Abstract
The lysosomal storage disorders encompass nearly fifty diseases provoked by lack or deficiency of enzymes essential for the breakdown of complex molecules and hallmarked by accumulation in the lysosomes of metabolic residues. Histochemistry and cytochemistry studies evidenced patterns of circadian variation of the lysosomal marker enzymes, suggesting that lysosomal function oscillates rhythmically during the 24-h day. The circadian rhythmicity of cellular processes is driven by the biological clock ticking through transcriptional/translational feedback loops hardwired by circadian genes and proteins. Malfunction of the molecular clockwork may provoke severe deregulation of downstream gene expression regulating a complex array of cellular functions leading to anatomical and functional changes. In this review we highlight that all the genes mutated in lysosomal storage disorders encode circadian transcripts suggesting a direct participation of the biological clock in the pathophysiological mechanisms underlying cellular and tissue derangements hallmarking these hereditary diseases. The 24-h periodicity of oscillation of gene transcription and translation could lead in physiological conditions to circadian rhythmicity of fluctuation of enzyme levels and activity, so that gene transfer could be envisaged to reproduce 24-h periodicity of variation of enzymatic dynamics and circadian rhythmicity could have an impact on the schedule of enzyme replacement therapy.
Electronic supplementary material
The online version of this chapter (doi:10.1007/8904_2014_354) contains supplementary material, which is available to authorized users.
Introduction
The lysosomal storage disorders (LSDs) comprise about fifty diseases that are caused by the absence or insufficiency of particular enzymes necessary for the breakdown of complex molecules and are hallmarked by amassing in the lysosomes of metabolic residues (Futerman and van Meer 2004; Ballabio and Gieselmann 2009). Lysosomes are cytoplasmic organelles found in nearly every eukaryotic cell, which take part crucially in fundamental aspects of cellular homeostasis such as membrane repair, autophagy, endocytosis, and protein metabolism. LSDs are inherited prevalently in an autosomal recessive manner and only in a minor part as X-linked recessive disorders; even though each of them affects a modest fraction of the population, as a group LSDs represent an important challenge for the health-care systems (Meikle et al. 1999). The genetic modifications responsible of the LSDs alter the correct working of the lysosomes through the shortage of enzymes catalyzing hydrolysis of molecular substrates, hydrolase activators, or transporters or hinder the vesicular transport in the endosomal/lysosomal system causing lysosomal storage of substrates specific for each disorder type (Journet et al. 2002; Platt and Walkley 2004; Bagshaw et al. 2005; Lübke et al. 2009). Based on these premises, the LSDs are classified in defects in glycan degradation, defects in lipid degradation, defects in protein degradation, defects in lysosomal transporters, and defects in lysosomal trafficking. The lysosomal accumulation leads to formation of large intracellular vacuoles and perturbs the proper functioning of the cell, causing functional and anatomic derangements at the tissue level in numerous organ systems that are responsible of LSDs’ clinical manifestations (Bellettato and Scarpa 2010). The biological processes underlying cell, tissue, and organ system function show fluctuations that may be rhythmic, and when the periodicity corresponds to approximately 24 h, the rhythm is defined circadian (from the Latin words circa, about, and diem, a day) (Mazzoccoli et al. 2011a). Subcellular organelles, such as endoplasmic reticulum structures, show nycthemeral fluctuations in the relative amounts and regional differences in their distribution (Chedid and Nair 1972). Accordingly, histochemistry and cytochemistry studies evidenced patterns of circadian variation of the lysosomal marker enzymes, suggesting that lysosomal function oscillates rhythmically during the 24-h day (Bhattacharya and von Mayersbach 1976; Uchiyama et al. 1981; Uchiyama and von Mayersbach 1981).
In this review article we meant to suggest a possible involvement of the biological clock in the physiopathology of lysosomal storage diseases. We have reviewed the scientific literature on this issue and reported information from comparison of publicly available databases to corroborate our proposal.
The Circadian Clock Circuitry
Circadian rhythmicity characterizes physiological phenomena of all living beings and is driven in mammals by the circadian timing system, comprising central and peripheral oscillators (Hastings et al. 2003; Dibner et al. 2010; Mazzoccoli et al. 2011b, 2012a, b). The central oscillators are located in the hypothalamus and are represented by the neurons of the suprachiasmatic nuclei, which are influenced by photic stimuli perceived by the retinal ganglion cells and conveyed by the retino-hypothalamic tracts (Kalsbeek et al. 2006; Challet 2007). The alternation of light and darkness related to Earth’s rotation around its axis entrains the ticking of the central oscillators in the SCN, which in turn drives autonomous and self-sustained oscillators in the peripheral tissues through neural pathways (autonomic nervous system fibers) and humoral mediators (cortisol, melatonin) (Mazzoccoli 2011; Cailotto et al. 2009). At the cellular level the oscillation of biological functions is driven by molecular clockworks ticking through transcriptional-translational feedback loops operated by a set of genes called clock genes, which encode transcripts oscillating with circadian rhythmicity (Ko and Takahashi 2006; Lowrey and Takahashi 2011). The positive limb of the loop is started by the transcription factors Clock (or its paralog Npas2) and Arntl, also called Bmal1 (or its homologue Arntl2/Bmal2), which heterodimerize and activate the transcription of the genes Period (Per 1–3) and Cryptochrome (Cry1-2) (Nagoshi et al. 2004). The circadian proteins Per 1–3 and Cry 1–2 heterodimerize, pass back into the nucleus, and hinder the transcriptional activity of Clock/Arntl heterodimer, closing the loop in approximately 24 h (Duguay and Cermakian 2009). The PER and CRY proteins are posttranslationally modified by different processes, represented by phosphorylation, sumoylation, ubiquitylation, acetylation, and deacetylation, which influence their activity and degradation (Eide et al. 2002; Cardone et al. 2005). Phosphorylation is operated by casein kinase (CK)Iδ and CKIε and glycogen synthase kinase (GSK) 3-β, which target circadian proteins for degradation and regulate their nuclear translocation (Agostino et al. 2009; Sahar et al. 2010). A particular role is played by AMP-dependent kinase (AMPK) whose activity is influenced by ATP/AMP ratio and works also as a nutrient sensor (Fulco and Sartorelli 2008; Cantó and Auwerx 2009; Lamia et al. 2009). Acetylation is operated by Clock that has histone-acetyltransferase activity (Doi et al. 2006), whereas deacetylation is operated by SIRT1, an NAD+-dependent protein deacetylase necessary for high-magnitude circadian transcription of several core clock genes (Asher et al. 2008). The activity of SIRT1 depends on the availability of its cofactor, NAD+, whose synthesis is regulated by NAMPT, encoded by a clock-controlled gene that steers the salvage pathway responsible for circadian oscillation of NAD+ levels and ultimately of SIRT1 activity gauging the energy status of the cell and driving mitochondrial oxidative metabolism (Rutter et al. 2001; Ramsey et al. 2009; Nakahata et al. 2008, 2009; Peek et al. 2013). Besides, Clock/Arntl heterodimer activates the expression of the nuclear receptors Rev-erbα (encoded by NR1D1 gene) and Rorα (encoded by Rora gene), which drive in that order negatively and positively the rhythmic transcription of Bmal1, through competition for binding to a response element on the promoter of the Bmal1 gene (Preitner et al. 2002; Burris 2008; Bugge et al. 2012; Mazzoccoli et al. 2012c; Cho et al. 2012; Jetten et al. 2013). A gene necessary for circadian clock function in Drosophila melanogaster is Timeless (Tim), which is maintained in mammals and interplaying with its partner. TIMELESS-interacting protein (TIPIN) plays a role to regulate DNA replication processes under both normal and stress conditions. TIMELESS and TIPIN are important for ataxia telangiectasia and Rad3-related (ATR)-checkpoint kinase (Chk)1 and ataxia telangiectasia mutated (ATM)-checkpoint kinase (Chk)2-mediated signaling and S-phase arrest (Unsal-Kaçmaz et al. 2007; Smith et al. 2009; Yang et al. 2010; Kemp et al. 2010) (Fig. 1).
Fig. 1.
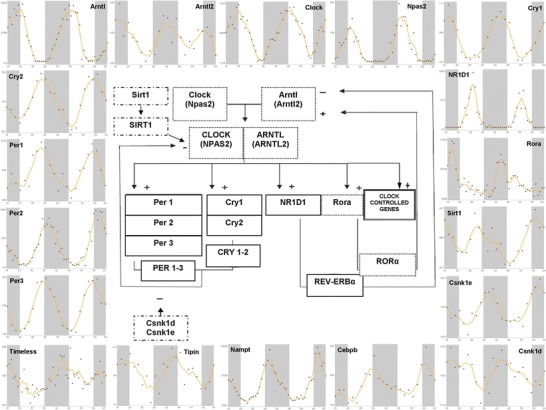
x–y plots showing the time-qualified profile of expression of core clock genes and clock-controlled genes (source: CircaDB, web addresses http://circadb.org, http://github.com/itmat/circadb, http://bioinf.itmat.upenn.edu/circa) and a scheme rendering the interactions among the cogwheels of the molecular clockwork. Continuous line boxes indicate transcriptional repressors such as PER, CRY, and REV-ERBα; dotted line boxes indicate transcriptional activators such as CLOCK, BMAL1, and RORα; dashed-dotted line boxes indicate posttranslational modifications catalyzed by SIRT1 and casein kinases such as CSNK1D and CSNK1E. On the x axis is represented time in hours, on the y axis are represented the mRNA relative expression levels in arbitrary units
The Biological Clock in Physiology and Pathology
The molecular clockwork drives the expression of thousands of genes, so that gene expression profiling studies performed with different algorithms and sampling frequency evidenced that approximately 5–20% of the transcriptome displays circadian rhythmicity (Asher and Schibler 2011). These genes are defined clock-controlled genes (CCGs) and steer cell processes, such as proliferation, differentiation, cell cycle, apoptosis, autophagy, and DNA damage response (Bozek et al. 2009). Tissue-specific output genes driven by the biological clock are responsible for various metabolic and homeostatic processes and distinctively control organ functions, synchronizing them to circadian environmental cues and making them available when mainly required at definite times during the 24-h day (Mazzoccoli et al. 2012d). In this way, the circadian outlines of metabolic gene expression may properly fit the swap between catabolic and anabolic phases corresponding with cycles of sleep/rest/fasting and wake/activity/feeding, respectively (Bass and Takahashi 2010; Bass 2012). This process is regulated by nuclear receptors expressed with 24-h periodicity in metabolically active tissues (liver, white and brown adipose tissues) (Yang et al; 2006), which recruit cofactors, coactivators, and corepressors and together with other processes, such as rhythmic histone methylation/demethylation, drive circadian waves of chromatin remodeling and epigenetic modifications impinging on transcriptional events and connect the molecular clockwork to metabolism integrating energy flux with varying physiological requirements during daytime and nighttime (Alenghat et al. 2008; Asher et al. 2010; Yin et al. 2010; Grimaldi et al. 2010; Berrabah et al. 2011; Di Tacchio et al. 2011; Dufour et al. 2011; Feng et al. 2011; Valekunja et al. 2013). The alteration of proper synchronization among physiological, behavioral, transcriptional, translational, and posttranslational modification rhythms underlies the pathological basis of metabolic, inflammatory, degenerative, and neoplastic diseases (Takahashi et al. 2008; Mazzoccoli et al. 2010; Maury et al. 2010; Anderson et al. 2013; Bonny et al. 2013; Vinciguerra et al., 2013; Tevy et al. 2013; De Cata et al. 2014; Mazzoccoli et al. 2014).
The Biological Clock and the LSDs: Toward and Beyond Lysosomes
Several experimental efforts using different methodologies have been carried out to identity lysosomal genes, and the categorization of the genes encoding structural, transport, and enzymatic proteins included in the lysosome allows a better understanding of the biology of this subcellular organelle. The Human Lysosome Gene Database (hLGDB) makes available a complete and reachable census of the 435 human genes encompassed in the lysosomal system (Brozzi et al. 2013) (Supplementary Table 1). We matched the genes stored in this database with those obtained by time-qualified genome-scale RNA profiling performed to identify and compare circadian transcripts from mouse liver (Hughes et al. 2009). A set of 2,892 sequence symbols (protein-coding genes, retained introns, pseudogenes, lincRNA, etc.) was refined through conversion of the symbols by using the Mouse Genome Informatics (v 5.17) resource (http://www.informatics.jax.org/batch) and obtaining current and old symbols, Ensembl Gene IDs, corresponding gene names, and feature type information. Among the 435 lysosomal genes listed in the hLGDB human database, 365 genes resulted expressed with circadian rhythmicity (http://circadb.org), and 70 did not show any 24-h periodicity (Fig. 2). On these premises, approximately 80% of the lysosomal genes turn out to oscillate with circadian rhythmicity and arguably are adequate to drive the nycthemeral changes of enzymatic activity and functional processes in the lysosome. Besides, a process tightly linked to lysosomal function is represented by autophagy (Schultz et al. 2011), which shows fluctuations with a pattern of circadian rhythmicity driven directly by the molecular clockwork through the transcription factor CEPB/β, whose expression is controlled by transcriptional activity of Bmal1 through binding at E-box sequences in its promoter, and in turn CEPB/β steers the circadian expression of genes encoding autophagy-related proteins (Ma et al. 2011; Ma and Lin 2012). Another mechanism involved in the pathogenesis of LSDs is represented by endoplasmic reticulum (ER) stress and unfolded protein response (UPR), an adaptive reaction universally preserved to handle the accumulation of unfolded proteins in this subcellular compartment, which eventually leads to apoptosis to preserve the organism in the case it is not sufficient. The amassing of unfolded proteins in the ER turns on IRE1a, PERK, and ATF6 pathways, causing the nuclear translocation of the transcription factors XBP1, ATF4, and ATF6, respectively, which induce the expression of genes encoding proteins involved in peptide folding and degradation to minimize the accumulation of unfolded proteins. UPR shows rhythmic oscillations with ultradian periodicity of approximately 12 h, and UPR-regulated genes are hallmarked by a rhythmic expression according to an ensuing 12-h period dependent on a functional circadian clock. This rhythmic activation of UPR-regulated genes seems to be the consequence of the activation of the IRE1a-XBP1 pathway, which is activated according to the same 12-h period rhythm (Cretenet et al. 2010). Secretory and transmembrane proteins fold in the ER into their native conformations and undergo posttranslational modifications, but alteration of these processes causes accumulation of misfolded proteins in the ER lumen and triggers the UPR. According to studies demonstrating UPR activation in fibroblasts from a large range of LSDs, this mechanism was recently advocated as a frequent mediator of apoptosis in LSDs. Accumulation of unfolded proteins can take place in response to changes in the ER environment, including nutrient starvation and reducing agents. UPR is also related to exhaustion of ER calcium stores and in LSDs has been found alteration of calcium homeostasis, suggesting that this pathway could be engaged in the pathological mechanisms set in motion in the diseases linked to lysosomal accumulation of unmetabolized substrates as well (Vitner et al. 2010). Besides, even if lysosome engulfment is the central derangement in LSDs, defective activity of lysosomal proteins prompts a number of pathogenic cascades, among which a leader role is played by improper activation of inflammation and immune response that may perpetuate inducing a chronic reaction (Vitner et al. 2010). These processes are linked to transcriptional circuits tightly controlled and temporally driven by the biological clock, so that inflammatory signaling pathways and immune-mediated responses are characterized by circadian rhythmicity of activity, rendered by nycthemeral variations of levels of humoral factors and cellular effectors, as well as phagocytic, complement, lysozyme, and peroxidase activity in innate immunity, and antibody and cytokine production, leukocyte trafficking, proliferation, and apoptosis in adaptive immunity (Cermakian et al. 2013; Vinciguerra et al. 2013, 2014).
Fig. 2.
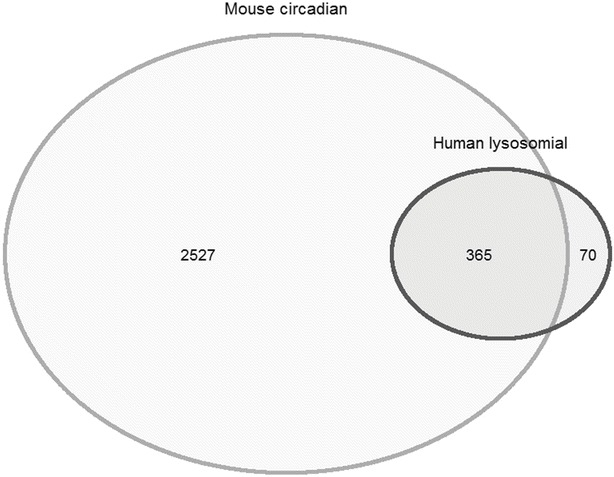
Comparison between 435 human lysosomal genes against 2,892 circadian protein-coding genes of Mus musculus. The mouse genes derive from refinement of a set of sequence symbols (protein-coding genes, retained introns, pseudogenes, lincRNA, etc., doi:10.1371/journal.pgen.1000442.s012). Refinement was done by converting the symbols by using the Mouse Genome Informatics (v 5.17) resource (http://www.informatics.jax.org/batch) and obtaining current and old symbols, Ensembl Gene IDs, corresponding gene names and feature type information
The Molecular Clockwork and Circadian Rhythmicity in LSDs
The hereditary diseases related to anomalous storage of molecules and intermediary metabolites in the lysosome are characterized by alteration of anatomical integrity and physiological function of many tissues and organ systems in the body underlying the clinical manifestations hallmarking the LSDs. Considering that the biological clock drives processes that are crucial for the maintenance of body homeostasis, it is tempting to speculate on the involvement of the circadian clock circuitry in the pathophysiological mechanism underlying these diseases. In line with this hypothesis, a severe deregulation of expression of clock genes and clock-controlled genes has been evidenced in a study that evaluated by whole transcriptome analysis through next-generation sequencing the expression of circadian genes in normal primary human fibroblasts and compared it to the circadian transcriptome of fibroblasts obtained from Hunter syndrome patients before and 24 h/144 h after iduronate-2-sulfatase treatment in vitro and evaluated also by qRT-PCR the time-related expression of core clock genes before and after 24 h of iduronate-2-sulfatase treatment upon synchronization by serum shock (Mazzoccoli et al. 2013). The expression of several core clock genes and clock-controlled genes was distorted and showed dynamic modifications 24 and 144 h after iduronate-2-sulfatase treatment. Besides, a semantic hypergraph-based analysis highlighted five gene clusters significantly associated to important biological processes or pathways and five genes, AHR, HIF1A, CRY1, ITGA5, and EIF2B3, proven to be central players in these pathways. The results imply a decisive contribution of deregulation of the clock gene machinery in the pathophysiological mechanisms underlying the derangement of cellular processes as well as the alteration of tissue function and anatomical integrity that hallmark the organ systems involved in the patients affected by Hunter syndrome (Mazzoccoli et al. 2013). More importantly, only circadian genes when mutated appear to be responsible of the altered lysosomal function and resulting anatomical and functional derangements that hallmark the LSDs. The enzyme iduronate-2-sulfatase is encoded by the IDS gene, whose expression oscillates with circadian rhythmicity, and this pattern of time-related variation features also the expression of the genes encoding the enzymes whose deficiency is responsible of the other LSDs known at present (source CircaDB, a data set of time course expression experiments from mice and humans deposited as publicly available microarray studies and highlighting circadian gene expression cycles, web addresses http://github.com/itmat/circadb, http://bioinf.itmat.upenn.edu/circa) (Pizarro et al. 2013). As listed in Table 1 and shown in Figs. 3 and 4, all the genes encoding lysosomal enzymes implicated in LSDs are customarily expressed with circadian rhythmicity, with the exception of GNPTG, encoding N-acetylglucosamine-1-phosphotransferase γ-subunit, whose mutation causes mucolipidosis III gamma (I-cell) and that is expressed rhythmically with a periodicity of 39 h. Accordingly, among the others the deregulation of circadian genes underlies the physiopathology of Niemann-Pick types A and B disease, caused by mutation of SGMS2 gene encoding sphingomyelin synthase 2, necessary for the transfer of phosphocholine from phosphatidylcholine onto ceramide to produce sphingomyelin, a major component of cell and Golgi membranes, as well as Niemann-Pick type C disease, caused by mutation of NPC1 gene, encoding Niemann-Pick C1, necessary for the intracellular trafficking of cholesterol from the late endosome to the trans-Golgi network (Panda et al. 2002; Hughes et al. 2009).
Table 1.
Circadian genes involved in lysosomal storage disorders
| Protein defect | Gene | Disease | OMIM | Chromosomal localization |
|---|---|---|---|---|
| Defects in glycan degradation | ||||
| Defects in glycoprotein degradation | ||||
| α-Sialidase | NEU1 | Sialidosis | 608272 | 6p21.33 |
| Cathepsin A | CTSA | Galactosialidosis | 256540 | 20q13.12 |
| α-Mannosidase | MAN2B | α-Mannosidosis | 248500 | 19p13.2 |
| β-Mannosidase | MANBA | β-Mannosidosis | 248510 | 4q24 |
| Glycosylasparaginase | AGA | Aspartylglucosaminuria | 208400 | 4q34.3 |
| α-Fucosidase | FUCA1 | Fucosidosis | 230000 | 1p36.11 |
| N-Acetyl-α-glucosaminidase | NAGA | Schindler disease, type III | 609421 | 22q13.2 |
| Defects in glycolipid degradation | ||||
| β-Galactosidase | GLB1 | GM1-gangliosidosis / MPS IVB | 230500 | 3p22.33 |
| β-Hexosaminidase α-subunit | HEXA | GM2-gangliosidosis (Tay-Sachs disease) | 606869 | 15q23 |
| β-Hexosaminidase β-subunit | HEXB | GM2-gangliosidosis (Sandhoff disease) | 606873 | 5q13.3 |
| GM2 activator protein | GM2A | GM2 gangliosidosis | 272750 | 5q33.1 |
| Glucocerebrosidase | GBA/GBA2 | Gaucher disease | 606463 | 1q22 |
| Saposins A, B, C, D | PSAP | Combined sphingolipid activator protein deficiency | 176801 | 10q22.1 |
| Defects in sulfatide degradation | ||||
| Arylsulfatase A | ARSA | Metachromatic leukodystrophy | 607574 | 22q13.33 |
| Saposin B | PSAP | Metachromatic leukodystrophy due to saposin B deficiency |
249900 | 10q22.1 |
| Formyl-glycin-generating enzyme | SUMF1 | Multiple sulfatase deficiency | 607939 | 3p26.1 |
| β-Galactosylceramidase | GALC | Globoid cell leukodystrophy (Krabbe disease) | 606890 | 14q31.3 |
| Defects in globotriaosylceramide degradation | ||||
| α-Galactosidase A | GLA | Fabry disease | 301500 | Xq22.1 |
| Defects in degradation of glycosaminoglycan (mucopolysaccharidoses, MPS) | ||||
| Degradation of heparan sulfate | ||||
| α-Iduronidase | IDUA | MPS I (Hurler, Scheie) | 607015 | 4p16.3 |
| Iduronate sulfatase | IDS | MPS II (Hunter) | 309900 | Xq28 |
| Heparan N-sulfatase | SGSH | MPS IIIa (Sanfilippo A) | 252900 | 17q25.3 |
| N-acetyl glucosaminidase | NAGLU | MPS IIIb (Sanfilippo B) | 252920 | 17q21.2 |
| Acetyl-CoA transferase | HGSNAT | MPS IIIc (Sanfilippo C) | 252930 | 8p11.21 |
| N-acetyl glucosamine 6-sulfatase | GNS | MPS IIId (Sanfilippo D) | 252940 | 12q14.3 |
| β-glucuronidase | GUSB | MPS VII (Sly) | 253220 | 7q11. 21 |
| Defects in degradation of other mucopolysaccharides | ||||
| N-Acetylgalactosamine 4-sulfatase | ARSB | MPS VI (Maroteaux-Lamy syndrome) | 253200 | 5q14.1 |
| Galactose 6-sulfatase | GALNS | MPS IVA (Morquio A) | 253000 | 16q24.3 |
| Hyaluronidase | HYAL1 | MPS IX | 601492 | 3p21.31 |
| Defects in glycogen degradation | ||||
| α-Glucosidase | GAA | Pompe disease | 232300 | 17q25.3 |
| Defects in lipid degradation | ||||
| Defects in sphingomyelin degradation | ||||
| Acid sphingomyelinase | SMPD1 | Niemann-Pick type A | 257200 | 11p15.4 |
| Acid sphingomyelinase | SMPD1 | Niemann-Pick type B (E and F) | 607616 | 11p15.4 |
| Acid ceramidase | ASAH1 | Farber lipogranulomatosis | 228000 | 8p22 |
| Defects in triglycerides and cholesteryl ester degradation | ||||
| Acid lipase | LIPA | Wolman/cholesteryl ester storage disease | 278000 | 10q23.31 |
| Defects in protein degradation | ||||
| Cathepsin K | CTSK | Pycnodysostosis | 601105 | 1q21.3 |
| Tripeptidyl peptidase | TPP1 | Ceroid lipofuscinosis 2 | 607998 | 11q15.4 |
| Palmitoyl-protein thioesterase | PPT1 | Ceroid lipofuscinosis, neuronal, 1 | 600722 | 1p34.2 |
| Defects in lysosomal transporters | ||||
| Cystinosin (cystin transport) | CTNS | Cystinosis | 606272 | 17p13.2 |
| Sialin (sialic acid transport) | SLC17A5 | Salla disease | 604322 | 6q13 |
| Defects in lysosomal trafficking proteins | ||||
| UDP- N -acetylglucosamine Phosphotransferase γ-subunit | GNPTG | Mucolipidosis III gamma (I-cell) | 607838 | 16p13.3 |
| Mucolipin-1(cation channel) | MCOLN1 | Mucolipidosis IV | 605248 | 19p13.2 |
| Lysosome-associated membrane protein 2 | LAMP2 | Danon | 309060 | Xq24 |
| Sphingomyelinase | NPC1/NPC2 | Niemann-Pick type C1 and D | 607623 | 11q11.2 |
| CLN3 protein (battenin) | CLN3 | Ceroid lipofuscinosis, neuronal, 3 | 204200 | 16p11.2 |
| Protein CLN6 | CLN6 | Ceroid lipofuscinosis, neuronal, 6 | 601780 | 15q23 |
| Protein CLN8 | CLN8 | Ceroid lipofuscinosis, neuronal, 8 | 607837 | 8p23.3 |
| Lysosomal trafficking regulator | LYST (CHS1) | Chediak-Higashi | 214500 | 1q42.3 |
| Myosin VA | MYO5A | Griscelli Type 1 | 214450 | 15q21.2 |
| RAB27 | RAB27A | Griscelli Type 2 | 607624 | 15q21.3 |
| Melanophilin | MLPH | Griscelli Type 3 | 609227 | 2q37.3 |
| HPS1 | HPS1 | Hermansky-Pudlak 1 | 203300 | 10q24.2 |
| AP3 β-subunit | AP3B1 | Hermansky-Pudlak 2 | 608233 | 5q14.1 |
In italic are indicated circadian transcripts, in bold is indicated a transcript characterized by 39 h periodicity
Source: CircaDB, web addresses http://circadb.org, http://github.com/itmat/circadb, http://bioinf.itmat.upenn.edu/circa
Fig. 3.

x–y plots showing the time-qualified profile of expression of clock-controlled genes whose mutation causes LSDs (source: CircaDB, web addresses http://circadb.org, http://github.com/itmat/circadb, http://bioinf.itmat.upenn.edu/circa). On the x axis is represented time in hours, and on the y axis are represented the mRNA relative expression levels in arbitrary units
Fig. 4.
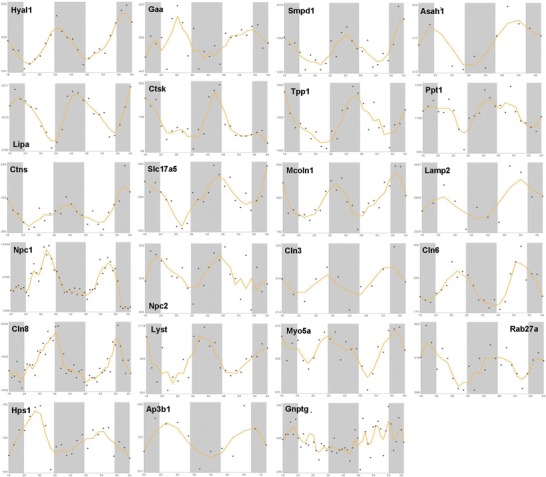
x–y plots showing the time-qualified profile of expression of clock-controlled genes whose mutation causes LSDs (source: CircaDB, web addresses http://circadb.org, http://github.com/itmat/circadb, http://bioinf.itmat.upenn.edu/circa). On the x axis is represented time in hours, and on the y axis are represented the mRNA relative expression levels in arbitrary units
The alteration of the circadian clock circuitry may be responsible also of changes of behavioral cycles of sleep/wake, rest activity, and fasting/feeding often evidenced in the patients affected by LSDs. A high prevalence of sleep disorders has been reported in patients affected by type III mucopolysaccharidosis, defined with the eponym Sanfilippo syndrome and representing the most frequent mucopolysaccharidosis, leading to neurodegeneration with habitually severe sleep and behavioral disturbance. Patients affected by Sanfilippo syndrome are characterized by alteration in the circadian rhythm of melatonin rendered by lower urinary concentrations of its metabolite 6-sulfatoxymelatonin at night and higher concentrations in the morning when compared to controls. Based on these reports, therapies aimed at circadian resynchronization such as behavioral treatment, light therapy, or melatonin administration rather than conventional hypnotics have been proposed (Fraser et al. 2002; Guerrero et al. 2006). Accordingly, a beneficial effect of melatonin administration was evidenced in a randomized, double-blind, placebo-controlled, parallel study conducted in a cohort of children with neurodevelopmental disorders and sleep impairment represented by difficulties in initiating and maintaining sleep (impossibility to fall asleep within 1 h of lights out or showing less than 6 h of continuous sleep). Compared to placebo, therapy with melatonin at escalating doses (from a starting dose of 0.5 mg through 2 mg and 6 mg to a maximal dose of 12 mg during the first month of treatment, at the end of which the child was maintained on the attained dose) improved sleep-onset latency and total nocturnal sleep time in a statistically significant way, although the increase of total nocturnal sleep time was not clinically significant (Appleton et al. 2012), which is a major downside of this approach and an example that biological pathways and statistical significance do not necessarily translate into tangible clinical benefit for the patient.
Conclusion
Genetically encoded oscillators maneuvered by transcriptional/translational feedback loops hardwired by circadian genes and proteins drive time-related variations of biological processes. The regular succession of intracellular phenomena is ordered by the biological oscillator ticking in every cell and steering the harmonization of crucial pathways and the compartmentalization in the temporal dimension of poorly compatible biochemical processes. Failure of time-of-day specific transcription of clock genes and clock-controlled genes caused by changes of working of the clock gene machinery may provoke severe deregulation of downstream gene expression regulating a complex array of cellular functions, such as molecule biosynthesis, posttranslational modification, processing, transport, conjugation, internalization and degradation, and cell processes such as cell cycle, autophagy, apoptosis, and DNA damage response. These changes cause perturbation of cellular homeostasis, cell dysfunction, and biochemical and structural derangements that may lead to cell death and tissue dysfunction. The key role played by the molecular clockwork in the control of lysosome function and the involvement of clock-controlled genes encoding circadian transcripts in the pathogenesis of LSDs suggest a direct involvement of the biological clock in the pathophysiological mechanisms underlying cellular and tissue derangements hallmarking these hereditary diseases, and that gene transfer or a proper timetable of enzyme replacement therapy could address the physiological fluctuations driven by the biological clock and appropriately outline circadian rhythmicity.
Electronic Supplementary Material
Acknowledgments
The study was supported by the “5x1000” voluntary contribution and by a grant (GM) from the Italian Ministry of Health through Department of Medical Sciences, Division of Internal Medicine and Chronobiology Unit (RC1201ME04, RC1203ME46, and RC1302ME31), IRCCS Scientific Institute and Regional General Hospital “Casa Sollievo della Sofferenza,” Opera di Padre Pio da Pietrelcina, San Giovanni Rotondo (FG), Italy.
Take-Home Message (Synopsis)
The lysosomal storage disorders are caused by mutation of genes whose expression is driven with 24-h periodicity by the biological clock, and the circadian pathways impact the pathophysiological mechanisms, implying the involvement of the temporal dimension in the pathogenesis of these hereditary diseases.
Compliance with Ethics Guidelines
Conflict of Interest Statement
Gianluigi Mazzoccoli, Tommaso Mazza, Manlio Vinciguerra, Stefano Castellana, and Maurizio Scarpa declare that there are no conflicts of interest with respect to the authorship and/or publication of this article.
Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Details of the Contributions of Individual Authors
GM conceived the purpose of the review and wrote the article, MV and MS wrote the article, and SC and TM performed bioinformatics analysis and represented scheme and figures.
Footnotes
Competing interests: None declared
Author contributed equally with all other contributors.
Contributor Information
Gianluigi Mazzoccoli, Email: g.mazzoccoli@operapadrepio.it.
Maurizio Scarpa, Email: m.scarpa@operapadrepio.it.
Collaborators: Johannes Zschocke, Matthias Baumgartner, K Michael Gibson, Marc Patterson, and Shamima Rahman
References
- Agostino PV, Harrington ME, Ralph MR, et al. Casein kinase-1-epsilon (CK1epsilon) and circadian photic responses in hamsters. Chronobiol Int. 2009;26:126–133. doi: 10.1080/07420520802675177. [DOI] [PubMed] [Google Scholar]
- Alenghat T, Meyers K, Mullican SE, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G, Beischlag TV, Vinciguerra M, Mazzoccoli G. The circadian clock circuitry and the AHR signaling pathway in physiology and pathology. Biochem Pharmacol. 2013;85:1405–1416. doi: 10.1016/j.bcp.2013.02.022. [DOI] [PubMed] [Google Scholar]
- Appleton RE, Jones AP, Gamble C et al (2012) The use of MElatonin in children with neurodevelopmental disorders and impaired sleep: a randomised, double-blind, placebo-controlled, parallel study (MENDS). Health Technol Assess 16:i-239 [DOI] [PubMed]
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, et al. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Bagshaw RD, Mahuran DJ, Callahan JW. Lysosomal membrane proteomics and biogenesis of lysosomes. Mol Neurobiol. 2005;32:27–41. doi: 10.1385/MN:32:1:027. [DOI] [PubMed] [Google Scholar]
- Ballabio A, Gieselmann V. Lysosomal disorders: from storage to cellular damage. Biochim Biophys Acta. 2009;1793:684–696. doi: 10.1016/j.bbamcr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian Integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellettato CM, Scarpa M. Pathophysiology of neuropathic lysosomal storage disorders. J Inherit Metab Dis. 2010;33:347–362. doi: 10.1007/s10545-010-9075-9. [DOI] [PubMed] [Google Scholar]
- Berrabah W, Aumercier P, Lefebvre P, et al. Control of nuclear receptor activities in metabolism by post-translational modifications. FEBS Lett. 2011;585:1640–1650. doi: 10.1016/j.febslet.2011.03.066. [DOI] [PubMed] [Google Scholar]
- Bhattacharya R, von Mayersbach H. Histochemistry of circadian changes of some lysosomal enzymes in rat liver. Acta Histochem Suppl. 1976;16:109–115. [PubMed] [Google Scholar]
- Bonny O, Vinciguerra M, Gumtz ML, Mazzoccoli G. Molecular bases of circadian rhythmicity in renal physiology and pathology. Nephrol Dial Transplant. 2013;28:2421–2431. doi: 10.1093/ndt/gft319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozek K, Relógio A, Kielbasa SM, et al. Regulation of clock-controlled genes in mammals. PLoS One. 2009;4(3):e4882. doi: 10.1371/journal.pone.0004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brozzi A, Urbanelli L, Germain PL, Magini A, Emiliani C (2013) hLGDB: a database of human lysosomal genes and their regulation. Database (Oxford) bat024 [DOI] [PMC free article] [PubMed]
- Bugge A, Feng D, Everett LJ, et al. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris TP. Nuclear hormone receptors for heme: REV-ERBalpha and REV-ERBbeta are ligand-regulated components of the mammalian clock. Mol Endocrinol. 2008;22:1509–1520. doi: 10.1210/me.2007-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cailotto C, Lei J, van der Vliet J, et al. Effects of nocturnal light on (clock) gene expression in peripheral organs: a role for the autonomic innervation of the liver. PLoS One. 2009;4(5):e5650. doi: 10.1371/journal.pone.0005650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone L, Hirayama J, Giordano F, et al. Circadian clock control by SUMOylation of BMAL1. Science. 2005;309:1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- Cermakian N, Lange T, Golombek D, et al. Crosstalk between the circadian clock circuitry and the immune system. Chronobiol Int. 2013;30:870–888. doi: 10.3109/07420528.2013.782315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challet E. Entrainment of the suprachiasmatic clockwork in diurnal and nocturnal mammals. Endocrinology. 2007;148:5648–5655. doi: 10.1210/en.2007-0804. [DOI] [PubMed] [Google Scholar]
- Chedid A, Nair V. Diurnal rhythm in endoplasmic reticulum of rat liver: electron microscopic study. Science. 1972;175:176–179. doi: 10.1126/science.175.4018.176. [DOI] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12 hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 2010;11(1):47–57. doi: 10.1016/j.cmet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- De Cata A, D’Agruma L, Tarquini R, Mazzoccoli G (2014) Rheumatoid arthritis and the biological clock. Expert Rev Clin Immunol 2014 May;10(5):687-95. doi:10.1586/1744666X.2014.899904 [DOI] [PubMed]
- Di Tacchio L, Le HD, Vollmers C, et al. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333:1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Dufour CR, Levasseur MP, Pham NH, et al. Genomic convergence among ERRα, PROX1, and BMAL1 in the control of metabolic clock outputs. PLoS Genet. 2011;7(6):e1002143. doi: 10.1371/journal.pgen.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguay D, Cermakian N. The crosstalk between physiology and circadian clock proteins. Chronobiol Int. 2009;26:1479–1513. doi: 10.3109/07420520903497575. [DOI] [PubMed] [Google Scholar]
- Eide EJ, Vielhaber EL, Hinz WA, et al. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iε. J Biol Chem. 2002;277:17248–17254. doi: 10.1074/jbc.M111466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331(6022):1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J, Wraith JE, Delatycki MB. Sleep disturbance in mucopolysaccharidosis type III (Sanfilippo syndrome): a survey of managing clinicians. Clin Genet. 2002;62:418–421. doi: 10.1034/j.1399-0004.2002.620512.x. [DOI] [PubMed] [Google Scholar]
- Fulco M, Sartorelli V. Comparing and contrasting the roles of AMPK and SIRT1 in metabolic tissues. Cell Cycle. 2008;7:3669–3679. doi: 10.4161/cc.7.23.7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futerman AH, van Meer G. The cell biology of lysosomal storage disorders. Nat Rev Mol Cell Biol. 2004;5:554–565. doi: 10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- Grimaldi B, Bellet MM, Katada S, et al. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 2010;12:509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero JM, Pozo D, Diaz-Rodriguez JL, Martinez-Cruz F, Vela-Campos F. Impairment of the melatonin rhythm in children with Sanfilippo syndrome. J Pineal Res. 2006;40:192–193. doi: 10.1111/j.1600-079X.2005.00294.x. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten AM, Kang HS, Takeda Y. Retinoic acid-related orphan receptors α and γ: key regulators of lipid/glucose metabolism, inflammation, and insulin sensitivity. Front Endocrinol (Lausanne) 2013;4:1. doi: 10.3389/fendo.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet A, Chapel A, Kieffer S, Roux F, Garin J. Proteomic analysis of human lysosomes: application to monocytic and breast cancer cells. Proteomics. 2002;2:1026–1040. doi: 10.1002/1615-9861(200208)2:8<1026::AID-PROT1026>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Palm IF, La Fleur SE, et al. SCN outputs and the hypothalamic balance of life. J Biol Rhythms. 2006;21:458–469. doi: 10.1177/0748730406293854. [DOI] [PubMed] [Google Scholar]
- Kemp MG, Akan Z, Yilmaz S, et al. Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J Biol Chem. 2010;285:16562–16571. doi: 10.1074/jbc.M110.110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS (2006) Molecular components of the mammalian circadian clock. Hum Mol Genet 15 (Spec No. 2):R271–R277 [DOI] [PubMed]
- Lamia KA, Sachdeva UM, Ditacchio L, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübke T, Lobel P, Sleat DE. Proteomics of the lysosome. Biochim Biophys Acta. 2009;1793:625–635. doi: 10.1016/j.bbamcr.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Lin JD. Circadian regulation of autophagy rhythm through transcription factor C/EBPβ. Autophagy. 2012;8:124–125. doi: 10.4161/auto.8.1.18081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Panda S, Lin JD. Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J. 2011;30:4642–4651. doi: 10.1038/emboj.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury E, Ramsey KM, Bass J. Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ Res. 2010;106:447–462. doi: 10.1161/CIRCRESAHA.109.208355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoccoli G. The timing clockwork of life. J Biol Regul Homeost Agents. 2011;25:137–143. [PubMed] [Google Scholar]
- Mazzoccoli G, Vendemiale G, De Cata A, Carughi S, Tarquini R. Altered time structure of neuro-endocrine-immune system function in lung cancer patients. BMC Cancer. 2010;10:314. doi: 10.1186/1471-2407-10-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoccoli G, Giuliani F, Sothern RB. A method to evaluate dynamics and periodicity of hormone secretion. J Biol Regul Homeost Agents. 2011;25:231–238. [PubMed] [Google Scholar]
- Mazzoccoli G, Sothern RB, Greco G, et al. Time-related dynamics of variation in core clock gene expression levels in tissues relevant to the immune system. Int J Immunopathol Pharmacol. 2011;24:869–879. doi: 10.1177/039463201102400406. [DOI] [PubMed] [Google Scholar]
- Mazzoccoli G, Cai Y, Liu S, et al. REV-ERBalpha and the clock gene machinery in mouse peripheral tissues: a possible role as a synchronizing hinge. J Biol Regul Homeost Agents. 2012;26:265–276. [PubMed] [Google Scholar]
- Mazzoccoli G, Francavilla M, Giuliani F, et al. Clock gene expression in mouse kidney and testis: analysis of periodical and dynamical patterns. J Biol Regul Homeost Agents. 2012;26:303–311. [PubMed] [Google Scholar]
- Mazzoccoli G, Francavilla M, Pazienza V, et al. Differential patterns in the periodicity and dynamics of clock gene expression in mouse liver and stomach. Chronobiol Int. 2012;29:1300–1311. doi: 10.3109/07420528.2012.728662. [DOI] [PubMed] [Google Scholar]
- Mazzoccoli G, Pazienza V, Vinciguerra M. Clock genes and clock controlled genes in the regulation of metabolic rhythms. Chronobiol Int. 2012;29:227–251. doi: 10.3109/07420528.2012.658127. [DOI] [PubMed] [Google Scholar]
- Mazzoccoli G, Tomanin R, Mazza T, et al. Circadian transcriptome analysis in human fibroblasts from Hunter syndrome and impact of iduronate-2-sulfatase treatment. BMC Med Genomics. 2013;6:37. doi: 10.1186/1755-8794-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoccoli G, Vinciguerra M, Oben J, Tarquini R, De Cosmo S (2014) Non-alcoholic fatty liver disease: the role of nuclear receptors and circadian rhythmicity. Liver Int. doi:10.1111/liv.12534 [DOI] [PubMed]
- Meikle PJ, Hopwood JJ, Clague AE, Carey WF. Prevalence of lysosomal storage disorders. JAMA. 1999;281:249–254. doi: 10.1001/jama.281.3.249. [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, et al. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/S0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Peek CB, Affinati AH, Ramsey KM, et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science. 2013;342:1243417. doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro A, Hayer K, Lahens NF, Hogenesch JB. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Res. 2013;41(Database issue):D1009–D1013. doi: 10.1093/nar/gks1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt FM, Walkley SU. Lysosomal defects and storage. In: Platt FM, Walkley SU, editors. Lysosomal disorders of the brain: recent advances in molecular and cellular pathogenesis and treatment. 1. New York: Oxford University Press; 2004. pp. 32–49. [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/S0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Ramsey K, Yoshino J, Brace CS, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J, Reick M, Wu LC, et al. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- Sahar S, Zocchi L, Kinoshita C, et al. Regulation of BMAL1 protein stability and circadian function by GSK3beta-mediated phosphorylation. PLoS One. 2010;5(1):e8561. doi: 10.1371/journal.pone.0008561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz ML, Tecedor L, Chang M, Davidson BL. Clarifying lysosomal storage diseases. Trends Neurosci. 2011;34:401–410. doi: 10.1016/j.tins.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Fu MA, Brown EJ. Tim-Tipin dysfunction creates an indispensible reliance on the ATR-Chk1 pathway for continued DNA synthesis. J Cell Biol. 2009;5:15–23. doi: 10.1083/jcb.200905006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, et al. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevy MF, Giebultowicz J, Pincus Z, Mazzoccoli G, Vinciguerra M. Aging signaling pathways and circadian clock-dependent metabolic derangements. Trends Endocrinol Metab. 2013;24:229–237. doi: 10.1016/j.tem.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama Y, von Mayersbach H (1981) Circadian changes of lysosomal enzyme activities in rat hepatocytes using ultracytochemistry. Prog Clin Biol Res 59C(00): 117–124.a [PubMed]
- Uchiyama Y, Groh V, von Mayersbach H (1981) Different circadian variations as an indicator of heterogeneity of liver lysosomes. Histochemistry 73:321–337 [DOI] [PubMed]
- Unsal-Kaçmaz K, Chastain PD, Qu PP, et al. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol. 2007;27:3131–3142. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valekunja UK, Edgar RS, Oklejewicz M, et al. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc Natl Acad Sci U S A. 2013;110:1554–1559. doi: 10.1073/pnas.1214168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra M, Borghesan M, Pazienza V, et al. The transcriptional regulators, the immune system and the circadian clock. J Biol Regul Homeost Agents. 2013;27:9–22. [PubMed] [Google Scholar]
- Vinciguerra M, Tevy MF, Mazzoccoli G (2014) A ticking clock links metabolic pathways and organ systems function in health and disease. Clin Exp Med. 2014 May;14(2):133-40. doi:10.1007/s10238-013-0235-8 [DOI] [PubMed]
- Vitner EB, Platt FM, Futerman AH. Common and uncommon pathogenic cascades in lysosomal storage diseases. J Biol Chem. 2010;285(27):20423–20427. doi: 10.1074/jbc.R110.134452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yang X, Wood PA, Hrushesky WJ. Mammalian TIMELESS is required for ATM-dependent CHK2 activation and G2/M checkpoint control. J Biol Chem. 2010;285:3030–3034. doi: 10.1074/jbc.M109.050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wu N, Lazar MA. Nuclear receptor Rev-erbalpha: a heme receptor that coordinates circadian rhythm and metabolism. Nucl Recept Signal. 2010;8:e001. doi: 10.1621/nrs.08001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


