Abstract
Mitochondrial DNA (mtDNA) is believed to be particularly susceptible to oxidative damage during aging, resulting in mtDNA point mutations, duplications, and deletions. Although mtDNA deletions have been reported in various human tissues, e.g., the brain, heart, and skeletal muscle, little is known about the occurrence in hair. Therefore, we screened for the presence of mtDNA 13162 bp, 10422 bp, 7663 bp, 7436 bp, 4989 bp, and 4977 bp deletions in 90 hair samples from subjects aged 5 days to 91 years by using polymerase chain reaction (PCR) and investigated the deletion load by TaqMan probe-based real-time PCR.
We detected the mtDNA 4977 bp deletion in hair samples, but none of the other deletions that were screened for. The proportion of mtDNA 4977 deletion carriers was 98.3% (89/90) and the deletion loads increased from 0 to 1.436 ± 0.2086% of the total mtDNA with an exponential increase with age (r = 0.677,p < 0.05). These results suggest that mtDNA 4977 bp deletion is a common phenomenon in hair and increases with age. These findings expand our understanding of the tissue-specific distribution of mtDNA deletions.
KEY WORDS: mtDNA 4977bp deletion, hair, ageing, mtDNA mutation
INTRODUCTION
Human mtDNA encodes 22 tRNAs, 2 rRNAs, and 13 polypeptides from a 16,549 bp circular genome. It is at high risk of being affected by free radical damage during cell respiration [1, 2]. Approximately 90% of cellular oxygen is metabolized in mitochondria; some of this oxygen is converted to reactive oxygen species (ROS) as a toxic by-product of electron transport system complexes. Located in close proximity to the source of ROS, mtDNA becomes a frequent target of endogenous ROS [3]. As a result, the deletion mutation rate is much higher in mtDNA compared with that in nuclear DNA [4]. mtDNA point mutations and deletion mutations were initially considered to be linked with mitochondrial diseases, such as Pearson's syndrome, Kearns–Sayre syndrome (KSS), Alzheimer's disease, and progressive external ophthalmoplegia [4, 5]. In recent years, there has been growing evidence of the involvement of large-scale deletions in the age-dependent decline of cell or tissue function, since the removal of large essential parts of coding sequences usually results in the loss of the corresponding gene products [3, 6]. mtDNA point mutations or deletion mutations may also lead to faulty assembly of the respiratory chain and functional impairment of oxidative phosphorylation [7]. To date, more than 100 deletion types have been identified in human mtDNA, some of which are involved in the aging process [5, 8]. In particular, several mtDNA mutants with large deletions are frequently detected molecules with steadily increased incidence through successive decades of life [4, 9-11]. Previous studies have showed that mtDNA deletion types and amounts may vary in different tissues even within the same individual [12-15]. However, most of these studies were focused on the brain, heart, and skeletal muscle [10, 11, 16, 17]. As yet, little is known about mtDNA deletions in hair. Therefore, in the present study, we aimed to investigate the presence of the representative aging-related markers, namely, mtDNA 13162 bp, 10422 bp, 7663 bp, 7436 bp, 4989 bp, and 4977 bp deletions [10, 11, 16, 17], in hair samples and to explore their possible relationships with age.
MATERIALS AND METHODS
Tissue collection
For this study, hair shafts without roots were examined from 90 unrelated Chinese subjects aged 5 days to 91 years. All the participants were without known mitochondrial diseases, such as Pearson's syndrome, KSS, and mitochondrial myopathies. Three heart tissue samples with certified multiple mtDNA deletions were used as positive controls for detecting mtDNA deletions. The present study was performed under the supervision of the Ethics Committee of Zhongshan Medical School of Sun Yat-Sen University. Informed consent was obtained from all subjects.
DNA extraction
Hair shafts were cleaned by a 5-min soak in 1 mL ethanol (95%) and washed with sterile water. Then, the samples were cut into 1- to 5-mm fragments with clean scissors. The hair samples were placed into a 50-mL tube with 400 μL TET solution (100 mmol/L Tris-HCl, 1 mmol/L EDTA, 1% TritonX-100), 25 μL DTT (1 mol/L), and 20 μL proteinase K (20 mg/mL). The solution was then incubated on a shaker at 150 r/min at 60°C for 24 h. The digestion product was extracted with phenol and chloroform. The aqueous phase was then transferred into a new tube, followed by the addition of a onetenth volume of 3 mol/L sodium acetate and 4 μL DNAmate (TaKaRa Biotechnology, Japan). The desired mtDNA was ethanol precipitated and dissolved in 30 μL of distilled water.
DNA amplification
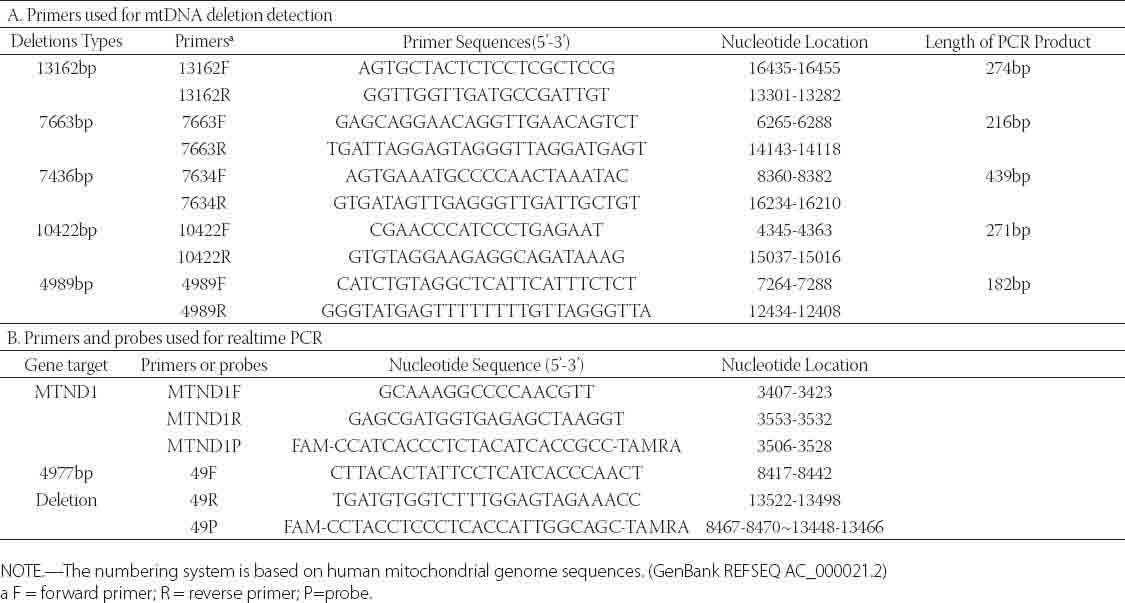
PCR was performed in a 25 μL reacti on mixture containing 2.5 μL 10× LA PCR Buffer II (Mg2+ Plus) (TaKaRa Biotechnology, Japan), 200 μM dNTPs, 0.5 μ;M primers (Table 1), 1.25 U Taq DNA polymerase, and 30 ng DNA template. Amplification comprised an initial denaturation at 94°C for 3 min, followed by 45 cycles of 30 s at 94°C, 30 s at 53°C, and 1 min at 72°C. After the reaction, 5 μL of the PCR product was separated on 2% agarose gel and visualized with SYBR Safe (Invitrogen, USA) with UV.
TABLE 1.
Primers used for mtDNA deletion detection and Real-time PCR
Plasmid construction for the standard curve
DNA fragments were amplified by PCR using the following primer sets: (1) for MTND1, forward 5’-GACGCCATAAAACTCTTCACC-3’ and reverse 5’-ATGAG- ATTGTTTGGGCTACT-G-3’; (2) for mtDNA 4977 bp deletion, forward 5’-AGTGA- AATGCCCCAACTAAATAC-3’ and reverse 5’-TGACCTGTTAGGG TGAGAAGA- AT-3’. Purified PCR products were cloned into the PMDT-19 vector (Ta-KaRa Biotechnology, Japan). The plasmids were further confirmed by DNA sequencing. Plasmid DNA concentration was determined by measuring the absorbance at 260 nm (A260).
TaqMan probe-based real-time PCR analysis
The primers and probes were designed according to the methods given in a previous report [18]. Real-time PCR amplifications were performed in 25 μL reaction volumes containing 30 ng DNA template, 300 nmol/L concentration of each primer, 150 nmol/L of each probe (Table. 1), and the TaqMan Universal Master Mix kit (Toyobo, Japan). The following cycling parameters were used for both assays: 1 min at 95°C, then 45 cycles of 15 s at 95°C and 1 min at 60°C. Each sample was repeated 3 times and a negative control was set. All real-time PCR assays were carried out on an ABI Prism 7500 Sequence Detector.
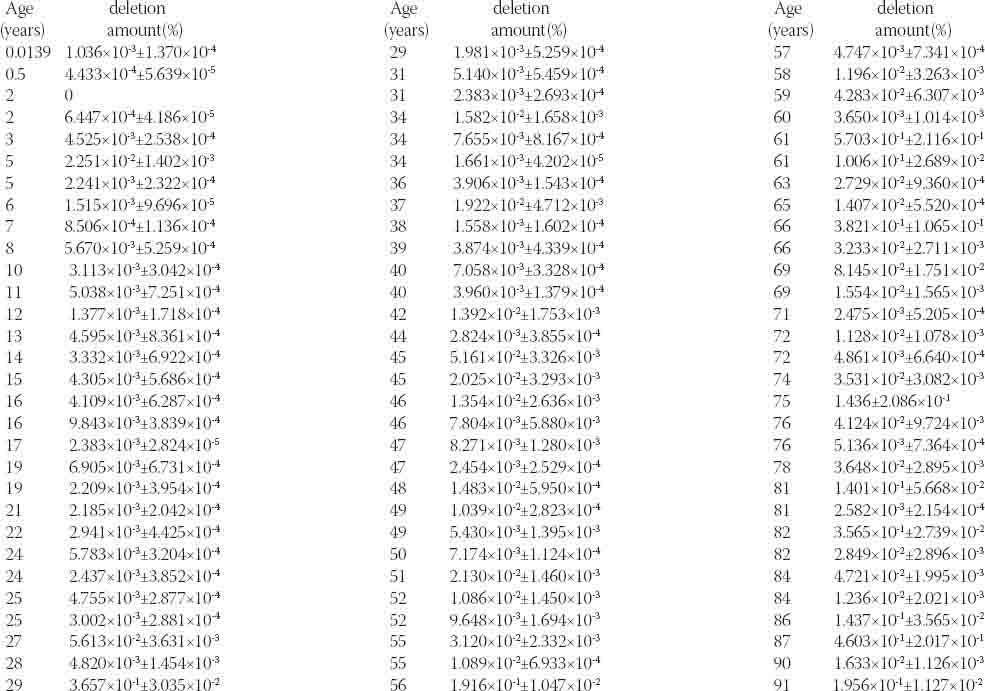
Supplementary Table.1.
MtDNA 4977bp deletion loads in hair samples
Statistical analysis
Comparison between groups was performed by one-way analysis of variance. The coefficient of variation was used to estimate the variability of sets of values. The association between age and mtDNA 4977 bp deletion load was analyzed by the Spearman rank correlation test. The regression equation was established by curve estimation. Statistical significance was set at p < 0.05.
RESULTS
Detection of mtDNA 13162 bp, 7663 bp, 7436 bp, 10422 bp, 4989 bp, and 4977 bp deletions in hair samples
mtDNA deletions were detected as previously described [19, 20]. Briefly, primer pairs were designed to match both ends of deletion points. In the absence of 13162 bp, 10422 bp, 7663 bp, 7436 bp, 4989 bp, and 4977 bp deletions in the mtDNA, the PCR product lengths would theoretically be 13436 bp, 10693 bp, 7879 bp, 7875 bp, 5171 bp, and 5278 bp, respectively. However, when the extension time is set at 1 min (sufficient time for amplification of only 1000 bp), there will not be any amplification fragment in the absence of these large deletions. Instead, amplification will only occur in the presence of those deletions, and the fragment sizes of the PCR products will be 273 bp, 216 bp, 440 bp, 271 bp, 182 bp, and 301 bp, respectively. Three heart tissue samples with certified multiple mtDNA deletions were used as positive controls. As expected, the lengths of amplification fragments were consistent with the theoretical sizes (Figure 1). In each amplification, negative controls were included in the mtDNA extraction and PCR reaction to detect any possible contamination. Interestingly, mtDNA 4977 bp deletions were detected in 89 of the 90 samples within 45 cycles of PCR amplification. The percentage of mtDNA 4977 deletion carriers was as high as 98.3%. However, amplification indicative of mtDNA 13162 bp, 10422 bp, 7663 bp, 7436 bp, and 4989 bp deletions was not observed in any samples in our PCR screening (Table 2). The mtDNA 4977 deletions from the positive PCR products were further confirmed by DNA sequencing (Figure 2). These results suggest that in hair the mtDNA 4977 bp deletion is common, but the mtDNA 13162 bp, 7663 bp, 7436 bp, 10422 bp, and 4989 bp deletions are not.
FIGURE 1.

Agarose gel electrophoresis of the PCR products. mtDNA and mtDNA 13162 bp, 10422 bp, 7663 bp, 7436 bp, 4989 bp, and 4977 bp deletions were detected in hair samples by PCR. Lane M is a DL1000 DNA ladder consisting of 100 bp, 200 bp, 300 bp, 400 bp, 500 bp, 700 bp, and 1000 bp markers. Lanes P to 6 represent positive control, negative control, and samples from individuals aged 3 years, 23 years, 45 years, 60 years, 72 years, and 91 years, respectively. (A) PCR analysis of mtDNA 13162 bp deletion. (B) PCR analysis of mtDNA 7663bp deletion. (C) PCR analysis of mtDNA 7436 bp deletion. (D) PCR analysis of mtDNA 10422 bp deletion. (E) PCR analysis of mtDNA 4989 bp deletion. (F) PCR analysis of mtDNA 4977 bp deletion.
TABLE 2.
Results of detecting mtDNA 13162bp, 10422bp, 7663bp, 7436bp, 4989bp and 4977bp deletions in hair samples
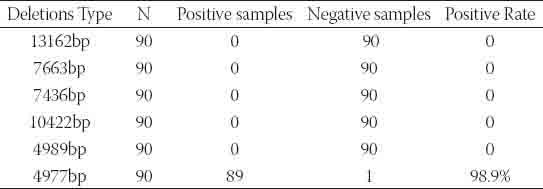
FIGURE 2.
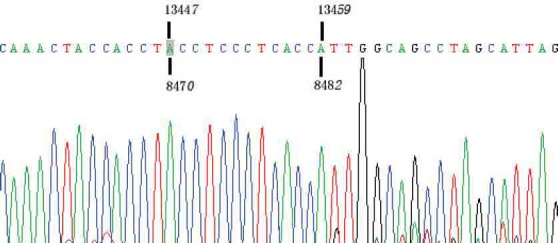
Schematic diagram of mtDNA 4977 bp deletion. Positive PCR products of mtDNA 4977 bp deletion were confirmed by DNA sequencing.
Reliability and reproducibility of the applied PCR assays
In the TaqMan probe-based real-time PCR approach, fluorescence was undetectable at the early amplification stage, but eventually exceeded a threshold value. The cycle number at which this value reached was defined as the cycle threshold (CT). The CT values were then plotted as a parameter of the DNA amount [18]. To test the reliability and reproducibility of the real-time PCR assay, calibration curves for the MTND1 (mtDNA total) and the mtDNA 4977 deletion were constructed. As shown in Figure 3, there was an excellent correlation between the cycle number and mtDNA amount, across 6 log-orders of magnitude (10-5¬−1 pg/μL) with correlation coefficients of 0.9977 for MTND1 and 0.9984 for the mtDNA 4977 deletion. It also demonstrated the sensitivity, linearization, and large dynamic range of the TaqMan probe-based real-time PCR approach used in this study.
FIGURE 3.
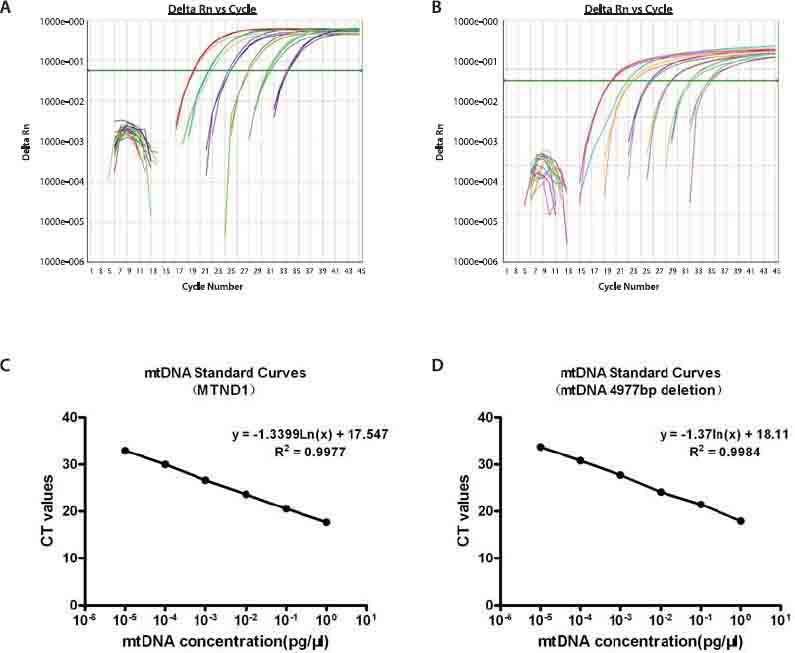
mtDNA amplification plots and standard curves. (A) Amplification plot of 10-fold serial dilutions (from 1 × 10-5 pg/μL to 1 pg/μL) of the MTDN1 plasmid. (B) Amplification plot of 10-fold serial dilutions (from 1 × 10-5 pg/μL to 1 pg/μL) of the mtDNA 4977 bp deletion plasmid. (C) Standard curves displaying average CT values from experiments with the MTND1 plasmid. (D) Standard curves displaying average CT values from experiments with the mtDNA 4977 bp deletion plasmid. CT values were reproducible in all 3 experiments.
Presence and amount of mtDNA 4977 bp deletion
For each sample, 2 different PCR reactions were performed, 1 for the intact mtDNA and another for the mtDNA4977 deletion. The MTDN1 gene, which is not generally subject to deletion, represents the total amount of mtDNA. The fragment that spanned the deletion junction represented the mtDNA 4977 bp deletion [18]. We measured the amount of mtDNA 4977 bp deletion in hair samples from subjects aged 5 days to 91 years. The detection rate of the mtDNA 4977 bp deletion was approximately 98.9% (89 out of 90). It was consistent with the result of standard PCR detection. The means and standard deviations of the samples are shown in Table 2. Measurement of the loads of the mtDNA 4977 bp deletion ranged from 0 to 1.436% (0.2086%) of total mtDNA (Figure 4). A large variability was detected, even between individuals of the same age decade. The association between age and the mtDNA 4977 bp deletion load was analyzed by the Spearman rank correlation test (r = 0.639,p < 0.05), which indicated that the 2 factors were positively correlated. The regression equation, developed by curve estimation, was y = 2E − 05e0 042x (R2 = 0.408, p < 0.05). Together, these results suggest that the mtDNA 4977 bp deletion is a common phenomenon in hair and increases with age.
FIGURE 4.

Correlation between mtDNA 4977 bp deletion load and age in 90 human hair samples. The regression equation, developed by curve estimation, was y = 2E − 05e0.042× (R2 =0.408, p < 0.05)
DISCUSSION
Large-scale mtDNA deletions have been reported to occur in numerous individuals and multiple tissues. There is increasing evidence that some of them are aging-related, including the mtDNA 13162 bp, 10422 bp, 7663 bp, 7436 bp, 4989 bp, and 4977 bp deletions [10, 12, 16-19, 21]. Although mtDNA 13162 bp, 10422 bp, 7663 bp, 7436 bp, 4989 bp, and 4977 bp deletions have been previously detected at high rates in the brain, heart, and muscle fibers [10, 12, 16-19, 21], we only detected the mtDNA 4977 bp deletion in the hair samples in the present study. Of these, 15 samples were from individuals aged 76 to 91 years. These results suggest that less mtDNA deletion types are present in hair compared with those that have been detected in the heart, brain, muscle, or skin in other studies [10, 12, 16-19, 21]. The present data also confirm and extend the previous observation that distribution of mutated mtDNA molecules can vary depending on the tissue type [13, 14]. In standard PCR analysis, several factors may result in false negative results. Although the theoretical detection limit is a single DNA copy, realistically, a higher amount of template DNA must be present [22]. The stabilities of reaction buffer and Taq polymerase are also principally important for PCR efficiency [23]. In this regard, we tried to re-amplify negative samples by improving the PCR reaction system by methods including applying two-round PCR, using more efficient DNA polymerases, and increasing Mg2+ concentrations. However, we still did not detect mtDNA 13162 bp, 10422 bp, 7663 bp, 7436 bp, and 4989 bp deletions in our hair samples. Previous studies have reported that the mtDNA 4977 bp deletion could hardly be detected in the heart and muscle fibers from subjects under 20 years of age [24, 25]. In contrast, this deletion was detected in 93.3% (14 out of 15) of hair samples from the 0–15-year age group in this study. We even observed a deletion rate of 1.036 × 10-3% (1.370 × 10-4%) of total mtDNA in the hair sample of a 5-day-old newborn. However, a hair sample from a 2-year-old subject showed negative amplification in 3 independent experiments with improved measures for amplification efficiency. In the present study, the mtDNA 4977 bp deletion showed an exponential increase with age. However, the correlation coefficient was weaker than those found in the heart tissue, muscle fiber, or brain tissue [10, 12, 17, 24]. In this study, strong variabilities of mtDNA 4977 bp deletion load were observed even within each age group. The highest deletion rate of 1.436% (0.2086%) was from a 75-year-old subject rather than the oldest subject (91-year-old). This deletion load was substantially lower than the rate of 2.93% measured in brain tissue in another study [12]. Interestingly, a deletion rate of 0.3657% (0.03035%) was found in a 29-year-old individual. This deletion amount was approximately 100 times higher than the lowest one from a subject in the same age decade. The sample belonged to a healthy man without clinical symptoms of heart diseases, Pearson's syndrome, KSS, Alzheimer's disease, or the other obvious diseases. But his maternal grandmother had died of Parkinson's disease, which has been recognized as a high mtDNA deletion disease [26]. Although genetic factors were not systematically analyzed in this study, these may be one of the determinants of variability in the rates of mtDNA 4977 bp deletion within groups of subjects with similar ages.
CONCLUSION
Our data suggest that the mtDNA 4977 bp deletion is a common phenomenon in hair and increases with age. These findings may lead to a better understanding of the distribution of mtDNA deletions in human tissues.
ACKNOWLEDGMENTS
We are indebted to all the volunteers who provided hair samples for this study. This research was partially supported by the Science & Research Program of Guangdong Province (NO. 2006B35502007), the Guangdong Province Science Foundation (NO. 04105351), and the summer student program of Sun Yat-Sen University. We are also grateful to Dr. Chaoquan Luo (Department of Biochemistry and Molecular Biology, Sun Yat-Sen University) for the useful advice on this paper. We also thank the editors of Editage Company for professional editing of the article.
DECLARATION OF INTEREST
Authors do not have any commercial affiliations, or potential conflicts of interest associated with this work submitted for publication
REFERENCES
- [1].Turchi C, Buscemi L, Giacchino E, Onofri V, Fendt L, Parson W, et al. Polymorphisms of mtDNA control region in Tunisian and Moroccan populations: an enrichment of forensic mtDNA databases with Northern Africa data. Forensic Sci Int Genet. 2009;3:166–172. doi: 10.1016/j.fsigen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- [2].Haigis MC, Yankner BA. The aging stress response. Mol Cell. 2010;40:333–344. doi: 10.1016/j.molcel.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Indo HP, Davidson M, Yen HC, Suenaga S, Tomita K, Nishii T, et al. Evidence of ROS generation by mitochondria in cells with impaired electron transport chain and mitochondrial DNA damage. Mitochondrion. 2007;7:106–118. doi: 10.1016/j.mito.2006.11.026. [DOI] [PubMed] [Google Scholar]
- [4].Rotig A, Poulton J. Genetic causes of mitochondrial DNA depletion in humans. Biochim Biophys Acta. 2009;1792:1103–1108. doi: 10.1016/j.bbadis.2009.06.009. [DOI] [PubMed] [Google Scholar]
- [5].Alberio S, Mineri R, Tiranti V, Zeviani M. Depletion of mtDNA: syndromes and genes. Mitochondrion. 2007;7:6–12. doi: 10.1016/j.mito.2006.11.010. [DOI] [PubMed] [Google Scholar]
- [6].Jou MJ, Peng TI, Wu HY, Wei YH. Enhanced generation of mitochondrial reactive oxygen species in cybrids containing. 4977-bp mitochondrial DNA deletion. Ann N Y Acad Sci. 2005;1042:221–228. doi: 10.1196/annals.1338.024. [DOI] [PubMed] [Google Scholar]
- [7].Ma YS, Wu SB, Lee WY, Cheng JS, Wei YH. Response to the increase of oxidative stress and mutation of mitochondrial DNA in aging. Biochim Biophys Acta. 2009;1790:1021–1029. doi: 10.1016/j.bbagen.2009.04.012. [DOI] [PubMed] [Google Scholar]
- [8].Wiesner RJ, Zsurka G, Kunz WS. Mitochondrial DNA damage and the aging process: facts and imaginations. Free Radic Res. 2006;40:1284–1294. doi: 10.1080/10715760600913168. [DOI] [PubMed] [Google Scholar]
- [9].Pavicic WH, Richard SM. Correlation analysis between mtDNA 4977-bp deletion and ageing. Mutat Res. 2009;670:99–102. doi: 10.1016/j.mrfmmm.2009.07.009. [DOI] [PubMed] [Google Scholar]
- [10].Pesce V, Cormio A, Fracasso F, Vecchiet J, Felzani G, Lezza AM, et al. Age-related mitochondrial genotypic and phenotypic alterations in human skeletal muscle. Free Radic Biol Med. 2001;30(11):1223–33. doi: 10.1016/s0891-5849(01)00517-2. [DOI] [PubMed] [Google Scholar]
- [11].Mohamed SA, Hanke T, Erasmi AW, Bechtel MJ, Scharfschwerdt M, Meissner C, et al. Mitochondrial DNA deletions and the aging heart. Exp Gerontol. 2006;41:508–517. doi: 10.1016/j.exger.2006.03.014. [DOI] [PubMed] [Google Scholar]
- [12].Meissner C, Bruse P, Mohamed SA, Schulz A, Warnk H, Storm T, et al. The 4977 bp deletion of mitochondrial DNA in human skeletal muscle, heart and different areas of the brain: a useful biomarker or more? Exp Gerontol. 2008;43:645–652. doi: 10.1016/j.exger.2008.03.004. [DOI] [PubMed] [Google Scholar]
- [13].Meissner C, Bruse P, Oehmichen M. Tissue-specific deletion patterns of the mitochondrial genome with advancing age. Exp Gerontol. 2006;41:518–524. doi: 10.1016/j.exger.2006.03.010. [DOI] [PubMed] [Google Scholar]
- [14].Fernandez-Vizarra E, Enriquez JA, Perez-Martos A, Montoya J, Fernandez-Silva P. Tissue-specific differences in mitochondrial activity and biogenesis. Mitochondrion. 2010;11:207–213. doi: 10.1016/j.mito.2010.09.011. [DOI] [PubMed] [Google Scholar]
- [15].Rock A, Irwin J, Dur A, Parsons T, Parson W. SAM: String-based sequence search algorithm for mitochondrial DNA database queries. Forensic Sci Int Genet. 2011;5:126–132. doi: 10.1016/j.fsigen.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zheng Y, Wang X, Yang M. A very large scale deletion mutation in human peripheral blood cells mitochondrial DNA. Acad Mil Med Univ. 2000;21:586–587. [Google Scholar]
- [17].Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, et al. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am J Hum Genet. 2006;79:469–480. doi: 10.1086/507132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pogozelski WK, Hamel CJ, Woeller CF, Jackson WE, Zullo SJ, Fischel-Ghodsian N, et al. Quantification of total mitochondrial DNA and the 4977-bp common deletion in Pearson's syndrome lymphoblasts using a fluorogenic 5’-nuclease (TaqMan) real-time polymerase chain reaction assay and plasmid external calibration standards. Mitochondrion. 2003;2:415–427. doi: 10.1016/S1567-7249(03)00033-3. [DOI] [PubMed] [Google Scholar]
- [19].Lim PS, Cheng YM, Wei YH. Large-scale mitochondrial DNA deletions in skeletal muscle of patients with end-stage renal disease. Free Radic Biol Med. 2000;29:454–463. doi: 10.1016/s0891-5849(00)00334-8. [DOI] [PubMed] [Google Scholar]
- [20].Kotake K, Nonami T, Kurokawa T, Nakao A, Murakami T, Shimomura Y. Human livers with cirrhosis and hepatocellular carcinoma have less mitochondrial DNA deletion than normal human livers. Life Sci. 1999;64:1785–1791. doi: 10.1016/s0024-3205(99)00117-4. [DOI] [PubMed] [Google Scholar]
- [21].von Wurmb-Schwark N, Schwark T, Caliebe A, Drenske C, Nikolaus S, Schreiber S, et al. Low level of the mtDNA(4977) deletion in blood of exceptionally old individuals. Mech Ageing Dev. 2010;131:179–184. doi: 10.1016/j.mad.2010.01.005. [DOI] [PubMed] [Google Scholar]
- [22].Alonso A, Martin P, Albarran C, Garcia P, Garcia O, de Simon LF, et al. Real-time PCR designs to estimate nuclear and mitochondrial DNA copy number in forensic and ancient DNA studies. Forensic Sci Int. 2004;139:141–149. doi: 10.1016/j.forsciint.2003.10.008. [DOI] [PubMed] [Google Scholar]
- [23].Alonso A, Martin P, Albarran C, Garcia P, Primorac D, Garcia O, et al. Specific quantification of human genomes from low copy number DNA samples in forensic and ancient DNA studies. Croat Med J. 2003;44:273–280. [PubMed] [Google Scholar]
- [24].Meissner C, von Wurmb N, Oehmichen M. Detection of the agedependent 4977 bp deletion of mitochondrial DNA. A pilot study. Int J Legal Med. 1997;110:288–291. doi: 10.1007/s004140050089. [DOI] [PubMed] [Google Scholar]
- [25].Dai P, Yang W, Jiang S, Gu R, Yuan H, Han D, et al. Correlation of cochlear blood supply with mitochondrial DNA common deletion in presbyacusis. Acta Otolaryngol. 2004;124:130–136. doi: 10.1080/00016480410016586. [DOI] [PubMed] [Google Scholar]
- [26].Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]


