Cell growth relies on Hrr25/CK1δ-directed phosphorylation of Ltv1, which allows its release from nascent 40S ribosomal subunits and promotes subunit maturation.
Abstract
Casein kinase 1δ/ε (CK1δ/ε) and their yeast homologue Hrr25 are essential for cell growth. Further, CK1δ is overexpressed in several malignancies, and CK1δ inhibitors have shown promise in several preclinical animal studies. However, the substrates of Hrr25 and CK1δ/ε that are necessary for cell growth and survival are unknown. We show that Hrr25 is essential for ribosome assembly, where it phosphorylates the assembly factor Ltv1, which causes its release from nascent 40S subunits and allows subunit maturation. Hrr25 inactivation or expression of a nonphosphorylatable Ltv1 variant blocked Ltv1 release in vitro and in vivo, and prevented entry into the translation-like quality control cycle. Conversely, phosphomimetic Ltv1 variants rescued viability after Hrr25 depletion. Finally, Ltv1 knockdown in human breast cancer cells impaired apoptosis induced by CK1δ/ε inhibitors, establishing that the antiproliferative activity of these inhibitors is due, at least in part, to disruption of ribosome assembly. These findings validate the ribosome assembly pathway as a novel target for the development of anticancer therapeutics.
Introduction
Ribosome biogenesis is required for cell growth. The assembly of ribosomal subunits involves the action of >200 assembly factors (AFs) including helicases, ATPases, GTPases, and kinases (Lafontaine and Tollervey, 2001; Granneman and Baserga, 2004; Hage and Tollervey, 2004; Zemp and Kutay, 2007; Henras et al., 2008; Strunk and Karbstein, 2009; Karbstein, 2011; Panse, 2011; Martin et al., 2013; Rodríguez-Galán et al., 2013; Thomson et al., 2013; Woolford and Baserga, 2013). These nonribosomal factors transiently associate with ribosome assembly intermediates to promote and regulate their assembly. AFs bound to late cytoplasmic precursors of both 40S and 60S subunits also prevent untimely translation initiation on immature subunits (Karbstein, 2013).
Defects in ribosome assembly and its regulation underlie many human diseases (Freed et al., 2010; Narla and Ebert, 2010; Armistead and Triggs-Raine, 2014). For example, a reduction in the production of functional ribosomes impairs translation, cell growth, and division, and provokes cell death. Conversely, a hallmark of human cancers is the up-regulation of the ribosome assembly pathway (Ruggero et al., 2003; Ruggero and Pandolfi, 2003; Stumpf and Ruggero, 2011).
We recently discovered a novel quality control mechanism during cytoplasmic 40S maturation that involves a translation-like cycle, where the translation initiation factor eIF5B promotes joining of 60S subunits to pre-40S subunits (Lebaron et al., 2012; Strunk et al., 2012). These studies also suggested that dissociation of the AF Ltv1 occurs before 60S subunit joining and that this event commits stable 40S assembly intermediates to the translation-like cycle (Strunk et al., 2012). Further, the cryogenic EM (cryo-EM) structure of a late pre-40S assembly intermediate indicates that Ltv1 must be released from a complex of the AF Enp1 and the ribosomal protein Rps3, which is located on the solvent side of the beak structure near the mRNA entry channel and blocks binding of Rps10 (Strunk et al., 2011).
The essential yeast casein kinase 1 (CK1) δ/ε homologue Hrr25 is required for 40S maturation and phosphorylates one or more components of the Enp1–Ltv1–Rps3 ternary complex (Schäfer et al., 2006). However, how Hrr25-mediated phosphorylation of this complex affects pre-40S maturation is not resolved. Further, Hrr25 has other roles in important processes, including cell cycle control (Butler et al., 1994; Mehlgarten and Schaffrath, 2003), tRNA modifications (Mehlgarten et al., 2009), 60S ribosome biogenesis (Ray et al., 2008), vesicle transport (Lord et al., 2011; Bhandari et al., 2013), DNA repair (Hoekstra et al., 1991; Ho et al., 1997), signaling (Kafadar et al., 2003), spindle formation during meiosis (Petronczki et al., 2006; Rumpf et al., 2010), and autophagy (Pfaffenwimmer et al., 2014; Tanaka et al., 2014). Thus, Hrr25-dependent control of the committed step in late 40S maturation may integrate ribosome assembly with other key cellular processes.
Like Hrr25, the human homologues CK1δ and CK1ε are components of pre-40S subunits and are required for cytoplasmic 40S maturation (Zemp et al., 2014). CK1δ and CK1ε also regulate multiple cellular processes, including the Wnt and Hedgehog signaling pathways (Price and Kalderon, 2002; Price, 2006), chromosome segregation, cell cycle and growth (Behrend et al., 2000; Stöter et al., 2005), DNA repair and microtubule dynamics (Knippschild et al., 1997; Li et al., 2004; Grozav et al., 2009; Ikeda et al., 2011), circadian rhythm (Eide et al., 2005; Gallego and Virshup, 2007), and vesicle trafficking (Wolff et al., 2006). Further, CK1δ expression is elevated in several tumor types and in Alzheimer’s and Parkinson’s disease (Ghoshal et al., 1999; Schwab et al., 2000; Yasojima et al., 2000; Knippschild et al., 2005b; Tsai et al., 2007; Brockschmidt et al., 2008; Perez et al., 2011; Hirner et al., 2012; Rodriguez et al., 2012; Knippschild et al., 2014; Rosenberg et al., 2015). Accordingly, CK1δ and CK1ε have been targets of drug design for more than a decade, and CK1δ/CK1ε inhibitors are active in preclinical models of these diseases (Badura et al., 2007; Brockschmidt et al., 2008; Sprouse et al., 2009, 2010; Walton et al., 2009; Meng et al., 2010; Li et al., 2011; Bryant et al., 2012; Perreau-Lenz et al., 2012; Toyoshima et al., 2012; Bibian et al., 2013; Smadja Storz et al., 2013; Kurihara et al., 2014). However, the molecular basis for the antiproliferative effects from CK1δ/CK1ε inhibitors has not been resolved.
Here we report that the essential function of Hrr25 is in ribosome biogenesis, where biochemical and genetic epistasis studies show that yeast cell growth relies on Hrr25-directed phosphorylation and release of Ltv1 from pre-40S. This circuit is necessary and sufficient for maturation of pre-40S subunits. Further, inhibiting Hrr25 and thus Ltv1 release has been shown to block the formation of 80S-like ribosomes that are required for maturation of pre-40S subunits. Finally, this circuit is operational in human breast cancer cells where knockdown of Ltv1 impairs the potency of CK1δ/CK1ε inhibitors, establishing the ribosome biogenesis pathway as an important drug target for anticancer therapeutics.
Results
Rps3 recruitment to ribosomes is energy independent but requires Ltv1 and Enp1
The delivery of the Rps3 ribosomal protein to pre-40S subunits requires the chaperone Yar1 (Koch et al., 2012), but how the handover of Rps3 from Yar1 to pre-40S subunits occurs is unclear. In pre-40S subunits, Rps3 forms a complex with the AFs Ltv1 and Enp1, and on pre-40S subunits this complex locks Rps3 in a position that differs from its locale in mature 40S subunits. In addition, the Ltv1–Enp1–Rps3 complex blocks Rps10 incorporation, preventing final assembly of the mRNA channel (Strunk et al., 2011).
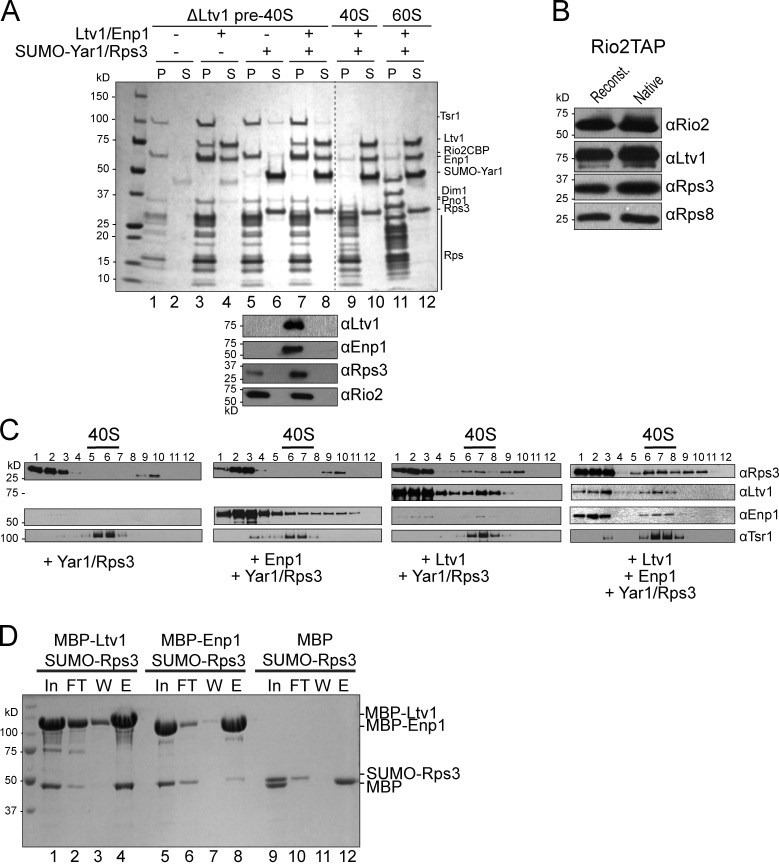
It has been suggested that both Hrr25-dependent phosphorylation and then dephosphorylation of Ltv1, Enp1, and Rps3 are required for 40S assembly (Schäfer et al., 2006). To test if phosphorylation is required for Rps3 recruitment to pre-40S subunits from Yar1, or if this process is spontaneous and is rather driven by higher affinity interactions between Rps3 and pre-40S subunits, we developed an in vitro system to assess Rps3 incorporation. Pre-40S ribosomes were isolated from Ltv1 knockout yeast cells (ΔLtv1). These ribosomes also lack Enp1 and Rps3, and have reduced levels of Rps20 and Rps29 (Strunk et al., 2011). As Rps3 is incorporated after Ltv1 and Enp1, we reasoned that addition of Enp1/Ltv1 would reconstitute the pre-40S assembly intermediate to which Rps3 is transferred in vivo. Co-sedimentation assays were used to assess incorporation of recombinant factors into pre-40S subunits (Fig. 1, A and B). Enp1/Ltv1/Yar1 and Rps3 were incubated with and without pre-40S ribosomes, and loaded on top of a sucrose cushion. After high-speed centrifugation, ribosomes and ribosome-bound proteins form a pellet, whereas soluble, free proteins remain in the supernatant. Control experiments demonstrated that Yar1, Rps3, Enp1, or Ltv1 do not sediment in the absence of pre-40S subunits (Fig. 1 A, lanes 10 and 12). In contrast, after incubation with pre-40S subunits, Enp1 and Ltv1 efficiently incorporate into the subunits (Fig. 1 A, lanes 3 and 7). This interaction is specific, as neither protein bound to mature 40S or 60S subunits. The addition of recombinant Yar1–Rps3 to pre-40S ribosomes reconstituted with Ltv1 and Enp1, but not to ΔLtv1 pre-40S ribosomes alone, leads to the incorporation of Rps3 and the release of Yar1 (Fig. 1 A). Further, the levels of Rps3 in the reconstituted pre-40S ribosomes mirror those found in purified pre-40S ribosomes from cells (Fig. 1 B).
Figure 1.
Recruitment of Rps3 to pre-40S ribosomes. (A) Coomassie-stained SDS-PAGE of the ribosome-bound (pellet [P]) and free (supernatant [S]) fractions. Binding to mature 40S or 60S subunits was evaluated as a control. The lower panel shows Western blots of lanes 5–8. Antibodies used are indicated on the right. The broken line indicates that intervening lanes have been spliced out. (B) Western blot analyses of native purified pre-40S and reconstituted pre-40S ribosomes establish that Rps3 levels are comparable in both. Ltv1 and Rps8 are the loading controls; Rio2 is the tagged component used for purification. (C) Western blot analyses of the gradient fractions from ribosomes reconstituted with the factors indicated. The positions of 40S ribosomes, as determined by absorbance at 254 nm, are indicated. (D) Coomassie-stained SDS-PAGE of protein binding assays on amylose beads. The pulled down fractions shown are: In, input; FT, flow-through; W, the final wash; and E, eluted.
To test if Ltv1 or Enp1 alone were sufficient to direct Rps3 recruitment, subunits were reconstituted, omitting one or the other component, and then assayed via sucrose gradient sedimentation (Fig. 1 C). This was necessary, as some precipitation of Enp1 was observed in the absence of Ltv1, leading to pelleting in the absence of ribosomes. In contrast, in gradient sedimentation experiments, 40S ribosomes do not pellet, avoiding this problem. In these studies, the position of pre-40S subunits is monitored by the sedimentation of the AF Tsr1, as well as by measuring RNA absorbance at 254 nm. As above, these studies showed that Rps3 does not bind to pre-40S subunits in the absence of Enp1 and Ltv1. Further, the addition of Ltv1 alone is sufficient to recruit Rps3, whereas Enp1 alone is not. These data are consistent with yeast two-hybrid interactions between Ltv1 and Rps3 (Ito et al., 2001; Merwin et al., 2014), and a direct interaction between Rps3 and Ltv1, as demonstrated by in vitro protein binding experiments (Fig. 1 D). Thus, transfer of Rps3 from Yar1 to pre-40S subunits is spontaneous but requires the presence of Ltv1, which directly interacts with Rps3.
Reconstituted preribosomes are indistinguishable from native preribosomes
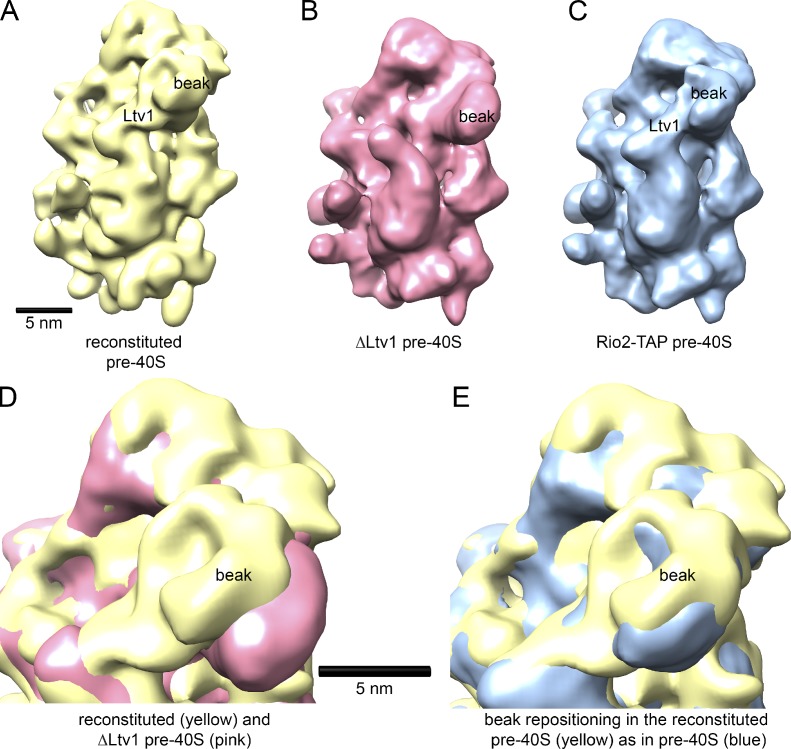
To test if reconstituted ribosomes accurately mimic native pre-40S ribosome intermediates purified from yeast, we used cryo-EM (Strunk et al., 2011). ΔLtv1 subunits were reconstituted with Enp1–Ltv1–Rps3 as above, before preparation of specimens for cryo-EM data acquisition (see Materials and methods). Comparison of the reconstituted pre-40S subunits with the ΔLtv1 subunits established additional density in the beak area where the Enp1–Ltv1–Rps3 complex is located (Figs. 2 and S1). Furthermore, a side view shows that the beak is retracted after the addition of Enp1–Ltv1–Rps3, as observed in the native purified pre-40S assembly intermediates (Strunk et al., 2011). Indeed, a comparison of the reconstituted pre-40S subunits with native pre-40S subunits shows that these structures are superimposable (Fig. 2). Thus, the in vitro delivery of Rps3 produces a molecule indistinguishable from native assembly intermediates, thereby validating the in vitro reconstitution.
Figure 2.
ΔLtv1 pre-40S ribosomes reconstituted with Yar1–Rps3 and Ltv1/Enp1 are structurally identical to native pre-40S ribosomes. (A) The solvent face of the reconstituted pre-40S ribosome shows the features expected from the presence of Enp1/Ltv1 and Rps3 near the beak. (B) Identical view of the ΔLtv1 pre-40S ribosomes used as a starting material in reconstitutions (EMD1924; Strunk et al., 2011). (C) Natively purified pre-40S ribosome (EMD1927; Strunk et al., 2011). (D) A superimposition of reconstituted (yellow) and ΔLtv1 pre-40S (pink) shows the repositioning of the beak upon addition of Enp1–Ltv1–Rps3, which suggests a structural recapitulation of the biochemically characterized pre-40S state. (E) Superimposition of reconstituted (yellow) and natively purified (blue) pre-40S ribosomes shows that reconstituted pre-40S ribosomes are structurally identical to purified native pre-40S ribosomes.
Release of Ltv1 requires Hrr25 kinase activity
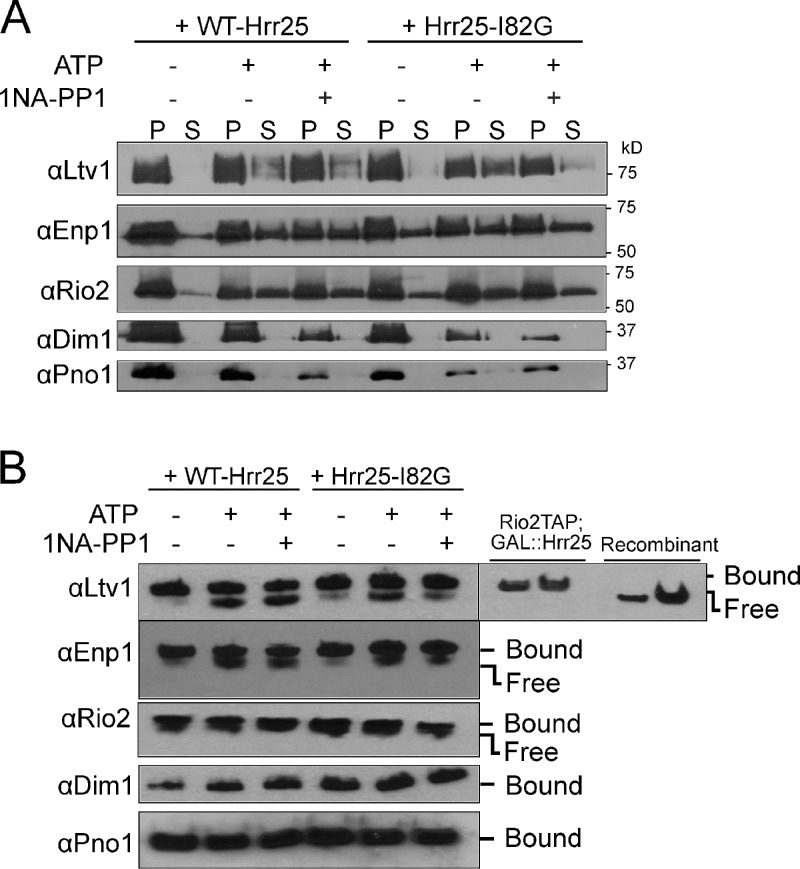
The kinase Hrr25 plays roles in pre-40S maturation, and Hrr25 depletion leads to reduced levels of phosphorylated Enp1, Ltv1, and Rps3 (Schäfer et al., 2006; Zemp et al., 2014). To test if Hrr25 kinase activity was required for release of Ltv1 or Enp1 from pre-40S subunits, we developed a release assay. Purified pre-40S ribosomes were incubated with nanomolar concentrations of ATP, and release of Ltv1 and Enp1 was monitored by sedimentation through a sucrose cushion as described above (Fig. 3 A) or by native gel shift analysis (Fig. 3 B). Both methods show that addition of ATP to pre-40S ribosomes leads to dissociation of Ltv1 and Enp1 (Fig. 3, A and B). The Rio2 ATPase is also dissociated, which is expected as its ATPase activity is linked to its dissociation from pre-40S ribosomes (Ferreira-Cerca et al., 2012). In contrast, other AFs remain bound.
Figure 3.
Hrr25-dependent release of Ltv1 from pre-40S ribosomes. (A) Co-sedimentation assays. Western blot analysis of AFs in the bound (pellet [P]) and released (supernatant [S]) fractions of the purified pre-40S ribosome from cells containing wild-type (WT-Hrr25) or 1NA-PP1-sensitive mutant (Hrr25-I82G) of Hrr25. (B) Native gel assay for Ltv1 release. Western blot analyses of native gels are shown. The position of ribosome-bound and free Ltv1 is identified by Rio2TAP-purified ribosomes and recombinant Ltv1.
To confirm that Ltv1 release arises from copurifying Hrr25 (Fig. S3A), we used a fully functional mutant of Hrr25 (Hrr25-I82G) that complements yeast depleted for Hrr25 in vivo (Fig. S2), but which has an enlarged ATP-binding pocket that renders Hrr25 sensitive to kinase inhibition by the bulky ATP analogue 1-(1,1-dimethylethyl)-3-(1-naphthalenyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine (1NA-PP1; Bishop et al., 2000). Notably, ATP-dependent dissociation of Ltv1 was blocked by addition of nanomolar concentrations of 1NA-PP1 to pre-40S ribosomes isolated from Hr225-I82G mutant cells, but Ltv1 dissociation from wild-type pre-40S ribosomes was not affected by 1NA-PP1 (Fig. 3). Thus, Ltv1 release from pre-40S subunits requires the Hrr25 kinase activity.
Enp1 is also released from reconstituted pre-40S ribosomes after the addition of ATP. However, its release (and the release of Rio2) is not efficiently blocked by 1NA-PP1, which indicates that Enp1 release is not (only) directed by Hrr25. This finding suggests that Enp1 remains behind in Ltv1-released, pre-40S subunits in vivo, in accord with the accumulation of Enp1 in 80S-like ribosomes (Strunk et al., 2012).
Release of Ltv1 is the essential function of Hrr25
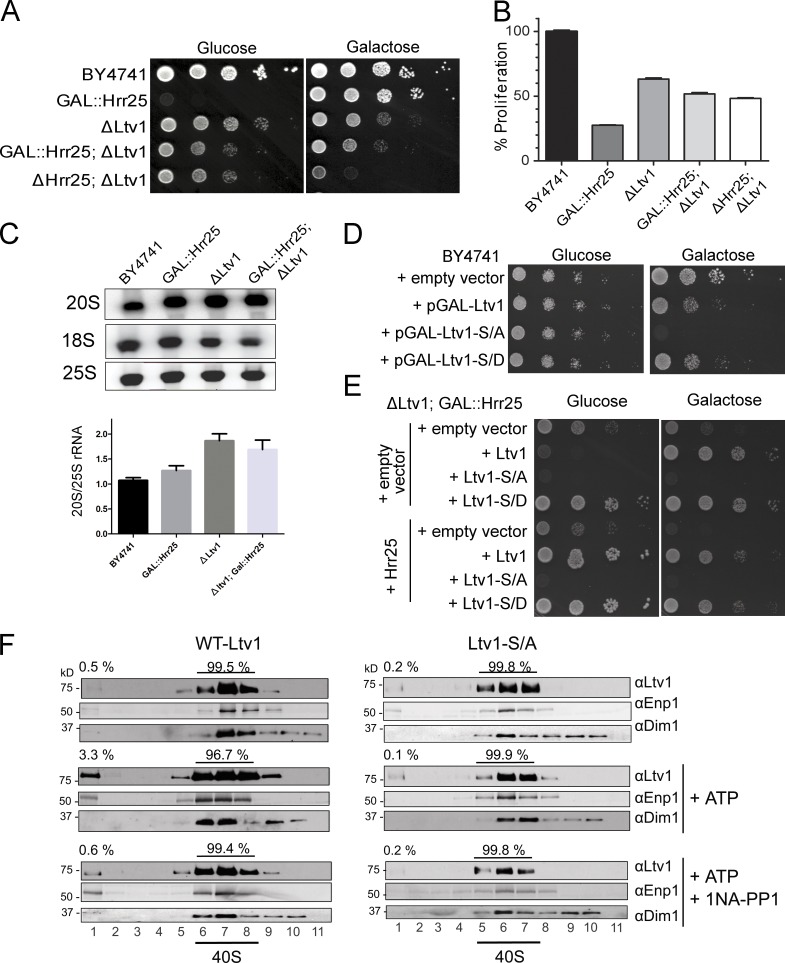
To confirm the role of Hrr25 in releasing Ltv1 in vivo, we used suppression analysis. While Hrr25 is an essential protein, Ltv1 is not (Fig. 4 A; Hoekstra et al., 1991; Winzeler et al., 1999; Seiser et al., 2006). Thus, we reasoned that if displacement of Ltv1 from pre-40S ribosomes was the essential function of Hrr25, then deletion of Ltv1 would be predicted to rescue the lethal phenotype of Hrr25 depletion. Depletion of Hrr25 in yeast cells engineered to express a galactose-inducible/glucose-repressible Hrr25 transgene (Gal::Hrr25) essentially abolished growth in glucose, as expected because Hrr25 is an essential gene (Fig. 4, A and B). Strikingly, however, loss of Ltv1 rescued growth of this mutant strain when grown in glucose (Fig. 4, A and B), and the ΔLtv1ΔHrr25 strain is viable (Fig. 4 A). Thus, the essential function of Hrr25 is linked to Ltv1. The more profound growth defect in the ΔLtv1ΔHrr25 cells relative to ΔLtv1 cells likely reflects roles of Hrr25 in other cellular processes as discussed above (Knippschild et al., 2005a). ΔLtv1ΔHrr25 cells also do not grow well on galactose, which indicates a role for Hrr25 in the regulation of galactose metabolism, consistent with previously observed interactions between Hrr25 and Gal10, as well as Sgm1 (Ho et al., 2002; Fasolo et al., 2011).
Figure 4.
Ltv1 release is the essential function of Hrr25. (A) Growth of wild-type yeast (BY4741), and those with galactose-inducible/glucose-repressible Hrr25 (GAL::Hrr25) or lacking Ltv1 (ΔLtv1), and of double mutant GAL::Hrr25;ΔLtv1 and ΔHrr25;ΔLtv1 cells, was compared. Fig. S2 B has a close-up to compare colony size. (B) Growth rates of wild-type, GAL::Hrr25,ΔLtv1, and GAL::Hrr25;ΔLtv1 yeast cells, as determined by their doubling times in YPD. All measurements were done in triplicate, and error bars show the standard deviation of these data. (C) Northern blot analysis of the 18S rRNA precursor (20S) and 25S rRNA. The quantitation shown below the data is the mean and standard deviation from four independent experiments. (D) Dominant-negative effects from Ltv1-S/A. Wild-type yeast (BY4741) were transformed with plasmids encoding wild-type Ltv1, Ltv1-S/A, or Ltv1-S/D under a galactose-inducible promoter. Induction of Ltv1 is weakly dominant-negative for wild-type Ltv1 and Ltv1-S/D, and strongly dominant-negative for Ltv1-S/A. (E) Growth of GAL::Hrr25;ΔLtv1 cells carrying plasmids of wild-type or phosphomutant Ltv1, with and without Hrr25, is compared on glucose (no endogenous Hrr25) and galactose-containing (endogenous Hrr25) plates. (F) Alanine mutations in the Ltv1 phosphosite block Enp1/Ltv1 release in vitro. Shown are Western blots of gradient fractions from ΔLtv1;Rio2TAP ribosomes, purified from cells containing Hrr25_I82G, and reconstituted on the calmodulin beads with Enp1–Yar1–Rps3 and wild-type Ltv1 or Ltv1-S/A. The positions of 40S ribosomes and free proteins are indicated.
Analysis of ribosomal RNA (rRNA) processing in yeast confirmed that depletion of Hrr25 and deletion of Ltv1 are epistatic; depletion of Hrr25 leads to a small but reproducible accumulation of 20S rRNA, the 18S rRNA precursor, and deletion of Ltv1 produces a more substantial accumulation of 20S rRNA. In contrast, depletion of Hrr25 in ΔLtv1 cells (the ΔLtv1/Gal::Hrr25 strain) reduces the accumulation of 20S rRNA that is manifest in the ΔLtv1 yeast (Fig. 4 C). In contrast, if Hrr25 and Ltv1 were independent, their combined depletion would increase levels of 20S rRNA.
Together, these data strongly support a model where Hrr25 directs the release of Ltv1 from pre-40S subunits in vivo. Even more importantly, they demonstrate that the essential function of Hrr25 is in ribosome assembly.
Hrr25 phosphorylates Ltv1
To test if Ltv1, Enp1, Rps3, or any other ribosomal protein were the target of the Hrr25 kinase, we included γ-[32P]ATP in the release assay and used SDS-PAGE analysis to identify radioactively labeled proteins. Both Ltv1 and Rio2 are significantly phosphorylated upon addition of γ-[32P]ATP to pre-40S ribosomes (Fig. S3 A). Further, Ltv1 phosphorylation, but not that of Rio2, is blocked by the addition of 1NA-PP1 to pre-40S ribosomes isolated from yeast that only express the Hrr25-I82G mutant. In contrast, 1NA-PP1 does not block Ltv1 phosphorylation in pre-40S ribosomes purified from cells expressing wild-type Hrr25. Thus, Ltv1 is the target of phosphorylation by Hrr25. These data were further confirmed by in vitro kinase assays using recombinant Hrr25, Ltv1, Enp1, and the Yar1–Rps3 complex. Whereas Hrr25 phosphorylates Ltv1 efficiently, no significant phosphorylation was observed for Enp1, Rps3, or Yar1 (Fig. S3 B).
Mutation of Ltv1 phosphosites blocks release of Ltv1
To confirm that Hrr25-directed phosphorylation of Ltv1 provokes its release from pre-40S ribosomes, we mapped its phosphorylation sites. Casein kinases like Hrr25 phosphorylate multiple adjacent sites on their substrates (Flotow et al., 1990). To examine which of the predicted phosphorylation sites are functionally relevant, we systematically mutated putative phosphorylation sites, prioritized based on conservation, and then tested their relevance in vivo and in vitro. Using this approach, we identified three highly conserved serine residues (S336, S339, and S342; Fig. S4). To test their importance in vivo and in vitro, we generated alanine mutants, which cannot be phosphorylated, as well as aspartate mutations, which are often used as phosphomimetics (Tarrant and Cole, 2009), as they mimic the negative charge of the phosphate. Importantly, mutation of these serines to alanine (Ltv1-S/A) or aspartate (Ltv1-S/D) residues reduces phosphorylation by Hrr25 (Fig. S3 C), which suggests that these constitute major phosphorylation sites. Further, the residual phosphorylation is more significant in Ltv1-S/D than Ltv1-S/A, as expected from the biochemistry of casein kinases, which are activated by prior phosphorylation events (Knippschild et al., 2005a), mimicked here by Ltv1-S/D.
To confirm the functional relevance of these Hrr25-dependent phosphorylation sites, we tested if Ltv1-S/A could be released from ribosomes. ΔLtv1 ribosomes from Hrr25-I82G cells were reconstituted on beads with recombinant wild-type Ltv1 and Ltv1-S/A (and Enp1, Yar1, and Rps3), unbound protein was washed out, and reconstituted ribosomes were eluted. While the addition of ATP to ribosomes containing wild-type Ltv1 leads to Ltv1 release (Fig. 4 F), as shown for native complexes above, Ltv1-S/A remained bound (Fig. 4 F). Addition of 1NA-PP1 confirms that release of wild-type Ltv1 is Hrr25 dependent. Release of Ltv1-S/D could not be tested as it does not bind pre-40S ribosomes in vitro (see “Phosphomimetic mutants bind weakly to pre-40S ribosomes in vivo and in vitro”). Interestingly, higher-resolution SDS-PAGE gels also demonstrate that an Ltv1 antibody–reactive band of lower electrophoretic mobility is observed upon ATP addition to pre-40S ribosomes containing wild-type Ltv1, but not Ltv1-S/A (Fig. S3 D). The electrophoretic mobility of Ltv1-S/D is akin to the lower mobility band of wild-type Ltv1, and also does not change upon ATP addition. These data further support the notion that Ltv1 residues S336, S339, and S342 are phosphorylated by Hrr25, leading to Ltv1 release from pre-40S ribosomes.
In vivo, Ltv1-S/A functions as a dominant-negative mutant, as expected from a defect in Ltv1 release (Fig. 4 D). In contrast, the Ltv1-S/D phosphomimetic mutant is Hrr25 independent, and grows nearly as well as cells containing wild-type Ltv1 and Hrr25 (Fig. 4 E). Importantly, the Ltv1-S/D mutant complements the growth defects manifest in ΔLtv1 yeast (Fig. 4 E), which indicates that Ltv1-S/D does bind to pre-40S ribosomes in the nucleus to promote their export (Seiser et al., 2006; Merwin et al., 2014). Collectively, these findings establish that Hrr25-dependent phosphorylation of Ltv1 is necessary and sufficient for its release from pre-40S ribosomes.
Phosphomimetic mutants bind weakly to pre-40S ribosomes in vivo and in vitro
Gradient ultracentrifugation was used to test if, as predicted by these findings, phosphorylation of Ltv1 impairs its ability to bind to pre-40S subunits, leading to Hrr25-independent release from pre-40S ribosomes. Analysis of extracts from yeast expressing wild-type Ltv1, Ltv1-S/A, or Ltv1-S/D shows a significantly increased peak of free 60S subunits and a reduced polysome fraction in cells expressing the Ltv1-S/A mutant, which is consistent with defects in 40S assembly (Fig. S5 A). Western blot analyses of these gradients demonstrates that wild-type Ltv1 and Ltv1-S/A are efficiently incorporated into pre-40S subunits. In contrast, the phosphomimetic Ltv1-S/D mutant bound very weakly to pre-40S subunits, as expected if phosphorylation leads to its release. We note however that Ltv1-S/D does complement the ΔLtv1 phenotype (Fig. 4 E), which indicates that Ltv1-S/D binds long enough to allow for pre-40S export to the cytoplasm.
Essentially identical results were obtained when ΔLtv1 ribosomes were reconstituted with Enp1 and wild-type Ltv1, Ltv1-S/A, or Ltv1-S/D in vitro. While Ltv1 and Ltv1-S/A cosediment with pre-40S ribosomes, Ltv1-S/D does not, and, correspondingly, Ltv1-S/D does not allow for Rps3 incorporation in vitro (Fig. S5 B), as Ltv1 is required for in vitro incorporation of Rps3 (Fig. 1 C).
We also note that Ltv1-S/A appears to be bound more weakly than wild-type Ltv1, as its peak in 40S is weaker, and it streaks toward the free peak, which is indicative of dissociation during the spin. This is not surprising, as the mutated alanine residues would be expected to be near or in contact with the 40S subunit, which would allow phospho-dependent regulation of binding.
These findings demonstrate that the Ltv1-S/D mutant, which mimics the phosphorylated state of Ltv1, readily dissociates from pre-40S ribosomes, supporting a model whereby Hrr25-dependent phosphorylation of Ltv1 triggers its release from pre-40S subunits.
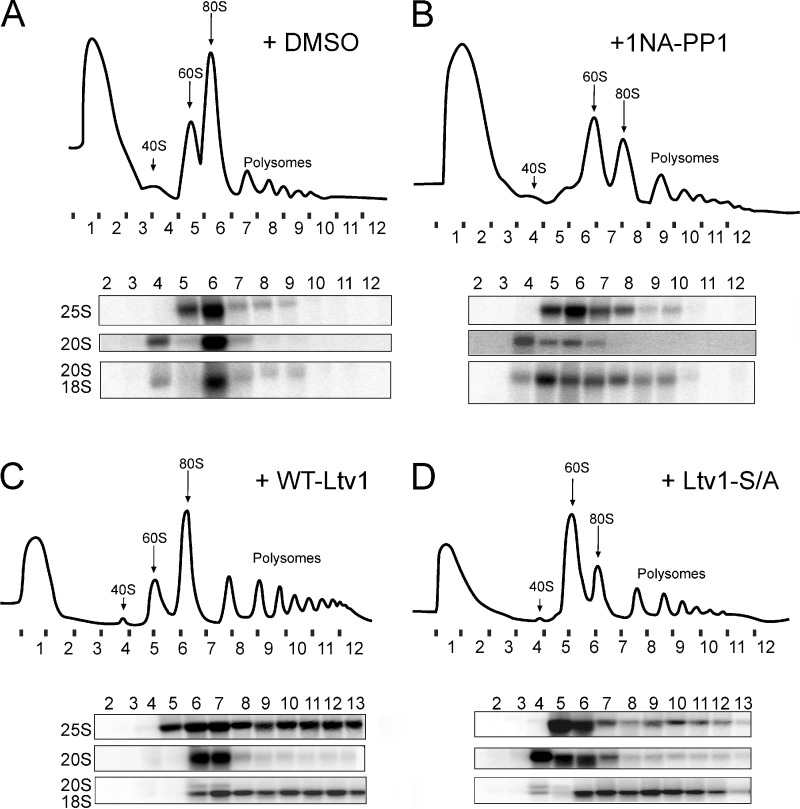
Inhibition of Ltv1 release blocks subunit joining
Maturation of pre-40S involves the joining of a large 60S ribosomal subunit and the formation of 80S-like ribosomes in a quality control cycle (Lebaron et al., 2012; Strunk et al., 2012). Depletion of the AF Fap7 leads to accumulation of 80S-like ribosomes. Hence, a block in the joining of 60S and pre-40S subunits can be monitored by the loss of 80S-like ribosomes when Fap7 is depleted (Strunk et al., 2012). To test the role of the Hrr25-Ltv1 circuit for entering this maturation cascade, we constructed a yeast strain where both Hrr25 and Fap7 are galactose-inducible and thus can be simultaneously depleted by growth in glucose. This strain was supplemented with a plasmid encoding the 1NA-PP1–sensitive mutant of Hrr25 (Hrr25-I82G). Inhibition of Hrr25 by addition of 1NA-PP1 to cultures grown in the presence of glucose resulted in loss of 80S-like assembly intermediates, whereas 80S-like ribosomes were observed in vehicle-treated cells (Fig. 5, A and B). Further, ΔLtv1/Gal::Fap7 cells supplemented with a plasmid encoding the dominant-negative Ltv1-S/A failed to form 80S-like ribosomes containing the 20S rRNA precursor in the absence of Fap7, whereas those with wild-type Ltv1 formed this key intermediate during 40S maturation (Fig. 5, C and D). These effects were not due to effects on export of pre-40S ribosomes from the nucleus, as in both experiments only the cytoplasmic fractions were analyzed (Fig. S5 C). Thus, Hrr25-mediated release of Ltv1 from pre-40S subunits is required for joining of 60S subunits and for entering into the translation-like cycle.
Figure 5.
Inhibition of Hrr25 or phosphosite mutations of Ltv1 block subunit joining. (A and B) 10–50% sucrose gradients of cytoplasmic extracts from GAL::Hrr25;GAL::Fap7 cells transformed with a plasmid carrying Hrr25-I82G, grown in glucose for 16 h, and treated with DMSO vehicle (A) or 1NA-PP1 (B). The quality of the nucleo-cytoplasmic separation is shown in Fig. S5 C. (C and D) Sucrose gradients of cytoplasmic extracts from ΔLtv1;GAL::Fap7 cells transformed with a plasmid carrying WT-Ltv1 (C) or Ltv1-S/A (D), grown in glucose for 16 h. Absorbance profiles at 254 nm and Northern blots for rRNAs and precursors are shown.
Human CK1δ/ε also has essential functions in 40S ribosome maturation
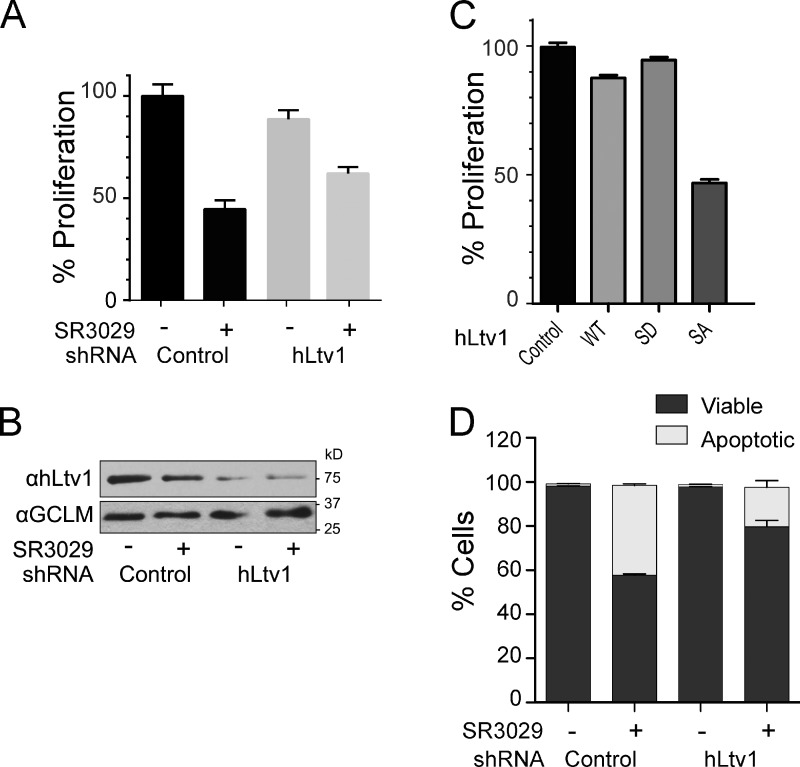
Human CK1δ and CK1ε have ∼65% sequence identity and 85% sequence similarity to Hrr25, and both have been implicated in 40S ribosome maturation (Zemp et al., 2014). To assess the effect of CK1δ/CK1ε inhibition on ribosome assembly and cell growth, we used the small molecule inhibitor SR-3029, a potent and highly specific inhibitor of CK1δ and CK1ε (Bibian et al., 2013). For our experiments, we used MDA-MB-231 triple-negative human breast cancer cells, which express high levels of CK1δ, are sensitive to knockdown of CK1δ (but not to silencing of CK1ε), and are highly sensitive to SR-3029 (Rosenberg et al., 2015). As expected, treatment of these cells with low concentrations of SR-3029 (30 nM) led to marked and rapid reductions in cell growth (Fig. 6 A). This is consistent with previous findings that both CK1δ and CK1ε have essential functions in human cells (Knippschild et al., 2014).
Figure 6.
The essential function of human CK1δ is in ribosome maturation. (A) Cell proliferation (as determined by doubling times) of control MDA-MB-231 cells, or of MDA-MB-231 cells where hLtv1 was silenced by Dox-directed induction of hLtv1 shRNA for 3 d, before addition of 30 nM SR3029 or vehicle for another 3 d. (B) Western blot analyses established ∼80% depletion of Ltv1 after 3 d of Ltv1 knockdown. (C) Proliferation of MDA-MB-231 cells engineered to inducibly overexpress wild-type hLtv1, hLtv1-S/D, or hLtv1-S/A after Dox treatment for 3 d. (D) Annexin V/DAPI staining combined with FACS analysis of control MDA-MB-231 cells and those depleted of hLtv1 in the presence or absence of 100 nM SR-3029. All measurements in A, C, and D were done in triplicate, and error bars show the standard deviation of these data.
We reasoned that if the essential role of CK1δ and CK1ε in human cells is also to direct Ltv1 release during ribosome assembly, then knockdown of human Ltv1 (hLtv1) should rescue the growth inhibition of cells treated with SR-3029. To test this hypothesis, we generated MDA-MB-231 cells stably expressing the fluorescence marker EGFP along with doxycycline (Dox)-inducible shRNAs that selectively silence hLtv1 or Renilla luciferase. Treatment of EGFP-sorted cells with Dox triggered knockdown of hLtv1 relative to the control cells (∼80% knockdown; Fig. 6 B). Knockdown of Ltv1 in the absence of SR-3029 had minor effects on cell growth (90% of the proliferation rate of wild-type cells; Fig. 6 A). While this might be an underestimation due to the incomplete knockdown of Ltv1, the small growth defect contrasts with the strong growth inhibitory phenotype upon Ltv1 deletion in yeast, where Ltv1 loss inhibits pre-40S export (Seiser et al., 2006; Fassio et al., 2010). However, the modest effects of hLtv1 knockdown are consistent with the presence of additional export adaptors for pre-40S subunits in human cells (Zemp et al., 2009; Carron et al., 2011).
Strikingly, the partial knockdown of Ltv1 partially rescued the growth inhibitory effects of SR-3029 (Fig. 6 A). Further, Ltv1 knockdown also impaired SR-3029–induced apoptosis of MDA-MB-231 breast cancer cells (Fig. 6 C). Thus, the antiproliferative effects observed after inhibition of Hrr25 or CK1δ/CK1ε in both yeast and human cells are conserved and are, in large part, due to effects on pre-40S ribosome maturation. Furthermore, alanine mutation of the serine residues homologous to the identified yeast phosphor-sites (hLtv1-S/A) also induces strong dominant-negative phenotypes (Fig. 6 C), again indicating that blocking the release of Ltv1 has the potential to inhibit human tumor cell growth. In contrast, and as also observed in yeast, forced overexpression of wild-type hLtv1 produces weak dominant-negative effects, and hLtv1-S/D is even more active than wild-type hLtv1.
Discussion
Ltv1 directs Rps3 incorporation into pre-40S subunits
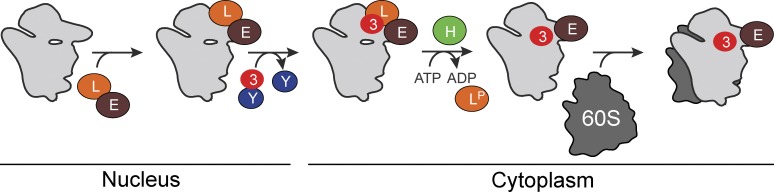
The data presented herein, along with previous studies, reveal a two-step mechanism for Rps3 recruitment into the beak structure of pre-40S subunits, where it forms the mRNA entry channel (Fig. 7). Yar1 delivers Rps3 to pre-40S subunits in the nucleus (Koch et al., 2012). Here we show that this is a spontaneous exchange driven by the stronger affinity interaction of Rps3 with pre-40S ribosome–bound Ltv1. In the resulting assembly, intermediate Rps3 is not bound at its mature binding site (Strunk et al., 2011), and can be salt-extracted from pre-40S subunits in a complex with the AFs Enp1 and Ltv1 (Schäfer et al., 2006). Here we show that Hrr25 phosphorylates Ltv1, leading to its release from pre-40S subunits. Enp1 is also released from pre-40S subunits, but the data do not provide strong support for a specific role of Hrr25 in this release. Nevertheless, we do note that Enp1 on pre-40S subunits is destabilized by the absence of Ltv1 (Strunk et al., 2011; Fig. 1 C). Ltv1 is required for efficient export of pre-40S ribosomes (Seiser et al., 2006; Merwin et al., 2014). Thus, Hrr25-dependent phosphorylation and release of Ltv1 must occur after nuclear export. How this presumed spatial regulation is achieved remains unknown, as there are pools of nuclear Hrr25 (Huh et al., 2003; Breker et al., 2014). It is possible that the nuclear pools of Hrr25 are not freely diffusible and instead interact with distinct structures, e.g. microtubules, which is consistent with Hrr25 having a role in spindle assembly (Petronczki et al., 2006; Rumpf et al., 2010). Alternatively, or additionally, it is possible that Hrr25-dependent Ltv1 release is slow relative to nuclear export, which is consistent with the observation that Ltv1 release is the rate-limiting step in 40S ribosome maturation (see “Kinase-mediated regulation of 40S ribosome assembly”). Finally, additional levels of regulation might be present, perhaps involving the recruitment or release of other AFs, that could regulate Hrr25 access or activity.
Figure 7.
Model for incorporation of Rps3 into pre-40S ribosomes. Enp1 (cyan) and Ltv1 (magenta) bind to pre-40S subunits in the nucleus. Yar1 (orange) delivers Rps3 (yellow) to pre-40S–bound Ltv1, in a process that is energy independent. Hrr25-dependent (green) phosphorylation of Ltv1 releases Ltv1 from pre-40S subunits to allow for subunit joining and entry into the translation-like maturation cycle.
The data also indicate that Ltv1 regulates the assembly of the beak structure in pre-40S subunits. Specifically, Ltv1 initially recruits Rps3, but blocks binding at its mature binding site. Presumably, Rps3 forms initial interactions with rRNA and ribosomal proteins in the presence of Ltv1, which are then remodeled after Ltv1 release. This is reminiscent of the step-wise manner in which other ribosomal proteins form interactions with rRNA, as revealed by hydroxyl radical footprinting data (Adilakshmi et al., 2008), with the exception that the time scale between these binding events is stretched out by Ltv1 binding.
Interestingly, recent work from bacteria indicates that the functions of Ltv1 in the formation of the beak structure and the mRNA entry channel are conserved between prokaryotes and eukaryotes, even though Ltv1 itself is not conserved (Clatterbuck Soper et al., 2013). In particular, small subunit assembly intermediates purified from bacteria lacking RimM do not contain the beak-binding proteins S3, S10, and S14, which are the homologues of Rps3, Rps20, and Rps29. All of these proteins are missing from pre-40S subunits purified from ΔLtv1 yeast (Strunk et al., 2011). Further, RimM has been suggested to bind to S19 (Lövgren et al., 2004), the homologue for Rps15, which interacts directly with Ltv1 (Campbell and Karbstein, 2011). Thus, we suggest that RimM and Ltv1 are functional homologues, though likely structurally unrelated.
Why does the beak area need the chaperone activities provided by Ltv1/RimM? Structure probing of the bacterial assembly intermediate reveals that the beak needs extensive refolding during the heat-activation step (RI→RI*), which allows S3, S10, and S14 to bind (Mizushima and Nomura, 1970; Held et al., 1974; Holmes and Culver, 2004). Among regions refolded during this transition, H42 and H43 of 16S rRNA are exposed in the absence of RimM in vivo and also form part of the Rps3 binding site. These analogies suggest that Ltv1, akin to RimM, prevents misfolding of the beak structure, which is consistent with the cold-sensitive phenotype manifest in ΔLtv1 yeast strains (Loar et al., 2004).
Ltv1 release regulates entry into the cytoplasmic maturation cascade
The data herein demonstrate that Hrr25-mediated phosphorylation and release of Ltv1 regulates entry into the 40S maturation cascade. Specifically, inhibition of Hrr25, or mutations in Ltv1 that block phosphorylation by Hrr25, prevent subunit joining to enter a translation-like cycle that is required for 40S maturation (Lebaron et al., 2012; Strunk et al., 2012). This was unanticipated, as the Ltv1 binding site on the small subunit is on the solvent side and not at the subunit interface. Thus, Ltv1 release appears to lead to global changes in the structure of pre-40S subunits, which are in turn required for joining of the large subunit.
Comparison of cryo-EM structures in the presence and absence of Ltv1 shows that Ltv1 dissociation leads to an 18° head rotation (Strunk et al., 2011). This rearrangement likely affects the stability of the intersubunit bridges located on the head, thereby weakening the formation of 80S-like ribosomes. We propose that interaction of Ltv1/Enp1 with 40S subunits prevents the formation of crucial subunit bridges on the head that are required for stabilizing the interaction of 60S with pre-40S. Future experiments are required to test this model.
Kinase-mediated regulation of 40S ribosome assembly
Why is Ltv1 dissociation a regulated step in 40S assembly? Due to the binding of seven AFs, which block all functional sites on the nascent 40S subunit, the 40S ribosome assembly intermediate that is exported from the nucleus is exceptionally stable and translationally inert (Strunk et al., 2011). In contrast, intermediates in the translation-like cycle are short-lived and subject to degradation (Strunk et al., 2012). Our data show that Ltv1 release is required for entry into the translation-like cycle, and therefore gates the conversion of a stable, translationally inert intermediate into one that is subject to quality control and degradation.
In addition, the data establish that the Hrr25-dependent release of Ltv1 is the rate-limiting step in cytoplasmic 40S maturation, and likely for all of 40S production, as the cytoplasmic precursor that contains Ltv1 and is the substrate for Hrr25 accumulates to substantial levels in wild-type yeast. These two properties render Hrr25-mediated Ltv1-relase well-suited for regulatory control. Further, we note that Ltv1 blocks incorporation of the ribosomal proteins lining the mRNA entry channel (Rps3 and Rps10), which indicates that this regulated step also controls a key maturation event.
Interestingly, Hrr25 has also been implicated in several cellular processes, including transcription, DNA repair, signaling, and meiosis (Knippschild et al., 2014). Thus, Hrr25-dependent control of the committed step in late 40S maturation could integrate ribosome assembly with these processes. Intriguingly, Hrr25 interacts genetically and physically with several other kinases that play important roles in nutrient-dependent signaling, including Gcn2, Pho80, Pcl10, Rim15, Rck1, and Ste20 (Ptacek et al., 2005; Gavin et al., 2006; Fiedler et al., 2009; Breitkreutz et al., 2010; Fasolo et al., 2011). As nutrient availability is a major modulator of ribosome assembly (Schmelzle and Hall, 2000; Li et al., 2006; Zoncu et al., 2011), it is tempting to speculate that one or more of these kinases might regulate Hrr25 in response to nutrient availability.
An essential function of CK1δ/CK1ε in 40S ribosome maturation
CK1δ/CK1ε are targets for the development of novel chemotherapeutics against cancer and neurodegenerative diseases (Knippschild et al., 2005b, 2014; Perez et al., 2011). Importantly, some CK1δ/CK1ε inhibitors have potent antiproliferative effects ex vivo, and lead to tumor regression and improved survival in preclinical animal cancer models (Knippschild et al., 2014). Nevertheless, because CK1δ/CK1ε have diverse roles (Knippschild et al., 2005a), it is not clear which of the pathways they control are the key vulnerabilities disabled by CK1δ/CK1ε inhibitors.
Our data show that the essential function of Hrr25 is in ribosome assembly versus in other biological processes, as deletion of Ltv1 rescues the lethal effects from Hrr25 depletion. Similarly, knockdown of Ltv1 also rescues the growth and promotes the survival of human MDA-MB-231 breast cancer cells that are treated with the CK1δ/CK1ε inhibitor SR-3029. As these cells rely exclusively in CK1δ for growth and survival (Rosenberg et al., 2015), these findings indicate that the CK1δ-hLtv1 circuit is operational and essential for ribosome assembly in higher eukaryotes, which is consistent with previous findings that demonstrate a role for CK1δ/CK1ε in 40S ribosome maturation (Zemp et al., 2014). Our findings are also consistent with previous observations that the ribosome assembly pathway is up-regulated in all cancers, which require marked increases in protein synthesis (Ruggero and Pandolfi, 2003; Stumpf and Ruggero, 2011). Finally, our findings validate the ribosome biogenesis machinery as an attractive and novel target for anticancer therapeutics.
Materials and methods
Yeast strains and cloning
Galactose-inducible and deletion strains were generated by PCR-based recombination (Longtine et al., 1998), confirmed by PCR and Western blotting (if antibodies were available), and are listed in Table S1. Yar1 was cloned into a pET28a-based plasmid (EMD Millipore) containing an N-terminal 6×His tag followed by a SUMO tag. Rps3 was cloned into the first multiple cloning site of pET-Duet1 (EMD Millipore). The Hrr25-I82G mutation and the phosphosite mutations of Ltv1 were introduced by site-directed mutagenesis and confirmed by sequencing. Plasmids used in this study are listed in Table S2.
Protein expression and purification
Tagged or untagged versions of Enp1 and Ltv1 were overexpressed from modified pET28a plasmids expressing an N-terminal His6 tag followed by an MBP tag. Overexpression of the proteins was induced by the addition of 1 mM IPTG to Rosetta cells (EMD Millipore) grown in 2×YT medium supplemented with appropriate antibiotics to an OD600 of 0.6. Cells were harvested after 16 h of growth at 18°C, and lysed by sonication in lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole, and 5 mM β-mercaptoethanol [β-ME]). The soluble fraction was applied to Ni2+-NTA resin (QIAGEN) equilibrated with lysis buffer. Resin was washed with 5 volumes of lysis buffer containing 20 mM imidazole. Proteins were eluted with lysis buffer supplemented with 300 mM imidazole. For cleavage of the MBP tag, eluates were mixed with TEV protease and dialyzed overnight at 4°C against 50 mM Tris, pH 7.5, 200 mM NaCl, and 2 mM DTT. The dialyzed samples were further purified by ion exchange chromatography over a Mono Q column (GE Healthcare) equilibrated with dialysis buffer. Proteins were eluted with a linear gradient of 100–500 mM NaCl over 30 column volumes. The final step of protein purification was carried over a size-exclusion chromatography column (Superdex 200; GE Healthcare) preequilibrated with 20 mM Tris, pH 7.5, 200 mM NaCl, and 2 mM DTT. Fractions with pure protein were pooled and concentrated using Amicon concentrators (EMD Millipore).
For copurification of Enp1 and Ltv1, cell pellets were mixed and resuspended in lysis buffer (50 mM Tris-NaOH, pH 7.5, 200 mM NaCl, and 5 mM β-ME) supplemented with 100 µg/ml lysozyme, and lysed by sonication. The cleared lysate was loaded onto amylose resin equilibrated with lysis buffer. Beads were washed with 5 volumes of lysis buffer before elution with lysis buffer supplemented with 15 mM maltose. Eluates were mixed with TEV protease and dialyzed overnight at 4°C against dialysis buffer (25 mM Tris-NaOH, pH 7.5, 200 mM NaCl, and 2 mM DTT), then purified by ion exchange chromatography on a MonoQ column equilibrated with dialysis buffer. Proteins were eluted with a linear gradient from 200 mM to 500 mM NaCl over 30 column volumes, and loaded on a Superdex200 size-exclusion column equilibrated with dialysis buffer. Fractions containing pure proteins were pooled and concentrated.
For copurification of Yar1 and Rps3, cells were transformed with pET-Duet1-Rps3 and pET-SUMO-Yar1 plasmids and grown at 37°C to OD600 of 0.6 in 2×YT media supplemented with antibiotics. Cells were transferred to 18°C, induced by addition of 0.3 mM IPTG, and harvested after 12 h. Cell pellets were resuspended in lysis buffer (20 mM Hepes-NaOH, pH 7.5, 200 mM NaCl, and 5 mM β-ME) supplemented with 100 µg/ml lysozyme, and lysed by sonication. The cleared lysate was loaded onto Ni2+-NTA resin equilibrated with lysis buffer. Beads were washed with 10 volumes of lysis buffer and 5 volumes of lysis buffer supplemented with 20 mM imidazole. Proteins were eluted with lysis buffer containing 300 mM imidazole, and further purified on a Superdex200 size-exclusion column preequilibrated with 20 mM Hepes-NaOH, pH 7.5, 200 mM NaCl, and 2 mM DTT. Fractions containing pure proteins were pooled and concentrated.
A plasmid encoding GST-Hrr25 was obtained from M.S. Cyert (Stanford University, Stanford, CA), and GST-Hrr25 was purified as described by Kafadar et al. (2003). In brief, BL21(DE3)pLys (EMD Millipore) cells carrying the pGEX-4T-3-Hrr25 plasmid were grown to an OD600 of 0.5 in 2×YT medium at 37°C, induced by addition of 0.5 mM IPTG, and harvested after 5 h of expression at 30°C. Cells were lysed by sonication in lysis buffer (50 mM Tris, pH 8.0, 1.5 M NaCl, 2 mM EDTA, 2 mM EGTA, and 2 mM DTT) supplemented with a complete protease inhibitor cocktail tablet (Roche). The lysate was then supplied with 0.1% Tween-20 and cleared by centrifugation. The soluble fraction was purified over Glutathione Sepharose resin (GE Healthcare) equilibrated with lysis buffer. The resin was washed with 5 column volumes of wash buffer (50 mM Tris, pH 8.0, 110 mM KOAc, 2 mM MgCl2, 2 mM DTT, and 0.1% Tween-20), 2 column volumes of wash buffer supplemented with 1 mM ATP, and 3 column volumes of wash buffer once again. Proteins were eluted in wash buffer supplemented with 15 mM reduced glutathione and dialyzed in 20 mM Hepes, pH 6.8, 150 mM KOAc, 250 mM sorbitol, 2 mM MgOAc, and 1 mM DTT.
TAP purification of pre-40S ribosomes
TAP-tagged yeast cells were grown at 30°C in yeast extract peptone dextrose (YP-dextrose) to an OD600 of 1 in a 10-liter fermenter. Affinity purification of TAP-tagged ribosomes was performed essentially as described previously (Strunk et al., 2011). In brief, lysed yeast cells were applied to IgG resin (GE Healthcare) equilibrated in IgG binding buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 10 mM MgCl2, and 0.075% NP-40) and washed with 100 column volumes of binding buffer, followed by 30 column volumes of TEV cleavage buffer (IgG binding buffer containing 0.5 mM EDTA and 2 mM DTT). Pre-40S ribosomes were collected after TEV cleavage and applied to calmodulin resin (Agilent Technologies) equilibrated in calmodulin binding buffer (IgG binding buffer supplemented with 2 mM CaCl2 and 1 mM imidazole). The resin was washed with 100 column volumes of binding buffer and ribosomes were eluted by the addition of calmodulin-binding buffer supplemented with 2 mM EGTA.
Cosedimentation/ribosome binding assays
2 pmol of pre-40S ribosomes from ΔLtv1 cells, purified through both steps of the TAP protocol, were incubated with 10 pmol of recombinant proteins in 50 µl RB buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 0.075% NP-40, and 10 mM β-ME). The mixture was incubated for 15 min on ice and then placed on 400 µl of a 20% sucrose cushion, and centrifuged for 4 h at 400,000 g in a TLA 100.1 rotor. After centrifugation, the supernatant was removed and acetone precipitated, and the pellet was resuspended in SDS-load dye. Supernatants and pellets were analyzed by SDS-PAGE followed by Western blotting.
Release assays
Pre-40S ribosomes used in release assays were purified only through the first step of the TAP protocol to allow for more copurifying Hrr25. No significant ATP-dependent release is observed using pre-40S purified through both steps. Rio2TAP pre-40S ribosomes were incubated with equimolar concentrations of purified ATP or with 1NA-PP1 plus ATP in RB buffer. The mixture was incubated for 10 min at room temperature, and then analyzed by cosedimentation analysis as described above, by 4% native gels, or by gradient sedimentation. Native gels were prepared as described previously (Acker et al., 2007). In brief, samples were mixed with native gel dye and loaded on a 4% acrylamide/bisacrylamide (37.5:1) gel prepared and run in THEM buffer at 4°C. For gradient centrifugation, the samples were loaded on 5–20% sucrose gradients in gradient buffer (20 mM Hepes, pH 7.4, 5 mM MgCl2, 100 mM NaCl, and 2 mM DTT), centrifuged for 4 h at 400,000 g in a SW41Ti rotor. Importantly, control experiments demonstrate that at higher concentrations 1-NAPP1 and/or the DMSO solvent destabilize pre-40S ribosomes.
Kinase assay
Kinase assays were performed using 1 µM Ltv1, Enp1, and SUMO-Yar1-Rps3 and 0.1 µM recombinant GST-tagged Hrr25, or 50 nM Rio2TAP or Tsr1TAP-purified preribosomes and no kinase. Reactions were set up in 10 µl RB buffer with 1 µM or 50 nM cold ATP and 10 µCi γ-[32P]ATP, incubated for 10 min at 30°C, stopped by the addition of SDS loading buffer, and analyzed by SDS-PAGE and autoradiography.
Protein interaction studies
3 µM MBP-Ltv1, MBP-Enp1, or MBP were mixed with 5 µM SUMO-Rps3 in binding buffer (50 mM Tris, pH 7.5, 200 mM NaCl, and 2 mM DTT) and incubated for 15 min at 4°C before the addition of 25 µl of equilibrated amylose resin (New England Biolabs, Inc.). The mixture was incubated on a rotating platform at 4°C for 30 min before flow-through fractions were collected. Resin was washed four times with 200 µl of binding buffer before being eluted with 25 µl of binding buffer supplemented with 20 mM maltose.
Sucrose density gradient analysis
Sucrose gradient centrifugation and analysis was performed as described previously (Strunk et al., 2012). In brief, cells were grown to mid log phase in YP-dextrose and harvested after addition of 0.1 mg/ml cycloheximide, washed, and lysed in ice-cold gradient buffer supplemented with 0.1 mg/ml cycloheximide and complete protease Inhibitor cocktail (Roche). Lysate was cleared by 10 min of centrifugation at 10,000 g, applied to 10–50% sucrose gradients in gradient buffer, and centrifuged for 2 h at 40,000 RPM in a SW41Ti rotor. Gradients were fractionated and scanned by UV 260 nm absorbance. Fractions were analyzed for their protein and RNA content by Western and Northern blotting. Cytoplasmic extraction was performed similar to Rieder and Emr (2001). In brief, cells were grown to mid log phase in YP-dextrose, harvested after addition of 0.1 mg/ml cycloheximide, and washed with TSD reduction buffer (100 mM Tris, pH 9.4, 10 mM DTT, and 0.1 mg/ml cycloheximide). Spheroplasts were prepared by incubation in buffer A (2% glucose, 1 M sorbitol, 1 mM DTT, and 5 mg/ml Zymolyase; Seikagaku) at 30°C for 20 min, washed twice with buffer A lacking Zymolyase, and lysed in ice-cold gradient buffer (supplemented with cycloheximide and protease inhibitors) using a Dounce homogenizer. Lysate was cleared of unbroken cells by centrifugation at 300 g. Cytoplasmic (C) and nuclear (N) fractions were separated by an additional centrifugation step at 13,000 g.
Antibodies
Antibodies against soluble recombinant Ltv1, Enp1, Tsr1, Dim1, Pno1, Rio2, Rrp5, and Rok1 were raised in rabbits by Josman LLP. HRP-conjugated anti–rabbit secondary antibody was obtained from Rockland Immunochemicals, Inc., the hLtv1 antibody (a gift from U. Kutay, ETH Zürich, Zürich, Switzerland) was raised in rabbits, the Rps3 antibody (a was gift from M. Seedorf, Zentrum für Molekulare Biologie der Universität Heidelberg, Heidelberg, Germany) was raised in rabbits, and Tif11 and Tif35 antibodies (a gift from A. Hinnebusch, National Institutes of Health) were raised in rabbits.
Cryo-EM analysis
4 µl of reconstituted pre-40S particles was applied to a holey carbon film (QuantiFoil 2.2; EMS) and prepared for EM imaging by plunge freezing on a Vitrobot Mark IV (FEI). Images were collected using Leginon (Suloway et al., 2005; Shrum et al., 2012) on a Titan Krios (FEI) operating at 120 kV with a nominal magnification of 59,000 and a defocus range of −1 to −3 µm. Images were recorded on an UltraScan 4000 camera (Gatan). 6,018 particles were selected using the manual particle-picking tool in Appion (Lander et al., 2009). To avoid any potential for model bias, we independently refined the data against two different initial models (Fig. S1 A). In the first case, a density map generated from the atomic coordinates of the 40S ribosome (Ben-Shem et al., 2011) and filtered to 50-Å resolution was used as the starting model. In the second case, the ΔLtv1 pre-40S (EMD1924), also filtered to 50-Å resolution, was used as an initial model. In both cases, after five rounds of projection matching in EMAN (Ludtke et al., 1999), the beak appears in the retracted position. After five additional refinement rounds, density for Ltv1 was clear. Relion version 1.2 was used for final refinement, resulting in a final 21-Å resolution structure (judging by the Fourier shell correlation [FSC] cutoff of 0.5) that clearly shows a ridge of density at the beak to hold it in the retracted position (Fig. 2 A and Fig. S1 B). Further multimodel 3D refinement and CTF correction in Relion (Scheres, 2012), which allows the data to separate into two distinct populations, did not reveal any underlying heterogeneity at the 20-Å resolution limit of the dataset. Importantly, we know that the changes between the ΔLtv1 and native pre-40S are visible within this resolution limit (Strunk et al., 2011).
Retroviral vector packaging and transduction
EcoPack (Takara Bio Inc.) cells were calcium phosphate transfected using the ProFection system (Promega) with RIEP, pRetroX-dsREDmirE(Ren)-GFP, pRetroX-dsREDmirE(hLtv1)-GFP, pRETROX-TIGHT_GFP-Tomato, pRETROX-TIGHT_GFP-hLtv1, pRETROX-TIGHT_GFP-hLtv1-S/A, or pRETROX-TIGHT_GFP-hLtv1-S/D. Retrovirus was collected twice, filtered, and concentrated using a 50,000 mol wt cut-off filter (VIVASPIN 20; Sartorius Stedim Biotech). MD-MBA-231 cells expressing the ecotropic receptor and the reverse tetracycline transactivator rtTA2 (MD-MBA-231-RIEP; obtained from C. Miething, Uniklinikum Freiburg, Freiburg im Breisgau, Germany) were generated using concentrated virus from RIEP and Ecotropic Receptor Booster (Takara Bio Inc.) according to the manufacturer’s protocol. Cells were selected with 0.5 µg/ml puromycin for several passages. MD-MBA-231-RIEP cells were then spin-infected (2,500 rpm, 30°C, 90 min) with concentrated virus from pRetroX-dsREDmirE(Ren)-GFP, pRetroX-dsREDmirE(hLtv1)-GFP, pRETROX-TIGHT_GFP-Tomato, pRETROX-TIGHT_GFP-hLtv1, pRETROX-TIGHT_GFP-hLtv1-S/A, or pRETROX-TIGHT_GFP-hLtv1-S/D in the presence of polybrene. Cells were expanded and FACS sorted for the presence of GFP. The expression of Renilla- or hLtv1-specific shRNAs and dsRED was induced by adding 0.3 µg/ml Dox. Expression of Tomato and wild-type or mutant hLtv1 (hLtv1-S/A and hLtv1-S/D) proteins was induced by addition of 0.3 µg/ml Dox and incubation for 3 d.
Quantification of apoptotic cells
Cells were washed twice with PBS and then incubated with 1× Annexin V binding buffer (BD) and Annexin V Alexa Fluor 647 (Biolegend) for 20 min. The cells were then washed with Annexin V binding buffer and resuspended in Annexin V binding buffer with DAPI. Cells were sorted using a flow cytometer (LSRII; BD) and analyzed using FlowJo software.
Online supplemental material
Fig. S1 shows supporting data for the EM analysis of in vitro reconstituted pre-40S ribosomes. Fig. S2 shows serial dilutions demonstrating that the Hrr25-I82G mutant is fully functional in the absence of 1NA-PP1. Fig. S3 shows that the target of the Hrr25 kinase activity in vitro is Ltv1. Fig. S4 shows a sequence alignment of residues 322–386 of yeast Ltv1 and its homologues. Fig. S5 shows binding of Ltv1-S/A and Ltv1-S/D mutants to pre-40S ribosomes in vitro and in vivo. Table S1 shows yeast strains used in this study. Table S2 shows plasmids used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201409056/DC1.
Supplementary Material
Acknowledgments
We thank U. Kutay, M. Seedorf, and A. Hinnebusch for antibodies against hLtv1, Rps3, and Tiff11/Tif35, respectively; M.S. Cyert (Stanford) for the Hrr25 expression plasmid; C. Miething (Uniklinikum Freiberg) for the RIEP plasmid; the flow cytometry core at Scripps Florida for cell sorting; and members of the Karbstein laboratory for comments on the manuscript.
This work was supported by National Institutes of Health (NIH) grants R01-GM086451 (to K. Karbstein) and CA154739 (to W.R. Roush and J.L. Cleveland), Molecular Library Screening Center Network grant U54MH074404 (chemistry portion, to W.R. Roush), National Cancer Institute Comprehensive Cancer Center grant P30-CA076292 (awarded to the H. Lee Moffitt Cancer Center & Research Institute), funding from the ThinkPink Kids Foundation (to J.L. Cleveland), and by grant 1149763 from the National Science Foundation (to M.E. Stroupe). H. Ghalei was supported by a PGA National Cancer Awareness Fellowship. F.X. Schaub was supported by the Swiss National Foundation (P300P3-147907).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- 1NA-PP1
- 1-(1,1-dimethylethyl)-3-(1-naphthalenyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine
- AF
- assembly factor
- CK1
- casein kinase 1
- cryo-EM
- cryogenic EM
- Dox
- doxycycline
- rRNA
- ribosomal RNA
References
- Acker M.G., Kolitz S.E., Mitchell S.F., Nanda J.S., and Lorsch J.R.. 2007. Reconstitution of yeast translation initiation. Methods Enzymol. 430:111–145 10.1016/S0076-6879(07)30006-2 [DOI] [PubMed] [Google Scholar]
- Adilakshmi T., Bellur D.L., and Woodson S.A.. 2008. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 455:1268–1272 10.1038/nature07298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armistead J., and Triggs-Raine B.. 2014. Diverse diseases from a ubiquitous process: the ribosomopathy paradox. FEBS Lett. 588:1491–1500 10.1016/j.febslet.2014.03.024 [DOI] [PubMed] [Google Scholar]
- Badura L., Swanson T., Adamowicz W., Adams J., Cianfrogna J., Fisher K., Holland J., Kleiman R., Nelson F., Reynolds L., et al. 2007. An inhibitor of casein kinase I epsilon induces phase delays in circadian rhythms under free-running and entrained conditions. J. Pharmacol. Exp. Ther. 322:730–738 10.1124/jpet.107.122846 [DOI] [PubMed] [Google Scholar]
- Behrend L., Stöter M., Kurth M., Rutter G., Heukeshoven J., Deppert W., and Knippschild U.. 2000. Interaction of casein kinase 1 delta (CK1δ) with post-Golgi structures, microtubules and the spindle apparatus. Eur. J. Cell Biol. 79:240–251 10.1078/S0171-9335(04)70027-8 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., and Yusupov M.. 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 334:1524–1529 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- Bhandari D., Zhang J., Menon S., Lord C., Chen S., Helm J.R., Thorsen K., Corbett K.D., Hay J.C., and Ferro-Novick S.. 2013. Sit4p/PP6 regulates ER-to-Golgi traffic by controlling the dephosphorylation of COPII coat subunits. Mol. Biol. Cell. 24:2727–2738 10.1091/mbc.E13-02-0114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibian M., Rahaim R.J., Choi J.Y., Noguchi Y., Schürer S., Chen W., Nakanishi S., Licht K., Rosenberg L.H., Li L., et al. 2013. Development of highly selective casein kinase 1δ/1ε (CK1δ/ε) inhibitors with potent antiproliferative properties. Bioorg. Med. Chem. Lett. 23:4374–4380 10.1016/j.bmcl.2013.05.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A.C., Ubersax J.A., Petsch D.T., Matheos D.P., Gray N.S., Blethrow J., Shimizu E., Tsien J.Z., Schultz P.G., Rose M.D., et al. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 407:395–401 10.1038/35030148 [DOI] [PubMed] [Google Scholar]
- Breitkreutz A., Choi H., Sharom J.R., Boucher L., Neduva V., Larsen B., Lin Z.Y., Breitkreutz B.J., Stark C., Liu G., et al. 2010. A global protein kinase and phosphatase interaction network in yeast. Science. 328:1043–1046 10.1126/science.1176495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breker M., Gymrek M., Moldavski O., and Schuldiner M.. 2014. LoQAtE—Localization and quantitation atlas of the yeast proteome. A new tool for multiparametric dissection of single-protein behavior in response to biological perturbations in yeast. Nucleic Acids Res. 42:D726–D730 10.1093/nar/gkt933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockschmidt C., Hirner H., Huber N., Eismann T., Hillenbrand A., Giamas G., Radunsky B., Ammerpohl O., Bohm B., Henne-Bruns D., et al. 2008. Anti-apoptotic and growth-stimulatory functions of CK1 delta and epsilon in ductal adenocarcinoma of the pancreas are inhibited by IC261 in vitro and in vivo. Gut. 57:799–806 10.1136/gut.2007.123695 [DOI] [PubMed] [Google Scholar]
- Bryant C.D., Parker C.C., Zhou L., Olker C., Chandrasekaran R.Y., Wager T.T., Bolivar V.J., Loudon A.S., Vitaterna M.H., Turek F.W., and Palmer A.A.. 2012. Csnk1e is a genetic regulator of sensitivity to psychostimulants and opioids. Neuropsychopharmacology. 37:1026–1035 10.1038/npp.2011.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A.R., White J.H., Folawiyo Y., Edlin A., Gardiner D., and Stark M.J.. 1994. Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol. Cell. Biol. 14:6306–6316 10.1128/MCB.14.9.6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M.G., and Karbstein K.. 2011. Protein-protein interactions within late pre-40S ribosomes. PLoS ONE. 6:e16194 10.1371/journal.pone.0016194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carron C., O’Donohue M.F., Choesmel V., Faubladier M., and Gleizes P.E.. 2011. Analysis of two human pre-ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals. Nucleic Acids Res. 39:280–291 10.1093/nar/gkq734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatterbuck Soper S.F., Dator R.P., Limbach P.A., and Woodson S.A.. 2013. In vivo X-ray footprinting of pre-30S ribosomes reveals chaperone-dependent remodeling of late assembly intermediates. Mol. Cell. 52:506–516 10.1016/j.molcel.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide E.J., Kang H., Crapo S., Gallego M., and Virshup D.M.. 2005. Casein kinase I in the mammalian circadian clock. Methods Enzymol. 393:408–418 10.1016/S0076-6879(05)93019-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasolo J., Sboner A., Sun M.G., Yu H., Chen R., Sharon D., Kim P.M., Gerstein M., and Snyder M.. 2011. Diverse protein kinase interactions identified by protein microarrays reveal novel connections between cellular processes. Genes Dev. 25:767–778 10.1101/gad.1998811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassio C.A., Schofield B.J., Seiser R.M., Johnson A.W., and Lycan D.E.. 2010. Dominant mutations in the late 40S biogenesis factor Ltv1 affect cytoplasmic maturation of the small ribosomal subunit in Saccharomyces cerevisiae. Genetics. 185:199–209 10.1534/genetics.110.115584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Cerca S., Sagar V., Schäfer T., Diop M., Wesseling A.M., Lu H., Chai E., Hurt E., and LaRonde-LeBlanc N.. 2012. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat. Struct. Mol. Biol. 19:1316–1323 10.1038/nsmb.2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler D., Braberg H., Mehta M., Chechik G., Cagney G., Mukherjee P., Silva A.C., Shales M., Collins S.R., van Wageningen S., et al. 2009. Functional organization of the S. cerevisiae phosphorylation network. Cell. 136:952–963 10.1016/j.cell.2008.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotow H., Graves P.R., Wang A.Q., Fiol C.J., Roeske R.W., and Roach P.J.. 1990. Phosphate groups as substrate determinants for casein kinase I action. J. Biol. Chem. 265:14264–14269. [PubMed] [Google Scholar]
- Freed E.F., Bleichert F., Dutca L.M., and Baserga S.J.. 2010. When ribosomes go bad: diseases of ribosome biogenesis. Mol. Biosyst. 6:481–493 10.1039/b919670f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M., and Virshup D.M.. 2007. Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 8:139–148 10.1038/nrm2106 [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L.J., Bastuck S., Dümpelfeld B., et al. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature. 440:631–636 10.1038/nature04532 [DOI] [PubMed] [Google Scholar]
- Ghoshal N., Smiley J.F., DeMaggio A.J., Hoekstra M.F., Cochran E.J., Binder L.I., and Kuret J.. 1999. A new molecular link between the fibrillar and granulovacuolar lesions of Alzheimer’s disease. Am. J. Pathol. 155:1163–1172 10.1016/S0002-9440(10)65219-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S., and Baserga S.J.. 2004. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296:43–50 10.1016/j.yexcr.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Grozav A.G., Chikamori K., Kozuki T., Grabowski D.R., Bukowski R.M., Willard B., Kinter M., Andersen A.H., Ganapathi R., and Ganapathi M.K.. 2009. Casein kinase I δ/ε phosphorylates topoisomerase IIα at serine-1106 and modulates DNA cleavage activity. Nucleic Acids Res. 37:382–392 10.1093/nar/gkn934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage A.E., and Tollervey D.. 2004. A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 1:9–15 10.4161/rna.1.1.932 [DOI] [PubMed] [Google Scholar]
- Held W.A., Ballou B., Mizushima S., and Nomura M.. 1974. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J. Biol. Chem. 249:3103–3111. [PubMed] [Google Scholar]
- Henras A.K., Soudet J., Gérus M., Lebaron S., Caizergues-Ferrer M., Mougin A., and Henry Y.. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65:2334–2359 10.1007/s00018-008-8027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirner H., Günes C., Bischof J., Wolff S., Grothey A., Kühl M., Oswald F., Wegwitz F., Bösl M.R., Trauzold A., et al. 2012. Impaired CK1 delta activity attenuates SV40-induced cellular transformation in vitro and mouse mammary carcinogenesis in vivo. PLoS ONE. 7:e29709 10.1371/journal.pone.0029709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Mason S., Kobayashi R., Hoekstra M., and Andrews B.. 1997. Role of the casein kinase I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 94:581–586 10.1073/pnas.94.2.581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 415:180–183 10.1038/415180a [DOI] [PubMed] [Google Scholar]
- Hoekstra M.F., Liskay R.M., Ou A.C., DeMaggio A.J., Burbee D.G., and Heffron F.. 1991. HRR25, a putative protein kinase from budding yeast: association with repair of damaged DNA. Science. 253:1031–1034 10.1126/science.1887218 [DOI] [PubMed] [Google Scholar]
- Holmes K.L., and Culver G.M.. 2004. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat. Struct. Mol. Biol. 11:179–186 10.1038/nsmb719 [DOI] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., and O’Shea E.K.. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Ikeda K., Zhapparova O., Brodsky I., Semenova I., Tirnauer J.S., Zaliapin I., and Rodionov V.. 2011. CK1 activates minus-end-directed transport of membrane organelles along microtubules. Mol. Biol. Cell. 22:1321–1329 10.1091/mbc.E10-09-0741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., and Sakaki Y.. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 98:4569–4574 10.1073/pnas.061034498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafadar K.A., Zhu H., Snyder M., and Cyert M.S.. 2003. Negative regulation of calcineurin signaling by Hrr25p, a yeast homolog of casein kinase I. Genes Dev. 17:2698–2708 10.1101/gad.1140603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K.2011. Inside the 40S ribosome assembly machinery. Curr. Opin. Chem. Biol. 15:657–663 10.1016/j.cbpa.2011.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K.2013. Quality control mechanisms during ribosome maturation. Trends Cell Biol. 23:242–250 10.1016/j.tcb.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knippschild U., Milne D.M., Campbell L.E., DeMaggio A.J., Christenson E., Hoekstra M.F., and Meek D.W.. 1997. p53 is phosphorylated in vitro and in vivo by the delta and epsilon isoforms of casein kinase 1 and enhances the level of casein kinase 1 delta in response to topoisomerase-directed drugs. Oncogene. 15:1727–1736 10.1038/sj.onc.1201541 [DOI] [PubMed] [Google Scholar]
- Knippschild U., Gocht A., Wolff S., Huber N., Löhler J., and Stöter M.. 2005a. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell. Signal. 17:675–689 10.1016/j.cellsig.2004.12.011 [DOI] [PubMed] [Google Scholar]
- Knippschild U., Wolff S., Giamas G., Brockschmidt C., Wittau M., Würl P.U., Eismann T., and Stöter M.. 2005b. The role of the casein kinase 1 (CK1) family in different signaling pathways linked to cancer development. Onkologie. 28:508–514 10.1159/000087137 [DOI] [PubMed] [Google Scholar]
- Knippschild U., Krüger M., Richter J., Xu P., García-Reyes B., Peifer C., Halekotte J., Bakulev V., and Bischof J.. 2014. The CK1 family: contribution to cellular stress response and its role in carcinogenesis. Front. Oncol. 4:96 10.3389/fonc.2014.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch B., Mitterer V., Niederhauser J., Stanborough T., Murat G., Rechberger G., Bergler H., Kressler D., and Pertschy B.. 2012. Yar1 protects the ribosomal protein Rps3 from aggregation. J. Biol. Chem. 287:21806–21815 10.1074/jbc.M112.365791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara T., Sakurai E., Toyomoto M., Kii I., Kawamoto D., Asada T., Tanabe T., Yoshimura M., Hagiwara M., and Miyata A.. 2014. Alleviation of behavioral hypersensitivity in mouse models of inflammatory pain with two structurally different casein kinase 1 (CK1) inhibitors. Mol. Pain. 10:17 10.1186/1744-8069-10-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D.L., and Tollervey D.. 2001. The function and synthesis of ribosomes. Nat. Rev. Mol. Cell Biol. 2:514–520 10.1038/35080045 [DOI] [PubMed] [Google Scholar]
- Lander G.C., Stagg S.M., Voss N.R., Cheng A., Fellmann D., Pulokas J., Yoshioka C., Irving C., Mulder A., Lau P.W., et al. 2009. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166:95–102 10.1016/j.jsb.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaron S., Schneider C., van Nues R.W., Swiatkowska A., Walsh D., Böttcher B., Granneman S., Watkins N.J., and Tollervey D.. 2012. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 19:744–753 10.1038/nsmb.2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Yin H., and Kuret J.. 2004. Casein kinase 1 delta phosphorylates tau and disrupts its binding to microtubules. J. Biol. Chem. 279:15938–15945 10.1074/jbc.M314116200 [DOI] [PubMed] [Google Scholar]
- Li H., Tsang C.K., Watkins M., Bertram P.G., and Zheng X.F.. 2006. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 442:1058–1061 10.1038/nature05020 [DOI] [PubMed] [Google Scholar]
- Li D., Herrera S., Bubula N., Nikitina E., Palmer A.A., Hanck D.A., Loweth J.A., and Vezina P.. 2011. Casein kinase 1 enables nucleus accumbens amphetamine-induced locomotion by regulating AMPA receptor phosphorylation. J. Neurochem. 118:237–247 10.1111/j.1471-4159.2011.07308.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loar J.W., Seiser R.M., Sundberg A.E., Sagerson H.J., Ilias N., Zobel-Thropp P., Craig E.A., and Lycan D.E.. 2004. Genetic and biochemical interactions among Yar1, Ltv1 and Rps3 define novel links between environmental stress and ribosome biogenesis in Saccharomyces cerevisiae. Genetics. 168:1877–1889 10.1534/genetics.104.032656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Lord C., Bhandari D., Menon S., Ghassemian M., Nycz D., Hay J., Ghosh P., and Ferro-Novick S.. 2011. Sequential interactions with Sec23 control the direction of vesicle traffic. Nature. 473:181–186 10.1038/nature09969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lövgren J.M., Bylund G.O., Srivastava M.K., Lundberg L.A., Persson O.P., Wingsle G., and Wikström P.M.. 2004. The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits. RNA. 10:1798–1812 10.1261/rna.7720204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke S.J., Baldwin P.R., and Chiu W.. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- Martin R., Straub A.U., Doebele C., and Bohnsack M.T.. 2013. DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol. 10:4–18 10.4161/rna.21879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlgarten C., and Schaffrath R.. 2003. Mutant casein kinase I (Hrr25p/Kti14p) abrogates the G1 cell cycle arrest induced by Kluyveromyces lactiszymocin in budding yeast. Mol. Genet. Genomics. 269:188–196. [DOI] [PubMed] [Google Scholar]
- Mehlgarten C., Jablonowski D., Breunig K.D., Stark M.J., and Schaffrath R.. 2009. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol. Microbiol. 73:869–881 10.1111/j.1365-2958.2009.06811.x [DOI] [PubMed] [Google Scholar]
- Meng Q.J., Maywood E.S., Bechtold D.A., Lu W.Q., Li J., Gibbs J.E., Dupré S.M., Chesham J.E., Rajamohan F., Knafels J., et al. 2010. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc. Natl. Acad. Sci. USA. 107:15240–15245 10.1073/pnas.1005101107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merwin J.R., Bogar L.B., Poggi S.B., Fitch R.M., Johnson A.W., and Lycan D.E.. 2014. Genetic analysis of the ribosome biogenesis factor Ltv1 of Saccharomyces cerevisiae. Genetics. 198:1071–1085 10.1534/genetics.114.168294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima S., and Nomura M.. 1970. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 226:1214–1218 10.1038/2261214a0 [DOI] [PubMed] [Google Scholar]
- Narla A., and Ebert B.L.. 2010. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 115:3196–3205 10.1182/blood-2009-10-178129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse V.G.2011. Getting ready to translate: cytoplasmic maturation of eukaryotic ribosomes. Chimia (Aarau). 65:765–769 10.2533/chimia.2011.765 [DOI] [PubMed] [Google Scholar]
- Perez D.I., Gil C., and Martinez A.. 2011. Protein kinases CK1 and CK2 as new targets for neurodegenerative diseases. Med. Res. Rev. 31:924–954 10.1002/med.20207 [DOI] [PubMed] [Google Scholar]
- Perreau-Lenz S., Vengeliene V., Noori H.R., Merlo-Pich E.V., Corsi M.A., Corti C., and Spanagel R.. 2012. Inhibition of the casein-kinase-1-epsilon/delta/ prevents relapse-like alcohol drinking. Neuropsychopharmacology. 37:2121–2131 10.1038/npp.2012.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Matos J., Mori S., Gregan J., Bogdanova A., Schwickart M., Mechtler K., Shirahige K., Zachariae W., and Nasmyth K.. 2006. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 126:1049–1064 10.1016/j.cell.2006.07.029 [DOI] [PubMed] [Google Scholar]
- Pfaffenwimmer T., Reiter W., Brach T., Nogellova V., Papinski D., Schuschnig M., Abert C., Ammerer G., Martens S., and Kraft C.. 2014. Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep. 15:862–870 10.15252/embr.201438932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.A.2006. CKI, there’s more than one: casein kinase I family members in Wnt and Hedgehog signaling. Genes Dev. 20:399–410 10.1101/gad.1394306 [DOI] [PubMed] [Google Scholar]
- Price M.A., and Kalderon D.. 2002. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell. 108:823–835 10.1016/S0092-8674(02)00664-5 [DOI] [PubMed] [Google Scholar]
- Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., et al. 2005. Global analysis of protein phosphorylation in yeast. Nature. 438:679–684 10.1038/nature04187 [DOI] [PubMed] [Google Scholar]
- Ray P., Basu U., Ray A., Majumdar R., Deng H., and Maitra U.. 2008. The Saccharomyces cerevisiae 60 S ribosome biogenesis factor Tif6p is regulated by Hrr25p-mediated phosphorylation. J. Biol. Chem. 283:9681–9691 10.1074/jbc.M710294200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder S.E., and Emr S.D.. 2001. Isolation of subcellular fractions from the yeast Saccharomyces cerevisiae. Curr. Protoc. Cell Biol. Chapter 3:Unit 3.8. [DOI] [PubMed] [Google Scholar]
- Rodriguez N., Yang J., Hasselblatt K., Liu S., Zhou Y., Rauh-Hain J.A., Ng S.K., Choi P.W., Fong W.P., Agar N.Y., et al. 2012. Casein kinase I epsilon interacts with mitochondrial proteins for the growth and survival of human ovarian cancer cells. EMBO Mol. Med. 4:952–963 10.1002/emmm.201101094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Galán O., García-Gómez J.J., and de la Cruz J.. 2013. Yeast and human RNA helicases involved in ribosome biogenesis: current status and perspectives. Biochim. Biophys. Acta. 1829:775–790 10.1016/j.bbagrm.2013.01.007 [DOI] [PubMed] [Google Scholar]
- Rosenberg L.H., Lafitte M., Grant W., Chen W., Bibian M., Noguchi Y., Fallahi M., Roush W.R., Cleveland J.L., and Duckett D.R.. 2015. A casein kinase 1δ-β-catenin circuit as an exploitable driver of human breast cancer. Sci. Transl. Med. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D., and Pandolfi P.P.. 2003. Does the ribosome translate cancer? Nat. Rev. Cancer. 3:179–192 10.1038/nrc1015 [DOI] [PubMed] [Google Scholar]
- Ruggero D., Grisendi S., Piazza F., Rego E., Mari F., Rao P.H., Cordon-Cardo C., and Pandolfi P.P.. 2003. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 299:259–262 10.1126/science.1079447 [DOI] [PubMed] [Google Scholar]
- Rumpf C., Cipak L., Dudas A., Benko Z., Pozgajova M., Riedel C.G., Ammerer G., Mechtler K., and Gregan J.. 2010. Casein kinase 1 is required for efficient removal of Rec8 during meiosis I. Cell Cycle. 9:2657–2662 10.4161/cc.9.13.12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T., Maco B., Petfalski E., Tollervey D., Böttcher B., Aebi U., and Hurt E.. 2006. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 441:651–655 10.1038/nature04840 [DOI] [PubMed] [Google Scholar]
- Scheres S.H.2012. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415:406–418 10.1016/j.jmb.2011.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T., and Hall M.N.. 2000. TOR, a central controller of cell growth. Cell. 103:253–262 10.1016/S0092-8674(00)00117-3 [DOI] [PubMed] [Google Scholar]
- Schwab C., DeMaggio A.J., Ghoshal N., Binder L.I., Kuret J., and McGeer P.L.. 2000. Casein kinase 1 delta is associated with pathological accumulation of tau in several neurodegenerative diseases. Neurobiol. Aging. 21:503–510 10.1016/S0197-4580(00)00110-X [DOI] [PubMed] [Google Scholar]
- Seiser R.M., Sundberg A.E., Wollam B.J., Zobel-Thropp P., Baldwin K., Spector M.D., and Lycan D.E.. 2006. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics. 174:679–691 10.1534/genetics.106.062117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrum D.C., Woodruff B.W., and Stagg S.M.. 2012. Creating an infrastructure for high-throughput high-resolution cryogenic electron microscopy. J. Struct. Biol. 180:254–258 10.1016/j.jsb.2012.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smadja Storz S., Tovin A., Mracek P., Alon S., Foulkes N.S., and Gothilf Y.. 2013. Casein kinase 1δ activity: a key element in the zebrafish circadian timing system. PLoS ONE. 8:e54189 10.1371/journal.pone.0054189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouse J., Reynolds L., Swanson T.A., and Engwall M.. 2009. Inhibition of casein kinase I epsilon/delta produces phase shifts in the circadian rhythms of cynomolgus monkeys. Psychopharmacology (Berl.). 204:735–742 10.1007/s00213-009-1503-x [DOI] [PubMed] [Google Scholar]
- Sprouse J., Reynolds L., Kleiman R., Tate B., Swanson T.A., and Pickard G.E.. 2010. Chronic treatment with a selective inhibitor of casein kinase I delta/epsilon yields cumulative phase delays in circadian rhythms. Psychopharmacology (Berl.). 210:569–576 10.1007/s00213-010-1860-5 [DOI] [PubMed] [Google Scholar]
- Stöter M., Bamberger A.M., Aslan B., Kurth M., Speidel D., Löning T., Frank H.G., Kaufmann P., Löhler J., Henne-Bruns D., et al. 2005. Inhibition of casein kinase I delta alters mitotic spindle formation and induces apoptosis in trophoblast cells. Oncogene. 24:7964–7975 10.1038/sj.onc.1208941 [DOI] [PubMed] [Google Scholar]
- Strunk B.S., and Karbstein K.. 2009. Powering through ribosome assembly. RNA. 15:2083–2104 10.1261/rna.1792109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk B.S., Loucks C.R., Su M., Vashisth H., Cheng S., Schilling J., Brooks C.L. III, Karbstein K., and Skiniotis G.. 2011. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 333:1449–1453 10.1126/science.1208245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk B.S., Novak M.N., Young C.L., and Karbstein K.. 2012. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 150:111–121 10.1016/j.cell.2012.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf C.R., and Ruggero D.. 2011. The cancerous translation apparatus. Curr. Opin. Genet. Dev. 21:474–483 10.1016/j.gde.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C.S., and Carragher B.. 2005. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151:41–60 10.1016/j.jsb.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Tanaka C., Tan L.J., Mochida K., Kirisako H., Koizumi M., Asai E., Sakoh-Nakatogawa M., Ohsumi Y., and Nakatogawa H.. 2014. Hrr25 triggers selective autophagy-related pathways by phosphorylating receptor proteins. J. Cell Biol. 207:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrant M.K., and Cole P.A.. 2009. The chemical biology of protein phosphorylation. Annu. Rev. Biochem. 78:797–825 10.1146/annurev.biochem.78.070907.103047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E., Ferreira-Cerca S., and Hurt E.. 2013. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 126:4815–4821 10.1242/jcs.111948 [DOI] [PubMed] [Google Scholar]
- Toyoshima M., Howie H.L., Imakura M., Walsh R.M., Annis J.E., Chang A.N., Frazier J., Chau B.N., Loboda A., Linsley P.S., et al. 2012. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc. Natl. Acad. Sci. USA. 109:9545–9550 10.1073/pnas.1121119109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai I.C., Woolf M., Neklason D.W., Branford W.W., Yost H.J., Burt R.W., and Virshup D.M.. 2007. Disease-associated casein kinase I δ mutation may promote adenomatous polyps formation via a Wnt/β-catenin independent mechanism. Int. J. Cancer. 120:1005–1012 10.1002/ijc.22368 [DOI] [PubMed] [Google Scholar]
- Walton K.M., Fisher K., Rubitski D., Marconi M., Meng Q.J., Sládek M., Adams J., Bass M., Chandrasekaran R., Butler T., et al. 2009. Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. J. Pharmacol. Exp. Ther. 330:430–439. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 285:901–906 10.1126/science.285.5429.901 [DOI] [PubMed] [Google Scholar]
- Wolff S., Stöter M., Giamas G., Piesche M., Henne-Bruns D., Banting G., and Knippschild U.. 2006. Casein kinase 1 delta (CK1δ) interacts with the SNARE associated protein snapin. FEBS Lett. 580:6477–6484 10.1016/j.febslet.2006.10.068 [DOI] [PubMed] [Google Scholar]
- Woolford J.L. Jr, and Baserga S.J.. 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 195:643–681 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasojima K., Kuret J., DeMaggio A.J., McGeer E., and McGeer P.L.. 2000. Casein kinase 1 delta mRNA is upregulated in Alzheimer disease brain. Brain Res. 865:116–120 10.1016/S0006-8993(00)02200-9 [DOI] [PubMed] [Google Scholar]
- Zemp I., and Kutay U.. 2007. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 581:2783–2793 10.1016/j.febslet.2007.05.013 [DOI] [PubMed] [Google Scholar]
- Zemp I., Wild T., O’Donohue M.F., Wandrey F., Widmann B., Gleizes P.E., and Kutay U.. 2009. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 185:1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I., Wandrey F., Rao S., Ashiono C., Wyler E., Montellese C., and Kutay U.. 2014. CK1δ and CK1ε are components of human 40S subunit precursors required for cytoplasmic 40S maturation. J. Cell Sci. 127:1242–1253 10.1242/jcs.138719 [DOI] [PubMed] [Google Scholar]
- Zoncu R., Efeyan A., and Sabatini D.M.. 2011. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12:21–35 10.1038/nrm3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.