The luminal domain of Nup210 that lacks NPC sorting signals is sufficient for myogenesis, which suggests that Nup210 may operate within the nuclear envelope/ER lumen during differentiation.
Abstract
Previously, we identified the nucleoporin gp210/Nup210 as a critical regulator of muscle and neuronal differentiation, but how this nucleoporin exerts its function and whether it modulates nuclear pore complex (NPC) activity remain unknown. Here, we show that gp210/Nup210 mediates muscle cell differentiation in vitro via its conserved N-terminal domain that extends into the perinuclear space. Removal of the C-terminal domain, which partially mislocalizes gp210/Nup210 away from NPCs, efficiently rescues the differentiation defect caused by the knockdown of endogenous gp210/Nup210. Unexpectedly, a gp210/Nup210 mutant lacking the NPC-targeting transmembrane and C-terminal domains is sufficient for C2C12 myoblast differentiation. We demonstrate that the endoplasmic reticulum (ER) stress-specific caspase cascade is exacerbated during Nup210 depletion and that blocking ER stress-mediated apoptosis rescues differentiation of Nup210-deficient cells. Our results suggest that the role of gp210/Nup210 in cell differentiation is mediated by its large luminal domain, which can act independently of NPC association and appears to play a pivotal role in the maintenance of nuclear envelope/ER homeostasis.
Introduction
Nuclear pore complexes (NPCs) are multiprotein transport channels that span the nuclear envelope (NE) and serve as exclusive communication conduits between the nucleus and the cytoplasm (Hoelz et al., 2011). NPCs are composed of multiple copies of ∼30 different proteins called nucleoporins or Nups (Allen et al., 2001; Cronshaw et al., 2002). Although the primary function of NPCs has been traditionally viewed to be nucleocytoplasmic transport, recent studies in eukaryotes ranging from yeast to mammals indicate that nucleoporins play roles in nuclear processes such as chromatin organization and gene regulation (Akhtar and Gasser, 2007; Brown and Silver, 2007; Liang and Hetzer, 2011). Moreover, accumulating evidence suggests that NPCs have cell type–specific composition (Raices and D’Angelo, 2012; Ori et al., 2013) and that single nucleoporin mutations often give rise to developmental defects and human diseases (Nakamura et al., 1996; Zhang et al., 2008; Lupu et al., 2008; Gomez-Cavazos and Hetzer, 2012). Altogether, these studies raise the exciting possibility that a subset of nucleoporins might act as critical regulators of gene expression programs during differentiation and development in a transport-independent manner.
One striking example of a NPC protein that exhibits cell type–specific expression is the transmembrane nucleoporin gp210/Nup210 (herein referred to as Nup210). Nup210 was the first nucleoporin to be discovered more than three decades ago, and as a result of its membrane association and close resemblance to viral fusogenic proteins, it was initially predicted to promote the fusion between the inner nuclear membrane and outer nuclear membrane during NPC biogenesis (Gerace et al., 1982; Wozniak et al., 1989). However, several studies have shown that Nup210 is absent in many tissue types and found to be dispensable for NPC assembly, maintenance, and distribution (Olsson et al., 1999, 2004; Eriksson et al., 2004; Stavru et al., 2006; Ori et al., 2013). This raises the important question of what Nup210’s function might be and whether it requires tethering to the NPC. We have recently shown that Nup210 is not detectable in myoblasts and embryonic stem cells but becomes expressed and incorporated into NPCs upon differentiation into myotubes and neuroprogenitor cells, respectively. Importantly, we found that the depletion of Nup210 by shRNA blocked differentiation in both cell lineages, resulting in increased cell death and down-regulation of genes essential for cell differentiation. Knockdown of either Pom121 or NDC1, two transmembrane NPC proteins known to participate in NPC assembly, did not affect myoblast differentiation, indicating that Nup210 function is distinct from other membrane-anchored nucleoporins (D’Angelo and Gomez-Cavazos et al., 2012).
Here, we extend our previous studies and show that a luminal fragment of Nup210 that lacks NPC sorting signals is sufficient to reconstitute myogenesis in differentiated cells lacking Nup210. This suggests that the role of Nup210 in differentiation can be functionally separated from its association with the NPC and may operate within the NE/ER lumen. Analysis of Nup210 knockdown cells undergoing apoptosis reveals activation of the ER stress-specific caspase cascade and up-regulation of ER stress-responsive proteins. Furthermore, inhibition of ER stress-mediated apoptosis restores differentiation of Nup210-depleted cells, suggesting that Nup210 plays an antiapoptotic role by regulating NE/ER homeostasis to promote cell fate determination in muscle cells.
Results and discussion
Overexpression of Nup210 lacking its C-terminal domain accelerates myotube formation
The topology of Nup210 within the NE is unique among transmembrane nucleoporins. Nup210 is a single pass transmembrane nucleoporin of 1,886 amino acids, of which only a short C-terminal tail of 57 residues faces the transport channel. The majority of the protein is composed of a 1,783-aa luminal domain that resides inside the perinuclear space, which is continuous with the ER lumen (Fig. 1 A; Wozniak et al., 1989). To understand how Nup210 functions in muscle differentiation, we analyzed its primary protein sequence conservation across metazoans (Fig. 1 B). Alignment of representative sequences from mammals, fish, echinoderms, flies, and worms revealed high conservation of Nup210 luminal domain and poor conservation of the C-terminal tail (Fig. 1 B). To directly test the role of the conserved luminal domain of Nup210 in muscle differentiation, we generated two truncation mutants fused to GFP, Nup210ΔLUMEN lacking the luminal domain, and Nup210ΔCT, which lacks the C-terminal domain (Figs. 1 C and S1 B), and stably expressed them in C2C12 cell lines (Fig. S1, A–C). We have previously shown that ectopic expression of full-length Nup210 in C2C12 myoblasts efficiently accelerates myotube formation (D’Angelo and Gomez-Cavazos et al., 2012). We found that C2C12 cells expressing Nup210ΔCT, but not Nup210ΔLUMEN, phenocopied full-length Nup210 overexpression, exhibiting a similar increase in myotube formation after differentiation was induced as characterized by an earlier appearance of myosin heavy chain (MHC)–positive cells and generation of larger myotubes (Fig. 1, D and E). These results suggest that the luminal domain of Nup210 is important for muscle differentiation.
Figure 1.
Overexpression of Nup210 lacking its C-terminal domain accelerates myotube formation in C2C12 cells. (A) Schematic representation of the topology of Nup210. S.S., signal sequence; TM, transmembrane domain; CT, C-terminal domain. (B) Conservation analysis of Nup210 domains across metazoans: H. sapiens, M. musculus, D. rerio, S. purpuratus, D. melanogaster, and C. elegans. 10 = conserved; 0 = not conserved. Red bars highlight residues that are 100% conserved across all metazoans analyzed. N, N terminus; C, C terminus. (C) Schematic representation of Nup210 GFP fusion constructs overexpressed in C2C12 cells. (D) C2C12 myoblasts were infected with retrovirus carrying GFP alone, Nup210-GFP, Nup210ΔLUMEN-GFP, and Nup210ΔCT-GFP and induced to differentiate. Immunofluorescence against MHC (red) and GFP (green) was performed at 48, 72, and 96 h after differentiation. Hoechst (blue) was used as a nuclear stain. Insets show a digital magnification (3×) of the GFP channel. Bar, 100 µm. (E) Quantification of nuclei in MHC-positive cells (two or more nuclei) at 48, 72, and 96 h after differentiation. Data were collected from three independent experiments and plotted. *, P < 0.05; and **, P < 0.01 indicate a significant difference between overexpression experiments carrying GFP and Nup210ΔCT-GFP by t test. Red bars and numbers represent means.
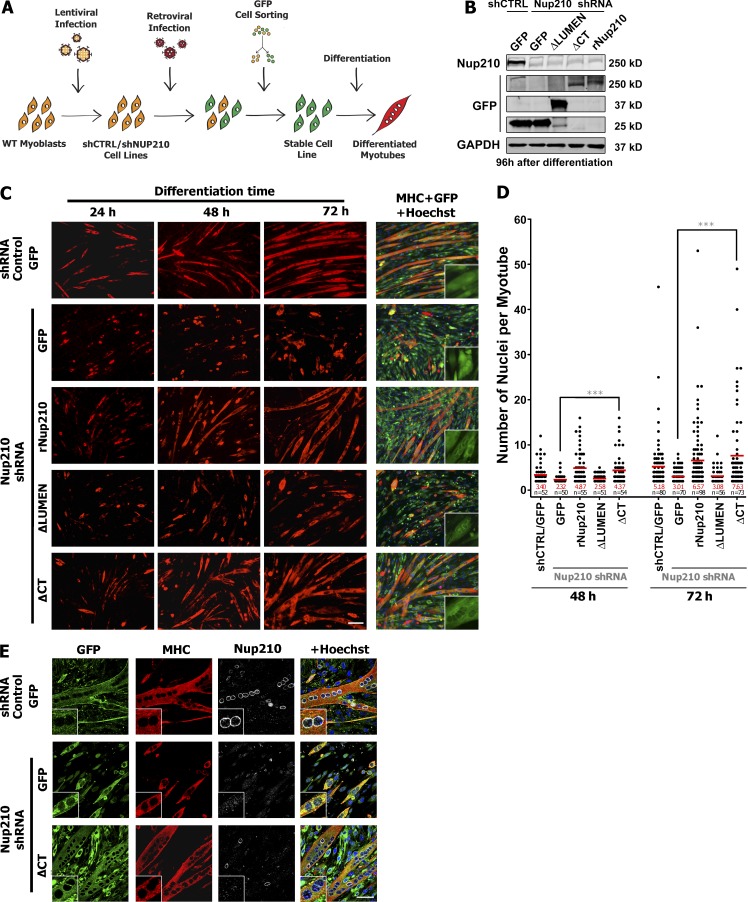
The Nup210ΔCT truncation mutant is sufficient to restore C2C12 differentiation
We have previously observed that the depletion of Nup210 in C2C12 inhibits differentiation, a phenotype that can be rescued by the overexpression of an shRNA-resistant Nup210 (rNup210; D’Angelo and Gomez-Cavazos et al., 2012). To determine whether the luminal domain of Nup210 is sufficient to mediate myogenesis, we expressed shRNA-resistant mutants Nup210ΔCT-GFP or Nup210ΔLUMEN-GFP as well as rNup210-GFP and GFP in C2C12 myoblasts carrying Nup210 shRNAs (Fig. 2, A and B; and Fig. S1 D). As expected, cells expressing shRNA-resistant full-length Nup210 restored myogenesis, whereas those expressing GFP alone did not (Fig. 2, C and D). Consistent with our overexpression data, cells expressing Nup210ΔCT-GFP, but not Nup210ΔLUMEN-GFP, were able to form multinucleated myotubes and rescue C2C12 differentiation (Fig. 2, C–E). Interestingly, cells expressing a Nup210ΔTMCT-GFP were not able to restore differentiation, implying a requirement for this domain to be anchored to the NE membrane (unpublished data). These findings demonstrate that the conserved luminal domain is the primary mediator of Nup210’s role during muscle differentiation.
Figure 2.
Nup210ΔCT is sufficient to restore C2C12 differentiation. (A) Schematic representation of rescue experiments methodology. C2C12 myoblasts were first infected with lentivirus carrying control and Nup210 shRNAs followed by retroviral transduction of GFP or Nup210 constructs. Cells were then GFP sorted to obtain high expressors. (B) Protein levels of endogenous Nup210 and GFP-tagged Nup210 fragments in reconstituted cell lines expressing scrambled or Nup210 shRNAs. GAPDH was used a loading control. shCTRL, control shRNA. (C) C2C12 myoblasts expressing scrambled or Nup210 shRNAs were infected with retrovirus carrying GFP alone or shRNA-resistant Nup210-GFP, Nup210ΔLUMEN-GFP, and Nup210ΔCT-GFP and induced to differentiate. Immunofluorescence against MHC (red) and GFP (green) was performed at 24, 48, and 72 h after differentiation. Insets show a digital magnification (3×) of GFP channel. Bar, 100 µm. (D) Quantification of nuclei in MHC-positive cells (two or more nuclei) at 48 and 72 h after differentiation from experiments shown in C. Data were collected from four independent experiments. ***, P < 0.001 indicates a significant difference between rescue experiments of GFP and Nup210ΔCT-GFP by t test. Red bars and numbers represent means. (E) Immunofluorescence against GFP (green), MHC (red), and Nup210 (C-terminal–specific antibody, white) in C2C12 cells carrying control or Nup210 shRNAs reconstituted with GFP or Nup210ΔCT-GFP 96 h after differentiation. Hoechst (blue) was used as a nuclear stain. Insets show a digital magnification (3×) of each respective channel. Bar, 50 µm.
Nup210ΔCT mutant is not efficiently targeted to the NE and NPCs
We have previously reported Nup210 to be exclusively located at NPCs in differentiated C2C12 cells (D’Angelo and Gomez-Cavazos et al., 2012). However, it remained unclear whether NPC association is important for Nup210’s function in cell differentiation. Targeting of Nup210 to the nuclear pore membrane has been shown to be dependent on its single transmembrane segment and its C-terminal tail (Wozniak and Blobel, 1992). To determine the localization of Nup210 fragments to the NE and NPCs in C2C12 cells, stable cell lines expressing Nup210-GFP, Nup210ΔLUMEN-GFP, and Nup210ΔCT-GFP were fixed and stained with mAb414 to visualize nuclear pores. Stable cell lines expressing Sec61-β–GFP and NDC1-GFP were used as experimental controls for ER and NE/NPC localization, respectively (Fig. 3, A–D). Using confocal microscopy and structured illumination microscopy, we were able to confirm that Nup210ΔLUMEN-GFP, which contains both previously characterized NE sorting signals, properly localized at the NE and NPCs similar to full-length Nup210 (60.2 ± 3.7% vs. 75.6 ± 6.7%; Fig. 3, A–D). In contrast, Nup210-ΔCT-GFP lost its ability to concentrate at the NE and NPCs (36.3 ± 8.3% vs. 75.6 ± 6.7%), being predominantly distributed throughout the ER (Fig. 3, A–D). These results confirm that the C-terminal domain, which faces the nuclear pore, plays a critical role in targeting Nup210 to the NPC.
Figure 3.
Nup210ΔCT truncation mutant fails to accumulate at the NE and is partially mislocalized from NPCs. (A) NE distribution of Sec61-β–GFP, NDC1-GFP, Nup210-GFP, Nup210ΔLUMEN-GFP, or Nup210ΔCT-GFP in stable myoblast cell lines. C2C12 cells were stained for GFP (green) and nuclear pores using mAb414 (red). Experiments were performed at least three times independently. Representative data for each condition is shown. Bar, 5 µm. (B) GFP and mAb414 signal profiles at NE cross sections were determined by ImageJ. (C) Localization of GFP and mAb414 signals at NE surfaces of C2C12 myoblasts stably expressing Sec61-β–GFP (n = 555), NDC1-GFP (n = 603), Nup210-GFP (n = 510), Nup210ΔLUMEN-GFP (n = 502), and Nup210ΔCT-GFP (n = 652) where n represents the number of NPCs quantified. C2C12 cells were stained for GFP and nuclear pores using mAb414. Experiments were repeated three times, and representative images are presented. Bar, 1 µm. (D) GFP and mAb414 signal profiles at the NE surface were determined by ImageJ. Colocalization percentage of nuclear pore signals with GFP signals was determined using Imaris.
Because Nup210-ΔCT-GFP still showed partial association with NPCs (Fig. 3, C and D), we wanted to determine whether the transmembrane domain was also involved in targeting Nup210 to NPCs in C2C12 cells. We generated a protein chimera in which the transmembrane domain of Nup210 was replaced with that of Sec61-β and compared the ability of this construct to associate with NPCs to Nup210Δ26–1,327aa-GFP, a fragment containing both the transmembrane and C-terminal domains (68.8 ± 6.3%; Fig. S1, E and F). We found that swapping the Nup210 transmembrane domain for that of Sec61-β impaired its localization to nuclear pores (31.0 ± 6.7%), despite containing an intact C-terminal domain. These findings indicate that the transmembrane domain of Nup210 is required for NPC targeting (Fig. S1, E and F). Consistently, the transmembrane and C-terminal domains of Nup210 are sufficient for nuclear pore localization, as fusion of these domains with the N-terminal domain of the ER resident protein Calnexin relocalized it to the NE and NPCs (CalnexinLUMEN+NUP210TM+CT-GFP [63.0 ± 9.6%] vs. Calnexin-GFP [15.7 ± 6.2%]; Fig. S1, G–J). These results confirm that the transmembrane and C-terminal domains both contribute to the targeting of Nup210 to the NE and NPCs. Given that the Nup210ΔCT truncation rescues myogenesis despite attenuated localization at the NPC suggests that the role of Nup210 during muscle differentiation may in fact be linked to the perinuclear space, where Nup210 may be able to function without NPC interaction.
Association of Nup210 to the NPC is dispensable for C2C12 differentiation
To directly test whether NPC association of Nup210 was required for C2C12 differentiation, we generated two Nup210 protein constructs that were no longer able to associate with NPCs, yet contained an intact luminal domain. To achieve this, we replaced the transmembrane domain of Nup210 in our Nup210ΔCT mutant with that of Sec61-β (ΔCTSec61-βTM) or Lap2-β (ΔCTLap2-βTM), the latter being an inner nuclear membrane protein known to be dispensable for C2C12 differentiation and to not associate with NPCs (Figs. 4 A and S2 A; Huber et al., 2009). First, we characterized NE and NPC localization in C2C12 cells stably overexpressing these constructs. Both proteins localized to NE/ER membranes and failed to associate with the NPCs (Fig. 4, B–D; and Fig. S2, B–D). Interestingly, differentiation of these cell lines led to rapid fusion of myotubes, similar to full-length Nup210 and Nup210ΔCT overexpression (Fig. S2, E and F).
Figure 4.
Nup210 mutant lacking NPC sorting signals rescues myotube formation. (A) Schematic representation of Nup210ΔCTLap2-βTM-GFP. N, N terminus; C, C terminus. (B) NE distribution of Nup210ΔCTLap2-βTM-GFP in myoblasts. C2C12 cells were stained for GFP and nuclear pores using mAb414. Bar, 5 µm. (C) Structured illumination microscopy in NE surfaces of C2C12 myoblasts stably expressing Nup210ΔCTLap2-βTM-GFP. Cells were stained for GFP and nuclear pores using mAb414. Bar, 1 µm. (D) Localization of GFP (green line) and mAb414 (red line) signals at the NE of Nup210ΔCTLap2-βTM-GFP (n = 547) was determined by using ImageJ. Colocalization percentage of nuclear pore signals with GFP signals was determined using Imaris. (E) C2C12 myoblasts expressing scrambled or Nup210 shRNAs were infected with retrovirus carrying GFP alone or shRNA-resistant Nup210ΔCTLap2-βTM-GFP and induced to differentiate. Immunofluorescence against MHC (red) and GFP (green) was performed at 24, 48, and 72 h after differentiation. Insets show a digital magnification (3×) of GFP channel. Bar, 100 µm. (F) Quantification of MHC-positive cells with two or more nuclei at 48 and 72 h after differentiation from experiments shown in E. Data were collected from three independent experiments. ***, P < 0.001; and **, P < 0.01 indicate a significant difference between rescue experiments of GFP and Nup210ΔCTLap2-βTM-GFP by t test. Red bars and numbers represent means. (G) Immunofluorescence against GFP (green), MHC (red), and Nup210 (C-terminal–specific antibody, white) in C2C12 cells carrying control or Nup210 shRNAs reconstituted with GFP or Nup210ΔCTLap2-βTM-GFP 96 h after differentiation. Hoechst (blue) was used as a nuclear stain. Insets show a digital magnification (3×) of each respective channel. Bar, 50 µm. (H) FRAP analysis in NE membranes of C2C12 myoblasts stably expressing Nup210ΔCTLap2-βTM-GFP (t1/2 = 64.64 ± 7.10 s; MFR = 0.715 ± 0.019; n = 10), Nup210-GFP (t1/2 = 108.5 ± 12.61 s; MFR = 0.476 ± 0.027; n = 25), and Sec61-β–GFP (t1/2 = 44.8 ± 2.48; MFR = 44.8 ± 2.48; n = 16). White boxes represent photobleached areas. Error bars represent standard deviation of the mean. Bar, 10 µm.
Second, we tested the ability of the non-NPC–associated Nup210ΔCTLap2-βTM-GFP construct to rescue C2C12 differentiation in myoblasts carrying Nup210 shRNAs. We found that despite exclusion from NPCs, C2C12 cells expressing Nup210ΔCTLap2-βTM-GFP were able to differentiate under Nup210 knockdown conditions (Fig. 4, E–G; Fig. S1 D; and Fig. S2 G). Importantly, Nup210ΔCTLap2-βTM-GFP was highly dynamic at the NE in C2C12 myoblasts, showing similar kinetics as the ER protein Sec61-β. This substantiates the idea that NPC-association of Nup210 at the NE is not required for differentiation (Fig. 4 H). Collectively, these data suggest that the transmembrane and C terminus of Nup210 are dispensable for differentiation and that the primary function of Nup210 in mediating C2C12 differentiation lies in the lumen of the NE/ER cisternae.
Depletion of Nup210 exacerbates the ER stress-specific caspase cascade during C2C12 differentiation
In recent years, there has been a growing appreciation that ER stress is triggered as a part of normal skeletal muscle development (Nakanishi et al., 2005; Alter and Bengal, 2011; Deldicque et al., 2012). During differentiation, moderate ER stress helps maintain the homeostasis of both calcium levels and protein synthesis in the ER, selecting these cells for myotube formation and enhancing resistance to apoptosis (Huppertz et al., 2001; Morishima et al., 2002; Nakanishi et al., 2005; Schöneich et al., 2014). Upon induction of C2C12 differentiation, a proportion of cells undergoes apoptosis and fails to differentiate into myotubes, a physiological response that has been attributed to the ER stress-specific caspase cascade, which involves caspase-12, -9, and -3 (Nakagawa et al., 2000; Morishima et al., 2002; Nakanishi et al., 2005). Intriguingly, we have found that differentiation of C2C12 myoblasts expressing Nup210 shRNA leads to a substantial increase in apoptosis as assessed by caspase-3 activation (Fig. S3 A; D’Angelo and Gomez-Cavazos et al., 2012). This raised the exciting possibility that Nup210 might play a role in modulating ER stress. Consistently, Nup210 protein is induced as early as 24 h after C2C12 differentiation (Fig. S3 B), the same time point at which we detect apoptotic cells when Nup210 is knocked down. To test whether apoptosis triggered by Nup210 depletion involved an ER stress response, we examined the ER-specific caspase cascade in Nup210-depleted cultures. Western blot analysis showed that procaspase-12 (50 kD) had been extensively processed to its active form (35 kD) in detached cells and remained largely unchanged in attached cells 48 h after differentiation (Fig. 5 A). This activated form of caspase-12 was only observed when control C2C12 myotubes were treated with tunicamycin, an inhibitor of N-linked glycosylation and a widely used inducer of ER stress (Fig. 5 A). After processing of caspase-12, the ER stress-specific cascade continues with the activation of caspase-9, which then catalyzes cleavage of procaspase-3 (Morishima et al., 2002). Activation of caspase-9 and -3 was also present in detached cells from Nup210 knockdown cultures (Fig. 5 A), indicating that depletion of Nup210 from differentiating myoblasts triggers the ER stress-specific caspase cascade and apoptosis. Consistently, we detected an increase in caspase-12, -9, and -3 activation in whole-cell lysates (attached and detached) of Nup210-depleted cell populations when compared with control (Fig. S3 A).
Figure 5.
Inhibition of C2C12 differentiation by Nup210 depletion leads to an ER stress profile and can be rescued by ER stress inhibitors. (A and B) Western blot analysis in attached and detached cells from Nup210 knockdown cultures 48 h after differentiation, showing scramble shRNA attached cells (negative control) and attached plus detached cells treated with 500 ng/ml tunicamycin (24 h, positive control). (A) Blot of caspase-12, -9, and -3 activation. (B) Blot of ATF6 activation, BiP, and PDI. (C) PDI immunofluorescence in differentiated cultures (attached + detached) 48 h after differentiation expressing scrambled or Nup210 shRNAs. Hoechst was used to stain nuclei. Images were taken at the confocal plane of detached cells. Insets show a digital magnification (3×) of red and blue channels. (D) Differentiation of myoblasts expressing control or Nup210 shRNAs in the presence of TUDCA (500 µg/ml) or vehicle (doubled-distilled H2O). Immunofluorescence against MHC (red) was performed at 24, 48, and 72 h after differentiation. (E) Quantification of nuclei in MHC-positive cells (two or more nuclei) at 24, 48, and 72 h after differentiation from experiments shown in D. Data were collected from three independent experiments. ***, P < 0.001 indicates a significant difference between Nup210 knockdown cells treated with TUDCA or vehicle by t test. Red bars and numbers represent means. (F) Immunofluorescence against Cleaved caspase-3 (green) on differentiated myoblasts expressing control (n = 6,086) or Nup210 shRNAs in the presence of TUDCA (500 µg/ml; n = 4,402), or vehicle (double-distilled H2O; n = 4,612) 72 h after differentiation. Percentages indicate the proportion of cells positive for Cleaved caspase-3 staining versus total number of cells (n). (G) Differentiation of C2C12 cells carrying Nup210 shRNAs expressing GFP alone or MAGE-A3–GFP. Immunofluorescence against MHC (red) and GFP (green) was performed at 24, 48, and 72 h after differentiation. (H) Quantification of nuclei in MHC-positive cells (two or more nuclei) at 48 and 72 h after differentiation from experiments shown in G. Data were collected from three independent experiments. ***, P < 0.001 indicates a significant difference between rescue experiments of GFP and MAGE-A3–GFP by t test. Red bars and numbers represent means. shCtrl, control shRNA. Bars: (C) 50 µm; (D, F, and G) 100 µm.
Among the three sensor proteins of ER stress, ATF6 (Activating Transcription Factor 6) has been previously proposed to be the key mediator of ER stress induced apoptosis during normal physiological conditions in C2C12 differentiation and to be responsible for inducing the expression of ER chaperones important to cope with ER stress (Nakanishi et al., 2005; Yamamoto et al., 2007; Morishima et al., 2011). We detected ATF6 processing as well as up-regulation of the ER-resident chaperones binding immunoglobulin protein (BiP) and protein disulfide isomerase (PDI) in detached cells from cultures carrying Nup210 shRNA (Fig. 5 B). In agreement with the activation of ER stress-induced apoptosis, Nup210-depleted cell populations exhibited a higher number of detached cells positive for PDI compared with control (Fig. 5 C). These findings indicate that Nup210 depletion leads to an increase in the ER stress response typically accompanying skeletal muscle differentiation and suggest a potential role for Nup210 in the regulation of NE/ER homeostasis during myoblast differentiation.
The chemical chaperone TUDCA reduces apoptosis and restores differentiation in Nup210-depleted cells
If the absence of Nup210 triggers substantial ER stress-mediated apoptosis, blocking ER stress response might compensate for the loss of Nup210. To test this, we used the chemical chaperone tauroursodeoxycholic acid (TUDCA), a bile acid derivative that has been shown to exhibit beneficial effects in numerous diseases associated with ER stress by reducing activation of the unfolded protein response and blocking the ER stress-mediated apoptotic pathway (Xie et al., 2002; Özcan et al., 2006; Amaral et al., 2009). Interestingly, myoblasts carrying Nup210 shRNAs induced to differentiate in the presence of TUDCA regained the ability to fuse and develop into mature and healthy myotubes, whereas cells treated with vehicle failed to differentiate (Fig. 5, D and E; and Fig. S3 C). We also detected a reduction in the number of caspase-3–positive cells in TUDCA-treated differentiated cultures carrying Nup210 shRNAs, which is consistent with inhibition of the ER stress-specific apoptotic cascade (Fig. 5 F). Similarly, TUDCA could promote survival of terminally differentiated myotubes depleted of Nup210 (Fig. S3, D and E), indicating that TUDCA can protect cells from Nup210 knockdown-induced cell death at both early and late stages of C2C12 differentiation.
Overexpression of the caspase-12 inhibitor MAGE-A3 rescues differentiation of Nup210-depleted cells
Finally, we wanted to test whether Nup210’s antiapoptotic activity is specific to ER stress. One of the mechanisms by which TUDCA is thought to protect cells from ER stress mediated apoptosis is through inhibition of caspase-12 (Xie et al., 2002). To determine whether inhibition of caspase-12 was sufficient to restore differentiation of Nup210-deficient cells, we generated cell lines stably expressing Nup210 shRNAs and overexpressing MAGE-A3, a known inhibitor of caspase-12 in C2C12 cells (Fig. S3 F; Morishima et al., 2002; Nakanishi et al., 2005). Cells overexpressing MAGE-A3–GFP recovered their ability to differentiate and fused into multinucleated myotubes (Fig. 5, G and H). Consistent with a reduction in apoptosis, MAGE-A3–GFP cultures had a decrease in active caspase-3 at 36 and 72 h after differentiation (Fig. S3 G). These results further support the finding that stress caused by Nup210 knockdown can be relieved by inhibiting caspase-12–mediated apoptosis.
Here, we provide first evidence for an antiapoptotic function of Nup210 during skeletal muscle differentiation. Our findings suggest that Nup210 regulates cell differentiation not by modulating NPC activity or gene activation directly but rather by a process that involves luminal NE/ER components and is critical for dampening a physiological response to NE/ER stress during cell fate adaptation.
Multiple nucleoporins, including Nup210, are in constant exchange from NPCs, residing at the pore only for a few seconds to a few hours (Rabut et al., 2004). Mobility of these nucleoporins could be attributed to various functions both on and off the NPC. One clear example is Nup98, a nucleoporin that has been shown to shuttle between the NPC and the nucleoplasm and to contribute to several processes including mRNA export and gene regulation (Griffis et al., 2002; Capelson et al., 2010). Intriguingly, unlike other transmembrane nucleoporins (i.e., POM121), the association of Nup210 with the NPC is short lived and highly dynamic (Rabut et al., 2004). However, the purpose of Nup210’s high mobility is poorly understood. Our data show that a Nup210 chimera with increased mobility at the NE membranes does not interfere with its role in myogenesis, highlighting the exciting possibility of an NPC-independent function of Nup210 during cell differentiation. Whether the primary role of the NPC is to serve as an anchor to concentrate and retain Nup210 within the NE lumen or whether Nup210 has yet an unknown NPC-dependent function remains unclear.
Currently, we can only speculate as to how Nup210 exerts its function. In this regard, it has previously been suggested that the luminal domain of Nup210 contains calcium-binding motifs known as EF-hands at amino acids 160–171, 424–435, 580–591, 1,413–1,424, and 1,610–1,621 (Greber and Gerace, 1995). We have found that that these regions are not required for C2C12 differentiation (Fig. S3, H–K). In fact, analysis of these regions reveals weak conservation across metazoans and no consensus sequence for EF-hands, indicating that these amino acid stretches are unlikely to play a role in Nup210 function during myogenic differentiation (unpublished data). These experiments do not exclude the possibility that Nup210 might still play a role in Ca2+ homeostasis. Using protein structure prediction software, we were able to identify eight bacterial-Ig–like domains in the luminal domain of Nup210 (unpublished data). Importantly, bacterial-Ig–like domains have been recently characterized as novel calcium-binding modules in bacteria (Mans et al., 2004; Raman et al., 2010; Wang et al., 2013). It will be important to test whether Nup210 has Ca2+ binding capacity and whether it can act as a Ca2+ buffer in the NE/ER lumen. This could explain why depletion of Nup210 leads to an increase in ER stress-induced cell death as perturbation of calcium homeostasis has been recently associated with ER stress-mediated apoptosis in differentiating myoblasts (Nakanishi et al., 2015).
Our data suggest a potential antiapoptotic role for Nup210 during cellular stress. Interestingly, a recent expression analysis of NE proteins in cancer samples identified Nup210 as one of the most consistently up-regulated NE proteins in numerous tumors (de las Heras et al., 2013). It would be interesting to test whether Nup210 has an antiapoptotic role in cancer cells and whether its overexpression promotes tumorigenesis. In summary, the finding that Nup210 functions via its luminal domain establishes a new link between the NPC and the NE lumen during muscle differentiation and provides new insights in our understanding of how this tissue-specific nucleoporin may function across different cell types.
Materials and methods
Cells
C2C12 cells were obtained from ATCC. Proliferating myoblasts were maintained in DMEM containing 20% FBS. Differentiation into myotubes was induced by shifting confluent myoblasts to media containing 2% horse serum. C2C12 stable cell lines were generated at least three times independently and kept under the same conditions during their generation, maintenance, and differentiation. Stable cell lines consisted of mixed populations (all selected and sorted cells) and not isolated subclones. Low and high GFP sorted cells were obtained to further validate phenotypic differences. For differentiation experiments, cell lines were counted and plated in 8-well plates (Ibidi) at the same density and induced to differentiate upon confluency.
Western blot analysis
For protein extracts, proliferating myoblasts or differentiated myotubes were lysed in 1% SDS buffer preheated to 95°C, incubated at 95°C for 5 min, and passed 5–10 times though a 27-gauge syringe. For analysis of C2C12 attached and detached populations, detached cells were harvested by collecting the floating and loosely attached cells in the media and recovered living cells by scraping the plates. Protein concentration was determined using the bicinchoninic acid reagent (Thermo Fisher Scientific) and normalized, and 6% β-mercaptoethanol plus 0.01% bromophenol blue was added. For Western blot analysis, 40–80 mg of protein was resolved in SDS-PAGE and transferred to Immobilon-FL membranes (EMD Millipore). Membranes were blocked with PBS-0.05% Tween 20 (PBS-T) plus 5% nonfat milk for 1 h, washed, and incubated with primary antibody for 1 h at RT or overnight at 4°C. The secondary antibody was added after three washes with PBS-T for 1 h at RT. Membranes were analyzed with an infrared imaging system (Odyssey; LI-COR Biosciences) or using ECL (Thermo Fisher Scientific).
Antibodies and chemicals
Anti-Nup210/rabbit (IQ294) purchased from Immunoquest was used for Western blot analysis (1:1,000), while anti-Nup210/rabbit (A301-795A) obtained from Bethyl Laboratories, Inc. was used for immunofluorescence (1:500). mAb414/mouse was purchased from Covance. Anti-MHC antibody/mouse (MF-20) was obtained from Developmental Studies Hybridoma Bank at the University of Iowa. Anti-GFP/rabbit used for Western blotting was obtained from Santa Cruz Biotechnology, Inc. (sc-8384; 1:1,000). Anti-GFP/rabbit used for immunofluorescence was purchased from Invitrogen (A-11122; 1:1,000). Anti-GAPDH/mouse (6C5) was obtained from Abcam (1:2,000). Anti-PDI/mouse used for immunofluorescence was obtained from Abcam (ab5484; 1:1,000) and for Western blot/rabbit from Cell Signaling Technology (mAb #3501; 1:1,000). Anti-BiP/rabbit was purchased from Cell Signaling Technology (mAb #3177). Anti-ATF6/rabbit was obtained from Santa Cruz Biotechnology, Inc. (sc-22799). Caspase-12/rat antibody was obtained from Sigma-Aldrich (#C7611). Anti–caspase-9/rabbit (#9504) and anti–activated caspase 3/rabbit (#9661) was obtained from Cell Signaling Technology. TUDCA dihydrate was obtained from Tokyo Chemical Industry Co.
Nup210 protein conservation analysis
Nup210 protein sequences from Homo sapiens, Mus musculus, Danio rerio, Strongylocentrotus purpuratus, Drosophila melanogaster, and Caenorhabditis elegans were retrieved from the NCBI data bank. Multiple sequence alignment of Nup210 amino acid sequences was performed using PRALINE online software (http://www.ibi.vu.nl/programs/pralinewww; Simossis and Heringa, 2005). PRALINE conservation values from each residue on Nup210 were plotted using Excel (Microsoft).
Immunofluorescence
Differentiated myotubes were cultured in 8-well plates (Ibidi). For MHC and GFP staining, cells were fixed in 4% PFA for 20 min at RT. For Nup210 staining, cells were fixed in −20°C methanol for 2 min and then permeabilized in PBS/1% Triton X-100 for 1–2 min. For PDI staining, C2C12 cultures were fixed in 4% PFA + 0.1% glutaraldehyde for 5 min at RT. Fixed cells were blocked using immunofluorescence buffer (PBS, 10 mg/ml BSA, 0.02% SDS, and 0.1% Triton X-100) and incubated with primary antibody in immunofluorescence buffer for 2 h at RT or overnight at 4°C. Cells were washed in immunofluorescence buffer and incubated with secondary antibody for 1 h at RT. Cells were incubated with Hoechst for 5 min and washed three times with PBS buffer at RT before imaging. Samples were kept on PBS and imaged immediately at RT. For C2C12 differentiation assays, images were taken with a motorized microscope (20×; Axio Observer.Z1; Carl Zeiss) using AxioVision Viewer 4.8 software (Carl Zeiss). Number of nuclei per myotube were counted using Photoshop (Adobe) from 8–12 separate fields of vision per condition (700 × 500 µm each). Data were plotted using Prism (GraphPad Software). For NE and nuclear pore distribution of Nup210 constructs, C2C12 myoblasts cell lines were grown on coverslips and fixed for 2 min in 4% PFA at RT. Immunofluorescence of C2C12 myoblasts was performed similarly as in myotubes using GFP and mAb414 antibodies. Samples were mounted in ProLong gold (Life Technologies). For NPC localization analysis, images were taken with a super-resolution microscope (63×; Elyra PS.1; Carl Zeiss) at RT. Reconstruction and chromatographic alignment was performed for all images using ZEN 2011 software (Carl Zeiss) with the structured illumination microscopy analysis module. Confocal images to examine NE distribution of Nup210 fragments were taken with a laser-scanning confocal microscope (63×; LSM 710; Carl Zeiss) at RT. Colocalization of fluorophore signals was analyzed by Imaris (Bitplane), and signal profiles were generated using RGB profiler plugin in ImageJ (National Institutes of Health).
FACS
C2C12 myoblasts were dissociated with trypsin for 5 min, centrifuged at 1,000 g, and resuspended in PBS plus 2% FBS. Cells exhibiting similar fluorescent levels were collected in 100% FBS and plated on DMEM with 20% FBS. Cell sorting was performed in the Flow Cytometry Core facility at the Salk Institute.
Constructs
Rat Nup210-GFP on cytomegalovirus (CMV)-pEGFP-C1 backbone was obtained from J. Ellenberg’s laboratory (European Molecular Biology Laboratory Heidelberg, Heidelberg, Germany; Rabut et al., 2004). To generate Nup210 retroviral constructs, Nup210-GFP (1–1,886 aa), Nup210ΔLUMEN-GFP (1–25 aa + 1,809–1,886 aa), Nup210ΔCT-GFP (1–1,829 aa), and all Nup210-derived fragments were first amplified from pEGFP-C1-shRNA–resistant Nup210-GFP by PCR, cloned into pDONR207 (pENTRY) and recombined into the destination vector CMV-pQCXIB (retroviral vector) using the Gateway Cloning System (Invitrogen). Generation of shRNA-resistant Nup210 was performed as previously described in D’Angelo and Gomez-Cavazos et al. (2012). In brief, site-directed mutagenesis was performed on Nup210-GFP (pEGFP-C1) with a site-directed mutagenesis kit (QuikChange; QIAGEN) following the manufacturer’s protocol. Mutations were targeted to the 21-bp Nup210 shRNA site changing the third nucleic acid of every codon in that region without altering the amino acid sequence. Human NDC1, Sec61-β, Calnexin, and MAGE-A3 were first cloned into CMV-pEGFP-N1 (C-terminal GFP tag), recloned into pDONR207, and recombined into CMV-pQCXIB. CalnexinLumen(1–480 aa) + Nup210TM+CT(1,809–1,886 aa)-GFP chimera was generated by fusion PCR, cloned into pDONR207, and recombined into CMV-pQCXIB. For transmembrane chimeras ΔCTSec61-βTM and ΔCTLap2-βTM, transmembrane domains of human Lap2-β (411–430 aa) and Sec61-β (68–90 aa) were added by extension PCR to the 3′ end of Nup210’s luminal domain (1–1,808 aa), respectively.
Lentiviral and retroviral production, infection, and selection
Lentivirus was packaged in 293T cells grown in DMEM (Invitrogen) containing 20% FBS and penicillin/streptomycin. 293T cells were transfected in 10-cm plates with 5.2 µg of shRNA vector and 2.8 µg of packaging mix using 24 µl of Lipofectamine 2000. Media were replaced 12 h after transfection, and virus was collected at 36 and 60 h. Proliferating myoblasts at 30% confluency were infected with the 36-h supernatant and with the 60 h, 24 h later. Myoblasts were then split and cultured in media containing 2 µg/ml puromycin for selection. Nontarget shRNA control (SHC002) was purchased from Sigma-Aldrich (sequence 5′-CAACAAGATGAAGAGCACCAA-3′). Nup210 (TRCN0000101938) shRNA lentiviral vector was obtained from GE Healthcare (sequence 5′-GCTGACAGATAAGCAACTGAA-3′). For retroviral production, 293T cells were transfected in 10-cm plates with 4 µg of retroviral vector and 4 µg of Ampho retroviral packaging vector using 24 µl of Lipofectamine 2000. Proliferating myoblasts were infected with retrovirus supernatant. Myoblasts were then split and cultured in media containing 10 µg/ml blasticidin for selection. For myotube lentiviral infections, cells were infected 36 h after differentiation was induced with a 2 ×108 transducing unit.
FRAP
C2C12 cells were plated on 8-well plates (Ibidi) and imaged in a laser-scanning confocal microscope (LSM 710). FRAP was performed on sections of the NE in C2C12 myoblasts stably expressing GFP-tagged constructs. To circumvent overexpression artifacts, only cells expressing low levels of these constructs were subjected to FRAP analysis. FRAP parameters used were five prebleach scans followed by ∼50 photobleaching iterations and 115 postbleach scans taken at 2.5 frames/s. Fluorescence intensity data were collected, normalized, and fitted for each sample to determine the maximum fluorescence recovery (MFR) and half-time of recovery (t1/2) using Prism.
Online supplemental material
Fig. S1 shows representative images of all stable cell lines used on this manuscript. Fig. S2 shows the acceleration of myotube formation by the overexpression of Nup210 chimeras lacking NPC sorting signals. Fig. S3 shows that TUDCA and MAGE-A3–GFP overexpression can restore differentiation of myoblasts expressing Nup210 shRNAs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201410047/DC1.
Supplementary Material
Acknowledgments
We thank the members of the Hetzer laboratory and Jennifer M. Higginbotham for critical discussion and reading of the manuscript. We also thank Dongyang Li for help with experiments not included in this manuscript.
M.W. Hetzer is supported by the National Institutes of Health grant RO1GM098749. The project described was supported by award number P30CA014118195 from the National Cancer Institute. J.S. Gomez-Cavazos is supported by UC-MEXUS CONACYT (University of California Institute for Mexico and the United States El Consejo Nacional de Ciencia y Tecnología) Fellowship.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BiP
- binding immunoglobulin protein
- CMV
- cytomegalovirus
- MFR
- maximum fluorescence recovery
- MHC
- myosin heavy chain
- NE
- nuclear envelope
- NPC
- nuclear pore complex
- PDI
- protein disulfide isomerase
- TUDCA
- tauroursodeoxycholic acid
References
- Akhtar A., and Gasser S.M.. 2007. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8:507–517 10.1038/nrg2122 [DOI] [PubMed] [Google Scholar]
- Allen N.P., Huang L., Burlingame A., and Rexach M.. 2001. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276:29268–29274 10.1074/jbc.M102629200 [DOI] [PubMed] [Google Scholar]
- Alter J., and Bengal E.. 2011. Stress-induced C/EBP homology protein (CHOP) represses MyoD transcription to delay myoblast differentiation. PLoS ONE. 6:e29498 10.1371/journal.pone.0029498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral J.D., Viana R.J., Ramalho R.M., Steer C.J., and Rodrigues C.M.. 2009. Bile acids: regulation of apoptosis by ursodeoxycholic acid. J. Lipid Res. 50:1721–1734 10.1194/jlr.R900011-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.R., and Silver P.A.. 2007. Transcriptional regulation at the nuclear pore complex. Curr. Opin. Genet. Dev. 17:100–106 10.1016/j.gde.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Capelson M., Liang Y., Schulte R., Mair W., Wagner U., and Hetzer M.W.. 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 140:372–383 10.1016/j.cell.2009.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw J.M., Krutchinsky A.N., Zhang W., Chait B.T., and Matunis M.J.. 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158:915–927 10.1083/jcb.200206106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo M.A., Gomez-Cavazos J.S., Mei A., Lackner D.H., and Hetzer M.W.. 2012. A change in nuclear pore complex composition regulates cell differentiation. Dev. Cell. 22:446–458 10.1016/j.devcel.2011.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de las Heras J.I., Batrakou D.G., and Schirmer E.C.. 2013. Cancer biology and the nuclear envelope: a convoluted relationship. Semin. Cancer Biol. 23:125–137 10.1016/j.semcancer.2012.01.008 [DOI] [PubMed] [Google Scholar]
- Deldicque L., Hespel P., and Francaux M.. 2012. Endoplasmic reticulum stress in skeletal muscle: origin and metabolic consequences. Exerc. Sport Sci. Rev. 40:43–49 10.1097/JES.0b013e3182355e8c [DOI] [PubMed] [Google Scholar]
- Eriksson C., Rustum C., and Hallberg E.. 2004. Dynamic properties of nuclear pore complex proteins in gp210 deficient cells. FEBS Lett. 572:261–265 10.1016/j.febslet.2004.07.044 [DOI] [PubMed] [Google Scholar]
- Gerace L., Ottaviano Y., and Kondor-Koch C.. 1982. Identification of a major polypeptide of the nuclear pore complex. J. Cell Biol. 95:826–837 10.1083/jcb.95.3.826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cavazos J.S., and Hetzer M.W.. 2012. Outfits for different occasions: tissue-specific roles of Nuclear Envelope proteins. Curr. Opin. Cell Biol. 24:775–783 10.1016/j.ceb.2012.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber U.F., and Gerace L.. 1995. Depletion of calcium from the lumen of endoplasmic reticulum reversibly inhibits passive diffusion and signal-mediated transport into the nucleus. J. Cell Biol. 128:5–14 10.1083/jcb.128.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E.R., Altan N., Lippincott-Schwartz J., and Powers M.A.. 2002. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell. 13:1282–1297 10.1091/mbc.01-11-0538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelz A., Debler E.W., and Blobel G.. 2011. The structure of the nuclear pore complex. Annu. Rev. Biochem. 80:613–643 10.1146/annurev-biochem-060109-151030 [DOI] [PubMed] [Google Scholar]
- Huber M.D., Guan T., and Gerace L.. 2009. Overlapping functions of nuclear envelope proteins NET25 (Lem2) and emerin in regulation of extracellular signal-regulated kinase signaling in myoblast differentiation. Mol. Cell. Biol. 29:5718–5728 10.1128/MCB.00270-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppertz B., Tews D.S., and Kaufmann P.. 2001. Apoptosis and syncytial fusion in human placental trophoblast and skeletal muscle. Int. Rev. Cytol. 205:215–253 10.1016/S0074-7696(01)05005-7 [DOI] [PubMed] [Google Scholar]
- Liang Y., and Hetzer M.W.. 2011. Functional interactions between nucleoporins and chromatin. Curr. Opin. Cell Biol. 23:65–70 10.1016/j.ceb.2010.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu F., Alves A., Anderson K., Doye V., and Lacy E.. 2008. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev. Cell. 14:831–842 10.1016/j.devcel.2008.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans B.J., Anantharaman V., Aravind L., and Koonin E.V.. 2004. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 3:1612–1637 10.4161/cc.3.12.1316 [DOI] [PubMed] [Google Scholar]
- Morishima N., Nakanishi K., Takenouchi H., Shibata T., and Yasuhiko Y.. 2002. An endoplasmic reticulum stress-specific caspase cascade in apoptosis. Cytochrome c-independent activation of caspase-9 by caspase-12. J. Biol. Chem. 277:34287–34294 10.1074/jbc.M204973200 [DOI] [PubMed] [Google Scholar]
- Morishima N., Nakanishi K., and Nakano A.. 2011. Activating transcription factor-6 (ATF6) mediates apoptosis with reduction of myeloid cell leukemia sequence 1 (Mcl-1) protein via induction of WW domain binding protein 1. J. Biol. Chem. 286:35227–35235 10.1074/jbc.M111.233502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Zhu H., Morishima N., Li E., Xu J., Yankner B.A., and Yuan J.. 2000. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 403:98–103 10.1038/47513 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Largaespada D.A., Lee M.P., Johnson L.A., Ohyashiki K., Toyama K., Chen S.J., Willman C.L., Chen I.M., Feinberg A.P., et al. 1996. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat. Genet. 12:154–158 10.1038/ng0296-154 [DOI] [PubMed] [Google Scholar]
- Nakanishi K., Sudo T., and Morishima N.. 2005. Endoplasmic reticulum stress signaling transmitted by ATF6 mediates apoptosis during muscle development. J. Cell Biol. 169:555–560 10.1083/jcb.200412024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K., Kakiguchi K., Yonemura S., Nakano A., and Morishima N.. 2015. Transient Ca2+ depletion from the endoplasmic reticulum is critical for skeletal myoblast differentiation. FASEB J. 29:1–13. [DOI] [PubMed] [Google Scholar]
- Olsson M., Ekblom M., Fecker L., Kurkinen M., and Ekblom P.. 1999. cDNA cloning and embryonic expression of mouse nuclear pore membrane glycoprotein 210 mRNA. Kidney Int. 56:827–838 10.1046/j.1523-1755.1999.00618.x [DOI] [PubMed] [Google Scholar]
- Olsson M., Schéele S., and Ekblom P.. 2004. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp. Cell Res. 292:359–370 10.1016/j.yexcr.2003.09.014 [DOI] [PubMed] [Google Scholar]
- Ori A., Banterle N., Iskar M., Andrés-Pons A., Escher C., Khanh Bui H., Sparks L., Solis-Mezarino V., Rinner O., Bork P., et al. 2013. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol. 9:648 10.1038/msb.2013.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan U., Yilmaz E., Özcan L., Furuhashi M., Vaillancourt E., Smith R.O., Görgün C.Z., and Hotamisligil G.S.. 2006. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 313:1137–1140 10.1126/science.1128294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G., Doye V., and Ellenberg J.. 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6:1114–1121 10.1038/ncb1184 [DOI] [PubMed] [Google Scholar]
- Raices M., and D’Angelo M.A.. 2012. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat. Rev. Mol. Cell Biol. 13:687–699 10.1038/nrm3461 [DOI] [PubMed] [Google Scholar]
- Raman R., Rajanikanth V., Palaniappan R.U., Lin Y.P., He H., McDonough S.P., Sharma Y., and Chang Y.F.. 2010. Big domains are novel Ca²+-binding modules: evidences from big domains of Leptospira immunoglobulin-like (Lig) proteins. PLoS ONE. 5:e14377 10.1371/journal.pone.0014377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneich C., Dremina E., Galeva N., and Sharov V.. 2014. Apoptosis in differentiating C2C12 muscle cells selectively targets Bcl-2-deficient myotubes. Apoptosis. 19:42–57 10.1007/s10495-013-0922-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simossis V.A., and Heringa J.. 2005. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 33(Suppl. 2):W289–W294 10.1093/nar/gki390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F., Nautrup-Pedersen G., Cordes V.C., and Görlich D.. 2006. Nuclear pore complex assembly and maintenance in POM121- and gp210-deficient cells. J. Cell Biol. 173:477–483 10.1083/jcb.200601002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Zhang J., Zhang X., Xu C., and Tu X.. 2013. Solution structure of the Big domain from Streptococcus pneumoniae reveals a novel Ca2+-binding module. Sci Rep. 3:1079 10.1038/srep01079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R.W., and Blobel G.. 1992. The single transmembrane segment of gp210 is sufficient for sorting to the pore membrane domain of the nuclear envelope. J. Cell Biol. 119:1441–1449 10.1083/jcb.119.6.1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R.W., Bartnik E., and Blobel G.. 1989. Primary structure analysis of an integral membrane glycoprotein of the nuclear pore. J. Cell Biol. 108:2083–2092 10.1083/jcb.108.6.2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Khaoustov V.I., Chung C.C., Sohn J., Krishnan B., Lewis D.E., and Yoffe B.. 2002. Effect of tauroursodeoxycholic acid on endoplasmic reticulum stress-induced caspase-12 activation. Hepatology. 36:592–601 10.1053/jhep.2002.35441 [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., and Mori K.. 2007. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev. Cell. 13:365–376 10.1016/j.devcel.2007.07.018 [DOI] [PubMed] [Google Scholar]
- Zhang X., Chen S., Yoo S., Chakrabarti S., Zhang T., Ke T., Oberti C., Yong S.L., Fang F., Li L., et al. 2008. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell. 135:1017–1027 10.1016/j.cell.2008.10.022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.