe than in ACG1B, in which only the upper part is usually ossified (paraglider shape); also, the spine is less ossified in ACG1B, with absent or only rudimentary calcification of the vertebral bodies. Other features of DLCD seem to overlap more with DTD, like the hitchhiker thumb, the increased distance between the first and the second toes, the equinovarus deformity of the feet, and the narrowing of the interpediculate distance of the lower lumbosacral spine. The pronounced hypoplasia of the ulna and fibula, which are reduced to triangular remnants (fig 2), was considered a specific feature of DLCD, distinguishing this condition from other lethal skeletal dysplasias. Later on, when more neonatal cases were classified as AO2, the discussion about whether this triangular shape of ulna and fibula was distinct from the limb bone hypoplasia seen in AO2 remained open among the experts. The radiological distinction of AO2 from DTD is not always evident and the criterion of perinatal lethality is used to classify patients with AO2; however, in our experience, AO2 rather than DTD must be considered when distinct hypoplasia of one or more long bones (humerus, ulna, radius, or fibula) is present. Based on this observation, the triangular ulna and fibula of DLCD is indeed a criterion of identity with AO2.
In the original description of DLCD,1 the first affected sibling (a boy) had in addition endocrine and haematological abnormalities, which were not present in the second affected sibling (a girl). In 1986 Whitley et al reported a third affected child (a girl) of the original family, whose pathology was described in detail and who also did not have the endocrine and haematological abnormalities of her older brother.2 We speculate that these peculiar features of the first sibling were probably not related to the skeletal dysplasia which appears to be the same in all three affected siblings.
The histopathological features of cartilage and bone in DLCD, particularly the lacunar halos observed around chondrocytes, are very similar to the typical findings in ACG1B; this observation induced Whitley et al2 to speculate that these two conditions may have the same molecular basis, which is indeed the case. This shows the importance of the histopathological data in both the interpretation of slight clinical and radiological differences between “novel, distinct” and known conditions and in directing the search for their genetic causes.
T512K is a missense mutation located in the sixth cytoplasmic domain of the sulfate transporter protein (Swiss-Prot protein knowledgebase http://www.expasy.org/sprot/, accession number P50443). The effect of this mutation in homozygosity is the severe DLCD phenotype, whereas the effect in compound heterozygosity with the IVS1+2T>C mutation is non-lethal diastrophic dysplasia. Thus, the affected residue appears to have an important structural or functional role, even if it is not located in a transmembrane domain. The observation that T512 is conserved in mammals supports this notion (supplemental fig 4) and the lack of residual activity of this mutant at sulfate uptake assay (fig 3) demonstrates its pathogenicity.
Our experience with more than 150 patients with sulfate transporter related dysplasias shows that T512K has been found neither in non-Finnish patients nor in 150 non-Finnish controls. Our analysis on 200 Finnish controls did not uncover T512K, thus indicating that T512K is not a common allele in this population. It appears therefore to be a rare pathogenic allele, restricted to the Finnish population. R492W, on the contrary, was found in seven out of 200 Finnish and five out of 150 non-Finnish controls in the heterozygous state. This relatively high frequency and the fact that in compound heterozygosity with a known pathogenic allele does not cause any phenotype clearly indicate that R492W is a polymorphic variant, despite its conservation through mammals (supplemental fig 4). The normal sulfate uptake activity in CRL-2245 cells transfected with the R492W mutant confirms that it is a polymorphic variant. Because of the change from arginine to tryptophan, which is a major amino acid substitution, and because it was believed that R492 was located in a transmembrane domain of the protein, R492W has been previously considered to be a pathogenic mutation.13 Indeed, the transmembrane structure predicted by Hästbacka et al8 included a transmembrane domain between amino acids 474 and 494, which would include R492; a more recent modelling of the sulfate transporter (Swiss-Prot accession number P50443) predicts a cytoplasmic loop between amino acids 476 and 523, thus reducing the structural importance of this region. In our past experience, three patients with DTD were diagnosed as compound heterozygotes for R492W and another known pathogenic mutation. We reviewed the sequences of these three patients and found that all three had indeed another rare mutation in cis phase with R492W (unpublished data), which is most probably the second pathogenic allele responsible for the diastrophic phenotype. Thus, in summary, the study of the original DLCD family revealed a second, rarer “Finnish” mutation in SLC26A2, confirmed that DLCD is part of the SLC26A2 dysplasia continuum, and unmasked a putative pathogenic mutation as a neutral polymorphism.
Supplementary Material
Key points.
-
►
De la Chapelle dysplasia is caused by a mutation in SLC26A2 and is allelic to the other sulfate transporter related dysplasias.
-
►
T512K is a novel, non-functional, SLC26A2 mutation specific to the Finnish population; it causes a severe bone dysplasia overlapping with atelosteogenesis type 2 when homozygous and diastrophic dysplasia when in compound heterozygosity with the milder common Finnish mutation.
-
►
R492W, previously considered a pathogenic SLC26A2 mutation, is a polymorphic variant with normal sulfate uptake activity.
Acknowledgements
We are grateful to the family who participated in the study. We thank Valerie Pittet for technical assistance.
Funding: This work has been supported by the Swiss National Research Foundation (grant 3100A0-100485 to LB and ASF), by the Telethon-Italy (grant GGP06076 to AR), by the European Community (FP6, “EuroGrow” project, LSHM-CT-2007-037471 to AR), and by the National Institutes of Health, USA (grant CA16058) to the Ohio State University Comprehensive Cancer Center. JH was supported by the Academy of Finland, The Sigrid Juselius Foundation and the Ulla Hjelt fund of the Finnish Pediatric Research Foundation.
Footnotes
Competing interests: None declared.
Ethics approval: Ethics committee approval has been obtained (from the University of Lausanne).
Patient consent: Parental/guardian consent obtained.
REFERENCES
- 1.De la Chapelle A, Maroteaux P, Havu N, Granroth G. A rare lethal bone dysplasia with recessive autosomic transmission. Arch Fr Pediatr. 1972;29:759–770. [PubMed] [Google Scholar]
- 2.Whitley CB, Burke BA, Granroth G, Gorlin RJ. de la Chapelle dysplasia. Am J Med Genet. 1986;25:29–39. doi: 10.1002/ajmg.1320250105. [DOI] [PubMed] [Google Scholar]
- 3.Sillence DO, Kozlowski K, Rogers JG, Sprague PL, Cullity GJ, Osborn RA. Atelosteogenesis: evidence for heterogeneity. Pediatr Radiol. 1987;17:112–118. doi: 10.1007/BF02388086. [DOI] [PubMed] [Google Scholar]
- 4.Schrander-Stumpel C, Havenith M, Linden EV, Maertzdorf W, Offermans J, van der Harten J. De la Chapelle dysplasia (atelosteogenesis type II): case report and review of the literature [corrected] Clin Dysmorphol. 1994;3:318–327. [PubMed] [Google Scholar]
- 5.Rossi A, van der Harten HJ, Beemer FA, Kleijer WJ, Gitzelmann R, Steinmann B, Superti-Furga A. Phenotypic and genotypic overlap between atelosteogenesis type 2 and diastrophic dysplasia. Hum Genet. 1996;98:657–661. doi: 10.1007/s004390050279. [DOI] [PubMed] [Google Scholar]
- 6.Superti-Furga A, Rossi A, Steinmann B, Gitzelmann R. A chondrodysplasia family produced by mutations in the diastrophic dysplasia sulfate transporter gene: genotype/phenotype correlations. Am J Med Genet. 1996;63:144–147. doi: 10.1002/(SICI)1096-8628(19960503)63:1<144::AID-AJMG25>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 7.Superti-Furga A. A defect in the metabolic activation of sulfate in a patient with achondrogenesis type IB. Am J Hum Genet. 1994;55:1137–1145. [PMC free article] [PubMed] [Google Scholar]
- 8.Hästbacka J, de la Chapelle A, Mahtani MM, Clines G, Reeve-Daly MP, Daly M, Hamilton BA, Kusumi K, Trivedi B, Weaver A, Coloma A, Lovett M, Buckler A, Kaitila I, Lander ES. The diastrophic dysplasia gene encodes a novel sulfate transporter: positional cloning by fine-structure linkage disequilibrium mapping. Cell. 1994;78:1073–1087. doi: 10.1016/0092-8674(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 9.Rossi A, Bonaventure J, Delezoide AL, Cetta G, Superti-Furga A. Undersulfation of proteoglycans synthesized by chondrocytes from a patient with achondrogenesis type 1B homozygous for an L483P substitution in the diastrophic dysplasia sulfate transporter. J Biol Chem. 1996;271:18456–18464. doi: 10.1074/jbc.271.31.18456. [DOI] [PubMed] [Google Scholar]
- 10.Rossi A, Bonaventure J, Delezoide AL, Superti-Furga A, Cetta G. Undersulfation of cartilage proteoglycans ex vivo and increased contribution of amino acid sulfur to sulfation in vitro in McAlister dysplasia/atelosteogenesis type 2. Eur J Biochem. 1997;248:741–747. doi: 10.1111/j.1432-1033.1997.t01-1-00741.x. [DOI] [PubMed] [Google Scholar]
- 11.Rossi A, Kaitila I, Wilcox WR, Rimoin DL, Steinmann B, Cetta G, Superti-Furga A. Proteoglycan sulfation in cartilage and cell cultures from patients with sulfate transporter chondrodysplasias: relationship to clinical severity and indications on the role of intracellular sulfate production. Matrix Biol. 1998;17:361–369. doi: 10.1016/s0945-053x(98)90088-9. [DOI] [PubMed] [Google Scholar]
- 12.Karniski LP. Mutations in the diastrophic dysplasia sulfate transporter (DTDST) gene: correlation between sulfate transport activity and chondrodysplasia phenotype. Hum Mol Genet. 2001;10:1485–1490. doi: 10.1093/hmg/10.14.1485. [DOI] [PubMed] [Google Scholar]
- 13.Rossi A, Superti-Furga A. Mutations in the diastrophic dysplasia sulfate transporter (DTDST) gene (SLC26A2): 22 novel mutations, mutation review, associated skeletal phenotypes, and diagnostic relevance. Hum Mutat. 2001;17:159–171. doi: 10.1002/humu.1. [DOI] [PubMed] [Google Scholar]
- 14.Esko JD, Elgavish A, Prasthofer T, Taylor WH, Weinke JL. Sulfate transport-deficient mutants of Chinese hamster ovary cells. Sulfation of glycosaminoglycans dependent on cysteine. J Biol Chem. 1986;261:15725–15733. [PubMed] [Google Scholar]
- 15.Forlino A, Piazza R, Tiveron C, Della Torre S, Tatangelo L, Bonafe L, Gualeni B, Romano A, Pecora F, Superti-Furga A, Cetta G, Rossi A. A diastrophic dysplasia sulfate transporter (SLC26A2) mutant mouse: morphological and biochemical characterization of the resulting chondrodysplasia phenotype. Hum Mol Genet. 2005;14:859–871. doi: 10.1093/hmg/ddi079. [DOI] [PubMed] [Google Scholar]
- 16.Spranger J. Bone dysplasia ‘families’. Pathol Immunopathol Res. 1988;7(1–2):76–80. doi: 10.1159/000157098. [DOI] [PubMed] [Google Scholar]
- 17.Nishimura G, Haga N, Kitoh H, Tanaka Y, Sonoda T, Kitamura M, Shirahama S, Itoh T, Nakashima E, Ohashi H, Ikegawa S. The phenotypic spectrum of COL2A1 mutations. Hum Mutat. 2005;26:36–43. doi: 10.1002/humu.20179. [DOI] [PubMed] [Google Scholar]
- 18.Passos-Bueno MR, Wilcox WR, Jabs EW, Sertie AL, Alonso LG, Kitoh H. Clinical spectrum of fibroblast growth factor receptor mutations. Hum Mutat. 1999;14:115–125. doi: 10.1002/(SICI)1098-1004(1999)14:2<115::AID-HUMU3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy J, Jackson GC, Barker FS, Nundlall S, Bella J, Wright MJ, Mortier GR, Neas K, Thompson E, Elles R, Briggs MD. Novel and recurrent mutations in the C-terminal domain of COMP cluster in two distinct regions and result in a spectrum of phenotypes within the pseudoachondroplasia – multiple epiphyseal dysplasia disease group. Hum Mutat. 2005;25:593–594. doi: 10.1002/humu.9342. [DOI] [PubMed] [Google Scholar]
- 20.Krakow D, Robertson SP, King LM, Morgan T, Sebald ET, Bertolotto C, Wachsmann-Hogiu S, Acuna D, Shapiro SS, Takafuta T, Aftimos S, Kim CA, Firth H, Steiner CE, Cormier-Daire V, Superti-Furga A, Bonafe L, Graham JM, Jr, Grix A, Bacino CA, Allanson J, Bialer MG, Lachman RS, Rimoin DL, Cohn DH. Mutations in the gene encoding filamin B disrupt vertebral segmentation, joint formation and skeletogenesis. Nat Genet. 2004;36:405–410. doi: 10.1038/ng1319. [DOI] [PubMed] [Google Scholar]
- 21.Ballhausen D, Bonafe L, Terhal P, Unger SL, Bellus G, Classen M, Hamel BC, Spranger J, Zabel B, Cohn DH, Cole WG, Hecht JT, Superti-Furga A. Recessive multiple epiphyseal dysplasia (rMED): phenotype delineation in eighteen homozygotes for DTDST mutation R279W. J Med Genet. 2003;40:65–71. doi: 10.1136/jmg.40.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.