CRISPR–Cas9-induced mutations in intraflagellar transport (IFT) motors reveal that IFT-specific dynein and cytoplasmic dynein have unique compositions but share components and regulatory mechanisms.
Abstract
Cilium formation and maintenance require intraflagellar transport (IFT). Although much is known about kinesin-2–driven anterograde IFT, the composition and regulation of retrograde IFT-specific dynein remain elusive. Components of cytoplasmic dynein may participate in IFT; however, their essential roles in cell division preclude functional studies in postmitotic cilia. Here, we report that inducible expression of the clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9 system in Caenorhabditis elegans generated conditional mutations in IFT motors and particles, recapitulating ciliary defects in their null mutants. Using this method to bypass the embryonic requirement, we show the following: the dynein intermediate chain, light chain LC8, and lissencephaly-1 regulate retrograde IFT; the dynein light intermediate chain functions in dendrites and indirectly contributes to ciliogenesis; and the Tctex and Roadblock light chains are dispensable for cilium assembly. Furthermore, we demonstrate that these components undergo biphasic IFT with distinct transport frequencies and turnaround behaviors. Together, our results suggest that IFT–dynein and cytoplasmic dynein have unique compositions but also share components and regulatory mechanisms.
Introduction
Cilia are microtubule-based structures that play diverse roles in motility, sensory perception, and signaling (Rosenbaum and Witman, 2002; Scholey, 2003; Eggenschwiler and Anderson, 2007; Ishikawa and Marshall, 2011). The assembly and maintenance of cilia require bidirectional intraflagellar transport (IFT), which ferries ciliary precursors bound to the IFT particle protein complex from the base of the ciliary axoneme to the tip (Rosenbaum and Witman, 2002; Scholey, 2003, 2013). Anterograde IFT is powered either by a single heterotrimeric kinesin-2 motor, which was first proposed in Chlamydomonas reinhardtii, or by the concerted action of a heterotrimeric kinesin-2 and a homodimeric OSM-3-kinesin in kinesin-2 family in Caenorhabditis elegans neuronal cilia (Rosenbaum and Witman, 2002; Scholey, 2003, 2013). Retrograde IFT is driven by a special cytoplasmic dynein that is proposed to recycle the anterograde motors, IFT particles, and breakdown products of ciliary turnover back to the cell body (Pazour et al., 1998, 1999; Porter et al., 1999; Signor et al., 1999; Rosenbaum and Witman, 2002; Scholey, 2003).
Cytoplasmic dyneins include cytoplasmic dynein 1 (DYNC1) and the IFT-specific cytoplasmic dynein 2 (DYNC2; Pfister et al., 2005). DYNC1 consists of a heavy chain homodimer, intermediate chains, light intermediate chains, and light chain dimers of the LC8, Tctex1, and Roadblock protein families (Kardon and Vale, 2009). These accessory subunits contribute to the structural integrity, motor regulation, and cargo specificity of the dynein complex. Moreover, cytoplasmic dynein motor activity appears to be governed by the Lis1–NudEL complex and the dynactin complex (Kardon and Vale, 2009; Roberts et al., 2013). In contrast, the composition and regulation of IFT–dynein remain unclear. Biochemical studies in C. reinhardtii revealed that IFT–dynein contains the heavy chain DHC1b, intermediate chains FAP133 and FAP163, light intermediate chain D1bLIC, and light chain LC8 (Pazour et al., 1998, 1999; Porter et al., 1999; Hou et al., 2004; Rompolas et al., 2007; Patel-King et al., 2013). A recent study using a temperature-sensitive allele of DHC1b demonstrated that retrograde IFT regulates flagellar assembly and function but not maintenance (Engel et al., 2012). Genetic research in Caenorhabditis elegans revealed that mutations of che-3 (DHC1b homologue) or xbx-1 (D1bLIC) caused defects in retrograde IFT (Signor et al., 1999; Schafer et al., 2003).
Identification of IFT–dynein components largely relied on biochemical fractionation and proteomic analysis of flagellar proteins in C. reinhardtii, and this strategy does not differentiate between the integral components and the cargo molecules of IFT–dynein, as axonemal dynein complexes share subunits with IFT and cytoplasmic dyneins (e.g., LC8; Pazour et al., 1998). Moreover, mass spectrometry may not be sensitive enough to detect IFT regulators present at substoichiometric concentrations. The C. elegans neuronal cilia provide a complementary system to study IFT–dynein using genetic and imaging approaches. However, the efficiency of RNAi in knocking down gene activity in C. elegans ciliated neurons has been very limited (Pastrana, 2010). Moreover, the use of RNAi may lead to variable penetrance of the phenotype due to the incompleteness of protein depletion, and may also be accompanied by off-target effects (Pastrana, 2010). Thus, new techniques that are efficient and robust are needed to access the role of the target gene in cilia.
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) system has been recently demonstrated as a powerful tool for genome editing (for review see Hsu et al., 2014). The specificity of Cas9 nuclease is determined by a synthetic guide RNA (sgRNA) that base-pairs with a G(N)19NGG “protospacer” DNA sequence in the genome. Cas9 generates double-strand breaks, which are often inaccurately repaired by the nonhomologous end joining pathway, causing frameshift insertions or deletions (indels) in the target locus. We have developed the somatic CRISPR–Cas9 technique in which CRISPR–Cas9 is expressed under the control of an inducible or tissue-specific promoter to generate conditional mutants in C. elegans (Shen et al., 2014). Here, we first demonstrate that somatic CRISPR–Cas9 efficiently creates conditional mutations in ciliated neurons. Using this platform, we bypass the embryonic requirement of dynein-associated chains and investigate their roles in the construction of C. elegans neuronal cilia.
Results and discussion
Generation of conditional mutants in ciliated neurons of C. elegans
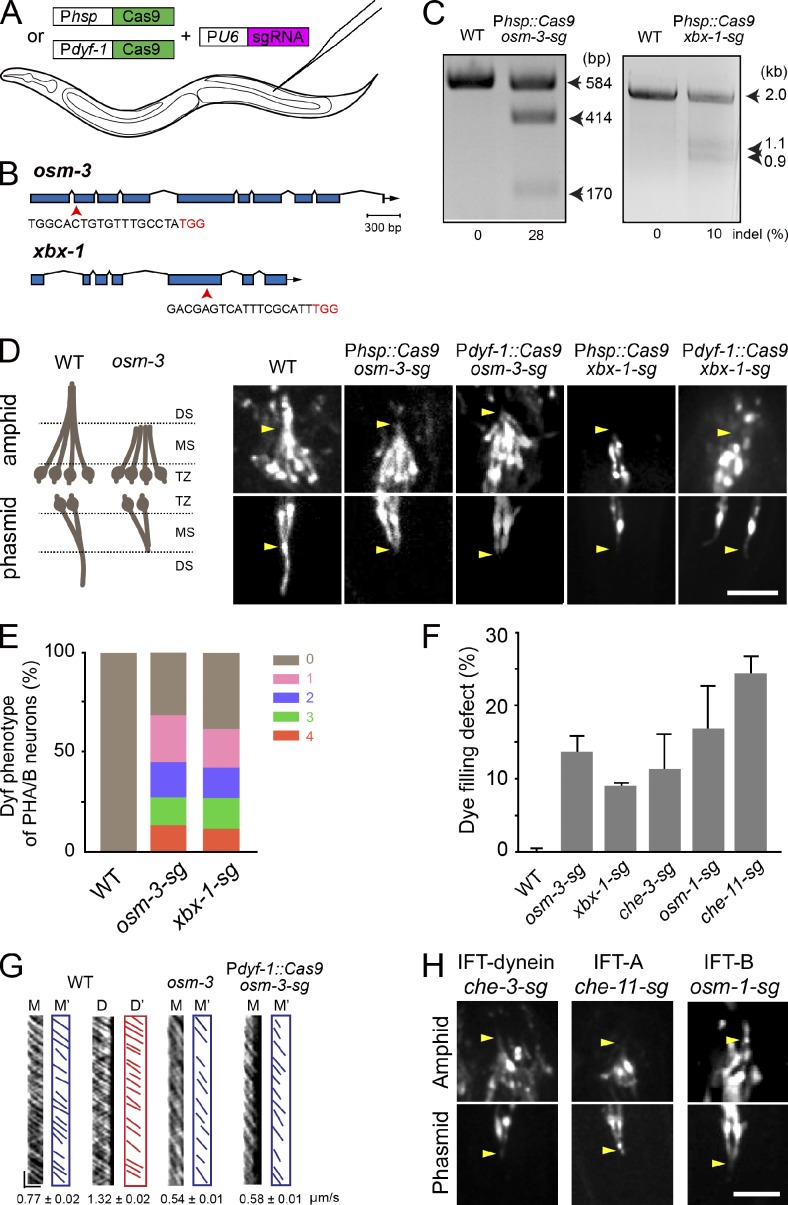
We generated transgenic animals that specifically express Cas9 under the control of either a heat-shock-inducible promoter (Phsp-16.2 or Phsp) or a ciliated neuron-specific promoter (Pdyf-1) and that ubiquitously express the sgRNA under the control of the U6 gene promoter (PU6; Fig. 1 A; Ou et al., 2005; Shen et al., 2014). We first designed sgRNA that was complementary to the coding sequence of the osm-3 gene or the xbx-1 gene (Fig. 1 B). osm-3 encodes a homodimeric kinesin-2 that is essential for building the ciliary distal segment (Snow et al., 2004), and xbx-1 encodes an IFT–dynein light intermediate chain (Schafer et al., 2003). We chose to target osm-3 and xbx-1 on the basis of their easily identifiable defects in the cilium structure, IFT, and uptake of a fluorescent dye (abnormal dye filling [Dyf] phenotype). To achieve temporally controlled mutation, the expression of Cas9 was controlled by Phsp (Fig. 1, B and C). We transformed a plasmid containing Cas9 and sgRNA into C. elegans, and in the resultant transgenic animals, we refer to the sgRNA targeting osm-3 or xbx-1 as osm-3-sg or xbx-1-sg, respectively. Because T7 endonuclease I (T7EI) recognizes and cleaves the imperfectly matched DNA (Hsu et al., 2014), we applied the T7EI assay to detect the indels in osm-3-sg and xbx-1-sg animals whose embryos were treated with heat shock. We first PCR-amplified the genomic DNA fragments containing the target sites from these transgenic animals. After T7EI digestion, these PCR fragments were cut into two small fragments of the expected sizes; however, the fragments were intact in wild-type (WT) animals (Fig. 1 C), which indicates that somatic CRISPR–Cas9 induced molecular lesions in the osm-3 and xbx-1 loci.
Figure 1.
The somatic CRISPR–Cas9 technique generates conditional mutations in IFT components in C. elegans ciliated sensory neurons. (A) Schematic illustrating the use of somatic CRISPR–Cas9 for creating conditional mutants. (B) osm-3 and xbx-1 gene models. Exons are in blue, and arrows indicate sgRNA sequences corresponding to exons (Table S4). The NGG (PAM) sequences are denoted in red. (C) Representative gels showing the T7EI assay results for osm-3 and xbx-1. Indels are indicated at the bottom. (D, left) Schematic of cilia in WT and osm-3 animals. DS, distal segments; MS, middle segments; TZ, transition zone. (D, right) The morphology of amphid (top) and phasmid (bottom) cilia visualized using OSM-6::GFP. Arrowheads indicate junctions between the middle and distal segments. The length of the cilia are 8.2 ± 0.1 µm (amphids [A]) or 7.9 ± 0.3 µm (phasmids [P]) in WT, 4.8 ± 0.1 µm (A) or 3.9 ± 0.2 (P) µm in Phsp::Cas9; osm-3-sg, and 4.5 ± 0.1 µm (A) or 3.5 ± 0.1 µm (P) in Pdyf-1::Cas9; osm-3-sg. n = 23–81. Mean ± SE. Bar, 5 µm. (E) Dye-filling defects under a 100× objective lens. 0–4 and their respective colors represent the number of PHA/B neurons defective in dye-filling assays. n = 26–51. (F) Dye-filling defects under a fluorescence stereoscope. n = 150–315. Mean ± SE (error bars). (G) Kymographs (M or D) and corresponding lines (M’ or D’) of OSM-6::GFP motility. Error bars indicate mean ± SE. Horizontal bar, 2 µm; vertical bar, 5 s. n = 120–323. (H) Cilium morphology in conditional mutants visualized by OSM-6::GFP. Arrowheads indicate the positions where the junctions between the middle and distal segments should be in WT animals. Bar, 5 µm.
We next used a compound microscope with a 100× objective lens to examine the Dyf phenotype (Fig. S1). By examining four phasmid ciliated neurons (PHA/B/L/R) of osm-3-sg and xbx-1-sg conditional mutants, we found that 69% and 62% of the animals showed defects in DiI uptake, respectively (n = 51 and 26; Figs. 1 E and S1 A). All neurons showed complete uptake of DiI in 100% of WT animals (n > 100; Fig. 1, E and F). We noticed that the mosaic Dyf phenotype ranged from 14% to 24% for osm-3-sg and from 12% to 19% for xbx-1-sg in one to four phasmid neurons (Fig. 1 E), which is a common feature in conditional mutants. We also assessed the Dyf phenotype using a fluorescence stereoscope and detected the lower Dyf penetrance (Fig. 1 F). This result may have occurred because the stereoscope is sensitive enough to detect fluorescence from a single neuron, and we only scored the Dyf phenotype in animals with phasmid neurons that completely failed to take up DiI (Figs. 1 E and S1, A and B).
To induce spatially controlled mutations, we used Pdyf-1 to express Cas9. Under a fluorescence stereoscope, we found that 92% (n = 50) and 56% (n = 27) of transgenic animals expressing Pdyf-1::Cas9; osm-3-sg and Pdyf-1::Cas9; xbx-1-sg showed the Dyf phenotype, respectively. Thus, the expression of Cas9 under the control of a heat-shock promoter or a ciliary-specific promoter allows efficient creation of conditional mutants of osm-3 and xbx-1. By comparing mutation efficacies and the penetrance of ciliary phenotypes between osm-3 and xbx-1, we noticed that the higher indel frequency (28% for osm-3, 10% for xbx-1) may correlate with the higher dye filling defect (15% for osm-3, 9% xbx-1; Fig. 1, C and F).
Next, we examined the defects in cilium structure and IFT in osm-3-sg and xbx-1-sg animals using the OSM-6/IFT52::GFP reporter, which undergoes biphasic IFT along the entire cilia (Fig. 1 D). The osm-3-sg animals lost their ciliary distal segments, and the cilium length was reduced from 8 µm to 4–5 µm (Fig. 1 D), phenocopying osm-3(p802) null mutants (Snow et al., 2004). We further performed live imaging analysis of IFT in osm-3-sg mutants and found that the IFT velocity was reduced from 0.77 ± 0.02 µm/s (mean ± SE, n = 213) in the WT animals to 0.58 ± 0.01 µm/s (n = 323) in the remaining middle segments, which is also comparable to the IFT speed in osm-3(p802) mutants (Fig. 1 G). In xbx-1-sg conditional mutants, OSM-6::GFP formed aggregates around the transition zone and no IFT was detected in the residual cilia (Fig. 1 D), which are similar to those observed in xbx-1(ok279) deletion mutants (Schafer et al., 2003). To extend our initial successes to other IFT components, we conditionally mutated the IFT–dynein heavy chain (che-3), IFT particle A (che-11), and IFT particle B (osm-1) components by using the heat-shock strategy (Signor et al., 1999; Scholey, 2003). We showed that these conditional mutants reproduced the defects in cilium structure caused by their corresponding null alleles, and that the Dyf penetrance ranged from 10% to 25% in four phasmid ciliated neurons (Fig. 1, F and H). Collectively, somatic CRISPR–Cas9 can be used to generate conditional mutations of IFT motors (osm-3, xbx-1, and che-3) and IFT particles (che-11 and osm-1).
The functions of cytoplasmic dynein components in ciliogenesis
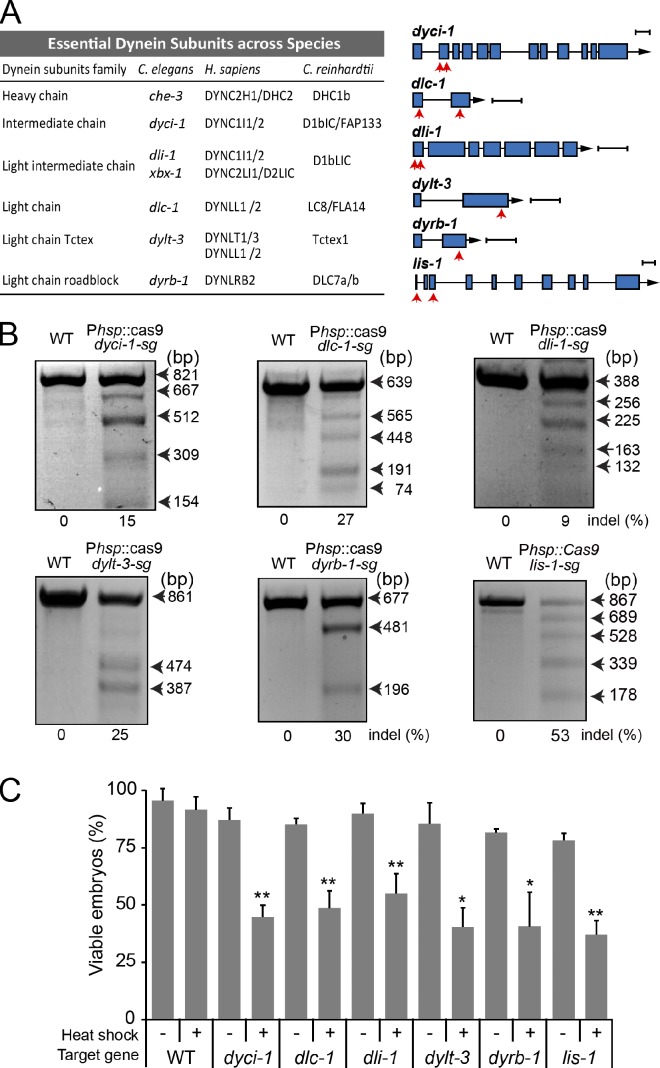
The C. elegans genome encodes 14 dynein-associated chains (Hao et al., 2011), five of which are essential for embryonic development (WormBase), including the intermediate chain (DYCI-1), light intermediate chain (DLI-1), light chain LC8 (DLC-1), Tctex-type light chain (DYLT-3), and roadblock-type light chain (DYRB-1; Fig. 2 A). We first generated transgenic animals that expressed Cas9 under the control of the Phsp promoter and ubiquitously expressed sgRNAs that target one site for dylt-3 and dyrb-1 or two sites for dyci-1, dlc-1, and dli-1 (Fig. 2 A). Our T7EI assays detected the expected molecular lesions (Fig. 2 B). Although the heat-shock treatment did not alter the embryonic viability of WT embryos, the conditional mutant embryos exhibited embryonic lethality with penetrances ranging from 40% to 55% after heat shock (Fig. 2 C). These data indicated that we generated conditional mutants of embryonically essential dynein subunits.
Figure 2.
Conditional mutations in six embryonically essential dynein components. (A, left) The essential dynein subunits and known IFT–dynein components across species. (A, right) Gene models of dyci-1, dlc-1, dli-1, dylt-3, dyrb-1, and lis-1. Exons are in blue and red arrows indicate one or two sgRNA sequences targeting dynein components (Table S4). Bars, 300 bp. (B) Representative gels showing the results of T7EI assays. One target site generated two small fragments. (C) Embryonic lethality was determined by quantifying viable embryos after heat-shock treatment. n = 97–242; mean ± SE (error bars); *, P < 0.05; **, P < 0.01.
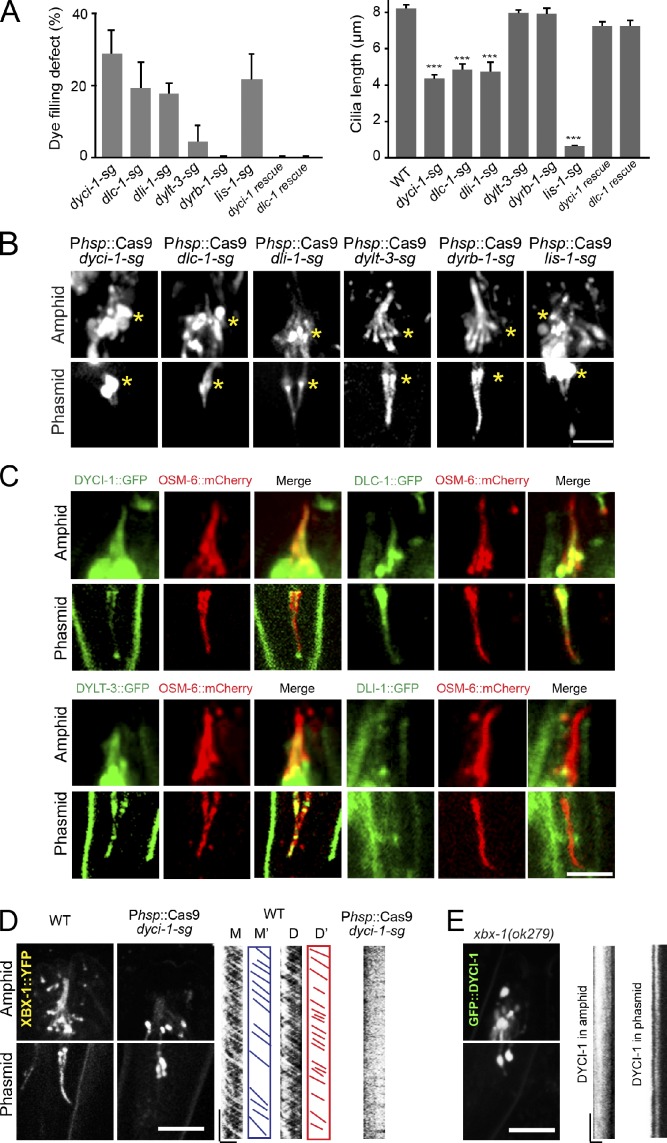
Next, we used these conditional mutants to study the contribution of the dynein components to cilium formation. We first showed that 29% (dyci-1), 19% (dlc-1), and 18% (dli-1) of the conditional mutants failed to take up DiI, whereas <4% of dylt-3 or none of dyrb-1 conditional mutants developed the Dyf phenotype (Fig. 3 A). Consistently, we found that the cilium length was significantly reduced from 8.3 µm in WT animals to 4.4 µm (dyci-1), 4.9 µm (dlc-1), and 4.8 µm (dli-1) in the conditional mutants but was not altered in dylt-3-sg or dyrb-1-sg animals (Fig. 3, A and B). IFT cargo includes IFT particles and ciliary receptors such as a transient receptor potential vanilloid channel OSM-9 (Qin et al., 2005). We did not find that OSM-6::GFP or OSM-9::GFP moved in the remaining cilia of the dyci-1-sg and dlc-1-sg conditional mutants (Figs. 3 B and S3 A). These ciliary defects are similar to those detected in che-3– and xbx-1–null mutants. To further test whether DYCI-1 and DLC-1 function with IFT–dynein, we determined the cellular localization of GFP-tagged DYCI-1 and DLC-1. We showed that they were distributed along the cilia and underwent the same biphasic IFT as XBX-1 (Fig. 3 C and see Fig. 5 B). Furthermore, we genetically introduced XBX-1::YFP into the dyci-1-sg conditional mutant and GFP::DYCI-1 into the xbx-1(ok279) mutant. We found that XBX-1::YFP and GFP::DYCI-1 formed aggregates around the transitional zone and were not motile in cilia (Fig. 3, D and E). The interdependence of XBX-1 and DYCI-1 in IFT further indicated that they may function together with IFT–dynein.
Figure 3.
Ciliary defects in conditional mutants of embryonically essential dynein components. (A) Defects of dye-filling assays (left) and the cilium length (right). Mean ± SE (error bars); n = 34–381 from three generations. ***, P < 0.001. (B) Amphid and phasmid cilia visualized by OSM-6::GFP. Asterisks indicate transition zones. Bar, 5 µm. (C) Ciliary localization of DYCI-1, DLC-1, DYLT-3, and DLI-1 (GFP) and OSM-6::mCherry (red). (D and E) Localization (left) and motility (kymographs, right) of XBX-1::YFP in WT and dyci-1 conditional mutants (D) or GFP::DYCI-1 in xbx-1 mutants (E). Micrograph bar, 5 µm; kymograph horizontal bar, 2 µm; vertical bar, 5 s.
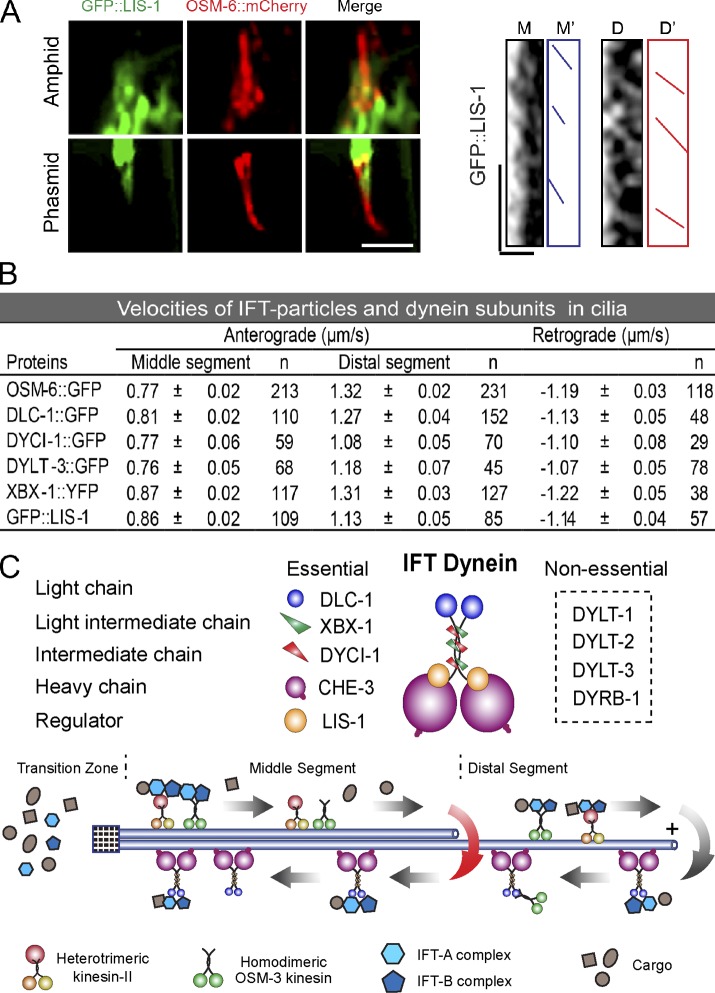
Figure 5.
IFT of LIS-1, IFT velocities, and the proposed model. (A) Colocalization of GFP::LIS-1 with OSM-6::mCherry (red) in cilia (left), and a kymograph of GFP::LIS-1 motility (right). Micrograph bar, 5 µm; kymograph horizontal bar, 2 µm; vertical bar, 5 s. (B) Velocities of IFT particle and dynein subunits (mean ± SE). (C) A proposed model of subunits in the IFT–dynein complex and multiple turnaround sites on the cilium.
We studied how DLI-1 contributes to ciliogenesis. We did not find that OSM-6::GFP formed aggregates within the cilia of dli-1-sg conditional mutants (Fig. 3 B), nor did we detect the ciliary localization of DLI-1::GFP in transgenic animals expressing the Pdli-1::dli-1::GFP or Pdyf-1::gfp::dli-1 reporter (Figs. 3 C and S2 A). Instead, we found that DLI-1::GFP was distributed along the dendrite in these neurons (Fig. S2 B) and that OSM-6::GFP formed aggregates in the dendrites of dli-1-sg conditional mutants (Fig. S2 C). To further examine the function of DLI-1 in the retrograde dendritic transport, we compared the transport velocities of OSM-6::GFP in the dendrites of WT and dli-1 conditional mutants. Consistent with the previous measurements (Signor et al., 1999), OSM-6::GFP moved along the dendrite at the anterograde speed of 0.75 ± 0.05 µm/s (n = 33) and at 0.90 ± 0.04 µm/s (n = 30) in WT animals. However, we did not detect the retrograde movement of OSM-6::GFP in the dendrites of the dli-1 conditional mutants, whereas we found that OSM-6::GFP moved at the anterograde speed of 0.68 ± 0.02 µm/s (n = 33) in these dendrites. These results indicate that DLI-1 indirectly contributes to cilium formation by regulating the retrograde dendritic transport.
Dynein subunits undergo diverse IFT
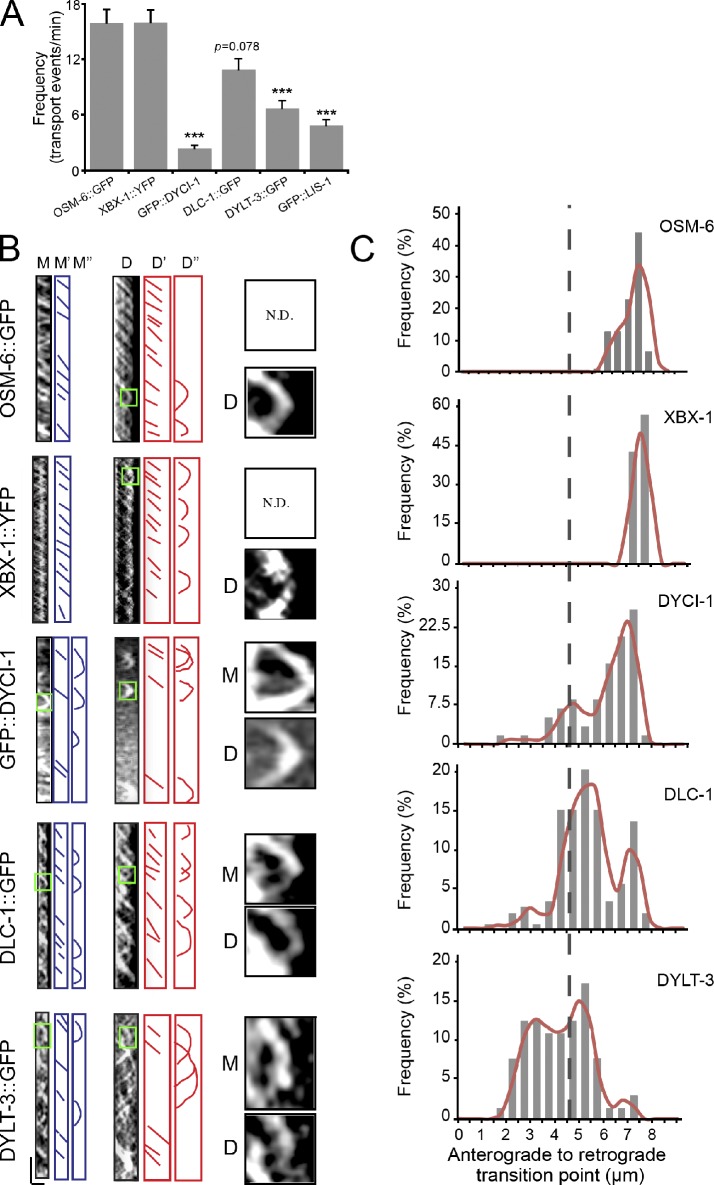
To understand the transport behavior of IFT–dynein subunits, we generated kymographs from time-lapse movies of IFT of GFP-tagged dynein subunits (Videos 1–4). We first quantified the transport frequency as the number of transport events per minute. OSM-6::GFP and XBX-1::YFP both showed a frequency of 16 events/min; however, DYCI-1, DLC-1, and DYLT-3 displayed frequencies of ∼3, 9, and 6 events/min, respectively (Fig. 4 A). We next examined their turnaround sites. The majority of OSM-6 and XBX-1 turned around at the distal tip, whereas DYCI-1, DLC-1, and DYLT-3 returned at the middle and distal segments (Fig. 4, B and C). 76% of the GFP::DYCI-1 puncta returned along the distal segment and very few reached the distal tip, and 24% of DYCI-1 turned around at the tip of the middle segment. 60% of the DLC-1::GFP puncta turned back at the distal segment, whereas 40% of DLC-1 returned at the middle tip. Finally, 69% of the DYLT-3::GFP puncta returned in the middle segment, not even reaching the middle tip (Fig. 4, B and C). These data suggest that multiple retrograde IFT pathways may operate within a single cilium.
Figure 4.
Transport behaviors of dynein subunits in cilia. (A) IFT frequencies of IFT particle and dynein subunits. n = 64–212; mean ± SE (error bars). ***, P ≤ 0.001. (B) Kymographs (left) of DYCI-1, DLC-1, DYLT-3, and XBX-1 in the middle (M) or distal segments (D). M″ and D″ indicate turnaround events. M and D (right) are 4× enlarged images of turnarounds in the green boxes on the left. Horizontal bar, 2 µm; vertical bar, 5 s. (C) Distribution of turnaround events along the cilia. The y axis indicates the percentage of turnaround events at the specific region of cilia among the total turnaround events. n = 58–140. The broken line shows the junction between the middle and distal segments.
Lissencephaly-1 regulates retrograde IFT
We studied the function of lissencephaly-1 in cilium formation. Lis1 is thought to function as a “clutch” to suppress cytoplasmic dynein motility and to cause the dynein to tightly bind microtubules (Huang et al., 2012). We generated lis-1-sg conditional mutants that exhibited the expected molecular lesions and embryonic lethality (Fig. 2, B and C). In lis-1-sg animals, we found that 22% of worms failed to take up DiI and reduced the cilium length, and that OSM-6::GFP did not move but rather aggregated along the residual cilia (Fig. 3, A and B), which indicates that the function of LIS-1 is similar to that of CHE-3 and XBX-1 in retrograde IFT. We then visualized GFP-tagged LIS-1 in ciliated neurons and showed that GFP::LIS-1 entered the middle segment of cilia and occasionally moved into the distal segment (Fig. 5, A and B). We detected sparse and biphasic IFT of GFP::LIS-1 puncta (Fig. 5, A and B). Thus, LIS-1 undergoes IFT and regulates retrograde IFT.
In conclusion, this study demonstrated that embryonically essential dynein-associated chains behave distinctly in C. elegans neuronal cilia: the intermediate chain DYCI-1, the light chain LC8/DLC-1, and the dynein regulator LIS-1 undergo biphasic IFT and are essential for retrograde IFT; the light intermediate chain DLI-1 regulates the dendritic transport of IFT particles; the light chain Tctex-type DYLT-3 and roadblock-type DYRB-1 are dispensable for ciliogenesis (Fig. 5, A and B). These findings suggest that cytoplasmic dynein and IFT–dynein have unique compositions but share essential components and regulatory mechanisms.
To visualize IFT of dynein subunits, this work relied on the GFP fusion technique, a standard protocol that has generated reliable results including ones related to IFT–dynein (Mello et al., 1991; Signor et al., 1999; Schafer et al., 2003). Despite the fact that we injected DNA with a low concentration (∼10 ng/µl), below which the GFP fluorescence is invisible, and that we did not notice any ciliary phenotype in these animals, this method can cause the overexpression of the transgene. Dynein is a holoenzyme complex with a strict stoichiometry, and the overexpression of subunits or GFP tagging could result in artifacts because some complexes would be disrupted or not fully assembled. We thus performed the rescue experiments to examine whether the overexpression of GFP-tagged dynein subunits could rescue ciliary phenotypes in the corresponding conditional mutants. We studied dyci-1 and dlc-1 as they regulate ciliogenesis. In these experiments, we first made all the possible synonymous mutations in the target sites of transgenes such that they would not be cleaved by CRISPR–Cas9 (Fig. S3 B). We transformed GFP-tagged mdyci-1 (m for synonymous mutation) and mdlc-1 into conditional mutants, and we did not detect any Dyf phenotype after inducing Cas9 expression by heat-shock treatment (GFP::mdyci-1, n = 59; mdlc-1::GFP, n = 87; Fig. 3 A). We further examined the ciliary structure and measured the cilium length in these animals, and we did not find any obvious abnormality compared with those in WT animals (Figs. 3 A and S3 C). These rescue results indicated that GFP fusion or overexpression of DYCI-1 and DLC-1 does not affect IFT–dynein’s function. Because the potential off-target effect is a major concern for Cas9, our results not only demonstrated the functionality of GFP fusion, but also alleviated the concern of off-target cleavages.
The GFP-tagged dynein subunits undergo IFT at the same speeds as other IFT motors (Fig. 5 B), which suggests that they are probably associating with the dynein complex. We did not detect DLI-1::GFP in the cilia using the same expression strategy, suggesting that the ciliary location of GFP-tagged dynein subunits can be specific (Fig. 3 C). It remains unknown why DLI-1 only functions within the dendrites but does not enter cilia. The emerging evidence suggests that a selective ciliary gate may regulate protein trafficking between cilia and the general cytoplasm (Ishikawa and Marshall, 2011), and DLI-1 may not be able to pass such a gate at the base of cilia.
Our results with IFT–dynein are consistent with biochemical data from other organisms, and also provide new insights. For example, LC8/DLC-1 was proposed as an essential component for retrograde IFT from C. reinhardtii (Pazour et al., 1998). However, LC8 can be the subunit or cargo—or both—of IFT–dynein, as it is a known component in axonemal dyneins. Our study from nonmotile cilia enables us to conclude that LC8 can be an IFT–dynein subunit. Similarly, a recent study used the biochemical approach to study IFT–dynein composition in vertebrates, and their data are also largely complementary to ours (Asante et al., 2014). However, Lis1 was not identified in IFT–dynein fraction but was localized in cilia by immunofluorescence (Asante et al., 2014). We found that GFP::LIS-1 was not associated with IFT–dynein all the time (Fig. 5 A), and the biochemical fractionation must be difficult to detect the transient interaction. Our data also indicate that DYCI-1 is an integral IFT–dynein component. The previous work using a transcriptional GFP reporter did not detect the expression of dyci-1 in C. elegans ciliated neurons (Hao et al., 2011). By increasing the length of the 5′ UTR regulatory sequence, we detected the expression of Pdyci-1::gfp in ciliated neurons (Fig. 3 C). Consistently, biochemical studies identified FAP133/DYCI-1 as an IFT–dynein component (Rompolas et al., 2007), and RNAi experiments in Trypanosome brucei showed that DIC5/DYCI-1 regulates retrograde IFT (Blisnick et al., 2014). Moreover, we showed that DYLT-3 and DYRB-1 do not regulate ciliogenesis (Fig. 3, A and B). Dynein-associated chains may complement each other in cilium formation, and the generation of double or multiple conditional mutations by somatic CRISPR–Cas9 may dissect their redundant roles in ciliogenesis. Interestingly, the single or double genetic deletions of C. elegans Tctex-type light chain DYLT-1 or DYLT-2 do not cause defects in cilium assembly, even though they undergo biphasic IFT (Hao et al., 2011). We cannot exclude the possibility that these dynein light chains regulate ciliogenesis, but they may also likely modulate currently unidentified physiological events in cilia.
Lis1 has been found to induce slow or nonmoving dynein (Huang et al., 2012). We showed that GFP::LIS-1 is distributed in the middle segment (Fig. 5 A), where it may inactivate IFT–dynein during anterograde IFT. We observed biphasic but infrequent IFT of LIS-1 in C. elegans (Fig. 5, A and B), which is consistent with the brief motility of Lis1 in Aspergillus nidulans (Egan et al., 2012). Because limited GFP::LIS-1 enters the ciliary distal segment where retrograde IFT commences, IFT–dynein motility may also require the detachment of LIS-1, and a substoichiometric amount of LIS-1 at the distal tip may initiate IFT. In Lis1 mutants, cytoplasmic dynein and its cargo move at normal speeds with reduced frequencies in A. nidulans (Egan et al., 2012; Huang et al., 2012). However, we did not observe retrograde IFT in the lis-1-sg conditional mutants, which suggests that LIS-1 may be more critical for the motility of IFT–dynein. Studies from mammalian motile cilia and C. reinhardtii flagella reported that Lis1 regulates the activity of the outer dynein arm (Pedersen et al., 2007), indicating that Lis1 plays manifold roles in the regulation of cytoplasmic dynein, IFT–dynein, and axonemal dynein.
In summary, the somatic CRISPR–Cas9 technique enables the functional analysis of embryonically essential dynein subunits in C. elegans ciliogenesis. These dynein-associated molecules may regulate the motor processivity or function as a switch to change the direction of motion, and single molecule biophysics studies should be pursued in the future to better understand the function of these dynein-associated chains. The present study focuses on the ciliary roles of embryonically essential dynein components, and these conditional mutants are also valuable resources for the study of other dynein-based processes. This method is also amenable to investigations of the ciliary function of other essential embryonic genes as well as the formation and function of other organelles.
Materials and methods
C. elegans strains, genetics, and DNA manipulations
C. elegans strains were raised on nematode growth media (NGM) plates seeded with the Escherichia coli strain OP50 at 20°C using standard methods. CRISPR–Cas9 constructs were produced according to a previously described method (Shen et al., 2014). In brief, to generate the conditional knockout vectors, CRISPR–Cas9 target sequences were added to CRISPR–Cas9 vectors by PCR using Phusion high-fidelity DNA polymerase (New England Biolabs, Inc.) with the primers listed in Table S3 followed by self-ligation and transformation into E. coli. GFP-tagged dyneins were constructed by insertion of genomic sequences (3 kb promoter plus coding region) into pPD95.77 (containing GFP and unc-54 3′ UTR) for endogenous expression or into pOG1264 (containing dyf-1 promoter, GFP, and unc-54 3′ UTR) for ciliated neuron specific expression, except that DYLT-3::GFP fusion was generated by PCR gene splicing by overlap extension (SOEing) of a dylt-3 genomic sequence (3 kb promoter plus coding region) with GFP::unc-54 3′ UTR DNA fragments. Transgenic C. elegans were generated by the germ line microinjection of DNA plasmids at 10–50 ng/µl and a selection marker pRF4(rol-6(su1006)), Podr-1::dsRed, WT unc-76 gene or Pegl-17::Myri-mCherry + Pegl-17::mCherry::his-24 into N2 or unc-76(e911) hermaphrodites. The strains, primers, and plasmids are listed in Tables S1–S3.
Molecular assays
Molecular lesions induced by CRISPR–Cas9 were detected according to previously described methods (Shen et al., 2014). In brief, worms were lysed with 10× PCR buffer (Mg2+) and were used as templates for amplifying DNA fragments containing CRISPR–Cas9 targets. PCR products were purified by QIAquick PCR Purification kit (QIAGEN) and digested by T7EI at 37°C for 30 min. The indel percentage was determined using the formula 100 × {1 – [1 – (b + c)/(a + b + c)]1/2}, where a is the integrated intensity of the intact band and b and c are the integrated intensities of the digested small bands.
Heat-shock treatment, dye-filling assay, and viability assay
Heat-shock treatment was performed at 33°C for 1 h at 8 h after egg laying. The dye-filling assay was performed according to a previously described protocol (Snow et al., 2004). In brief, young adult worms were collected into ∼200 µl M9 solution, and mixed with ∼200 µl dyes (DiI 1,1′-dioctadecyl-3,3,3′,3′,-tetramethylindo-carbocyanine perchlorate, or DiO 3,3′-dioctadecyloxacarbocyanine perchlorate) at a working concentration (20 µg/ml). After incubation at room temperature in a dark place for ∼30–60 min, animals were transferred to a seeded NGM plate, and examined for dye uptake 1 h later. For the viability assay, the number of viable adult worms was divided by the number of originally laid eggs.
Live cell imaging
C. elegans were anesthetized with 0.1 mmol/liter levamisole in M9 buffer, mounted on 2% agar pads, and maintained at 22°C. Our imaging system includes an Axio Observer Z1 microscope (Carl Zeiss) equipped with a 100×, 1.45 NA objective lens, an EM CCD camera (iXon+ DU-897D-C00-#BV-500; Andor Technology), and the 488 nm and 568 nm lines of a Sapphire CW CDRH USB Laser System (Coherent) with a spinning disk confocal scan head (CSU-X1 Spinning Disk Unit; Yokogawa Electric Corporation). Time-lapse images were acquired with an exposure time of 200 ms by µManager (www.micro-manager.org). We used ImageJ software (http://rsbweb.nih.gov/ij/) to process the images.
Statistical analysis
Student’s t test and χ2 analysis were used to examine significant differences in ciliary phenotypes between WT and mutant animals, as indicated in the figure legends.
Online supplemental material
Fig. S1 (related to Fig. 1), shows the Dyf phenotype in osm-3-sg or xbx-1-sg animals under a 100× objective lens and a fluorescence stereoscope, respectively. Fig. S2 (related to Fig. 3) shows DLI-1 localization and function in ciliated neurons. Fig. S3 (related to Fig. 3 and Fig. 4) indicates that GFP fusion or overexpressed DYCI-1 and DLC-1 do not affect IFT–dynein’s function. Table S1 lists strains used in this study. Table S2 and S3 list all PCR products and plasmids for construction of transgenic C. elegans. Table S4 lists the target sequence of each gene. Table S5 gives the primers for molecular analysis. Videos 1–4 (related to Fig. 4) show the IFT movements of embryonically essential dynein subunits in cilia (GFP::DYCI-1, DLC-1::GFP, DYLT-3::GFP, and GFP::LIS-1). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201411041/DC1.
Supplementary Material
Acknowledgments
G. Ou thanks Drs. J. Scholey, R. Vale, A. Carter, S. Reck-Peterson, G. Goshima, and E. Griffith for their support and discussions regarding this project.
This work was supported by the National Basic Research Program of China (973 Program, grants 2013CB945602 and 2012CB945002), the National Natural Science Foundation of China (grants 31222035, 31301134, 31171295, and 31190063), and the Junior Thousand Talents Program of China.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Cas
- CRISPR-associated
- CRISPR
- clustered regularly interspaced short palindromic repeats
- Dyf
- abnormal dye filling
- IFT
- intraflagellar transport
- sgRNA
- synthetic guide RNA
- T7EI
- T7 endonuclease I
- WT
- wild type
References
- Asante D., Stevenson N.L., and Stephens D.J.. 2014. Subunit composition of the human cytoplasmic dynein-2 complex. J. Cell Sci. 127:4774–4787 10.1242/jcs.159038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blisnick T., Buisson J., Absalon S., Marie A., Cayet N., and Bastin P.. 2014. The intraflagellar transport dynein complex of trypanosomes is made of a heterodimer of dynein heavy chains and of light and intermediate chains of distinct functions. Mol. Biol. Cell. 25:2620–2633 10.1091/mbc.E14-05-0961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan M.J., Tan K., and Reck-Peterson S.L.. 2012. Lis1 is an initiation factor for dynein-driven organelle transport. J. Cell Biol. 197:971–982 10.1083/jcb.201112101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler J.T., and Anderson K.V.. 2007. Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 23:345–373 10.1146/annurev.cellbio.23.090506.123249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel B.D., Ishikawa H., Wemmer K.A., Geimer S., Wakabayashi K., Hirono M., Craige B., Pazour G.J., Witman G.B., Kamiya R., and Marshall W.F.. 2012. The role of retrograde intraflagellar transport in flagellar assembly, maintenance, and function. J. Cell Biol. 199:151–167 10.1083/jcb.201206068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L., Efimenko E., Swoboda P., and Scholey J.M.. 2011. The retrograde IFT machinery of C. elegans cilia: two IFT dynein complexes? PLoS ONE. 6:e20995 10.1371/journal.pone.0020995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Pazour G.J., and Witman G.B.. 2004. A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol. Biol. Cell. 15:4382–4394 10.1091/mbc.E04-05-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Lander E.S., and Zhang F.. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 157:1262–1278 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Roberts A.J., Leschziner A.E., and Reck-Peterson S.L.. 2012. Lis1 acts as a “clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell. 150:975–986 10.1016/j.cell.2012.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., and Marshall W.F.. 2011. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 12:222–234 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Kardon J.R., and Vale R.D.. 2009. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 10:854–865 10.1038/nrm2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C.C., Kramer J.M., Stinchcomb D., and Ambros V.. 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10:3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G., Blacque O.E., Snow J.J., Leroux M.R., and Scholey J.M.. 2005. Functional coordination of intraflagellar transport motors. Nature. 436:583–587 10.1038/nature03818 [DOI] [PubMed] [Google Scholar]
- Pastrana E.2010. A toolset for the proficient geneticist. Nat. Methods. 7:488–489 10.1038/nmeth0710-488a [DOI] [PubMed] [Google Scholar]
- Patel-King R.S., Gilberti R.M., Hom E.F., and King S.M.. 2013. WD60/FAP163 is a dynein intermediate chain required for retrograde intraflagellar transport in cilia. Mol. Biol. Cell. 24:2668–2677 10.1091/mbc.E13-05-0266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Wilkerson C.G., and Witman G.B.. 1998. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol. 141:979–992 10.1083/jcb.141.4.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Dickert B.L., and Witman G.B.. 1999. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144:473–481 10.1083/jcb.144.3.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L.B., Rompolas P., Christensen S.T., Rosenbaum J.L., and King S.M.. 2007. The lissencephaly protein Lis1 is present in motile mammalian cilia and requires outer arm dynein for targeting to Chlamydomonas flagella. J. Cell Sci. 120:858–867 10.1242/jcs.03374 [DOI] [PubMed] [Google Scholar]
- Pfister K.K., Fisher E.M., Gibbons I.R., Hays T.S., Holzbaur E.L., McIntosh J.R., Porter M.E., Schroer T.A., Vaughan K.T., Witman G.B., et al. 2005. Cytoplasmic dynein nomenclature. J. Cell Biol. 171:411–413 10.1083/jcb.200508078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M.E., Bower R., Knott J.A., Byrd P., and Dentler W.. 1999. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell. 10:693–712 10.1091/mbc.10.3.693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Burnette D.T., Bae Y.K., Forscher P., Barr M.M., and Rosenbaum J.L.. 2005. Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr. Biol. 15:1695–1699 10.1016/j.cub.2005.08.047 [DOI] [PubMed] [Google Scholar]
- Roberts A.J., Kon T., Knight P.J., Sutoh K., and Burgess S.A.. 2013. Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol. 14:713–726 10.1038/nrm3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P., Pedersen L.B., Patel-King R.S., and King S.M.. 2007. Chlamydomonas FAP133 is a dynein intermediate chain associated with the retrograde intraflagellar transport motor. J. Cell Sci. 120:3653–3665 10.1242/jcs.012773 [DOI] [PubMed] [Google Scholar]
- Rosenbaum J.L., and Witman G.B.. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3:813–825 10.1038/nrm952 [DOI] [PubMed] [Google Scholar]
- Schafer J.C., Haycraft C.J., Thomas J.H., Yoder B.K., and Swoboda P.. 2003. XBX-1 encodes a dynein light intermediate chain required for retrograde intraflagellar transport and cilia assembly in Caenorhabditis elegans. Mol. Biol. Cell. 14:2057–2070 10.1091/mbc.E02-10-0677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey J.M.2003. Intraflagellar transport. Annu. Rev. Cell Dev. Biol. 19:423–443 10.1146/annurev.cellbio.19.111401.091318 [DOI] [PubMed] [Google Scholar]
- Scholey J.M.2013. Kinesin-2: a family of heterotrimeric and homodimeric motors with diverse intracellular transport functions. Annu. Rev. Cell Dev. Biol. 29:443–469 10.1146/annurev-cellbio-101512-122335 [DOI] [PubMed] [Google Scholar]
- Shen Z., Zhang X., Chai Y., Zhu Z., Yi P., Feng G., Li W., and Ou G.. 2014. Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev. Cell. 30:625–636 10.1016/j.devcel.2014.07.017 [DOI] [PubMed] [Google Scholar]
- Signor D., Wedaman K.P., Orozco J.T., Dwyer N.D., Bargmann C.I., Rose L.S., and Scholey J.M.. 1999. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J. Cell Biol. 147:519–530 10.1083/jcb.147.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow J.J., Ou G., Gunnarson A.L., Walker M.R., Zhou H.M., Brust-Mascher I., and Scholey J.M.. 2004. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat. Cell Biol. 6:1109–1113 10.1038/ncb1186 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.