Abstract
Objectives:
To investigate 15 respiratory viruses in children with acute respiratory tract infections (ARTIs) using multiplex reverse-transcriptase polymerase chain reaction (RT-PCR), and to analyze the clinical and epidemiological features of these viruses.
Methods:
In a cross-sectional study, 135 children, ≤5 years of age who presented with ARTIs in Najran Maternity and Children Hospital, Najran, Saudi Arabia between October 2012 and July 2013 were included. The clinical and sociodemographic data, and the laboratory results were recorded using a standardized questionnaire. Two nasopharyngeal swabs were collected from each child: one for bacteriological examination, and the second for viral detection using multiplex RT-PCR.
Results:
A single viral pathogen was detected in 76 patients, viral coinfections in 9, and mixed viral and bacterial pathogens in 15. Respiratory syncytial virus was isolated in 33 patients, human rhinovirus (hRV) in 22, adenovirus (AdV) in 19, human metapneumovirus in 13, influenza virus in 10, parainfluenza virus in 7, human corona virus (hCoV) in 4, and human bocavirus in one.
Conclusion
Respiratory syncytial virus, hRV, and AdV were the most frequent viruses, accounting for more than two-thirds of the cases. Other viruses, such as MPV, hCoV NL63, and hCoV OC43, may play a role in pediatric ARTIs. Of significance is the potential use of multiplex RT-PCR to provide epidemiological and virological data for early detection of the emergence of novel respiratory viruses in the era of the Middle East respiratory syndrome coronavirus.
A cute respiratory tract infections (ARTI's) are a leading cause of infectious disease-related morbidity and mortality among children under 5 years of age worldwide, particularly in the developing countries.1 The ARTI's can be caused by bacteria and fungi, but viral infections are the most common causes. In developing countries, viruses represent a considerable proportion of the pathogens associated with ARTI's, varying from 40-90% across studies.2-4 The most frequently implicated viruses among children are respiratory syncytial virus (RSV), influenza A and B viruses (IAV and IBV), parainfluenza viruses (PIVs), adenoviruses (AdV), human rhinoviruses (hRV), and human enteroviruses (hEV). Several recently discovered viruses, such as human metapneumovirus (hMPV), human bocavirus (hBoV), and the human coronaviruses (hCoVs) (NL63 and OC43), have been identified as potential respiratory pathogens. In addition, 2 novel human polyomaviruses (Washington University virus [WU] and Karolinska Institute virus [KI]) have been detected in patients with respiratory infections.5-7 Lower respiratory tract infections (LRTI's) are the most severe forms of infection and cause of approximately 1.9 million annual deaths among children,8 while upper respiratory tract infections (URTI's) are the most frequent presentation of respiratory infections. An estimated 50% of all illnesses in all age groups, and approximately 75% of illnesses in young children are viral URTI's.9 Proper diagnosis of viral ARTI's has been shown to reduce the misuse of antibiotics and shorten the length of hospital stay. However, current challenges in detecting respiratory viruses include inconsistent clinical manifestations of viral RTI's and variable sensitivity and specificity of available diagnostic assays. The traditional diagnostic methods for the respiratory viruses including cell culture and direct immunofluorescence assays are time consuming and technically demanding.10 Several types of molecular biological methods, including reverse transcription PCR (RT-PCR), PCR-hybridization, and real time PCR, have been introduced as more rapid and sensitive detection methods for respiratory infections.3,5,11-13 Most studies in Saudi Arabia focused on a small number of viral pathogens, and information on the relative contribution of each pathogen to clinical forms of ARTI's is incomplete.14-16 Hence, the aims of our study were to investigate the distribution of 15 respiratory viruses in children with ARTI's using multiplex RT-PCR, and to evaluate the presenting clinical, demographic, and epidemiological features of these different viral infections.
Methods
This study was conducted in Najran Maternity and Children Hospital, a 200-bed, tertiary-care hospital in Najran, southwestern Saudi Arabia between October 2012 and July 2013. Included in the study were children ≤5 years of age who presented with signs and symptoms of upper and/or lower respiratory tract infection seen in the pediatric emergency room, or admitted to the pediatric ward, or the pediatric intensive care unit (PICU). Written consent was obtained from their parents. Inclusion criteria were as follows: a temperature of ≥38°C, and at least one of the following: rhinitis, pharyngitis, cough, earache, hoarseness of voice, rhonchi, crepitations, or wheezy chest. The diagnosis for each patient was performed by the attending pediatrician based on the World Health Organization (WHO) standard clinical criteria.17 The clinical and socio-demographic data and routine laboratory tests results were recorded using a standardized questionnaire, including the age, gender, underlying medical conditions, signs and symptoms at presentation, findings on physical examination, laboratory findings (complete blood count [CBC], C-reactive protein [CRP], and bacterial cultures), and definitive clinical diagnosis.
Sampling
Two nasopharyngeal swaps were collected for each patient under strict infection control guidance using rayon-budded swabs with 2 ml of virus transport medium (Virocult, Wiltshire, UK); one for bacteriological examination and the other for respiratory virus detection using RT-PCR multiplex. The specimens were transported in an icebox to the Microbiology Department, College of Medicine, Najran University for further processing.
Bacteriological examination
Each nasopharyngeal sample from each case was inoculated on blood and MacConkey agar media (Difco Laboratories, Detroit, MI, USA). The plates were incubated at 37⁰C for 24-48 hours. Suspected colonies were identified as pathogenic organisms using standard laboratory procedures according to the biochemical and serological techniques recommended by the WHO.18
Multiplex RT-PCR analysis
Viral nucleic acids were extracted from the nasopharyngeal samples using QIAamp viral mini kit (Qiagen, Maryland, USA) according to the manufacturers’ instructions. The total RNAs extracted from the clinical samples were used for the synthesis of the first-strand cDNAs (Revert AidKit, Fermentas, Canada) according to the manufacturer's instructions. The sample cDNAs were tested in a 3-tube reaction following the protocol supplied by the manufacturer (Qiagen, Maryland, USA). Tube-1 contained primers set A targeting AdV, hCoV 229E/NL63, and PIV1-3. Tube-2 contained primers set B targeting hCoV OC43/HKU1, hRV A/B/C, RSV A/B, and IAV. Tube-3 contained primers set C targeting hBoV 1/2/3/4, IBV, hMPV, PIV4, and hEV. Positive controls included a mixture of all 15 virus clones. The negative control contained only double distilled water. The PCR amplification was performed using 10 µl of Seeplex RV master mix (Seegene, Seoul, Korea), 4 µl of 5X multiplex primer sets, 3 µl of 8-MOP solution, and 3 µl of newly synthesized first-strand cDNA (1 µg/3 µl). The Seeplex RV contained A, B, and C sets of primers designed from highly conserved regions of genetic sequences for the 15 respiratory viruses. An initial pre-PCR step of 94°C for 15 minutes was performed in a thermocycler 9600 (Perkin-Elmer, Ohio, USA), followed by a total of 35 PCR cycles under the following conditions: 94°C for 30 sec, 60°C for 1.5 min, and 72°C for 1.5 min. The final cycle was followed by an extension step at 72°C for 10 min. The amplified PCR products were separated on 2% agarose gel and stained with ethidium bromide. The type of respiratory virus was identified by comparison with the reference band size provided by the manufacturer.
Statistical analysis
Values were expressed as percentages for discrete variables, or as mean and standard deviation for continuous variables. Clinical characteristics and laboratory variables were compared using the chi-square or the Student t-test or Fisher's exact test, as appropriate. Statistical significance was defined as a p-value less than 0.05. All analyses were performed with the Statistical Package for the Social Sciences (SPSS), version 15.0 (SPSS Inc., Chicago, IL, USA).
Results
A total of 135 patients were included in the study period. The median age of patients was 12 months, and the number of males was 80 (63.3%). A single viral pathogen was detected in 76 (56.3%); viral co-infections in 9 (6.7%); and mixed viral and bacterial pathogen in 15 (11.1%) patients. The specific pathogens were RSV in 33 patients, hRV in 22, AdV in 19, hMPV in 13, IFV in 10, PIV's in 7, hCoV in 4, and hBoV in one) (Table 1). Children were divided into 5 age-groups: ≤12 months (n=73; 54.1%), 13-24 months (n=32; 23.7%), 25-36 months (n=9; 6.7%), 37-48 months (n=9; 6.7%), and 49-60 months (n=12; 8.9%). The infection rate in children ≤12-month age was significantly higher than in the other age groups. Interestingly, we observed a significantly larger number of positive cases for RSV infection in patients aged <24 months (25/34 versus 9/34, p=0.01). Moreover, hMPV and hCoV infections were detected only in children <24 months of age, p=0.001 (Figure 1).
Table 1.
Distribution of viral etiologic agents and co-infections.
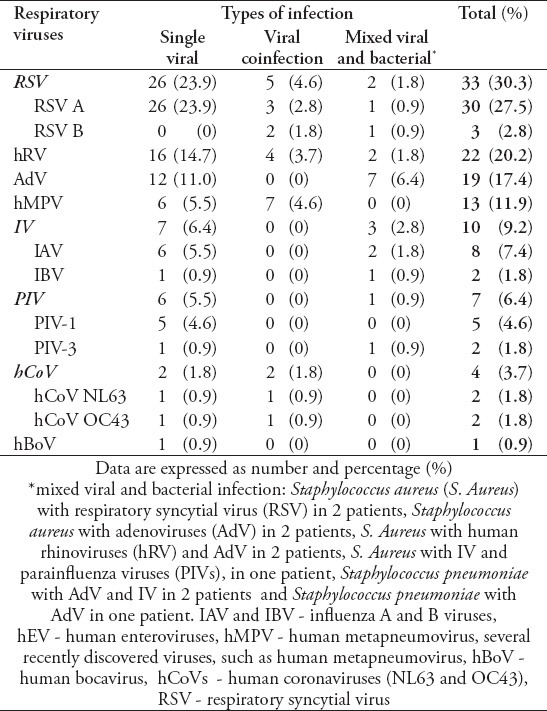
Figure 1.
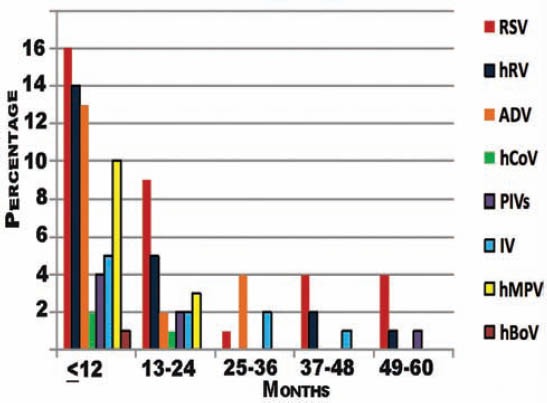
The distribution of viral agents according to age among the studied patients.
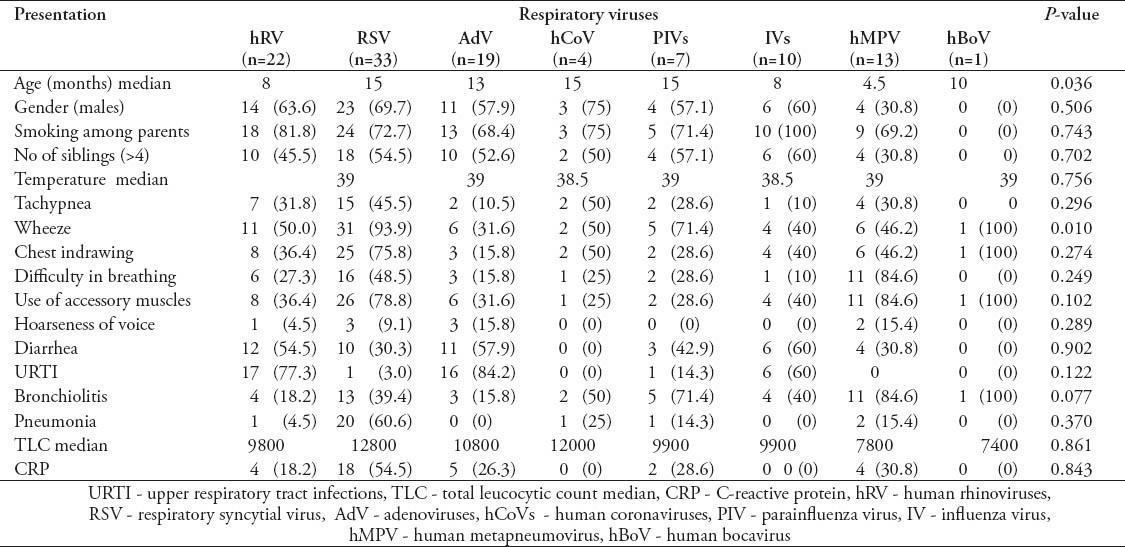
Demographic data and clinical presentation of patients were compared to general pathogens identified (Table 2) and the individual viral pathogens (Table 3). Generally, there were no significant differences in the clinical presentation, as well as, the gender of patients for the various pathogens isolated. However, the median age of children with hMPV was significantly higher than that of children infected with the other viruses (p=0.036). Wheeze was more frequently associated with viral infections (p=0.005) mainly RSV (p=0.01)
Table 2.
Demographic and clinical presentations of the studied patients.
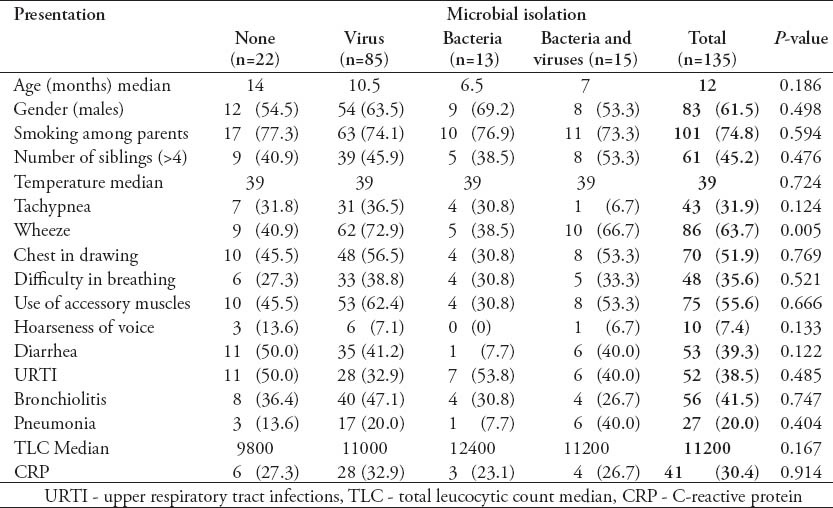
Table 3.
Demographic and clinical presentations of the studied patients infected with individual respiratory viruses.
Discussion
Viral infection is a major cause of respiratory illness among children. The frequency of respiratory virus detection in 135 children with ARTI's, in this study, was 80.7%. This finding is in support of previous studies, which reported viral detection rates of 47-95% in children.2-5,19 Possible explanations for the wide differences in detection rates in the literature include heterogeneity in study populations, differences in presenting respiratory symptoms, number of respiratory pathogens tested, method used for detection and genetic variability between populations.2,20 Overall, in 6.7% of the patients, viral co-infections were found. Previous studies using RT PCR techniques reported viral co-infection rates of 5-10%, with RSV, hRV, PIVs, and hMPV being the most commonly implicated viruses in cases of mixed infections.12,19-21 In this study, the high detection rate of RSV with hMPV (4/9; 44.4%) and hRV with hCoV (2/9, 22.2%) in viral co-infections was an interesting finding. Huijskens et al3 found that in 67% of the hRV-positive patients and 42% of the RSV cases, hRV and RSV were co-detected with another virus.
It has been reported that the unique characteristic of RSV facilitates infection with a second respiratory virus.22,23 Previous studies reported that hRV in URTI's could serve as a clinical illness promotion factor, functioning additively or synergistically in the pathogenesis of lower respiratory syndromes such as bronchiolitis.9,231,24 However, identification of 2 or more viruses in a patient may be due to prolonged viral shedding or asymptomatic persistence of viruses.5 Further, work would be needed to clarify this situation. Quantitative identification of the viral genome may help in explaining co-infections.
As expected, most of the respiratory pathogens in this study were detected in children <1 year. The higher detection rate of respiratory pathogens among infants and young children has been ascribed to a higher infection rate, lower viral clearance rate due to underdeveloped immune system, and higher load of the infectious agent associated with living conditions such as crowding.2,3,20 Furthermore, the parents of younger children may seek healthcare earlier in the course of disease due to parental anxiety. Our results confirm that RSV and hRV play a key role in RTI's in children; RSV was the most frequent respiratory virus detected, followed by hRV. The predominance of RSV in this study is in accordance with the assertion that this virus is the single most frequent lower respiratory tract pathogen in infants and young children in Saudi Arabia and worldwide.4,5,15,20 It would be important for local pediatricians to use antibiotics cautiously when children are hospitalized with ARTI's. The hRV has been known to be responsible for URTI's and some LRTI's in children.19,24 Our findings showed that the presence of hRV in all age groups. Most hRVs (17/22; 77.3%) were associated with URTI's and 22.7% of hRV were detected in bronchiolitis (4 cases), or pneumonia (one case).
Adenovirus was reported to be responsible for 5-10% of ARTI's in children, and has been largely associated with bronchiolitis obliterans and acute wheezing episodes.25-27 In our study, ADVs were found in 17.4% of all patients, being the third most frequent viral pathogen, and most ADVs (16/19; 84.2%) were associated with URTI's. The hMPV is one of the causes of upper and lower RTI's, especially in preschool and older children. In our study, hMPV was the fourth most frequent viral pathogen accounting for 11.9% of all cases. In a previous Saudi study,14 hMPV was identified in 8.3% of 489 children with ARTI's. In contrast to some studies,12,26,28 all hMPV-positive cases in this study were below 2 years of age and almost half of the cases (7/13; 54%) were co-infected with other viruses. The detection rates for IVs and PIVs in our study were comparable with other studies, reporting rates of 0.8-12.6% for IV's and 2.8-19.4% for PIV's.3,5,13,19,20,23,26
In our study, hCoVs were detected in 3.7% of all patients including 2 cases with bronchiolitis and 2 cases with pneumonia. In a previous Saudi study,14 hCoVNL63 type was detected in 2.8% of 489 children.14 However, in our study 2 hCoV types (hCoV NL63 and hCoV OC43) were reported. In September 2012, a novel human coronavirus, called the Middle East respiratory syndrome coronavirus (MERS-CoV), was first identified in samples obtained from a Saudi Arabian businessman who died from acute respiratory failure.29 As of May 2013, a total of 49 confirmed cases of MERS-CoV infection with 26 deaths have been reported to the WHO, including 37 in Saudi Arabia with 21 deaths.30 These figures highlight the characteristic distribution of hCoVs emerging types circulating in Saudi Arabia.
The similar clinical presentations of patients infected by various respiratory viruses and some bacterial pathogens make etiological diagnoses difficult when decisions are based only on clinical presentation.3-5,12 In our study, except the strong association between RSV infection and chest wheeze, the clinical manifestations of other viral infections were largely nonspecific with fever and cough as the main symptoms.
One limitation of this study is the inability to describe the seasonal distribution of respiratory viruses due to the short duration of the study. Further, molecular-based studies of longer surveillance duration are necessary to elucidate the seasonal patterns and disease burden associated with respiratory viral pathogens.
In conclusion, our study provided background information concerning the respiratory viral etiology in Najran, southwestern Saudi Arabia. The RSV, hRV and AdV were the most frequent pathogens, accounting for over than two-thirds of cases with ARTI's. However, other emerging viruses as hMPV, hCoV NL63 and hCoV OC43 seem to play an important role in pediatric LRTI's. Moreover, the use of multiplex RT-PCR is needed to provide not only epidemiological and virological data, but also the opportunity to understand the emergence of novel respiratory viral pathogens ahead of time.
Footnotes
Disclosure.
References
- 1.Bellos A, Mulholland K, O’Brien KL, Qazi SA, Gayer M, Checchi F. The burden of acute respiratory infections in crisis-affected populations: a systematic review. Confl Health. 2010;4:3. doi: 10.1186/1752-1505-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.der Zalm MM, van Ewijk BE, Wilbrink B, Uiterwaal CS, Wolfs TF, van der Ent CK. Respiratory pathogens in children with and without respiratory symptoms. J Pediatr. 2009;154:396–400. doi: 10.1016/j.jpeds.2008.08.036. 400.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huijskens EG, Biesmans RC, Buiting AG, Obihara CC, Rossen JW. Diagnostic value of respiratory virus detection in symptomatic children using real-time PCR. Virol J. 2012;9:276. doi: 10.1186/1743-422X-9-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwofie TB, Anane YA, Nkrumah B, Annan A, Nguah SB, Owusu M. Respiratory viruses in children hospitalized for acute lower respiratory tract infection in Ghana. Virol J. 2012;9:78. doi: 10.1186/1743-422X-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bicer S, Giray T, Çöl D, Erdağ GÇ, Vitrinel A, Gürol Y, et al. Virological and clinical characterizations of respiratory infections in hospitalized children. Ital J Pediatr. 2013;39:22. doi: 10.1186/1824-7288-39-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sloots TP, Whiley DM, Lambert SB, Nissen MD. Emerging respiratory agents: new viruses for old diseases? J Clin Virol. 2008;42:233–243. doi: 10.1016/j.jcv.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albuquerque MC, Pena GP, Varella RB, Gallucci G, Erdman D, Santos N. Novel respiratory virus infections in children, Brazil. Emerg Infect Dis. 2009;15:806–808. doi: 10.3201/eid1505.081603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudan I, Boschi-Pinto C, Biloglav Z, Mulholland K, Campbell H. Epidemiology and etiology of childhood pneumonia. Bull World Health Organ. 2008;86:408–416. doi: 10.2471/BLT.07.048769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fashner J, Ericson K, Werner S. Treatment of the common cold in children and adults. Am Fam Physician. 2012;86:153–159. [PubMed] [Google Scholar]
- 10.Advani S, Sengupta A, Forman M, Valsamakis A, Milstone AM. Detecting respiratory viruses in asymptomatic children. Pediatr Infect Dis J. 2012;31:1221–1226. doi: 10.1097/INF.0b013e318265a804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bibby DF, McElarney I, Breuer J, Clark DA. Comparative evaluation of the Seegene Seeplex RV15, and real-time PCR for respiratory virus detection. J Med Virol. 2011;83:1469–1475. doi: 10.1002/jmv.22125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Renois F, Talmud D, Huguenin A, Moutte L, Strady C, Cousson J, et al. Rapid detection of respiratory tract viral infections and coinfections in patients with influenza-like illnesses by use of reverse transcription-PCR DNA microarray systems. J Clin Microbiol. 2010;48:3836–3842. doi: 10.1128/JCM.00733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Culebras E, Betriu C, Vázquez-Cid E, López-Varela E, Rueda S, Picazo JJ. Detection and genotyping of human respiratory viruses in clinical specimens from children with acute respiratory tract infections. Rev Esp Quimioter. 2013;26:47–50. [PubMed] [Google Scholar]
- 14.Al Hajjar S, Al Thawadi S, Al Seraihi A, Al Muhsen S, Imambaccus H. Human metapneumovirus and human coronavirus infection and pathogenicity in Saudi children hospitalized with acute respiratory illness. Ann Saudi Med. 2011;31:523–527. doi: 10.4103/0256-4947.84633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdel-Moneim AS, Kamel MM, Al-Ghamdi AS, Al-Malky MI. Detection of bocavirus in children suffering from acute respiratory tract infections in Saudi Arabia. PLoS One. 2013;8:e55500. doi: 10.1371/journal.pone.0055500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Majhdi FN, Al-Jarallah A, Elaeed M, Latif A, Gissmann L, Amer HM. Prevalence of respiratory syncytial virus infection in Riyadh during the winter season 2007-2008 and different risk factors impact. Internat J Virol. 2009;5:154–163. [Google Scholar]
- 17.Geneva (CH): World Health Organization; 2000. World Health Organization. WHO integrated management of childhood illness hand book. [Google Scholar]
- 18.Vandepitte J, Verhaegen J, Engbaek K, Rohner P, Piot P, Heuck CC. 2nd ed. Geneva (CH): World Health Organization; 2003. Basic Laboratory Procedures in Clinical Microbiology. [Google Scholar]
- 19.Huang G, Yu D, Mao N, Zhu Z, Zhang H, Jiang Z, et al. Viral etiology of acute respiratory infection in Gansu Province, China 2011. PLoS One. 2013;8:e64254. doi: 10.1371/journal.pone.0064254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suryadevara M, Cummings E, Bonville CA, Bartholoma N, Riddell S, Kiska D, et al. Viral etiology of acute febrile respiratory illnesses in hospitalized children younger than 24 months. Clin Pediatr (Phila) 2011;50:513–517. doi: 10.1177/0009922810394834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasman H, Pachucki CT, Unal A, Nguyen D, Devlin T, Peeples ME, et al. Aetiology of influenza-like illness in adults includes parainfluenzavirus type 4. J Med Microbiol. 2009;58(Pt 4):408–413. doi: 10.1099/jmm.0.006098-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin ET, Kuypers J, Wald A, Englund JA. Multiple versus single virus respiratory infections: viral load and clinical disease severity in hospitalized children. Influenza Other Respir Viruses. 2012;6:71–77. doi: 10.1111/j.1750-2659.2011.00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JK, Jeon JS, Kim JW, Rheem I. Epidemiology of respiratory viral infection using multiplex rt-PCR in Cheonan, Korea (2006-2010) J Microbiol Biotechnol. 2013;23:267–273. doi: 10.4014/jmb.1212.12050. [DOI] [PubMed] [Google Scholar]
- 24.Greenberg SB. Update on rhinovirus and coronavirus infections. Semin Respir Crit Care Med. 2011;32:433–446. doi: 10.1055/s-0031-1283283. [DOI] [PubMed] [Google Scholar]
- 25.Jansen RR, Wieringa J, Koekkoek SM, Visser CE, Pajkrt D, Molenkamp R, et al. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J Clin Microbiol. 2011;49:2631–2636. doi: 10.1128/JCM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.García-García ML, Calvo C, Falcón A, Pozo F, Pérez-Breña P, De Cea JM, et al. Role of emerging respiratory viruses in children with severe acute wheezing. Pediatr Pulmonol. 2010;45:585–591. doi: 10.1002/ppul.21225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tregoning JS, Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev. 2010;23:74–98. doi: 10.1128/CMR.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panda S, Mohakud NK, Pena L, Kumar S. Human metapneumovirus: review of an important respiratory pathogen. Int J Infect Dis. 2014;25:45–52. doi: 10.1016/j.ijid.2014.03.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 30.Brookline (MA): ProMed, International Society for Infectious Diseases; 2013. ProMED. MERS-CoV-Eastern Mediterranean (06) [Google Scholar]


