Abstract
Leukemia stem cells (LSCs) are found in most aggressive myeloid diseases and contribute to therapeutic resistance. Leukemia cells exhibit a dysregulated developmental program as the result of genetic and epigenetic alterations. Overexpression of the RNA-binding protein Musashi2 (MSI2) has been previously shown to predict poor survival in leukemia. Here, we demonstrated that conditional deletion of Msi2 in the hematopoietic compartment results in delayed leukemogenesis, reduced disease burden, and a loss of LSC function in a murine leukemia model. Gene expression profiling of these Msi2-deficient animals revealed a loss of the hematopoietic/leukemic stem cell self-renewal program and an increase in the differentiation program. In acute myeloid leukemia patients, the presence of a gene signature that was similar to that observed in Msi2-deficent murine LSCs correlated with improved survival. We determined that MSI2 directly maintains the mixed-lineage leukemia (MLL) self-renewal program by interacting with and retaining efficient translation of Hoxa9, Myc, and Ikzf2 mRNAs. Moreover, depletion of MLL target Ikzf2 in LSCs reduced colony formation, decreased proliferation, and increased apoptosis. Our data provide evidence that MSI2 controls efficient translation of the oncogenic LSC self-renewal program and suggest MSI2 as a potential therapeutic target for myeloid leukemia.
Introduction
A dysregulated developmental epigenetic program is considered to be a general feature of many cancers, and mutations or chromosomal translocations with histone methyltransferases contribute to transformation in myeloid leukemias (1, 2). One of the most aggressive subtypes of acute myeloid leukemia (AML) is characterized by the presence of a mixed-lineage leukemia gene (MLL) translocation and is associated with a poor response to therapy and a worse overall survival (3, 4). Translocations that include MLL have a wide spectrum of more than 60 fusion partners (5). The most common of these fusion partners is the MLL-AF9 translocation t(9;11)(p22;q23). The MLL gene encodes for a protein that methylates lysine 4 on histone 3 (H3K4), and this activity is usually inhibited or altered in leukemia (6, 7). MLL belongs to a family that includes other epigenetic regulators that, through methylation, acetylation, and other modifications, establish a developmental epigenetic program. Thus, translocations or mutations in these epigenetic regulators can contribute to leukemia, resulting in enhanced self-renewal and a block in differentiation (4, 6).
Several studies have implicated the Musashi (MSI) family of RNA-binding proteins as translation regulators contributing to a variety of cancers (8–10). Genetic and functional studies show that expression of MSI1 is upregulated in pediatric brain tumors and metastatic breast cancer and knockdown of MSI1 reduces the proliferation of different solid tumor cell lines (8–10). Other genetic alterations resulting in MSI2 overexpression are observed in blast crisis chronic myelogenous leukemia (CML-BC), which harbors chromosomal translocations that fuse MSI2 with HOXA9 and, most recently, MSI2 with TTC40 (11, 12). MSI2 expression levels can predict survival in B cell acute lymphoid leukemia (B-ALL) (13), and patients with higher MSI2 expression in CML-BC have been shown to have worse prognosis (14, 15). These findings suggest that increased expression of MSI2 may predict aggressiveness in a variety of cancers. In summary, MSI2 expression was found to be an independent adverse prognostic marker in AML and is a critical regulator of HSC maintenance, which contributes to the pathogenesis of leukemia when dysregulated (14–19).
It is widely accepted that the disruption of genetic and epigenetic mechanisms alters signaling networks that mediate cancer progression, and these types of alterations are commonly studied at the level of transcriptional regulation. It remains to be established how processes affecting post-transcriptional and translational regulation influence leukemia development (20, 21). The wide discrepancy between the transcriptome and the proteome highlights the importance of such regulation. RNA-binding proteins are involved in the post-transcriptional and cotranslational regulation of mRNAs. In the hematopoietic system mutations in proteins involved in RNA metabolism, including DKC1, RPS19, or splicing regulators, have been associated with congenital hematologic diseases and myeloid malignancies (22–24). Interestingly, the translation machinery itself can become altered through oncogenic signaling as a result of overexpression of MYC, ERK, or mutational activation of the PI3K/AKT pathway (25–28). Furthermore, mTOR pathway activation in cancer results in altered translation that can be clinically targeted with the addition of inhibitors such as rapalogs (25). Nevertheless, the specific contribution of translational regulation to leukemia progression and its role in leukemia stem cell (LSC) function remain poorly defined. Moreover, the link between RNA-binding proteins and epigenetic controllers is not known. Our study uncovers the requirement of the MSI2 RNA-binding protein for maintenance of the self-renewal program in myeloid leukemia.
Results
MSI2 direct mRNA targets are enriched for MLL-regulated genes.
We have previously demonstrated that MSI2 is highly expressed in patients with a worse clinical prognosis in CML-BC and in AML (15). Additionally, shRNA depletion of MSI2 in human AML cell lines expressing the BCR-ABL or MLL-AF9 translocations resulted in reduced proliferation and increased differentiation (15). Although these studies suggested that MSI2 is important for leukemic maintenance in vitro, its role in leukemic transformation and stem cell function remains unknown.
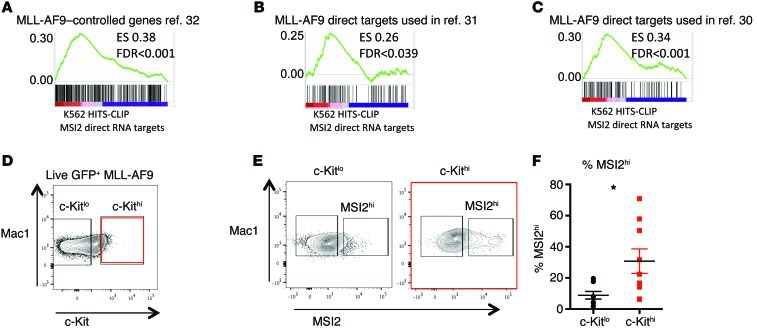
We previously mapped the direct MSI2 RNA binding targets by high-throughput sequencing and cross-linking immunoprecipitation (HITS-CLIP) using a FLAG-tagged version of MSI2 or a control vector in K562 (CML-BC) cells (16). As we reported, MSI2 binds to cellular targets that are associated with a variety of cellular pathways, including metabolism, mRNA regulation, and MYC. Among these pathways, we identified that MSI2’s direct RNA targets were associated with the MLL-AF9 self-renewal signature that is shared between leukemic granulocyte monocyte progenitors (GMPs) and HSCs (29). Therefore, we overlapped the rank list (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI78440DS1) of MSI2’s HITS-CLIP targets with additional gene sets associated with MLL-regulated genes (Figure 1, A–C, and refs. 30–32). These data suggested that the MSI2 direct mRNA targets were enriched in MLL-AF9 transcriptional target genes that have been previously defined (30–32).
Figure 1. MSI2-bound mRNAs are enriched for experimentally derived MLL transcriptional target genes.
(A) GSEA plot shows MSI2 rank list of HITS-CLIP targets overlapping with MLL-AF9–controlled genes (FLAG-MSI2–bound mRNAs overlapped with indicated gene sets as performed in ref. 16). (B) GSEA plot with MLL-AF9 direct transcriptional chromatin immunoprecipitated targets, same as A. (C) GSEA of MLL-AF4 targets overlapped with HITS-CLIP targets same as in A. (D) MLL-AF9 primary leukemia cells were stained with indicated antibodies and gated. (E) Representative flow cytometric plot showing MSI2 intracellular staining in gating of c-Kithi and c-Kitlo cells. (F) Frequency of MSI2hi cells in c-Kithi and c-Kitlo population. Experiments (D–F) include samples from 2 independent transplants. n = 9 mice. Means and SEM, *P < 0.05., unpaired Student’s t test.
These results prompted us to test whether MSI2 protein was expressed within the murine LSC-enriched compartment in MLL-AF9 transformed leukemia cells. In this model, the c-Kithi cells represent a population enriched for LSC activity (33). We identified a threefold increase in the frequency of cells highly expressing MSI2 in the c-Kithi population compared with the c-Kitlo MLL-AF9 transformed leukemia cells (Figure 1, D–F).
Msi2 delays leukemia initiation and progression.
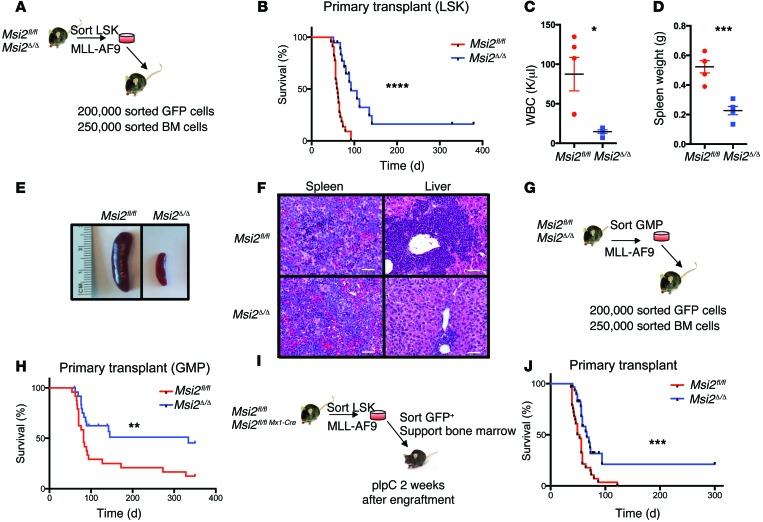
The elevated levels of MSI2 within the LSC population led us to investigate the requirement for Msi2 in leukemic initiation and transformation. To establish a model to study Msi2 and its contribution to myeloid leukemia, we have used the Msi2 conditional knockout mice that we previously crossed (Msi2fl/fl) into an Mx1-Cre background to generate the Msi2Δ/Δ allele after treatment with polyinositol-polycytosine (pIpC) allowing for conditional deletion within the hematopoietic compartment (16). In order to test whether Msi2 is critical for MLL-AF9–mediated initiation, we transduced control Msi2fl/fl and Msi2Δ/Δ Lin-Sca-1+c-Kit+ cells (LSKs) with MLL-AF9–expressing retroviruses coexpressing GFP (Figure 2A). GFP+ transformed LSK cells were sorted and serially replated for 5 rounds in a colony-forming assay. The deletion of Msi2 resulted in an approximately 50% reduction in colony formation, indicating that Msi2 quantitatively contributed to self-renewal in vitro (Supplemental Figure 1A).
Figure 2. Msi2 is required for efficient MLL leukemogenesis.
(A) Experimental scheme for LSK-derived MLL-AF9 initiation leukemia transplantation model. (B) Survival analysis of experiment scheme shown in A from indicated genotypes, n = 22 Msi2fl/fl and n = 19 Msi2Δ/Δ mice combined from 3 independent transplants. ****P < 0.0001, log-rank test. (C) White blood cell counts (WBCs) at 60 days post-transplantation as schemed in A were taken from n = 5 mice of each genotype. (D) Spleen weights at day 60 of indicated animals in C. Means and SEM, *P < 0.05, ***P < 0.005, unpaired Student’s t test. (E) Representative micrograph of indicated spleens from mice at 60 days post-transplantation. (F) H&E staining of spleen and liver sections from moribund leukemic mice (representative image from 3 independent animals). Original magnification, ×100. Scale bars: 20 μm. (G) Experimental scheme for GMP-derived MLL-AF9 leukemia transplantation model. (H) Survival analysis combined from 4 independent experiments with indicated genotypes, n = 24 Msi2fl/fl and n = 25 Msi2Δ/Δ, is shown. **P < 0.01, log-rank test. Msi2Δ/Δ mice identified as not deleted were censored from the analysis (Supplemental Table 2 and Supplemental Figure 2, G and I). (I) Experimental scheme for the Msi2 MLL-AF9 maintenance leukemia transplantation model. (J) Survival analysis of data combined from 3 independent maintenance transplants with indicated genotypes, n = 28 Msi2fl/fl and n = 29 Msi2Δ/Δ. ***P < 0.001, log-rank test. Unexcised Msi2 mice were censored from the survival curves (Supplemental Table 2 and Supplemental Figure 2, G and J).
To determine whether Msi2 was required for leukemic initiation in vivo, we injected MLL-AF9–transduced Msi2fl/fl and Msi2Δ/Δ LSK cells into lethally irradiated mice with support bone marrow cells. The mice that were injected with Msi2fl/fl leukemia cells began to develop signs of leukemia and were sacrificed because of a lethal myeloid disease with a median latency of 60 days post-transplantation (Figure 2B). In contrast, the majority of the mice injected with Msi2Δ/Δ leukemia cells lacked overt disease at 60 days with normal levels of white blood cells and dramatically reduced spleen size and weight compared with mice injected with Msi2fl/fl cells (Figure 2, C–E). Although the Msi2Δ/Δ leukemias arose after a delay of about 32 days, their disease burden was reduced compared with that of the Msi2fl/fl mice as assessed by their spleen and liver weights and a diminished leukemic infiltration (Figure 2F and Supplemental Figure 1, B and C).
As Msi2 is functionally required in LSCs transformed from LSKs, we wanted to determine whether it is also necessary in leukemias generated from GMPs. Therefore, Msi2fl/fl and Msi2Δ/Δ GMPs were sorted, transduced with MLL-AF9–expressing virus, and transplanted into lethally irradiated mice with support bone marrow (Figure 2G). Half of the Msi2Δ/Δ GMP transformed MLL-AF9 leukemias were delayed in their disease onset, and the other half remained disease free (Figure 2H). Therefore, Msi2 is also critical for maintaining leukemia that originated from committed myeloid progenitor cells known to express low levels of Msi2 compared with stem cell–enriched cells (15, 18, 19). The contribution of Msi2 to leukemia initiation led us to investigate the role of Msi2 in LSC maintenance. We transplanted Msi2fl/fl MLL-AF9 transformed LSK cells before pIpC excision, and after pIpC treatment we found a significant delay in leukemia progression with reduced disease burden in the spleen and liver (Figure 2, I and J, and Supplemental Figure 1, D and E).
Msi2 is required for leukemic stem cell survival.
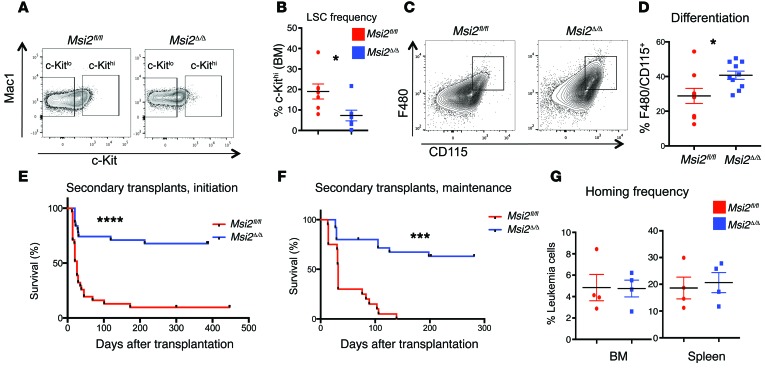
We found a significant reduction in the c-Kithi population in the Msi2Δ/Δ leukemias compared with the Msi2fl/fl (Figure 3, A and B). This corresponded to increased differentiation in the Msi2-deficient leukemia cells (Figure 3, C and D, and Supplemental Figure 2, A and B). However, we found no difference in cell cycle or increase in apoptosis (Supplemental Figure 2, C and D). The Msi2Δ/Δ leukemia cells formed approximately half the number of colonies compared with the controls, similar to the defect observed before transplantation (Supplemental Figure 2, E and F). We performed intracellular staining of MSI2 to determine the level of deletion in the initiation experiments from LSK-derived leukemia and found the deletion efficiency to be 62% (Supplemental Table 2 and Supplemental Figure 2, G and H). In the mice that succumbed to leukemia from the GMP-derived leukemias, we found a lower deletion rate of 37%, similar to the maintenance experiments (40%) (Supplemental Table 2 and Supplemental Figure 2, I and J). Despite the selection against deletion in some of the mice, these data suggested that leukemias could form without Msi2.
Figure 3. Msi2 maintains LSC function in MLL leukemia.
(A) Leukemic mice from Figure 2A analyzed for indicated surface markers and quantified in B. (B) LSC frequency from experiments gated in A, average from n = 7 Msi2fl/fl and n = 7 Msi2Δ/Δ. (C) Representative flow plot for differentiation markers F480 and CD115 in leukemic cells of mice from maintenance experiment. (D) Frequency of F480 and CD115 of leukemic cells from mice of indicated genotypes, n = 9 Msi2fl/fl and n = 10 Msi2Δ/Δ, from initiation LSK experiments. (E) Survival analysis of LSK-derived leukemia cells that were secondarily transplanted from the initiation experiment (Figure 2A) into sublethally irradiated mice, n = 31 Msi2fl/fl and n = 31 Msi2Δ/Δ, combined from transplant experiments in Supplemental Figure 3A. (F) Survival analysis of leukemia cells from the maintenance experiment (Figure 2J and one Cre-ER–expressing donor line of primary transplanted mice that were injected with tamoxifen), which were secondarily transplanted into sublethally irradiated mice, n = 20 Msi2fl/fl and n = 25 Msi2Δ/Δ, combined from individual transplants (Supplemental Figure 3). (G) The homing efficiency from the same donor cells assessed in E, measuring GFP+ cells in bone marrow (BM) or spleen at 18 hours post-transplantation. Experiments are from n = 4 Msi2fl/fl and n = 4 Msi2Δ/Δ. Means and SEM, *P < 0.05, unpaired Student’s t test for B and D. ***P < 0.001, ****P < 0.0001, log-rank test for E and F.
To determine the functional activity of the LSCs, we performed secondary transplantation experiments from both the initiation and maintenance derived primary leukemia. Msi2Δ/Δ leukemia cells from the initiation and maintenance experiments were predominantly incapable of engrafting efficiently in secondarily transplanted mice (7 of the 9 donors encompassing 7 independent transplants; Figure 3, E and F, and Supplemental Figure 3, A–G). Notably, we also observed a significant difference in survival in a secondary transplant from Msi2-deficient leukemias derived from Rosa-Cre-estrogen receptor–expressing (Cre-ER–expressing) animals that were injected with tamoxifen after engraftment in the primary recipients (Supplemental Figure 3G). Interestingly, homing of the leukemia cells to the bone marrow or spleen was not affected in the Msi2Δ/Δ compared with the control leukemia cells injected into mice (Figure 3G). Together these data suggested that Msi2 is required to maintain the LSC population. In the majority of the leukemias that formed in the secondary transplants, we were still able to detect efficient deletion (Supplemental Figure 3H).
MSI2 sustains MAPK signaling in LSCs.
Previous studies demonstrated that MSI2 depletion in human AML cell lines resulted in reduced MAPK signaling and overexpression of MSI2 in the LSKs increased pERK (15, 34). MAPK signaling is required for maintenance of HSCs (35) and preleukemic LSK self-renewal (36), and activating RAS results in a more aggressive leukemia (37). We used serum-starved leukemia cells taken from primary transplanted mice to test their ability to respond to growth factor receptor signaling by measuring their ERK activation after IL-3, IL-6, and SCF stimulation. We found that the Msi2Δ/Δ c-Kithi leukemia cells were able to respond, but at a significantly lower amplitude compared with the Msi2fl/fl c-Kithi leukemia cells (Supplemental Figure 4, A–C). In addition to ERK phosphorylation, we also found reduced phosphorylation of S6 in Msi2Δ/Δ c-Kithi cells (Supplemental Figure 4D). These results indicate that Msi2Δ/Δ LSCs have impaired signaling in MAPK and mTOR pathways in response to cytokine stimulation. To determine whether other growth factor signaling pathways were defective, we examined activation of JAK/STAT signaling by measuring phosphorylation of STAT5. In contrast to MAPK or S6, STAT5 was not reduced in the Msi2Δ/Δ LSCs, suggesting that Msi2 loss results in specific changes to MAPK output (Supplemental Figure 4E).
MSI2 controls self-renewal in MLL-AF9 leukemia.
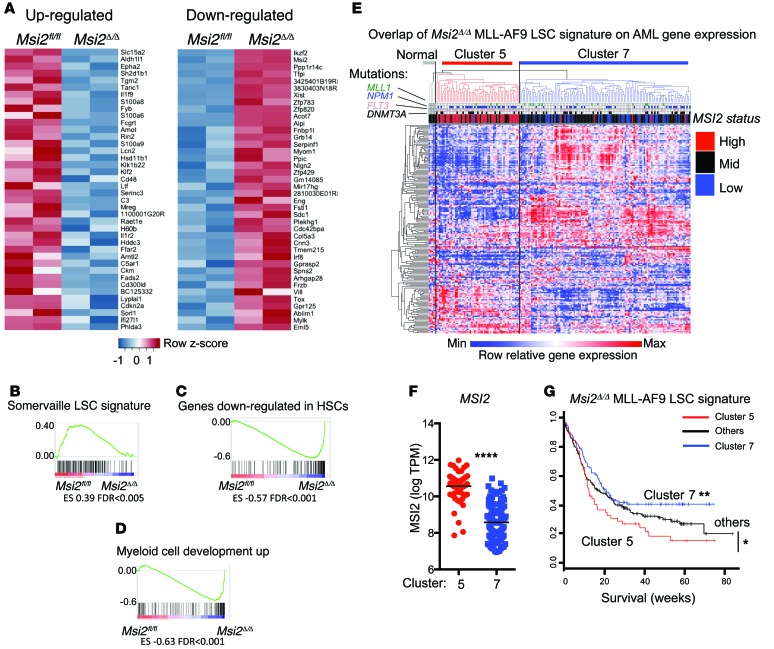
In order to further understand how Msi2 ablation results in reduced LSC function, we performed RNA sequencing on sorted c-Kithi leukemia cells from Msi2Δ/Δ and Msi2fl/fl mice. The Msi2Δ/Δ LSCs were significantly altered transcriptionally; we found 214 genes that were differentially expressed, of which 135 were downregulated and 79 were upregulated (log2 fold change > 0.75, false discovery rate [FDR] < 0.05; Supplemental Table 3). We generated a heat map to visualize the pronounced differences in the top 40 downregulated and upregulated genes (ranked by fold change in Figure 4A). We then functionally annotated our RNA sequencing analysis by performing gene set enrichment analysis (GSEA) (38) on all curated gene sets in the Molecular Signatures Database (http://www.broadinstitute.org/msigdb; 3,256 gene sets) combined with an additional set of relevant gene sets (92 gene sets from our experimentally derived or published hematopoietic self-renewal and differentiation signatures) (16, 38). We found 50 gene sets that were enriched for downregulated genes in Msi2Δ/Δ LSCs and 233 gene sets that were enriched in genes upregulated in Msi2Δ/Δ LSCs (Supplemental Tables 4 and 5). The top ranked gene sets in the Msi2Δ/Δ LSCs included enrichment in genes that are downregulated in LSCs (33) and HSCs (39) and genes upregulated during myeloid differentiation (ref. 40 and Figure 4, B–D). To determine whether the gene expression program in MLL-AF9 Msi2-deficient leukemias resembles that in patients with AML, we overlapped the RNA-seq gene signature corresponding to 177 genes from our LSCs in which Msi2 was deleted (human homologs and >6 mean log intensity; Supplemental Table 3) with the gene expression obtained from patients in the Eastern Cooperative Oncology Group E1900 trial (363 patients; GEO GSE24505) (41, 42). We found that an unsupervised clustering of the Msi2 signature was able to identify 9 distinct clusters in the AML patients (Supplemental Figure 5A). Patients with elevated MSI2 expression were grouped within cluster 5, while cluster 7 contained patients with reduced expression (Supplemental Figure 5 and Figure 4, E and F). These clusters also separated the patients with DNMT3A/FLT3-ITD/NPM1 mutations, suggesting that the Msi2 LSC signature could further distinguish this subtype of AML (Supplemental Figure 5, B and C). We found that patients within cluster 5 were statistically more likely to have FLT3-ITD and NPM1 mutations (Fisher’s exact test, FLT3-ITD or NPM1; P < 0.0001), but not DNMT3A (P = 0.08 or the triple mutant). The Msi2 LSC signature correlated with a poor clinical prognosis compared with the survival in the other clusters and in cluster 7 alone (Figure 4G). Overall, these data suggested that the MLL-AF9 Msi2 LSC gene signature distinguishes patients on the basis of their MSI2 expression and predicts their survival.
Figure 4. Msi2 maintains the HSC gene expression program and predicts survival in patients with AML.
(A) Gene expression heat map of the top 40 upregulated and downregulated genes from RNA sequencing analysis of n = 2 Msi2fl/fl and n = 2 Msi2Δ/Δ LSCs. (B–D) GSEA plot showing gene expression signature of (B) LSC-expressed genes downregulated in MLL-AF9 Msi2Δ/Δ, (C) HSC downregulated genes involved in self-renewal that are upregulated in MLL-AF9 Msi2Δ/Δ, and (D) genes upregulated in the MLL-AF9 Msi2Δ/Δ cells enriched in the gene set containing genes that are increased during myeloid development. (E) Msi2 differentially expressed genes (Msi2Δ/Δ MLL-AF9 LSC signature) corresponding to human homologs of >6 mean log intensity (177 genes) were overlapped with microarray data from AML patients obtained from the Eastern Cooperative Oncology Group E1900 trial, and unsupervised clustering was performed (363 patients; see Supplemental Figure 5 for all 9 clusters; cluster 5, n = 57, and cluster 7, n = 117, are shown). MSI2 expression was above 1 SD of the median in patients labeled as “High,” below in “Low,” and between in “Mid.” (F) Corresponding clusters from E were plotted for MSI2 expression. ****P < 0.0001, unpaired Student’s t test. (G) Overall survival analysis from patients in cluster 5, cluster 7, and others (based on E and Supplemental Figure 5). *P < 0.05, **P < 0.01, log-rank test.
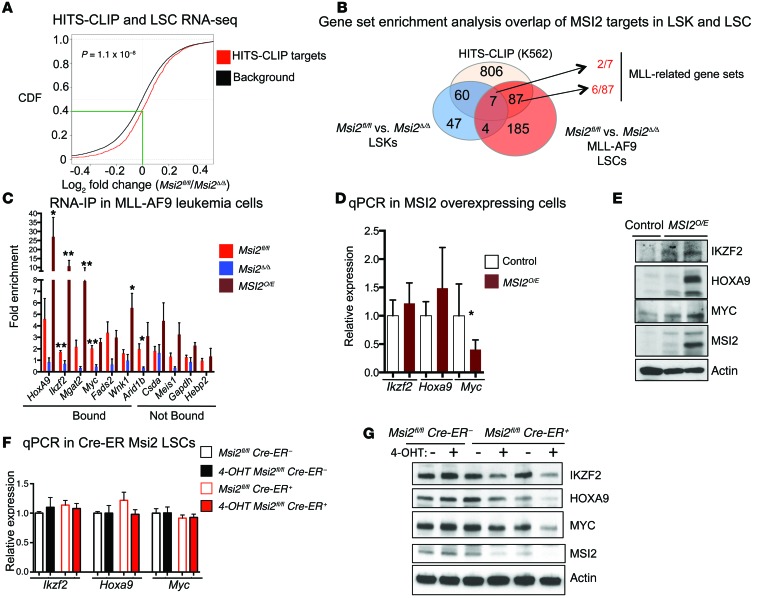
Our recent studies identified a relationship between the direct mRNA targets from the HITS-CLIP in K562 cells and the expression of these genes in hematopoietic stem and progenitor cells (16). Similarly, using a cumulative distribution function, we found a statistically significant difference in the mRNA level of HITS-CLIP targets compared with the background gene expression (genes expressed in MLL-AF9 leukemia cells that are not HITS-CLIP targets) in the Msi2Δ/Δ LSCs compared with the control (right shift) (P value = 1.1 × 10–8) (Figure 5A).
Figure 5. MSI2 binds and regulates the expression of Hoxa9, c-Myc, and Ikzf2.
(A) Cumulative distribution function (CDF) showing mRNA fold enrichment of HITS-CLIP targets in the RNA sequencing data of Msi2fl/fl and Msi2Δ/Δ LSCs, Kolmogorov–Smirnov test. (B) Functional pathway overlap analysis from HITS-CLIP and RNA sequencing data from Msi2fl/fl and Msi2Δ/Δ LSCs and microarray data from Msi2Δ/Δ LSKs. MLL-related gene sets shared among LSKs and LSCs (Supplemental Table 6). (C) RNA immunoprecipitation of MSI2 showing binding to selected MLL targets in Msi2fl/fl (n = 4–7 independent RNA-IPs and clones), Msi2Δ/Δ (n = 3 clones), and MSI2O/E (n = 4 clones; MLL-AF9 leukemia cells derived from LSK transformed cells from MSI2 doxycycline transgenic mice and induced for 2 days with doxycycline) (fold change calculated over IgG for each gene SEM). Means and SEM, *P < 0.05, **P < 0.01, unpaired Student’s t test. Hebp2 was not assessed. (D) Quantitative PCR for selected MLL target genes in control and MSI2O/E leukemic cells at 48 hours of MSI2 induction. Data are the mean of 3 independent experiments. Control, n = 3; MSI2O/E, n = 6. Means and SEM, *P < 0.05, unpaired Student’s t test. (E) Immunoblot analysis for MLL targets and MSI2 in control and MSI2O/E leukemic cells, from 2 independent experiments. (F) Quantitative PCR after acute deletion of Msi2 at 68 hours after 4-hydroxytestosterone (4-OHT) treatment in Msi2fl/fl Cre-ER– and Msi2fl/fl Cre-ER+ LSCs (n = 3 Msi2fl/fl Cre– and n = 4 Msi2fl/fl Cre+), average and SEM. (G) Immunoblot analysis for MLL targets and deletion of Msi2 at 68 hours after 4-OHT treatment in Msi2fl/fl Cre-ER– and Msi2fl/fl Cre-ER+ leukemic cells, representative of 2 independent experiments.
To further define MSI2’s role in LSCs, we tested the functional pathway overlap using the significantly enriched gene sets from the HITS-CLIP, microarray from normal LSKs deficient in Msi2 (16), and the RNA-seq in the LSCs (Figure 5B and Supplemental Table 5). Interestingly, in the Msi2-deficient LSCs only 3.8% (11/283) overlapped with normal LSK gene sets. However, of the overlapping gene sets, 7 gene sets were shared between MSI2’s direct targets, LSKs, and LSCs, and 2 of them were MLL-associated (Wang’s L-GMP/HOXA9-MEIS1 [ref. 43] and Krivtsov’s self-renewal [ref. 29]). Thirty-one percent (87/283) of the gene sets were found to be uniquely shared between LSCs and MSI2’s direct targets, and among these overlapping gene sets, 6 additional genes sets were also MLL-related (32), which includes gene sets from the self-renewal program, MLL-related subsets in human AML(44), HOXA9-MEIS1 targets genes (45) and genes enriched in LSCs (ref. 33, Figure 5B, and Supplemental Table 6). A possible explanation for a shared program among non-MLL transformed cells is that the endogenous MLL is required for both HSC function and leukemogenesis (46–49). Despite the lack of MLL oncogenes in K562, it has been demonstrated that c-MYB binds MLL through menin and brings it to the HOXA9 locus in CML-BC cell lines (50). Overall, these data further support the model that MSI2 binds to many of the downstream targets that are associated with the MLL self-renewal program found in both normal and leukemic cells.
To further support that MSI2 binds to the downstream targets within the MLL-controlled program, we performed RNA immunoprecipitation (IP) experiments in the murine MLL-AF9 transformed leukemia cells. To determine which MLL targets to test, we overlapped MLL direct target gene lists (30–32) with the top MSI2 HITS-CLIP targets (1,097 genes) that we previously identified (16), shown in Supplemental Tables 1 and 7, generating 137 genes (Supplemental Table 8). To prioritize the genes, we selected targets that (a) were demonstrated to be MLL transcriptional targets in 2 or more studies and overlapped as HITS-CLIP targets in K562 cells (Mgat2, Wnk1, Arid1b, and Csda); (b) were differentially expressed in the Msi2-deficient LSCs and MLL transcriptional targets (Ikzf2, most downregulated in RNA-seq; Figure 4A and Supplemental Figure 6; and Fads2 and Hebp2); or (c) were strongly associated with the MLL program (Hoxa9, Myc, and Meis1). We found enrichment for MSI2 binding to the mRNA transcripts Hoxa9, Ikzf2, Myc, Fads2, Wnk1a, Mgat2, and Arid1b (>1.5-fold over IgG control), but not Csda, Meis1, Hebp2, or Gapdh (Figure 5C). To further validate whether this binding is specific, we performed an RNA-IP experiment in the Msi2-deficient leukemia cells and found a statistically significant reduction of binding in Ikzf2, Myc, and Arid1b and also reduced binding in the other enriched targets (Hoxa9 and Fads2). We wanted to determine whether overexpression of MSI2 could result in enhanced binding. We then transformed LSKs from our previously characterized doxycycline-inducible MSI2 transgenic mouse model and developed a leukemia cell line (15). After 2 days of induction, we performed an additional RNA-IP and found statistically significant increased binding of Hoxa9, Ikzf2, Wnk1, and Mgat2. These data further suggest that MSI2 can bind to MLL-related targets in MLL-AF9 transformed leukemia cells.
On the basis of our RNA-IP results and previously published studies, we focused on Hoxa9, Ikzf2, and Myc. Myc has previously been demonstrated to be important in maintaining the self-renewal program in LSCs (32, 33, 45, 51–54). Ikzf2 encodes a transcription factor that regulates lymphocyte development by controlling quiescence and MAPK activation (55, 56). It has also been found to be frequently mutated, resulting in a loss of expression in hypodiploid B-ALL, and alternative isoforms were found in T cell ALL suggesting a dominant-negative function (57). HOXA9 is considered to be one of the most critical downstream targets of MLL-related oncogenes. Moreover, MSI2 was identified in chromosomal translocations with HOXA9 (11), and Msi2’s promoter could be bound by HOXA9 (14). To further determine whether MSI2 could directly alter translation, MSI2 overexpression resulted in an increase in IKZF2, MYC, and HOXA9 proteins without significantly increasing mRNA levels (Figure 5, D and E). Moreover, we detected equivalent mRNA stability of Ikzf2, Myc, and Hoxa9 after addition of actinomycin D to block transcription (Supplemental Figure 7A). These data support the concept that MSI2 expression increases protein translation of its direct targets.
To determine whether Msi2-deficient leukemias were reduced for MSI2 direct targets, we performed quantitative PCR on MLL-AF9 LSC-enriched cells and found a reduction in the mRNA levels for Ikzf2, Myc, and Hoxa9 in Msi2Δ/Δ cells compared with the controls (Supplemental Figure 7B). We then performed immunostaining for MYC in Msi2Δ/Δ LSCs and control Msi2fl/fl cells and found a reduction of MYC protein in the Msi2Δ/Δ LSCs (Supplemental Figure 7C). These data imply that MSI2 binding to Myc mRNA results in reduced expression in the LSCs. We also verified the reduced IKZF2 protein levels in the Msi2Δ/Δ LSCs compared with Msi2fl/fl LSCs by performing intracellular staining followed by flow cytometric analysis (Supplemental Figure 7D). Consistent with these results using single-cell approaches, we found reductions of MYC, HOXA9, and IKZF2 in Msi2-deficient leukemia cells by immunoblot analysis (Supplemental Figure 7E). A reduced mRNA and protein level of MYC, IKZF2, and HOXA9 in Msi2-deficient leukemia cells does not distinguish whether MSI2 binding alters translation or is a consequence of secondary effects. Therefore, to test the acute effect of Msi2 deletion, we used a Cre-ER–mediated deletion system. We examined the mRNA and protein levels in c-Kit–sorted LSCs and found reduced protein levels of IKZF2, MYC, and HOXA9 with equivalent mRNA expression 68 hours after tamoxifen-mediated deletion of Msi2 (Figure 5, F and G). Deletion of Msi2 and reduction of MYC and IKZF2 were also verified in intracellular staining in the c-Kithi cells compared with the c-Kitlo cells (Supplemental Figure 7, F–H). Taken together with the overexpression system, our data support a model where MSI2 maintains the translation of MLL downstream targets.
Ikzf2 maintains leukemia colony-forming ability.
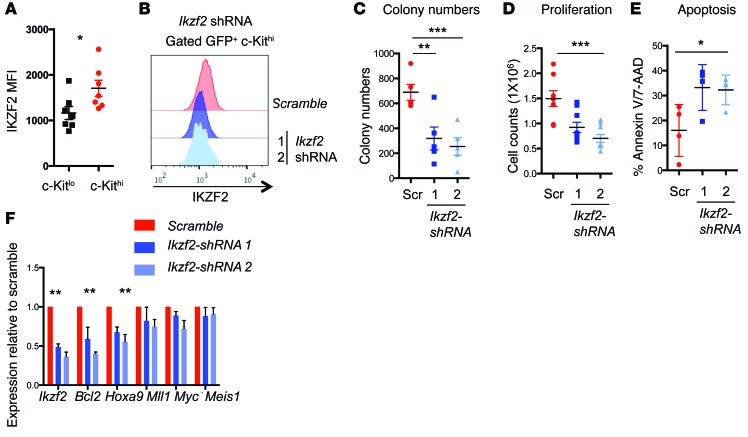
On the basis of our data that MSI2 directly binds to Ikzf2 mRNA and is starkly reduced in Msi2Δ/Δ LSCs, we hypothesized that it may be required for MLL-driven myeloid leukemia. Similar to MSI2, we found that IKZF2 was also detected at a higher abundance in the LSCs compared with the bulk cells (Figure 6A). To determine whether Ikzf2 contributes to leukemia survival, we performed shRNA-mediated depletion with 2 independent hairpins, which were validated for knockdown efficiency (Figure 6B). Leukemia cells with Ikzf2 depletion had a 60% reduction in colony formation, decreased proliferation, and increased apoptosis (Figure 6, C–E). Furthermore, forced reduction of Ikzf2 decreased Hoxa9 and Bcl2, but not Mll, Myc, or Meis1 (Figure 6F). In support of a role for IKZF2 in MLL-AF9 leukemia, we found that IKZF2 was reactivated and increased in secondary leukemias that had Msi2 deleted and that lacked a difference in survival (Supplemental Figure 8, A and B), compared with the leukemias that demonstrated a delay in their survival, which had reduced IKZF2 (Supplemental Figure 8C).
Figure 6. Ikzf2 contributes to the survival of myeloid leukemia cells.
(A) Median fluorescence intensity of IKZF2 in the c-Kithi and c-Kitlo MLL-AF9 leukemic cells (n = 7, combined from 2 independent experiments). (B) Representative flow cytometric plot for intracellular staining of IKZF2 showing knockdown in c-Kithi cells derived from Msi2fl/fl leukemia cells transduced with scramble and 2 Ikzf2 shRNAs (1 and 2) from 3 experiments. (C) Colony formation assay of MLL-AF9 leukemic cells transduced with scramble or Ikzf2 shRNAs, same as in B, plated in methylcellulose and counted after 5 days; n = 5. (D) Proliferation at 6 days post-transduction; n = 4. (E) Apoptosis at 6 days post-transduction; n = 4. (F) Quantitative PCR of MLL-related genes in MLL-AF9 leukemic cells transduced with scramble or Ikzf2 shRNAs. Results are means and SEM of n = 4 for scramble and Ikzf2 shRNA2 and n = 3 for Ikzf2 shRNA1. *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test for A to F.
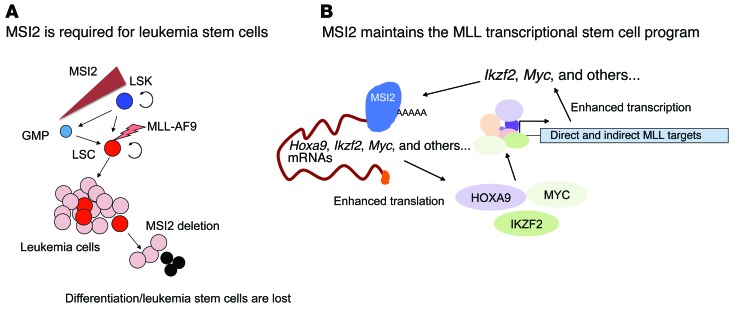
On the basis of these results, we demonstrate that, irrespective of the cell of origin, Msi2 is required for LSC function, as depletion results in loss of self-renewal and increased differentiation (Figure 7A). Additionally, we propose a working model where MSI2 binds and enhances translation of multiple MLL downstream target mRNAs including Hoxa9, Myc, and Ikzf2 and thus maintains a positive-feedback loop sustaining the MLL self-renewal program (Figure 7B).
Figure 7. MSI2 binds and regulates the MLL epigenetic self-renewal program.
(A) Schematic indicating the requirement for Msi2 in LSC function and reduced MSI2 levels in GMPs. Circular arrows indicate self-renewal in LSKs and LSCs. MLL-AF9 transduction results in transformation. (B) MSI2 binds to the mRNA transcripts associated with the MLL self-renewal program, including Hoxa9, Myc, and Ikzf2, and then enhances the protein expression of these genes. Increased HOXA9, MYC, and IKZF2 and other MLL-associated targets can control the transcription of the MLL self-renewal network, thus supporting a positive-feedback loop for sustaining LSC function.
Discussion
By combining our Msi2 conditional knockout model with gene expression and MSI2 target profiling, we have demonstrated that Msi2 is required for LSC maintenance through regulation of the MLL stem cell–associated program. In our global analysis using UV cross-linking and RNA immunoprecipitation to identify MSI2’s RNA targets, we have found that MSI2 binds to many targets associated with the MLL self-renewal program.
Using the MLL-AF9 model of AML, deletion of Msi2 resulted in a striking delay in leukemogenesis and decreased disease burden. Msi2 maintains the frequency and function of phenotypic LSCs. This was demonstrated by an increase in disease latency, reduced disease burden, and a strong selective advantage for unexcised Msi2 leukemias. The importance of understanding the cellular context of transformation was recently demonstrated by the obtaining of the expression signatures from LSK- versus GMP-derived leukemias, which could then predict the aggressiveness of human AML (58). On the basis of the low MSI2 expression in normal GMPs and a lack of a defect in Msi2-deficient GMPs, it was surprising to find that Msi2 was required for leukemias derived from GMPs (15, 18, 19). These data suggest a differential requirement for Msi2 in leukemic-GMPs compared with GMPs, and thus for maintaining LSCs derived from committed progenitors.
Microarray analysis indicated that MSI2 was not preferentially enriched in MLL-PTD (where PTD indicates partial tandem duplication) patients compared with other subgroups of AML (15). Despite these data, we report here that MSI2 is expressed in the MLL-related leukemia, as we have measured elevated expression of MSI2 within the LSC-enriched populations. Moreover, in our previous studies that included human AML cell lines NOMO-1 and THP-1 (both harbor MLL-AF9 translocations), we demonstrated that shRNA depletion of MSI2 resulted in reduced proliferation and increased differentiation (15). These data further suggest that measuring bulk expression in primary leukemias may not provide insight into the requirement for MSI2 within a particular patient. This point is strengthened by immunohistochemical examination of protein expression of MSI2 in AML patient bone marrow biopsies, in which demonstration of elevated MSI2 in as little as >1% of the cell population could still predict an unfavorable outcome (17). Therefore, it will be important to determine whether protein-based approaches to measure MSI2 combined with known stem cell markers can be used to predict survival or alter the therapeutic strategy in the various subtypes and genetic alterations that characterize AML. Additionally, our studies and others have previously demonstrated that MSI2 expression is elevated in some patients with AML and is enriched in the poor prognostic subset of patients who harbor FLT3-ITD, NPM1, and DNMT3A mutations (15, 59). The Msi2-deficient LSC signature could identify this cohort of patients, but these mutations did not define the Msi2 signature.
In our study we identify that MSI2 maintains the translation of MLL direct and indirect targets including Ikzf2, Myc, and Hoxa9. In previous studies it has been suggested that the MSI family of RNA-binding proteins can also act as translational repressors by binding to its consensus sequence and blocking the interaction of the poly(A) binding protein with eIF4E/G (60). Other studies have suggested that MSI proteins can alter polyadenylation in Xenopus laevis oocytes, resulting in the increase in translation (61). In line with our findings that MSI can maintain translation, a recent study in embryonic cortical neural stem cells (E12.5) found that although small changes occurred at the mRNA level there were profound effects on several genes in their ribosome occupancy (62). There were genes that demonstrated increased translation, and other genes had decreased translation, confirming a dual role for MSI2 function. Presumably the cellular context of other RNA-binding proteins may dictate whether MSI binding results in translation repression or activation.
Targeting the dysregulated epigenetic program in myeloid leukemia has been recently highlighted as an attractive strategy for therapeutic intervention in AML. For example, inhibitors of IDH2, DOT1L, KDM1A, and BET domain–containing proteins have demonstrated efficacy of clinical targeting of the epigenetic network (31, 32, 63–66). Additionally, NF-κB was recently linked to the MYC and MLL epigenetic network (52). Integrin and Syk signaling has also recently been associated with maintaining the undifferentiated program in MLL-AF9 leukemia (67, 68). The roles for RNA-binding proteins in myeloid leukemias are less understood. Besides the identification of an MLL/MYC/LIN28 axis that blocks miR150 in MLL leukemia (69), it is unclear whether RNA-binding proteins can control LSC function. It remains unclear how all these various overlapping factors contribute to the MLL self-renewal program. Moreover, our data demonstrate that MSI2 directly controls critical transcription factors and epigenetic modulators such as HOXA9, IKZF2, and MYC. The LSC self-renewal program encompasses a Myc/E2F (70) target gene signature that also depends on Jmjd1c (71) and Myb (45) and other genes including Nfkb and Brd4 (52, 72). It is difficult to dissociate all of these pathways, as they integrate many of the same signals to maintain an undifferentiated state.
We found that MSI2 HITS-CLIP targets overlapped with MLL direct and indirect transcriptional targets associated with MLL-AF9 LSCs. We validated these findings by demonstrating that MSI2 binds to one of the most downregulated genes in Msi2-deficient LSCs, Ikzf2, which has been shown to be commonly mutated in pediatric B-ALLs and is also a direct target of MLL fusions (30, 55). In contrast to its potential role as a tumor suppressor in pediatric B-ALL, it is normally expressed in common lymphoid progenitors, and we found it to be expressed in MLL-AF9 transformed LSCs. In pediatric B-ALLs, patients with deletions in IKZF2 had increased phosphorylation of ERK and S6, suggesting its role as a negative regulator of MAPK signaling (55). We found that loss of Msi2 resulted in decreased MAPK output in LSCs with reduced IKZF2 expression at the mRNA and protein level. It is unclear whether the reduced MAPK in the Msi2-deleted LSCs is a result of the loss of Ikzf2. However, we did observe a reduction in Bcl2 and Hoxa9 mRNA, suggesting that IKZF2 could be directly or indirectly involved in maintaining the survival of the undifferentiated LSCs by affecting these genes. Additionally, shRNA-mediated depletion of Ikzf2 resulted in reduced colony formation, suggesting that IKZF2 acts as a positive regulator of MLL leukemia cell growth and survival. One potential explanation for these results is that the loss of Msi2 results in a failure to maintain the normal feedback mechanisms in MAPK signaling and thus results in the loss of self-renewal. Nevertheless, these data suggest opposing roles for IKZF2 in B-ALL compared with myeloid leukemia. Future studies will be needed to dissect its role in human AML.
Although clinically targeting RNA-binding proteins remains challenging, our study provides a platform to identify inhibitors that potentially sensitize cells to Msi2 deletion. Moreover, this model supports the requirement for MSI2 in maintaining myeloid LSCs. In summary, our experiments provide evidence that the MSI2 RNA-binding protein can directly alter the MLL-associated self-renewal program.
Methods
Generation of MLL-AF9 leukemia and bone marrow transplants.
Bone marrow cells from 6- to 10-week-old Msi2fl/fl or Msi2Δ/Δ mice (16) in the C57BL/6 strain were processed for c-Kit enrichment by incubation with 20 μl of MACS CD117/c-Kit beads per mouse and then run on an AutoMACs (Miltenyi Biotec) following the manufacturer’s instructions. Cells were stained with Lineage antibody cocktail (CD3, CD4, CD8, Gr1, B220, CD19, TER119 conjugated with PeCy5), Sca-Pac Blue, CD34-FITC, SLAM-APC, CD48-PE, and c-KIT-APC-Cy7. Lin-Sca+Kit+ cells were sorted using a BD FACS Aria II instrument. Sorted LSKs were grown overnight in SFEM medium with 10 ng/ml IL-3, 10 ng/ml IL-6, 50 ng/ml SCF, 10 ng/ml thrombopoietin (TPO), and 20 ng/ml FLT3L. Cells were spinoculated twice with supernatant containing virus expressing MLL-AF9 together with GFP construct (a gift from Scott Armstrong, Memorial Sloan Kettering Cancer Center) on retronectin-coated plates. The cells were expanded for a week in M3434 methylcellulose medium (Stem Cell Technologies) and sorted for GFP+ cells. Two hundred thousand GFP+ cells were injected retro-orbitally with 250,000 bone marrow support cells into lethally irradiated 6-week-old C57BL/6 mice. For secondary transplants, 1 million fresh bone marrow leukemic cells were injected retro-orbitally into sublethally irradiated 6-week-old C57BL/6 mice.
Proliferation assay.
Mouse leukemic bone marrow cells were infected with virus expressing scramble and hairpins for Ikzf2 by spinfection of cells in RPMI medium containing 10% FBS, 30 ng/ml IL-3, 50 ng/ml SCF, and 500 ng/ml IL-6 together with respective viral supernatant. Cells were incubated overnight, and after 48 hours of infection, cells were treated with 5 μg/ml puromycin. Two days after puromycin selection, cells were plated at 250,000 cells/ml for proliferation assay. Cells were counted 2 days after plating (6 days after infection). All cytokines were purchased from PeproTech, and puromycin was manufactured by Sigma-Aldrich.
Colony assay.
Sorted LSKs were transduced with MLL-AF9–expressing virus and resorted for GFP, and then 10,000 cells were serially plated on M3434 methylcellulose medium. Colonies were counted every 5 days. For bone marrow cells from leukemic mice, 1,000 cells were used for colony assays.
Flow cytometry.
Bone marrow leukemic cells were stained for Mac1-PB, Gr1-PE, and c-Kit-PE-Cy7 and analyzed on a BD FACS LSR Fortessa instrument. For intracellular staining, cells were fixed with 1.5% paraformaldehyde for 15 minutes at room temperature and permeabilized with ice-cold methanol. Cells were washed 3 times with PBS and incubated with MSI2 (ab76148; Abcam) or IKZF2 antibody Pacific Blue (137220; BioLegend) in 2% FBS in PBS for 1 hour at room temperature. For MSI2 staining, cells were washed twice and incubated with secondary antibody conjugated with Alexa Fluor 647 (Molecular Probes).
For measuring apoptosis, cells were washed with PBS and incubated with anti–annexin V-PE (BD Biosciences) in the annexin V binding buffer (10 mM HEPES, pH 7.4, 140 mM NaCl, 4 mM KCl, 0.75 mM MgCl2, 1 mM CaCl2) together with 2 μl of 7-AAD in the reaction volume of 100 μl for 15 minutes as recommended by the manufacturer. The sample was measured using a BD Fortessa instrument.
Immunoblot analysis.
c-Kithi Msi2fl/fl Cre-ER– and Msi2fl/fl Cre-ER+ leukemic cells were sorted and were left untreated or treated with 600 nM 4-OH Tamoxifen (Sigma-Aldrich) for 68 hours in medium containing SCF (10 ng/ml), IL-3 (10 ng/ml), and IL-6 (10 ng/ml). One hundred thousand cells were counted and washed once with PBS and lysed in Laemmli sample buffer. Control or MSI2O/E cells were induced with 0.5 μg/ml of doxycycline for 48 hours. Lysates were run on 4%–15% gradient SDS-PAGE and transferred to nitrocellulose membrane. Membranes were blotted for IKZF2 (sc-9864; Santa Cruz), HOXA9 (07-178; Millipore), MYC (5605S; Cell Signaling), MSI2 (ab76148; Abcam), and actin (A3854; Sigma-Aldrich).
RNA immunoprecipitation.
Thirty million Msi2fl/fl, Msi2Δ/Δ, or MSI2O/E leukemia cells were used for RNA-IP using the Magna RIP RNA-binding protein immunoprecipitation kit (03-115; Millipore). First, cells were washed with cold PBS and then lysed according to the manufacturer’s directions. Anti-rabbit antibody or anti-MSI2 antibody (part of the kit 03-115; Millipore) incubated with magnetic beads was used to immunoprecipitate endogenous MSI2. After washing of the immunoprecipitated complexes, they were then treated with proteinase K. The phenol/chloroform method was used for RNA extraction, and the RNA was converted to cDNA using the Verso cDNA kit (Thermo Scientific). Quantitative PCR was used for validating candidates bound by MSI2.
Quantitative RT-PCR.
Total RNA from sorted c-Kithi MLL-AF9 leukemic bone marrow cells was isolated using TRIzol (Life Technologies) and RNeasy RNA extraction kit (Qiagen). RNA was reverse transcribed into cDNA with iSCRIPT (BioRad), and quantitative PCR was performed with primers for Bcl2 (TaqMan probe; ABI), Meis1 (forward: GCCATACAAGTGTTAAGGTTTCATC; reverse: CCTCCTTCTCTATCATCTATCACAA), Ikzf2 (forward: CATCACTCTGCATTTCCAGC; reverse: TGACCTCACCTCAAGCACAC), Myc (forward: ACGGAGTCGTAGTCGAGGTC; reverse: AGAGCTCCTCGAGCTGTTTG), and Hoxa9 (forward: GTAAGGGCATCGCTTCTTCC; reverse: ACAATGCCGAGAATGAGAGC).
RNA sequencing and GSEA.
Sorted c-Kithi cells from Msi2fl/fl mice (n = 2) and Msi2Δ/Δ mice (n = 2) were processed for RNA extraction using TRIzol and RNeasy RNA extraction kit. Samples were prepared with a standard Illumina kit using the TruSeq RNA SamplePrep Guide (Illumina). mRNA fragments with a length of 200–300 bp were selected for library construction. Sequencing was performed on a HiSeq 2000 platform using a standard paired-end protocol. Paired-end RNA-seq reads were first processed with Trimmomatic (73) to remove TruSeq adaptor sequences and bases with quality scores below 20, and reads with less than 30 remaining bases were discarded. Trimmed reads were then aligned to mm9 genome with the STAR spliced-read aligner (74). For each gene from the RefSeq annotations, the number of uniquely mapped reads overlapping with the exons was counted with HTSeq (http://www-huber.embl.de/users/anders/HTSeq/). Genes with fewer than 200 read counts were considered as not expressed and filtered out. For the rest of the genes, we used DESeq (75) to evaluate the differences in read counts between 2 Msi2Δ/Δ and 2 Msi2fl/fl mouse samples, which were represented by log2 fold-change and Benjamini-Hochberg corrected P values. These RNA-seq data have been deposited at the NCBI Gene Expression Omnibus (GSE64545). The rank list of log2 (Msi2fl/fl/Msi2Δ/Δ) was generated and GSEA was performed against the Molecular Signatures Database (http://www.broadinstitute.org/msigdb) combined with an additional set of relevant gene sets (92 gene sets from our experimentally derived or published hematopoietic self-renewal and differentiation signatures) (16, 38). GSEAs for the HITS-CLIP were selected based on the following criteria: (FDR < 0.01, P < 0.01) Msi2Δ/Δ negative; upregulated genes and positive enrichment; downregulated genes (FDR < 0.3 and P < 0.01). Microarray and mutation data obtained from (363 patients), (41) were normalized with raw read counts and converted to log TPM (tags per million). All original microarray data were deposited in the NCBI’s Gene Expression Omnibus (GEO GSE62115). The LSC gene signature was converted to human homologs (177 genes with >6 mean log intensity), which were filtered and row extracted. The genes from 363 patients underwent unsupervised clustering with the following parameters: agglomerative with a column and row distance metric of 1 minus Pearson correlation and an average linkage method with the default parameters in GeneE software from the Broad Institute. The 9 clusters were manually defined based on apparent breaks in the tree hierarchy, and clusters 5 and 7 were then further selected based on the largest clusters with the most differential expression of MSI2.
Statistics.
The 2-tailed Student’s t test was used for significance testing in the bar graphs, except where stated otherwise. P values less than 0.5 were considered significant. Error bars reflect the SEM, except where stated otherwise. Survival probabilities were estimated using the Kaplan-Meier method and compared with the log-rank test. All statistical analyses were carried out using GraphPad Prism 4.0 and the R statistical environment.
Study approval.
All animal procedures were approved by the Institutional Animal Care and Use Committee at Memorial Sloan Kettering Cancer Center. For patient sample data analysis, data from 363 AML samples were obtained from patients enrolled in the Eastern Cooperative Oncology Group’s E1900 clinical trial (41, 76). Institutional review board approval was obtained at Weill Cornell Medical College and Memorial Sloan Kettering Cancer Center.
Supplementary Material
Acknowledgments
We would like to thank Stephen Nimer, Scott Armstrong, and Neal Rosen for their critical advice and helpful suggestions. We would also like to thank Aly Azeem Khan, Agnes Viale, and the Memorial Sloan Kettering Cancer Center sequencing core for all their support. M.G. Kharas was supported by an NIH NIDDK Career Development Award (K01DK084261-01), NIDDK NIH R01-DK101989-01A1, a Louis V. Gerstner Young Investigator Award, an American Society of Hematology Junior Scholar Award, a Kimmel Scholar Award, and a V Scholar Award. L. Vu is supported by the Damon Runyon-Sohn Pediatric Cancer Fellowship Award. E. Paietta was supported by NIH grants U24-CA114737 and U10-CA180827. C.J. Lengner was supported by R01 CA16865 from the National Cancer Institute, and a fellowship from the W.W. Smith Charitable Trust. This clinical study was conducted by the ECOG-ACRIN Cancer Research Group (Robert L. Comis and Mitchell D. Schnall, group co-chairs) and supported in part by Public Health Service grants CA180820, CA180794, CA180791, CA189859, and CA180801, and by the National Cancer Institute, the NIH, and the Department of Health and Human Services. Its content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(3):1286–1298. doi:10.1172/JCI78440.
References
- 1.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339(6127):1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ezoe S. Secondary leukemia associated with the anti-cancer agent, etoposide, a topoisomerase II inhibitor. Int J Environ Res Public Health. 2012;9(7):2444–2453. doi: 10.3390/ijerph9072444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7(11):823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 5.Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009;23(8):1490–1499. doi: 10.1038/leu.2009.33. [DOI] [PubMed] [Google Scholar]
- 6.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10(10):500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell. 2004;6(3):437–443. doi: 10.1016/S1534-5807(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 8.Hemmati HD, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100(25):15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sureban SM, et al. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134(5):1448–1458. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 10.Oskarsson T, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nature. 2011;17(7):867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbouti A, et al. A novel gene, MSI2, encoding a putative RNA-binding protein is recurrently rearranged at disease progression of chronic myeloid leukemia and forms a fusion gene with HOXA9 as a result of the cryptic t(7;17)(p15;q23). Cancer Res. 2003;63(6):1202–1206. [PubMed] [Google Scholar]
- 12.Saleki R, et al. A novel TTC40-MSI2 fusion in de novo acute myeloid leukemia with an unbalanced 10;17 translocation. Leuk Lymphoma. doi: 10.3109/10428194.2014.947611. [published online ahead of print August 20, 2014]. doi: 10.3109/10428194.2014.947611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mu Q, et al. High expression of Musashi-2 indicates poor prognosis in adult B-cell acute lymphoblastic leukemia. Leuk Res. 2013;37(8):922–927. doi: 10.1016/j.leukres.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Ito T, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466(7307):765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kharas MG, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16(8):903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park S-M, et al. Musashi-2 controls cell fate, lineage bias, and TGF-β signaling in HSCs. J Exp Med. 2014;211(1):71–87. doi: 10.1084/jem.20130736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byers RJ, Currie T, Tholouli E, Rodig SJ, Kutok JL. MSI2 protein expression predicts unfavorable outcome in acute myeloid leukemia. Blood. 2011;118(10):2857–2867. doi: 10.1182/blood-2011-04-346767. [DOI] [PubMed] [Google Scholar]
- 18.de Andrés-Aguayo L, et al. Musashi 2 is a regulator of the HSC compartment identified by a retroviral insertion screen and knockout mice. Blood. 2011;118(3):554–564. doi: 10.1182/blood-2010-12-322081. [DOI] [PubMed] [Google Scholar]
- 19.Hope KJ, et al. An RNAi screen identifies Msi2 and Prox1 as having opposite roles in the regulation of hematopoietic stem cell activity. Cell Stem Cell. 2010;7(1):101–113. doi: 10.1016/j.stem.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3(3):179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 21.Kolle G, et al. Deep-transcriptome and ribonome sequencing redefines the molecular networks of pluripotency and the extracellular space in human embryonic stem cells. Genome Res. 2011;21(12):2014–2025. doi: 10.1101/gr.119321.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devlin EE, Dacosta L, Mohandas N, Elliott G, Bodine DM. A transgenic mouse model demonstrates a dominant negative effect of a point mutation in the RPS19 gene associated with Diamond-Blackfan anemia. Blood. 2010;116(15):2826–2835. doi: 10.1182/blood-2010-03-275776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Draptchinskaia N, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21(2):169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 24.Narla A, Ebert BL. Translational medicine: ribosomopathies. Blood. 2011;118(16):4300–4301. doi: 10.1182/blood-2011-08-372250. [DOI] [PubMed] [Google Scholar]
- 25.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485(7396):55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holland EC. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol Cell. 2003;12(4):889–901. doi: 10.1016/S1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- 27.Parsa AT, Holland EC. Cooperative translational control of gene expression by Ras and Akt in cancer. Trends Mol Med. 2004;10(12):607–613. doi: 10.1016/j.molmed.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10(4):254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 29.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 30.Guenther MG, et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22(24):3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernt KM, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20(1):66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuber J, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25(15):1628–1640. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Somervaille TCP, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10(4):257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, et al. Musashi2 modulates K562 leukemic cell proliferation and apoptosis involving the MAPK pathway. Exp Cell Res. 2014;320(1):119–127. doi: 10.1016/j.yexcr.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Chan G, Gu S, Neel BG. Erk1 and Erk2 are required for maintenance of hematopoietic stem cells and adult hematopoiesis. Blood. 2013;121(18):3594–3598. doi: 10.1182/blood-2012-12-476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, et al. Oncogenic Nras has bimodal effects on stem cells that sustainably increase competitiveness. Nature. 2013;504(7478):143–147. doi: 10.1038/nature12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuber J, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009;23(7):877–889. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaatinen T, et al. Global gene expression profile of human cord blood-derived CD133+ cells. Stem Cells. 2006;24(3):631–641. doi: 10.1634/stemcells.2005-0185. [DOI] [PubMed] [Google Scholar]
- 40.Brown AL, et al. Genetic regulators of myelopoiesis and leukemic signaling identified by gene profiling and linear modeling. J Leukoc Biol. 2006;80(2):433–447. doi: 10.1189/jlb.0206112. [DOI] [PubMed] [Google Scholar]
- 41.Patel JP, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, et al. The Wnt/β-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327(5973):1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mullighan CG, et al. Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia. 2007;21(9):2000–2009. doi: 10.1038/sj.leu.2404808. [DOI] [PubMed] [Google Scholar]
- 45.Hess JL, et al. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108(1):297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Artinger EL, Ernst P. Cell context in the control of self-renewal and proliferation regulated by MLL1. Cell Cycle. 2013;12(18):2969–2972. doi: 10.4161/cc.26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1(3):324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thiel AT, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17(2):148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Artinger EL, et al. An MLL-dependent network sustains hematopoiesis. Proc Natl Acad Sci U S A. 2013;110(29):12000–12005. doi: 10.1073/pnas.1301278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin S, Zhao H, Yi Y, Nakata Y, Kalota A, Gewirtz AM. c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. J Clin Invest. 2010;120(2):593–606. doi: 10.1172/JCI38030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.King B, et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 2013;153(7):1552–1566. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuo HP, et al. Epigenetic roles of MLL oncoproteins are dependent on NF-κB. Cancer Cell. 2013;24(4):423–437. doi: 10.1016/j.ccr.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi J, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27(24):2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo H, Li Q, O’Neal J, Kreisel F, Le Beau MM, Tomasson MH. c-Myc rapidly induces acute myeloid leukemia in mice without evidence of lymphoma-associated antiapoptotic mutations. Blood. 2005;106(7):2452–2461. doi: 10.1182/blood-2005-02-0734. [DOI] [PubMed] [Google Scholar]
- 55.Holmfeldt L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45(3):242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grzanka J, et al. FoxP3, Helios, and SATB1: roles and relationships in regulatory T cells. Int Immunopharmacol. 2013;16(3):343–347. doi: 10.1016/j.intimp.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 57.Fujimoto R, Ozawa T, Itoyama T, Sadamori N, Kurosawa N, Isobe M. HELIOS-BCL11B fusion gene involvement in a t(2;14)(q34;q32) in an adult T-cell leukemia patient. Cancer Genet. 2012;205(7):356–364. doi: 10.1016/j.cancergen.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 58.Krivtsov AV, et al. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia. 2013;27(4):852–860. doi: 10.1038/leu.2012.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thol F, et al. Prognostic significance of expression levels of stem cell regulators MSI2 and NUMB in acute myeloid leukemia. Ann Hematol. 2013;92(3):315–323. doi: 10.1007/s00277-012-1637-5. [DOI] [PubMed] [Google Scholar]
- 60.Kawahara H, Imai T, Imataka H, Tsujimoto M, Matsumoto K, Okano H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J Cell Biol. 2008;181(4):639–653. doi: 10.1083/jcb.200708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rutledge CE, et al. Efficient translation of Dnmt1 requires cytoplasmic polyadenylation and Musashi binding elements. PLoS One. 2014;9(2):e88385. doi: 10.1371/journal.pone.0088385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katz Y, et al. Musashi proteins are post-transcriptional regulators of the epithelial-luminal cell state. Elife. doi: 10.7554/eLife.03915. [published online ahead of print November 7, 2014]. doi: 10.7554/eLife.03915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Daigle SR, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20(1):53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daigle SR, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122(6):1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schenk T, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med. 2012;18(4):605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harris WJ, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21(4):473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 67.Miller PG, et al. In Vivo RNAi screening identifies a leukemia-specific dependence on integrin β3 signaling. Cancer Cell. 2013;24(1):45–58. doi: 10.1016/j.ccr.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hahn CK, et al. Proteomic and genetic approaches identify Syk as an AML target. Cancer Cell. 2009;16(4):281–294. doi: 10.1016/j.ccr.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiang X, et al. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22(4):524–535. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sroczynska P, et al. shRNA screening identifies JMJD1C as being required for leukemia maintenance. Blood. 2014;123(12):1870–1882. doi: 10.1182/blood-2013-08-522094. [DOI] [PubMed] [Google Scholar]
- 72.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fernandez HF, et al. Anthracycline dose intensification in acute myeloid leukemia. N Engl J Med. 2009;361(13):1249–1259. doi: 10.1056/NEJMoa0904544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.