Abstract
Aims
The mesenchymal cushions lining the early embryonic heart undergo complex remodelling to form the membranous ventricular septum as well as the atrioventricular and semilunar valves in later life. Disruption of this process underlies the most common congenital heart defects. Here, we identified a novel role for Slit-Robo signalling in the development of the murine membranous ventricular septum and cardiac valves.
Methods and results
Expression of Robo1 and Robo2 receptors and their ligands, Slit2 and Slit3, was present in or adjacent to all cardiac cushions/valves. Loss of Robo1 or both Robo1 and Robo2 resulted in membranous ventricular septum defects at birth, a defect also found in Slit3, but not in Slit2 mutants. Additionally, Robo1;Robo2 double mutants showed thickened immature semilunar and atrioventricular valves as well as highly penetrant bicuspid aortic valves. Slit2 mutants recapitulated the semilunar phenotype, whereas Slit3 mutants displayed thickened atrioventricular valves. Bicuspid aortic cushions were already observed at E12.5 in the Robo1;Robo2 double mutants. Expression of Notch- and downstream Hey and Hes genes was down-regulated in Robo1 mutants, suggesting that reduced Notch signalling in mice lacking Robo might underlie the defects. Luciferase assays confirmed regulation of Notch signalling by Robo.
Conclusion
Cardiac defects in mutants for Robo or Slit range from membranous ventricular septum defects to bicuspid aortic valves. These ligands and receptors have unique functions during development of specific cardiac cushion derivatives, and the Slit-Robo signalling pathway likely enforces its role by regulating Notch signalling, making these mutants a valuable new model to study cardiac valve formation.
Keywords: Heart development, Slit-Robo signalling pathway, Notch signalling pathway, Membranous ventricular septum defect, Bicuspid aortic valves
1. Introduction
Cardiac septation and valve formation are complex processes requiring precise gene regulation, and even the smallest disruption can lead to congenital defects such as ventricular septum defects or malformed and malfunctioning valves. The membranous interventricular septum and the atrioventricular and semilunar valves develop from primitive jelly-like cushions lining the early myocardial heart tube. The endothelial cells covering the cushions invade the underlying matrix by epithelial-to-mesenchymal transformation, where they proliferate and reshape the primitive cushions into relatively thick valve-like cellular structures by mid-gestation. Subsequently, the cushions remodel to form the thin mature leaflets of the atrioventricular and semilunar valves and the upper membranous part of the ventricular septum.1–3
In addition to the epithelial-to-mesenchymal transformation, parts of the cardiac cushions, including the future membranous ventricular septum, receive contributions from neural crest cells invading the arterial pole of the heart, adding to the complexity of the region.1 Correct formation and alignment of the cushions requires accurate signalling from within as well as from neighbouring tissues such as the myocardium, endocardium, second heart field, and neural crest. Major signalling pathways, such as Notch, Bmp, and Wnt, are crucial for cushion development and, in turn, regulate and are regulated by a plethora of transcription factors such as the Tbox and Sox families.1,2–5
Here, we describe a new candidate involved in membranous ventricular septum and valve formation, the Slit-Robo signalling pathway. The Roundabout (Robo) transmembrane receptors and their Slit ligands, initially identified in Drosophila,6,7 became known for their roles in axonal guidance in the embryonic nervous system. However, since this discovery, many new roles for the Slit-Robo signalling pathway have been identified, mainly in cancer and embryo development.8,9 In Drosophila and zebrafish, Slit-Robo signalling plays key roles in cell adhesion during cardiac cell polarization, morphogenesis, migration, and lumen formation.10–13 During murine heart development, roles for Slit-Robo signalling have now been described in cardiac chamber formation14 and cardiac neural crest migration and adhesion.15,16 Disrupted signalling results in partial absence of the pericardium and abnormal venous connections to the heart.16 However, knowledge of the pathway during mammalian heart development is limited. Its identified functions during neural crest migration and adhesion combined with the known expression patterns of Slit/Robo in the neural crest, outflow tract, and atrioventricular cushions14–16 suggest an additional role for this signalling pathway in the formation of these areas.
We have now identified a broad spectrum of cardiac defects in mutants for Robo and Slit, ranging from membranous ventricular septum defects to bicuspid aortic valves. We show that these ligands and receptors have unique functions during the development of the different cardiac cushion derivatives and that the Slit-Robo signalling pathway likely enforces its role by regulating Notch signalling. This is the first study indicating the involvement of this pathway in the development of membranous ventricular septum defects and bicuspid aortic valve disease, and the high penetrance of bicuspid aortic valves can make the Robo1/2 double mutant a valuable new tool to study the aetiology of this common human congenital disorder.
2. Methods
Transgenic mice and experimental procedures for in situ hybridization, immunohistochemistry, cell counts, three-dimensional reconstruction, volume and length measurements, qPCR, luciferase assays, and statistical analyses are provided in the Supplementary material online. All experimental procedures were performed in accordance with the UK Animals (Scientific Procedures) Act 1986 and institutional guidelines.
3. Results
3.1. Slit and Robo expression in the outflow tract and atrioventricular region
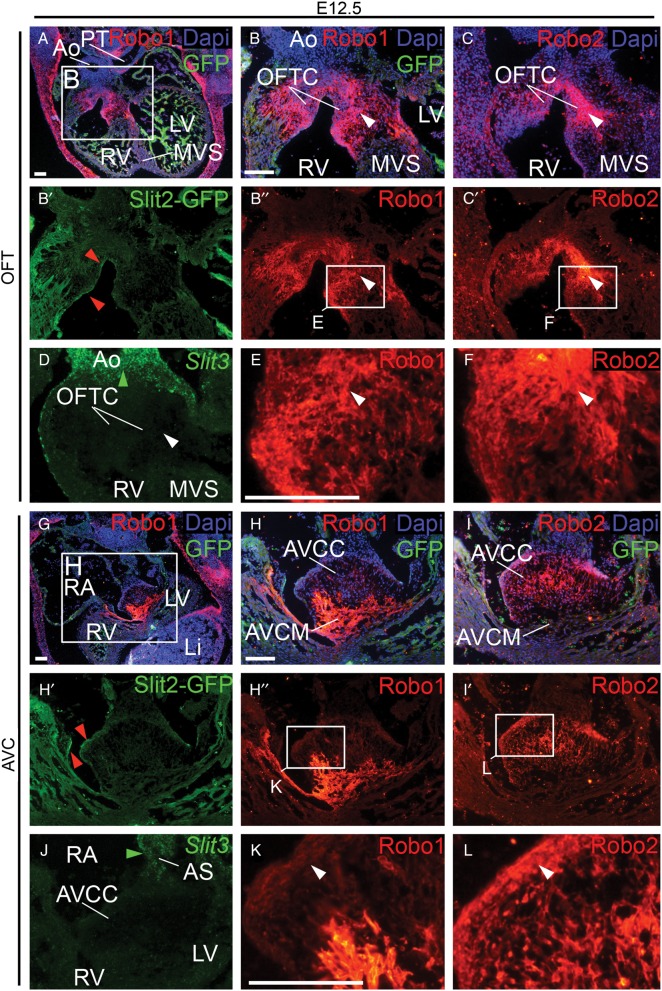
We have previously reported the overall expression patterns of Robo1, Robo2, Slit2 and Slit3 in the murine heart and migrating cardiac neural crest.16 The presence of these genes in and surrounding the cardiac cushions prompted us to study their expression in these regions in more detail. As we have previously shown that Robo3 is not expressed inside the heart and Robo4 only in the coronary circulation, we excluded these receptors from our study. Robo1 was expressed in the outflow tract and atrioventricular cushions, and subsequently valves, as well as in the atrioventricular canal myocardium throughout embryonic development (Figure 1A, B, E, G, H, and K; see Supplementary material online, Figure S1A–H),16 the only exception being its disappearance from specifically the aortic semilunar valves just before birth (data not shown). Robo2 was never observed in the myocardium but was present in both the outflow tract and atrioventricular cushions, and later valves, all through development (Figure 1C, F, I, and L; see Supplementary material online, Figure S1A–H). In the outflow tract around E12.5, expression of Robo2 was highest in the cushion area contributing to the separation of the aortic and pulmonary outflow within the heart (Figure 1C and F; see Supplementary material online, Figure S1C).
Figure 1.
Slit and Robo expression patterns in and surrounding the cardiac cushions. (A–L) immunohistochemistry (Robo1, Robo2, and Slit2-GFP), DAPI, and in situ hybridization (Slit3) staining at E12.5 in the outflow tract and atrioventricular cushion regions. Red arrowheads indicate the expression of Slit2 in the endocardium lining the cushions. (E, F, K, and L) show details of Robo1 and 2 expression in the indicated cushion regions. White arrowheads point to the region where both Robo1 and Robo2 are expressed. (D and J) Green arrowheads point to Slit3 expression in the outflow tract vessels and atrial septum, while Slit3 is not detectable in the cushions. Per stage for all genes analysed, n ≥ 3 embryos. Ao, Aorta; AVC, atrioventricular canal; AVCC, atrioventricular cushion; AVCM, atrioventricular canal myocardium; GFP, green fluorescent protein; Li, liver; OFT, outflow tract; OFTC, outflow tract cushion; MVS, membranous ventricular septum; PT, pulmonary trunk; RA, right atrium; R/LV, right/left ventricle. Scale bars depict 100 µm.
Slit1 was not expressed in the heart at any stage (see Supplementary material online, Figure S1A and data not shown) and was not analysed further. Besides its most evident expression in the ventricular trabecular myocardium, Slit2 was present in the endocardium lining both the Robo1- and Robo2-positive outflow tract and atrioventricular cushions and valves, and in the aortic semilunar valves just prior to birth (Figure 1A, B, G, and H; see Supplementary material online, Figure S1A–H). In contrast, Slit3 was not expressed in the cardiac cushions or valves (Figure 1D and J; see Supplementary material online, Figure S1A–H) but was found in the outflow tract and atrial myocardium adjacent to the cardiac cushions.
3.2. Absence of Slit-Robo signalling results in membranous ventricular septum defects
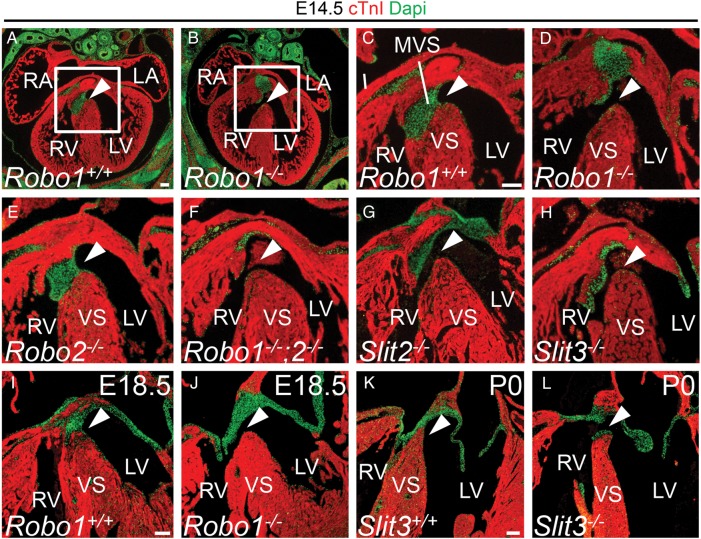
These expression patterns, combined with the identified roles of Slit-Robo during neural crest migration and adhesion, imply that defective signalling might result in cardiac valve and membranous septum defects. Therefore, we first analysed mice mutant for Robo1 and/or Robo2 receptor for septum defects. At E14.5, we noticed absence of the membranous ventricular septum combined with an overriding aorta in 60% of embryos lacking Robo1 (Figure 2A–D; Table 1), whereas the ventricular septum was closed in all littermate controls at this stage. The membranous ventricular septum closes the communication between the right and left ventricles by fusion of the outflow tract cushions with the atrioventricular cushions and is normally completed at E14.5. Robo2 mutant mice did not show any defects, whereas Robo1;Robo2 double mutants showed a phenotype similar to Robo1 lacking mice, indicating that Robo1 is the main Robo receptor required for the development of the region (Figure 2E and F; Table 1). All double mutants analysed exhibited a septum defect, suggesting an additional role for Robo2, although this difference might be caused by the lower numbers of double mutants analysed. The membranous septum defect was still present in 30% of Robo1 mutants at E18.5 (Figure 2I and J; Table 1), suggesting in one-third of mutants closure is merely delayed as the Mendelian ratio at birth is normal.17
Figure 2.
Disrupted Slit-Robo signalling results in membranous ventricular septum defects. (A–L) immunohistochemistry for cardiac Troponin I (cTnI) and DAPI on Robo1+/+ (A, C, and I), Robo1−/− (B, D, and J), Robo2−/− (E), Robo1−/−;Robo2−/− (F), Slit2−/− (G), Slit3+/+ (K) and Slit3−/− (H and L) hearts. The valves and the membranous ventricular septum are visible as green DAPI staining. White arrowhead points to the presence or absence of the membranous ventricular septum (see Table 1 for numbers of embryos analysed). VS, (muscular) ventricular septum. For other abbreviations, see the legend of Figure 1. Scale bars depict 100 µm.
Table 1.
The prevalence of membranous ventricular septum defects and bicuspid aortic valves in the absence of genes of the Slit-Robo signalling pathway
| Membranous ventricular septal defects |
Bicuspid aortic valves |
|||||
|---|---|---|---|---|---|---|
| E14.5 | E18.5 | E18.5 | ||||
| Embryo | +/+;+/− | −/− | +/+;+/− | −/− | +/+;+/− | −/− |
| Robo1 | 0/9 | 6/10 | 0/10 | 3/10 | 0/10 | 0/10 |
| Robo2 | 0/5 | 0/5 | 0/6 | 0/8 | 0/6 | 0/8 |
| Robo1;Robo2 | 0/3a | 3/3a | na | na | 0/3a | 3/3a |
| Slit2 | 0/6 | 2/6 | 0/8 | 0/7 | 0/8 | 1/7 |
| Slit3 | 0/5 | 2/5 | 0/4b | 1/4b | 0/4b | 0/4b |
The prevalence of membranous septum defects and bicuspid aortic valves in Robo1, Robo2, Robo1;Robo2, Slit2, and Slit3 mutants at the indicated developmental stages.
aRobo1;Robo2 was analysed at E15.5.
bSlit3 was analysed at P0; na means not analysed.
We next analysed mice mutant for Slit2 and Slit3 ligands. Interestingly, both mutants showed membranous ventricular septum defects at E14.5 (Figure 2G and H). However, the prevalence was lower compared with Robo1 mutants (Table 1). The septum defects in Slit2 lacking animals were relatively small, whereas the Slit3 knock-outs showed a phenotype more similar to Robo1 mutants. Of the ligand mutants, only mice lacking Slit3 showed a septum defect at P0 (Figure 2K and L; Table 1), indicating that Slit3-Robo1 interaction is most crucial for membranous ventricular septum development, but Slit2 and Robo2 are also involved.
To identify the cause of the ventricular septum defects, we first analysed cushion development at earlier stages. At E12.5, we did not observe defects in the cushions of any of the mutant mice analysed (data not shown), except for Robo1;Robo2 mutants. The size of the cushions was unaltered between the double mutants and wild-type littermates at E12.5 (Figure 3A). However, we observed reduced closure of outflow tract cushion regions that were already fused in wild-type littermates at this stage (Figure 3B and C; see Supplementary material online, Figure S2A and B). This region of impeded closure corresponded exactly to the outflow tract cushion region highly expressing both Robo1 and Robo2 (Figure 1A–L; see Supplementary material online, Figure S1C). As a result, the double mutants displayed a persistent connection between the right and left ventricle, while this connection was already closed in wild-type littermates at this stage (see Supplementary material online, Figure S2C and D).
Figure 3.
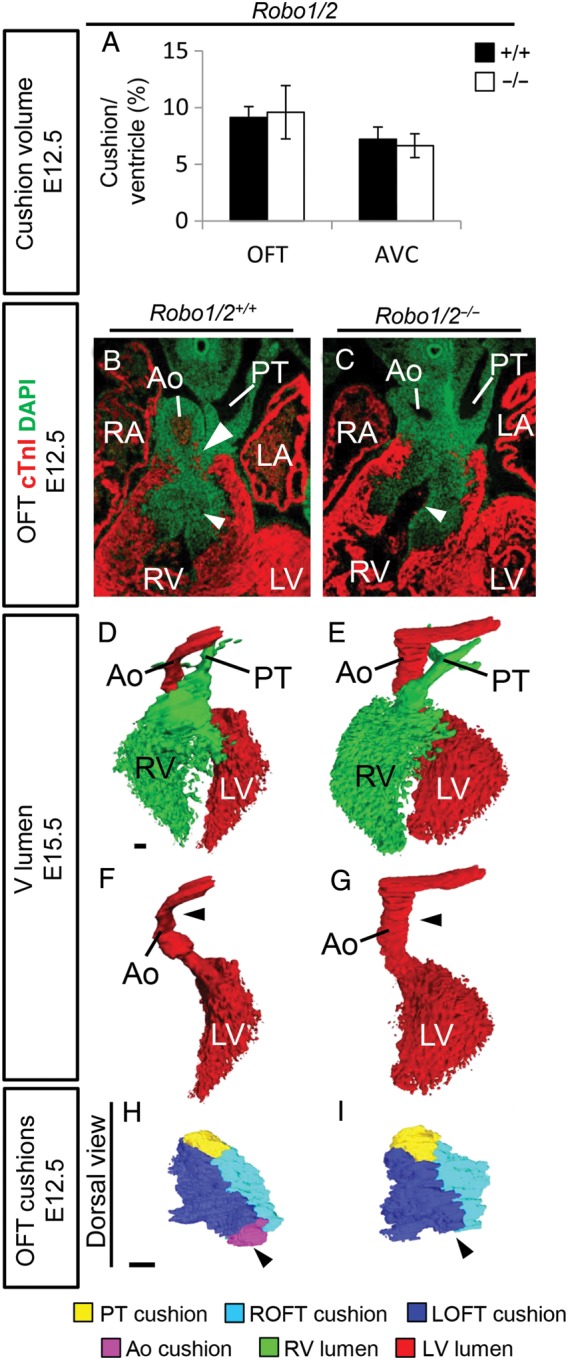
Defects in cushion formation in Robo1/2 double mutants. (A) Measurements of the OFT and AVC cushion volume corrected for ventricular volume (n = 3; OFT, P = 0.51; AVC, P = 0.27; Mann–Whitney U test). (B) Immunohistochemistry for cardiac Troponin I (cTnI) and DAPI on Robo1+/+;Robo2+/+, and Robo1−/−;Robo2−/− mutants. White arrowheads indicate increased space between OFT cushions in the mutant at E12.5. (D–G) Three-dimensional reconstructions of the ventricular lumen and outflow tract vessels of E15.5 Robo1+/+;Robo2+/+ and Robo1−/−;Robo2−/− hearts showing reduced rotation of the aorta in the mutant (black arrowheads), most clearly visible after removing the right ventricle and pulmonary trunk (F–G). (H–I) Three-dimensional reconstructions of the outflow tract cushions of E12.5 Robo1+/+;Robo2+/+ and Robo1−/−;Robo2−/− embryos showing the absence of the aortic posterior cushion in the Robo1/2 mutant. n = 3. For abbreviations, see the legend of Figure 1. Scale bars depict 100 µm.
Membranous ventricular septum defects can be caused by reduced contribution of the cardiac neural crest or second heart field to the outflow tract, resulting in aberrant alignment of the outflow tract vessels,4,5 or defects in endothelial-to-mesenchymal transformation and maturation of the cardiac cushions.1,2 Robo1, Robo2, and Slit3 are all expressed in the cardiac neural crest, while Slit2 is present in the neighbouring endoderm.16 As we previously demonstrated that increased apoptosis in the neural crest underlies the pericardial defects observed in Robo1 mutants,16 as well as the fact that the normally highly Robo1 and Robo2-expressing outflow tract cushion region showing reduced closure in the double mutant is neural crest derived (see Supplementary material online, Figure S3A), we started with analysing neural crest contribution to the outflow tract cushions in Robo1;Wnt1cre;R26REYFP embryos. We did not observe increased apoptosis over the different time points between E10.5 and E12.5, and correspondingly neural crest contribution to the outflow tract was not significantly reduced at E11.5 (see Supplementary material online, Figure S3B–D). Also, no increase in apoptosis was observed in Slit3 mutants (E12.5; see Supplementary material online, Figure S3E). These results do not completely rule out involvement of the neural crest in the defective formation of this region, but at least suggest additional mechanisms, likely second heart field defects, as Robo1 as well as Slit2 and 3 are expressed in the Isl1-positive second heart field at E10.5 (see Supplementary material online, Figure S4A). As both second heart field and neural crest defects can cause alignment defects of the outflow tract vessels, resulting in septum defects, we next analysed the arterioventricular alignment in Robo1 mutants at E14.5. The aorta and pulmonary trunk were normally separated in all mutants (see Supplementary material online, Figure S4B–D; data not shown). Although we did not observe any major defects, we noticed a difference in outflow tract vessel alignment, with the aorta slightly more to the right of the pulmonary trunk (see Supplementary material online, Figure S4B and C). Three-dimensional analysis of Robo1 mutant hearts further indicated reduced rotation of the outflow tract vessels and mainly the aorta (black arrowheads; see Supplementary material online, Figure S4D). This seemed to correspond to a narrower, longer ventricle, although the volume of the ventricular myocardium was unchanged. The muscular interventricular septum length was increased in Robo1 mutants, despite the presence of ventricular septum defects (see Supplementary material online, Figure S4E–G). The reduced outflow tract rotation was even more pronounced in the Robo1;Robo2 mutants (Figure 3D–G), possibly explaining the higher incidence of ventricular septum defects. These defects in outflow tract alignment are likely to underlie the ventricular septum defects in the absence of Robo1 or both Robo1 and Robo2.
3.3. A spectrum of valve malformations in Robo and Slit mutants
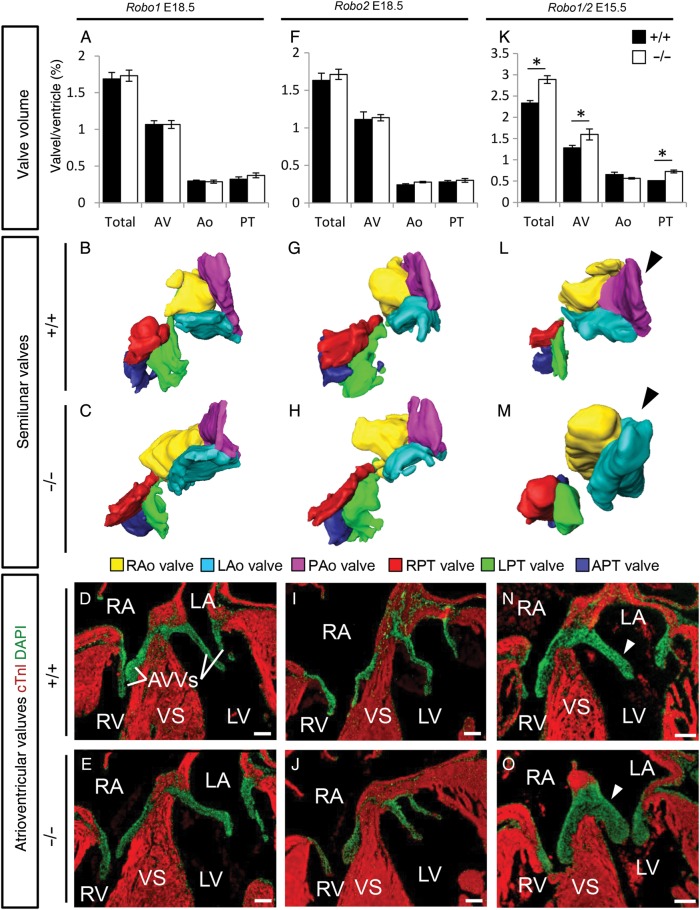
To investigate a role for defective cushion maturation in the formation of the ventricular septum defects, we next analysed the volume of the different cardiac cushion regions. We did not observe any differences in morphology or volume of the outflow tract or atrioventricular cushions in Robo1, Robo2, Slit2, or Slit3 mutant embryos at E14.5 (data not shown). However, in all E12.5 Robo1;Robo2 mutants, although cushion volume was not changed, the just forming aortic posterior cushion seemed absent (Figure 3H and I). In contrast, the pulmonary anterior cushion was forming normally. At E15.5, all Robo1;Robo2 double mutants showed thickening of both the semilunar and atrioventricular valves, in combination with now clearly visible bicuspid aortic valves (Figure 4K–O; see Supplementary material online, Figure S2E and F). While the overall aortic valve volume was similar between Robo1;Robo2 double mutants and wild-type littermates, this was caused by the absence of the posterior non-coronary aortic valve; the two remaining valves were thicker than their wild-type counterparts (Figure 4K–M). At E18.5, single Robo1 or Robo2 mutant valves remained indistinguishable from their wild-type littermates (Figure 4A–J). In contrast, E18.5 mice mutant for Slit2 showed thickening of both the aortic and pulmonary semilunar valves, similar to the Robo1;Robo2 double mutants; however, the atrioventricular valves retained a normal size (Figure 5A–E). As only one Slit2 mutant recapitulated the bicuspid aortic valves as seen in the absence of both Robo1 and Robo2, total aortic valve volume was significantly increased in mice lacking Slit2 (Figure 5A–C). The thickness of the semilunar valves in Slit3 mutants was relatively normal, but now the atrioventricular valves were strikingly thicker (Figure 5F–J). The posterior aortic semilunar leaflet, missing in the Robo1;Robo2 double mutants, was highly hypoplastic in the absence of Slit3, but never completely absent (Figure 5G and H). These data indicate a role for Slit-Robo signalling during valve maturation and functional redundancy and requirement of Robo1 and Robo2 in both the outflow tract and the atrioventricular valve regions, whereas the Slit ligands show region-specific functions. Furthermore, these results underline the possible multi-causal origin of the ventricular septum defects.
Figure 4.
A spectrum of valve malformations in Robo mutants. (A–O) Analysis of the cardiac valves at the indicated developmental stages in Robo1+/+ (A, B, and D) Robo1−/− (A, C, and E) Robo2+/+ (F, G, and I) Robo2−/− (F, H, and J) Robo1+/+;Robo2+/+ (K, L, and N), and Robo1−/−;Robo2−/− (K, M, and O) embryos. (A, F, and K) Measurements of the total valve (Total), atrioventricular valve (AV), aortic valve (Ao), and pulmonary trunk valve (PT) volume corrected for ventricular volume. (A) Total, n = 5, P = 0.47; AV, P = 0.60; Ao, P = 0.75; PT, P = 0.35. (F) Total, n = 5, P = 0.52; AV, P = 0.47; Ao, P = 0.08; PT, P = 0.92. (K) Total, WT n = 5, KO n = 3, P = 0.025; AV, P = 0.025; Ao, P = 0.30; PT, P = 0.025; Mann–Whitney U test. Note the overall increased valve volume in Robo1−/−;Robo2−/−. (B, C, G, H, L, and M) Example three-dimensional reconstructions as used for the volume measurements of the semilunar valves, seen from the ventricular side. Black arrowhead, note the absence of the posterior aortic valve in the Robo1−/−;Robo2−/− (M) embryo. (D, E, I, J, N, and O) Examples from immunohistochemistry sections (cTnI and DAPI) used for the measurements, showing the atrioventricular valves. White arrowheads, Robo1−/−;Robo2−/− (O) embryos show thickened valves. R/L/PAo, right/left/posterior aortic valves; R/L/APT, right/left/anterior pulmonary trunk valve; AVVs, atrioventricular valves. *P < 0.05. For other abbreviations, see the legend of Figures 1 and 2. Scale bars depict 100 µm.
Figure 5.
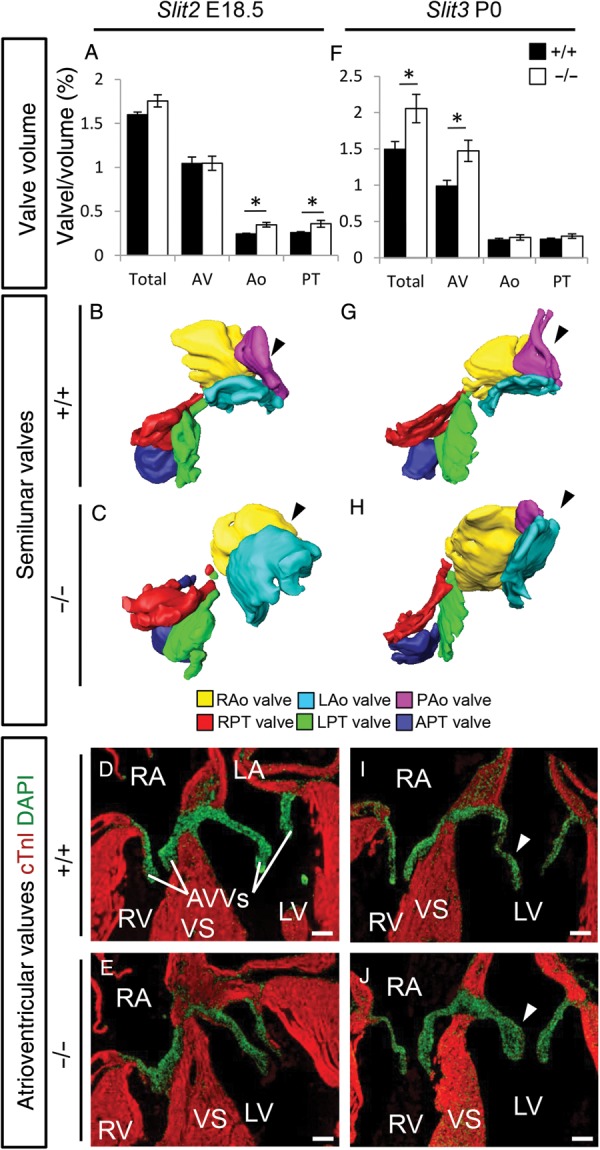
A spectrum of valve malformations in Slit mutants. (A–J) Analysis of the cardiac valves at the indicated developmental stages in Slit2+/+ (A, B, and D), Slit2−/− (A, C, and E), Slit3+/+ (F, G, and I), Slit3−/− (F, H, and J) embryos. (A and F) Measurements of the total valve (Total), atrioventricular valve (AV), aortic valve (Ao), and pulmonary trunk valve (PT) volume corrected for ventricular volume. (A) Total, n = 5, P = 0.08; AV, P = 0.92; Ao, P = 0.016; PT, P = 0.025. (F) Total, WT n = 5, KO n = 3, P = 0.025; AV, P = 0.025; Ao, P = 0.46; PT, P = 0.10; Mann–Whitney U test. Note that while there is overall increased valve volume in Robo1−/−;Robo2−/− mutants, Slit2−/− mutants only show increased semilunar (A) and Slit3−/− only increased total and atrioventricular valve volume (F). (B, C, G, and H) Three-dimensional reconstructions of the semilunar valves, seen from the ventricular side. Black arrowhead, note the absence of the posterior aortic valve in the Slit2−/− (C) while this valve is hypoplastic in the Slit3−/− (H) embryos. (D, E, I, and J) Immunohistochemistry sections (cTnI and DAPI) showing the atrioventricular valves. White arrowheads, Slit3−/− (J) embryos show thickened valves. R/L/PAo, right/left/posterior aortic valves; R/L/APT, right/left/anterior pulmonary trunk valve; AVVs, atrioventricular valves. *P < 0.05. For other abbreviations, see the legend of Figure 1. Scale bars depict 100 µm.
3.4. Deregulation of Robo2 and Slit3 in the absence of the Robo1 receptor
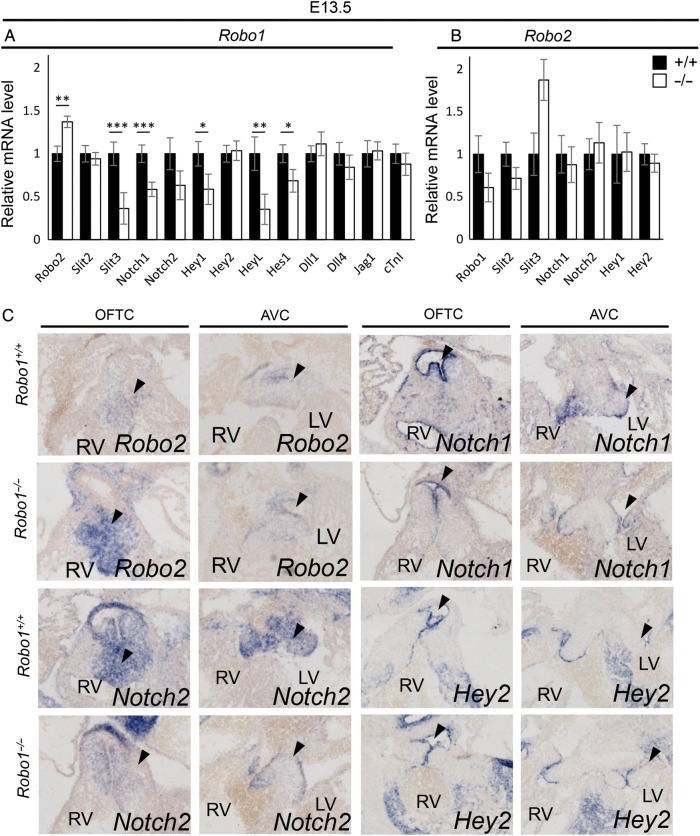
To address the possibility of functional redundancy and/or compensation of Slit/Robo genes in Robo1−/− mice, we isolated mRNA from E13.5 Robo1 mutant and their wild-type littermate hearts of which the ventricles and atria were removed, leaving the regions containing the outflow tract with the atrioventricular canal. In the absence of Robo1, qPCR indicated an increase in Robo2 expression levels. Slit2 levels were normal, while the amount of Slit3 mRNA was greatly reduced (Figure 6A). Additionally, we confirmed the absence of Slit1 and Robo3 expression in the heart, while Robo4 expression was unaltered (data not shown). These results were confirmed by in situ hybridization. While the reduction in Slit3 expression was much less obvious with in situ hybridization than when assessed by qPCR (data not shown), Robo2 expression was visibly increased, particularly in the outflow tract cushions and forming membranous interventricular septum, where normally Robo1 is highly expressed (Figure 6C; see Supplementary material online, Figure S5A and B). In the absence of Robo2, there was no significant change in Robo1 or Slit2 expression; in some, but not all mutants, Slit3 expression was much higher compared with the wild types (Figure 6B). These data suggest that most of the phenotype in Robo1 mutants is rescued by the increase of Robo2 expression and further suggests Slit3 as the main ligand for Robo1. While the increase in outflow tract Robo2 expression seemed to rescue semilunar valve malformations in Robo1 mutants, it failed to rescue the membranous septum defects or it may even underlie these defects.
Figure 6.
Down-regulation of Notch and downstream targets in the absence of Robo1. (A and B) Quantitative PCR on E13.5 Robo1+/+ and Robo1−/− (A, n = 6) and Robo2+/+ and Robo2−/− (B, n = 6) mRNA isolated from outflow tract with atrioventricular canal regions. All mutant levels visualized in relative levels to the wild-type expression set at 1. (A) Robo2, P = 0.007; Slit2, P = 0.62; Slit3, P ≤ 0.001; Notch1, P ≤ 0.001; Notch2, P = 0.09; Hey1, P = 0.026; Hey2, P = 0.82; HeyL, P = 0.003; Hes1, P = 0.029; Dll1, P = 0.53; Dll4, P = 0.39; Jag1, P = 0.86; cTnI, P = 0.45. (B) Robo1, P = 0.13; Slit2, P = 0.11; Slit3, P = 0.09; Notch1, P = 0.67; Notch2, P = 0.68; Hey1, P = 0.95; Hey2, P = 0.51; Student's t-test. (C) In situ hybridization on E13.5 Robo1+/+ and Robo1−/− embryos for the indicated genes showing up-regulation of Robo2, down-regulation of Notch1 and Notch2, but not Hey2 in the absence of Robo1. Black arrow heads indicate the areas of differential expression in the cushions. n ≥ 3 embryos per gene. *P < 0.05, **P < 0.01 and ***P < 0.001. For abbreviations, see the legend of Figure 1. Scale bars depict 100 µm.
3.5. Robo receptors are required for normal expression levels of the Notch signalling pathway
We have recently discovered the down-regulation of Notch2 and Hey1 during cortical development in Robo1 mutants.18 Additionally, another group reported regulation of Hes1 by Robo2 in the same system.19 These observations, combined with the type of defects found in the Slit and Robo mutants, suggested Robo-Notch interaction in the heart as well.3 Thus, using qPCR, we tested the expression levels of a number of Notch and downstream target genes in E13.5 Robo1 mutant and wild-type littermate mRNA isolated from the outflow tract and atrioventricular region. In agreement with our previous results in the developing cerebral cortex, Robo1 mutant hearts showed decreased expression of downstream Notch targets Hey1 and Hes1, although the decrease in Notch2 did not reach significance. Hes2 expression was below detection level in all samples (Figure 6A; data not shown). In addition, we found down-regulation of both Notch1 and HeyL, suggesting broad regulation of the Notch signalling pathway (Figure 6A). The expression of Notch ligands Dll1, Dll4, and Jag1 was unchanged, suggesting Robo does not regulate or influence Notch ligand presentation in interacting cells. The expression levels of Notch1, Notch2, Hey1, and Hey2 were unaltered in Robo2 mutants. The down-regulation of Notch1, and Hey1, but interestingly, most clearly Notch2 in the absence of Robo1 was confirmed by in situ hybridization (Figure 6C; see Supplementary material online, Figure S5A–H; data not shown); however, NICD1 expression was not visibly affected (see Supplementary material online, Figure S6A and B). Interestingly, Hey2 expression was unaltered in Robo1 mutants, both by qPCR and in situ hybridization (Figure 6A–C; see Supplementary material online, Figure S5G and H).
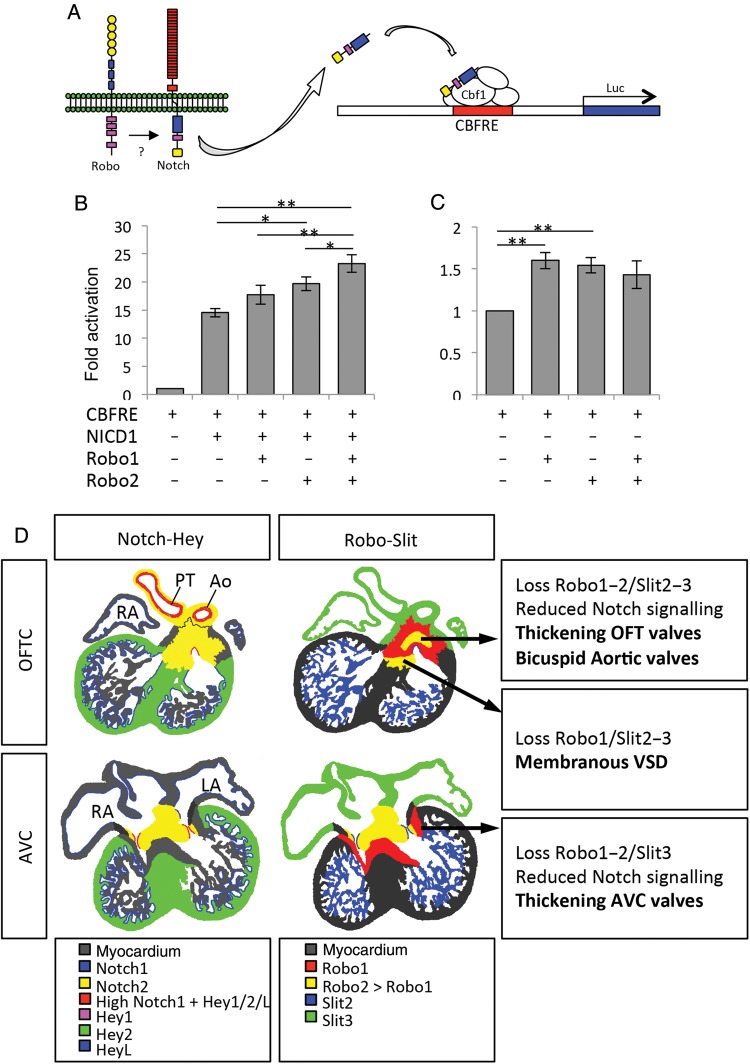
Next, we tested whether Robo receptors might directly enhance Notch signalling in vitro. Thus, we performed luciferase assays on COS7 cells, which were co-transfected with a plasmid containing four Notch-responsive Cbf1-binding elements and the basal simian virus 40 (SV40) promoter upstream of a luciferase cassette (CBFRE-luc), a plasmid with the Notch1 intracellular domain (NICD1) and/or Robo receptor expression plasmids (Figure 7A–C). In the control experiments, the presence of NICD1 expression strongly stimulated Notch-responsive CBFRE-luciferase activity (Figure 7B). This confirmed that the Notch-responsive element is activated by NICD1, with a 14-fold increase over basal expression levels found with an identical plasmid lacking NICD1 (Figure 7B). Co-transfection of Robo1 or Robo2-expressing plasmid and NICD1 further increased luciferase activation (respectively 18- and 20-fold), albeit Robo1 never reached significant levels. These results indicate that Robo2 is more important for regulating Notch signalling than Robo1. However, co-transfection of both Robo1 and Robo2 with NICD1 resulted in further increase in luciferase activity by 23-fold. This suggests that Notch signalling in the heart is spatially regulated by Robo expression, with maximum regulation when both Robo1 and Robo2 are present. Both Robo1 and Robo2 also minimally activated CBFRE-luciferase activity in the absence of NICD1 (1.6-, 1.5-, or 1.4-fold, respectively, Figure 7C). This might be caused by low levels of endogenous NICD1 present in COS7 cells or by direct regulation of Cbf1-responsive elements by Robo receptors. However, our data indicate that Robo receptors mainly function synergistically with NICD1, possibly by stabilizing N1ICD in the nucleus, although the nature of interaction, be it direct or indirect, will need to be further examined. Combined with the qPCR data, these results show that Robo1 and Robo2 receptors are capable of activating Notch signalling, suggesting that reduced activation of the Notch signalling pathway might underlie the defects observed in the mutants for Slit or Robo.
Figure 7.
Robo activates CBFRE-luciferase activity. (A) Robo and Notch are both membrane-bound receptors with intracellular signalling domains. Cleavage and binding of the intracellular domain of Notch to Cbf1 activates the CBFRE-luciferase reporter construct. (B and C) CBFRE-luciferase activation in the presence of the indicated transfected genes in COS7 cells. (B) P = 0.65, P = 0.045, P = 0.01 compared with NICD1 alone, respectively. Robo1 and 2 with NICD1 compared with Robo1 with NICD1 P = 0.007, to Robo2 with NICD1 P = 0.041, n = 5. (C) P = 0.004, P = 0.004, P = 0.14 compared with NICD1 alone, respectively, n = 5; one-way ANOVA with Bonferroni's multiple comparison test. (D) Scheme showing the similarity of the expression patterns between the Slit-Robo and Notch-Hey genes, with an overview of the observed defects. Luc, luciferase. *P < 0.05, **P < 0.01. For other abbreviations, see the legend of Figure 1.
4. Discussion
4.1. The Slit-Robo pathway as a new player in cardiac cushion development
Slit-Robo signalling has been implicated in heart tube assembly and lumen formation in Drosophila and zebrafish development,10–13 and in cardiac chamber formation,14 pericardial development,16 and cardiac neural crest migration and adhesion during murine heart development.15,16 However, a role in cardiac cushion development has not yet been described. We have shown here how genes of the Slit-Robo signalling pathway locally regulate membranous ventricular septum and valve formation. Both Robo1 and Robo2 receptors and their ligands Slit2 and Slit3 were expressed in or surrounding the cushions, and later valves, throughout development. The Robo1 receptor seemed the most crucial during heart development, which may be caused by the fact that Robo1, but not Robo2, can interact with Neuropilin1, a receptor required for outflow tract cushion development and membranous ventricular septum formation.20,21 Additionally, at least part of the difference between the phenotype in the Robo1 and Robo1;Robo2 mutants might be explained by rescue of Notch levels by an increase in Robo2 expression in the Robo1 mutant.
Bicuspid aortic valves were present in all Robo1;Robo2 double mutants analysed and were recapitulated, albeit with lower penetrance, in the absence of Slit2 and hypoplastic in the absence of Slit3, suggesting the requirement of all four genes for proper formation of the aortic semilunar valves. Thickening of the atrioventricular valves was only observed in Robo1;Robo2 and Slit3 mutants. This is interesting, as Slit2 is expressed in both locations in the endocardium covering the valves, whereas Slit3 is expressed in the neighbouring myocardium. This raises the question as to the role of Slit2 in this region but, most importantly, how does Slit3 reach the atrioventricular cushions as it is expressed in the atrial and outflow tract myocardium, but not in or directly surrounding the cushions? The expression level of Slit3 might be below detection level with in situ hybridization; however, Slit ligands have also been described able to act over long distances,22 indicating Slit3 might be able to reach the cushions from the relatively close by myocardium. Furthermore, some of the observed defects might be secondary to other abnormalities and not primary defects, possibilities that can only be investigated using tissue-specific conditional mutants. Taken together, our data demonstrate the local tight control of valve and membranous ventricular septum formation by the selective spatial interaction of genes of the Robo signalling pathway.
4.2. Defective Slit3-Robo1 signalling results in membranous ventricular septum defects
Recently, we have shown a role for Slit-Robo signalling in the development of both the systemic venous return to the heart as well as the pericardium. These pericardial defects seemed to be caused by abnormal localization of the caval veins combined with ectopic pericardial cavity formation, brought about by increased cell death and impaired adhesion and migratory responsiveness of the cardiac neural crest.16 In contrast, neural crest cell contribution to the outflow tract cushions was normal in Robo1 mutants. However, as the area of impaired outflow tract cushion closure in Robo1/2 double mutants was located in the neural crest derived part of the cushion, this still indicates a possible other role for the neural crest in the development of the membranous septum defects. More importantly, the expression of Robo1 as well as Slit2 and 3 in the second heart field suggests that disruptions in second heart field contribution to the outflow tract are another likely cause of the observed defects. The exact role of Slit-Robo signalling in the second heart field will still need to be further examined. Defects in both the second heart field and cardiac neural crest are linked to malrotation of the aorta and pulmonary trunk at their connection to the ventricle,4,5 as found to some degree in the Robo1 and Robo1/2 mutants, which is a known cause of membranous ventricular septum defects.
In addition to the septum defects, the neural crest has been linked with providing instructive signals for remodelling of the semilunar valves. Correct signalling and tissue–tissue interactions among second heart field, neural crest, and endocardial cushion mesenchyme are required for normal valve formation.23 The expression patterns of Robo1 and Slit3 in the second heart field and neural crest, Robo1 and Robo2 in the cushions, and Slit2 in the endocardium also suggest signalling between these cell populations.
4.3. Several downstream effectors of Notch are affected by disrupted Slit-Robo signalling
Notch intercellular signalling has been implicated in multiple aspects of heart development including cushion development, trabecular growth, atrioventricular patterning, neural crest differentiation, and outflow tract development.3 The Notch signalling pathway is known to regulate both the endocardial-to-mesenchymal transformation underlying early cushion development and their subsequent remodelling into the thin mature leaflets of the atrioventricular and semilunar valves.3,24
Bicuspid aortic valves are among the most common of congenital defects affecting 1–2% of the population25,26 and are associated with ventricular septum defects and arterial malformations such as aortic coarctation, aneurisms of the descending aorta, and carotid and vertebral artery defects. Bicuspid aortic valves have only two complete leaflets, while the third leaflet is either absent or incomplete.26 To date, NOTCH1 is the only transcriptional regulator linked to bicuspid aortic valve disease in humans in a limited number of familial cases and ∼4% of sporadic cases.27–29 While mice ubiquitously mutant for Notch1 die early during development due to severe defects, tissue-specific Notch inhibition has shown to mimic the defects seen in humans with mutations in NOTCH1.30 The defects observed in Slit-Robo mutants, in which Notch signalling is down-regulated, also closely resemble defects caused by human NOTCH1 mutations.
Notch signalling exerts its function through activation of effector genes such as the Hey and Hes families. The observed decrease of Notch1 and Notch2 in Robo1 mutants subsequently resulted in down-regulation of the downstream targets Hey1, HeyL, and Hes1, although not Hey2. Combined loss of Hey1 and HeyL results in membranous ventricular septum defects and dysplastic atrioventricular and pulmonary valves, similar to the defects observed in the absence of Slit-Robo signalling.31 Hey2 expression was unaltered in Robo1 mutants, in line with previous findings which suggested that aberrations in Notch signalling do not always result in changes in Hey2 expression.32 Consequently, the cardiac defects reported in Hey2 mutants are largely different from those seen in Robo1;Robo2 animals.33 The dextra-posed aorta phenotype present in Hes1 mutants was partially recapitulated in Robo1 mutants, consistent with the fact that Hes1 is down-regulated, but not absent in these mice.34,35 The extensive range of pharyngeal arch artery defects in the absence of Hes1 warrants further detailed investigation in Slit and Robo mutants in future studies.
As both Robo and Notch are transmembrane receptors, the question is how Robo regulates Notch signalling. There can be direct interaction at the membrane level, interaction of the intracellular domains either at the membrane or in the nucleus, or the intracellular domain of Robo independently binds to Cbf1 or Notch-responsive elements. Modulation of transcription by Robo signalling has been suggested in other systems, although no direct targets were identified.36,37 Transcriptional regulation by the intracellular domain of Robo is similar to a mechanism described in a recent study on the regulation of Hes1 by Robo2 during cortical development.19 In contrast to the latter study, however, we found that Notch expression is down-regulated in the absence of Robo, suggesting that Robo might activate Notch signalling instead of directly regulating Notch-responsive elements independently. The fact that the luciferase experiments suggest a more important role for Robo2 than Robo1 in the regulation of Notch signalling, in contrast to the mutant data, also needs further attention. This difference might be caused by the in vitro conditions, for example by a different response in COS7 cells, the lack of Slit ligands in vitro, or point to the involvement of other factors. The luciferase experiments do confirm maximal regulation in the presence of both receptors as found in the mutant analysis. An interesting observation is the lack of requirement for Slit ligand in the luciferase experiments, whereas the Slit2 and Slit3 mutant data indicate the requirement of Slit-Robo binding. This might be caused by the strong overexpression of Robo receptors in these experiments. The genes of the Slit-Robo and Notch-Hey/Hes pathways show strikingly similar expression patterns in the developing heart (Figures 1, 6, and 7D).3 Notch1 overlaps with the expression of Slit2 and Hey genes in the endothelium surrounding the cushions, while Notch2 is expressed in the cushions overlapping with Robo1 and Robo2. Hey2 and Slit3 show complementary gene expression in ventricle and atrium with outflow tract myocardium, respectively (Figure 7D). These similar patterns with different combinations of Notch-Hey and Slit-Robo genes in different cell types might explain the observed difference in phenotype between Slit and Robo mutants. The identification of the exact cellular interactions and signals required for the modulation of Notch by the Slit-Robo signalling pathway will be the focus of future studies.
Here, we uncovered a novel signalling pathway controlling membranous ventricular septum and cardiac valve formation, the Slit-Robo pathway. Further study of the identified range of valve defects, and in particular the already early in development recognizable bicuspid aortic valves, might help understand the aetiology of common congenital valve defects found in patients.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This work was supported by the Wellcome Trust (Programme Grant 089775) and the British Heart Foundation (PG/12/39/29626 to M.T.M.M.). M.T.M.M. was a recipient of an EMBO Long-Term Fellowship grant (ALTF 441-2010) and Netherlands Organisation for Scientific Research Rubicon grant (825.10.025). Funding to pay the Open Access publication charges for this article was provided by the Welcome Trust.
Acknowledgements
We thank Mary Rahman and Eleni Doumou for technical support and Alain Chédotal, Athena R. Ypsilanti and Pavol Zelina for providing the Robo1;Robo2 and Slit3 mutant embryos.
Conflict of interest: none declared.
References
- 1.Combs MD, Yutzey KE. Heart valve development: regulatory networks in development and disease. Circ Res 2009;105:408–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garside VC, Chang AC, Karsan A, Hoodless PA. Co-ordinating Notch, BMP, and TGF-beta signaling during heart valve development. Cell Mol Life Sci 2013;70:2899–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenthal N, Harvey RP. Heart Development and Regeneration. Amsterdam: Academic Press; 2010. [Google Scholar]
- 4.Rochais F, Mesbah K, Kelly RG. Signaling pathways controlling second heart field development. Circ Res 2009;104:933–942. [DOI] [PubMed] [Google Scholar]
- 5.Keyte A, Hutson MR. The neural crest in cardiac congenital anomalies. Differentiation 2012;84:25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seeger M, Tear G, Ferres-Marco D, Goodman CS. Mutations affecting growth cone guidance in Drosophila: genes necessary for guidance toward or away from the midline. Neuron 1993;10:409–426. [DOI] [PubMed] [Google Scholar]
- 7.Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell 1999;96:795–806. [DOI] [PubMed] [Google Scholar]
- 8.Andrews WD, Barber M, Parnavelas JG. Slit-Robo interactions during cortical development. J Anat 2007;211:188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ypsilanti AR, Zagar Y, Chedotal A. Moving away from the midline: new developments for Slit and Robo. Development 2010;137:1939–1952. [DOI] [PubMed] [Google Scholar]
- 10.Qian L, Liu J, Bodmer R. Slit and Robo control cardiac cell polarity and morphogenesis. Curr Biol 2005;15:2271–2278. [DOI] [PubMed] [Google Scholar]
- 11.Medioni C, Astier M, Zmojdzian M, Jagla K, Semeriva M. Genetic control of cell morphogenesis during Drosophila melanogaster cardiac tube formation. J Cell Biol 2008;182:249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santiago-Martinez E, Soplop NH, Patel R, Kramer SG. Repulsion by Slit and Roundabout prevents Shotgun/E-cadherin-mediated cell adhesion during Drosophila heart tube lumen formation. J Cell Biol 2008;182:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fish JE, Wythe JD, Xiao T, Bruneau BG, Stainier DY, Srivastava D, Woo S. A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development 2011;138:1409–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medioni C, Bertrand N, Mesbah K, Hudry B, Dupays L, Wolstein O, Washkowitz AJ, Papaioannou VE, Mohun TJ, Harvey RP, Zaffran S. Expression of Slit and Robo genes in the developing mouse heart. Dev Dyn 2010;239:3303–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calmont A, Ivins S, Van Bueren KL, Papangeli I, Kyriakopoulou V, Andrews WD, Martin JF, Moon AM, Illingworth EA, Basson MA, Scambler PJ. Tbx1 controls cardiac neural crest cell migration during arch artery development by regulating Gbx2 expression in the pharyngeal ectoderm. Development 2009;136:3173–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mommersteeg MT, Andrews WD, Ypsilanti AR, Zelina P, Yeh ML, Norden J, Kispert A, Chedotal A, Christoffels VM, Parnavelas JG. Slit-roundabout signaling regulates the development of the cardiac systemic venous return and pericardium. Circ Res 2013;112:465–475. [DOI] [PubMed] [Google Scholar]
- 17.Long H, Sabatier C, Ma L, Plump A, Yuan W, Ornitz DM, Tamada A, Murakami F, Goodman CS, Tessier-Lavigne M. Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron 2004;42:213–223. [DOI] [PubMed] [Google Scholar]
- 18.Yeh ML, Gonda Y, Mommersteeg MT, Barber M, Ypsilanti AR, Hanashima C, Parnavelas JG, Andrews WD. Robo1 modulates proliferation and neurogenesis in the developing neocortex. J Neurosci 2014;34:5717–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borrell V, Cardenas A, Ciceri G, Galceran J, Flames N, Pla R, Nobrega-Pereira S, Garcia-Frigola C, Peregrin S, Zhao Z, Ma L, Tessier-Lavigne M, Marin O. Slit/Robo signaling modulates the proliferation of central nervous system progenitors. Neuron 2012;76:338–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernandez-Miranda LR, Cariboni A, Faux C, Ruhrberg C, Cho JH, Cloutier JF, Eickholt BJ, Parnavelas JG, Andrews WD. Robo1 regulates semaphorin signaling to guide the migration of cortical interneurons through the ventral forebrain. J Neurosci 2011;31:6174–6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J, Pashmforoush M, Sucov HM. Endothelial neuropilin disruption in mice causes DiGeorge syndrome-like malformations via mechanisms distinct to those caused by loss of Tbx1. PLoS ONE 2012;7:e32429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson JH, Kidd T, Bland KS, Goodman CS. Short-range and long-range guidance by slit and its Robo receptors. Robo and Robo2 play distinct roles in midline guidance. Neuron 2000;28:753–766. [DOI] [PubMed] [Google Scholar]
- 23.Jain R, Engleka KA, Rentschler SL, Manderfield LJ, Li L, Yuan L, Epstein JA. Cardiac neural crest orchestrates remodeling and functional maturation of mouse semilunar valves. J Clin Invest 2011;121:422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacGrogan D, Luna-Zurita L, de la Pompa JL. Notch signaling in cardiac valve development and disease. Birth Defects Res A Clin Mol Teratol 2011;91:449–459. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol 2002;39:1890–1900. [DOI] [PubMed] [Google Scholar]
- 26.Siu SC, Silversides CK. Bicuspid aortic valve disease. J Am Coll Cardiol 2010;55:2789–2800. [DOI] [PubMed] [Google Scholar]
- 27.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature 2005;437:270–274. [DOI] [PubMed] [Google Scholar]
- 28.McKellar SH, Tester DJ, Yagubyan M, Majumdar R, Ackerman MJ, Sundt TM., III Novel NOTCH1 mutations in patients with bicuspid aortic valve disease and thoracic aortic aneurysms. J Thorac Cardiovasc Surg 2007;134:290–296. [DOI] [PubMed] [Google Scholar]
- 29.Mohamed SA, Aherrahrou Z, Liptau H, Erasmi AW, Hagemann C, Wrobel S, Borzym K, Schunkert H, Sievers HH, Erdmann J. Novel missense mutations (p.T596M and p.P1797H) in NOTCH1 in patients with bicuspid aortic valve. Biochem Biophys Res Commun 2006;345:1460–1465. [DOI] [PubMed] [Google Scholar]
- 30.High FA, Zhang M, Proweller A, Tu L, Parmacek MS, Pear WS, Epstein JA. An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation. J Clin Invest 2007;117:353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer A, Steidl C, Wagner TU, Lang E, Jakob PM, Friedl P, Knobeloch KP, Gessler M. Combined loss of Hey1 and HeyL causes congenital heart defects because of impaired epithelial to mesenchymal transition. Circ Res 2007;100:856–863. [DOI] [PubMed] [Google Scholar]
- 32.Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua-Belmonte JC, de la Pompa JL. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev 2004;18:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donovan J, Kordylewska A, Jan YN, Utset MF. Tetralogy of fallot and other congenital heart defects in Hey2 mutant mice. Curr Biol 2002;12:1605–1610. [DOI] [PubMed] [Google Scholar]
- 34.Rochais F, Dandonneau M, Mesbah K, Jarry T, Mattei MG, Kelly RG. Hes1 is expressed in the second heart field and is required for outflow tract development. PLoS ONE 2009;4:e6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Bueren KL, Papangeli I, Rochais F, Pearce K, Roberts C, Calmont A, Szumska D, Kelly RG, Bhattacharya S, Scambler PJ. Hes1 expression is reduced in Tbx1 null cells and is required for the development of structures affected in 22q11 deletion syndrome. Dev Biol 2010;340:369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grieshammer U, Le M, Plump AS, Wang F, Tessier-Lavigne M, Martin GR. SLIT2-mediated ROBO2 signaling restricts kidney induction to a single site. Dev Cell 2004;6:709–717. [DOI] [PubMed] [Google Scholar]
- 37.Rhee J, Buchan T, Zukerberg L, Lilien J, Balsamo J. Cables links Robo-bound Abl kinase to N-cadherin-bound beta-catenin to mediate Slit-induced modulation of adhesion and transcription. Nat Cell Biol 2007;9:883–892. [DOI] [PubMed] [Google Scholar]