Abstract
Deregulation of the apoptotic pathway, one of the hallmarks of tumor growth and -progression, has been shown to have prognostic value for tumor recurrence in rectal cancer. In order to develop clinically relevant bio-markers, we studied the methylation status of promoter regions of key apoptosis genes in rectal cancer patients, using methylation-sensitive restriction enzymes. DNA was extracted from fresh-frozen tumor tissues of 49 stage I-III rectal cancer patients and 10 normal rectal tissues. The results of this pilot study were validated in 88 stage III tumor tissues and 18 normal rectal tissues. We found that methylation of the intrinsic apoptotic pathway genes Apaf1, Bcl2 and p53 correlated with the apoptotic status (M30) of the tumor. Combined survival analyses of these three genes, based on the number of genes showing high methylation (all low, 1 high, 2 high or all high), showed shorter patient survival and recurrence-free periods with an increasing number of methylated markers. Multivariate analyses showed significant differences for overall survival (p = 0.01; HR = 0.28 (0.09–0.83)), cancer-specific survival (p = 0.004; HR = 0.13 (0.03–0.67)) and distant recurrence-free survival (p = 0.001; HR = 0.22(0.05–0.94)). The shortest survival was observed for patients showing low methylation of all markers, which—as was expected—correlated with high apoptosis (M30), but also with high proliferation (Ki-67). The study of epigenetic regulation of apoptosis genes provides more insight in the tumorigenic process in rectal cancer and might be helpful in further refining treatment regimens for individual patients.
Keywords: Rectal cancerl, Apoptosisl, DNA methylationl, Epigeneticsl, Promoter/enhancer analysisl, Clinical outcome
Introduction
Apoptosis is one of the major pathways frequently deregulated in cancer [1–3]. Deregulation of this pathway provides the tumor cell with a survival advantage and thereby promotes tumor growth and -progression. The apoptotic process is complicated and can be activated by stress or damage to the cell (intrinsic pathway), or by external factors (initiation by the immune system; extrinsic pathway), with both pathways converging at the level of the caspase cascade. This cascade eventually leads to cleavage of key proteins for cell structure and -function, causing fragmentation of the DNA, membrane blebbing and ultimately removal of the destructed cell by macrophages [4]. Previous studies have demonstrated that high levels of apoptosis in rectal cancer specimens correlated with low levels of local tumor recurrence [5–7]. A malfunctioning apoptotic pathway could also explain a poor response to anti-cancer treatment strategies such as pre-operative radiotherapy (RT).
Current treatment regimens of rectal cancer patients include radical removal of the primary tumor including all regional tumor cell deposits according to the total mesorectal excision (TME) technique. In addition to TME surgery, the majority of stage I and II patients and all stage III patients receive preoperative RT in order to reduce the local recurrence rate [8], based on data provided by several large randomized clinical trials [9–11]. Unfortunately, treating all rectal cancer patients with preoperative RT results in overtreatment of many individuals, as only a small number of patients—those who would develop a local recurrence—potentially benefit from this treatment and not all of these patients will respond to the therapy. However, most of the patients treated with preoperative RT will suffer from the side-effects such as increased risks of poor anal and sexual function, small bowel toxicity with obstruction and secondary malignancies [12–14].
The goal of this study was to investigate the regulation of the apoptotic pathway through DNA methylation, in order to better understand the biological processes underlying tumor growth and -progression in rectal cancer. Epigenetic mechanisms, responsible for regulation of gene transcription, have been shown to be deregulated in many cancers [15–18], thereby altering the expression levels of many genes in tumor cells. We aim to develop biomarkers that will assist in treatment decisions in rectal cancer patients. For this study we chose to focus on the apoptosis genes Apaf1, Bcl2, p53, Fas (CD95), and TrailR2, as a review of the current literature indicated these apoptotic proteins to have prognostic value in cancer [19–27]. We hypothesized that methylation of the promoter region of these genes would represent deregulation of the apoptotic pathway and, therefore, would correlate with patient survival and tumor recurrence in rectal cancer. Methylation assays were performed on DNA extracted from frozen tumor tissues of patients enrolled in the Dutch TME trial [28, 29] using a methylation-sensitive restriction enzyme-based approach [30]. We also assessed tumor cell proliferation status in this study, as a delicate balance may exist between apoptosis and proliferation in determining clinical outcome.
Materials and methods
Patient selection
Patients were selected from the study population of the non-irradiated (TME surgery only) arm of the Dutch TME trial [28, 29]. We selected patients with no evidence of disease after surgical resection of the tumor and of whom frozen tissue blocks were available (n = 137) [28, 29, 31]. Samples were collected between 1996 and 1999 and stored at −80 °C. The median follow-up time in this study cohort was 6 years. Trial eligibility criteria and follow-up protocols have been described previously [31–33]. Informed consent for the use of tumor specimens was obtained from all patients enrolled in the TME trial and the study has been approved by the Medical Ethical Committee of the Leiden University Medical Center. For the pilot study, frozen tumor tissues of 49 patients with stage I, II, or III rectal cancer were collected. In addition, normal colorectal tissues, taken at least 5 cm away from the tumor, were collected from 10 patients included in this study. The validation study consisted of a set of 88 frozen tumor tissues of patients with stage III rectal cancer of whom sufficient frozen tissue was available, and 18 normal rectal tissue samples. Clinicopathological parameters of all patients included in this study have been summarized in Table 1. This study was performed according to the REMARK criteria [34].
Table 1.
Patient characteristics of the pilot and validation study cohort
| Stage I-III patients non-irradiated arm TME trial (n = 685) |
Pilot study |
p value | Stage III patients non-irradiated arm TME trial (n = 281) |
Validation study |
p value | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| (n = 49) | (n = 88) | |||||||||
| N | (%) | n | (%) | N | (%) | n | (%) | |||
| Gender | ||||||||||
| Male | 432 | 63.1 | 35 | 71.4 | 181 | 64.4 | 62 | 70.5 | ||
| Female | 253 | 36.9 | 14 | 28.6 | 0.24 | 100 | 35.6 | 26 | 29.5 | 0.30 |
| Age at randomization | ||||||||||
| Mean | 64.3 | 66.0 | 63.37 | 62.59 | ||||||
| Standard error | 0.43 | 1.58 | 0.31 | 0.67 | 1.30 | 0.58 | ||||
| TNM stage | ||||||||||
| I | 209 | 30.5 | 14 | 28.6 | ||||||
| II | 195 | 28.5 | 25 | 51.0 | ||||||
| III | 281 | 41.0 | 10 | 20.4 | 0.13 | 281 | 100 | 88 | 100 | n.a. |
| Tumor location | ||||||||||
| Rectum | 607 | 88.6 | 41 | 83.3 | 246 | 89.5 | 84 | 95.4 | ||
| Anal region | 48 | 12.4 | 8 | 16.7 | 0.45 | 21 | 10.5 | 4 | 4.5 | 0.06 |
| Circumferential margin | ||||||||||
| Negative | 555 | 81.0 | 36 | 73.5 | 187 | 66.5 | 67 | 76.1 | ||
| Positive | 129 | 19.0 | 13 | 26.5 | 0.34 | 93 | 33.5 | 21 | 23.9 | 0.10 |
Patient characteristics are shown for both the pilot and validation study groups. The pilot study included stage I–III rectal cancer patients (n = 49), the validation study stage III patients only (n = 88). Variables listed are standard clinicopathological factors used in multivariate analyses, i.e. gender, age at randomization, TNM stage, tumor location, and circumferential margin involvement. p-values representing the difference between the TME trial patients and the pilot or validation study cohort, respectively, were calculated using a Student’s t test
DNA extraction and enzyme digestion
DNA was extracted from frozen tissues using a Trizol-based protocol according to manufacturer’s recommendations (Life Technologies Corp, Bleiswijk, the Netherlands). DNA was dissolved in Tris–EDTA buffer pH 8.0 and quantity was measured using a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE). To analyze the methylation status of specific promoter regions, we used methylation-sensitive restriction isoschizomers MspI (New England Biolabs, Ipswich, MA; R0106L) and HpaII (New England Biolabs, Ipswich, MA; R0171L). The isoschizomeric restriction enzymes share restriction site C^CGG but whereas MspI cuts the DNA irrespective of DNA methylation, HpaII is blocked by methyl groups at CpG dinucleotides. Reactions were optimized according to the manufacturer’s procotol (New England Biolabs, protocol “Optimizing restriction endonuclease reactions”). 250 ng of DNA was used for each digestion reaction. Per reaction, 25 units of HpaII and 100 units of MspI were used in combination with their respective buffers (5 µl) in a total reaction volume of 50 µl. Mock digestions were included for every sample, substituting the restriction enzymes with 5 µl 50 % glycerol. Overnight incubation for 16 h at 37 °C of all reaction mixtures was followed by heat inactivation for 20 min at 65 °C, and subsequent cooling down of the samples to 4 °C. Incubation times and conditions were optimized using the active (non-methylated) housekeeping gene β2-microglobulin (B2m) and the silenced (methylated) gene Myogenic differentiation 1 (MYOD1) that is only activated in muscle tissue. Methylated (universally methylated DNA, UMC; Millipore, Billerica, MA) and unmethylated (DNA from peripheral blood leukocytes, PBL, 5 different patients) controls were included in every digestion assay and on every PCR plate. We verified for every digestion procedure that >75 % of the DNA participated in the digestion reactions, measured as Ct >2 between the MspI- and mock-treated samples, and that the PCR product after digestion with HpaII was less than the product in the mock-treated samples.
Real-time PCR
PCR was performed in duplicate for apoptosis markers Apaf1, Bcl2, TrailR2, p53 and Fas (CD95) using genomic DNA primers surrounding (but not including) at least two enzyme restriction sites per amplicon. PCR was performed using 20 ng DNA, 2 pmol/µl primers and PerfeCTa® SYBR® Green SuperMix for iQ™ (Quanta Biosciences, Gaithersburg, MD, USA) in a final volume of 10 µl. Melting curves were used to ensure a single PCR product for each of the markers, and PCR products were run on a 1 % gel to ensure for correct size of the products. Quantitative PCR reactions were run on a 96 well CFX thermal cycler (BioRad, Benicia, CA). Methylated (UMC) and unmethylated controls (DNA from PBL) were included on every PCR plate. Thermal cycling reactions were as follows: hotstart for 3 min at 95 °C, followed by 40 cycles of denaturing for 10 s at 95 °C and annealing/extension for 30 s at the optimal melting temperatures for each primer set, as indicated below. Primer sequences and melting temperatures per primer set were as follows: Apaf1 Forward 5′- TTGACTGCTCCGCTGTC −3′; Apaf1 Reverse 5′- TCCCCACCTCTGGTTCT −3′ (Tm 63 °C); Fas Forward 5′- CCAACTTCCCAGGTTGAA −3′; Fas Reverse 5′- GCACAAATGGGCATTCC −3′ (Tm 63 °C); p53 Forward 5′- GTATCTACGGCACCAGGTC −3′; p53 Reverse 5′- CATGACAAGTAAGGGCAACT −3′ (Tm 62 °C); Bcl2 Forward 5’- GGTCCCGTGGATAGAGAT −3′; Bcl2 Reverse 5′- GCAGATGAATTACAATTTTCAG −3′ (Tm 56 °C); TrailR2 Forward 5′- CCTGGGAAGGGGAGAAGAT −3′; TrailR2 Reverse 5′- AGTTGAGGGAGGCACTTGG −3′ (Tm 60 °C). Primer sequences for methylation controls B2m and MYOD1 were as follows: B2m Forward 5′- GCCTTCTTA AACATCACGAG −3′; B2m Reverse 5′- CCAGCCAATCA GGACAA −3′ (Tm 58 °C); MYOD1 Forward 5′- TACAGC CGCTCTACCCAT −3′; MYOD1 Reverse 5′- CTCCAACA CCCGACTGC −3′ (Tm 60 °C). Methylation percentages were calculated as follows: methylation percentage = 2 − (Ct HpaII-treated samples) × 100 %. The amount of product detected after digestion with MspI was used to calculate the percentage of the DNA that was digested by the restriction enzymes, using the following formula: as 2 − (Ct HpaII-treated samples – Ct mock-treated samples) × 100 %.
Immunohistochemical staining and scoring
Whole tumor tissue sections (4 µm) of 117 patients of whom enough paraffin-embedded tumor tissue was available were IHC stained using a primary M30 antibody (Roche Diagnostics, Germany), staining for caspase-cleaved cytokeratin 18 [7]. Whole tissue sections (4 µm) of 40 patients in the validation study, representative of the complete series of stage III patients, were IHC stained at predetermined optimal concentrations using anti-Apaf1 (ab53152; Abcam, Cambridge, UK), anti-Bcl2 (ab7973; Abcam, Cambridge, UK), or anti-p53 (M7001; Dako, Glostrup, Denmark). A tissue microarray (TMA) was constructed including 495 patients from the non-irradiated arm of the Dutch TME trial. Three 1 mm tumor tissue cores were punched from each tumor block and transfered to a recipient block using a TMA Master (3D Histech, Budapest, Hungary). TMA sections, including 119 patients of our study cohort, were IHC stained at a predetermined optimal concentration with primary Ki-67 antibodies (Dako, Glostrup, Denmark; clone MIB-1). For all IHC stainings, tissue sections were incubated with the respective primary antibodies overnight (16 h). IHC staining was visualized using the Dako REAL™ EnVision™ Detection System, Peroxidase/DAB+, Rabbit/Mouse (Dako, Glostrup, Denmark). The level of apoptosis was scored as the number of M30-positive cells per mm2, as described previously [5]. Patients were classified into high or low apoptosis groups, based on the median number of M30-positive cells. The level of proliferation (Ki-67) was scored as the percentage of Ki-67-positive tumor cells. The average percentage positive cells per patient (three tumor cores) was used to classify patients into either high or low proliferation groups, based on the median percentage of Ki-67-positive tumor cells. For Apaf1, Bcl2 and p53, the percentage of positive tumor cells was scored in three different randomly chosen fields within the tumor tissue (similar to three tissues cores on a tissue microarray). For each tumor field, the percentages of negative, weak, moderate and strong staining in tumor cells was scored. For each of these categories, an average percentage was calculated over the three tumor fields. For each marker and for each patient separately, a histoscore—as a measure of marker expression—was calculated as follows: histoscore = (0 × mean percentage negative tumor cells) + (1 × mean percentage weak positive tumor cells) + (2 × mean percentage moderate positive tumor cells) + (3 × mean percentage strong positive tumor cells).
Statistical analysis
Only samples with a difference of ≥2 Ct between MspI-and mock-treated samples and <1 Ct difference between the supplicate PCR reactions were considered for statistical analyses. The distribution of data of each individual methylation marker was tested for normality using the Shapiro–Wilk test [35]. Methylation levels of each marker were defined as high or low methylation based on the median methylation percentage. Univariate and multivariate Cox proportional hazard models were used to statistically test the differences between the groups. χ2 tests were performed to compare the level of apoptosis (M30 staining) and the methylation percentage of individual markers. Interpolation plots were made to visualize the correlation between methylation and protein expression (IHC staining) data for Apaf1, Bcl2 and p53. We then studied combinations of two markers, both of the intrinsic and the extrinsic apoptosis pathways (Apaf1 and Bcl2, Apaf1 and p53, Bcl2 and p53, Fas and TrailR2). The patient groups were divided into three groups: 2 high (both markers showing high methylation), 1 high and 0 high (both markers showing low methylation). To obtain more power for statistical analyses, markers Apaf1, Bcl-2 and p53 were combined into a new variable. Data of all three markers were available for 78 patients in the validation cohort. The combined-marker set was divided into four groups: all low (low methylation in all three markers), 1 high (1 out of 3 markers high methylation), 2 high (2 out of 3 markers high methylation) and all high (high methylation in all three markers). Univariate and multivariate Cox proportional hazard analyses were performed to assess the correlation of the combined marker patient groups with overall survival (OS), cancer-specific survival (CSS), local recurrence-free survival (LRFS) and distant recurrence-free survival (DRFS). Multivariate analyses included covariates age, gender, circumferential margin and tumor location. Kaplan–Meier curves were used to visualize differences between the combined marker patient groups. Cumulative incidence curves were calculated for CSS, LRFS and DRFS, accounting for death due to other causes [36]. For all survival analyses we used a pre-established patient group, the patient group with the expected shortest survival and recurrence-free periods (the “all high” patient group), as the reference group. We assessed both apoptosis (M30) and proliferation (Ki-67) in the tumor specimens of the validation cohort. Only patients with both M30 and Ki-67 data available (n = 76) were included in these analyses. Combining both apoptosis and proliferation based on high versus low level of IHC staining resulted in four patient groups: low apoptosis and low proliferation (n = 16), low apoptosis and high proliferation (n = 13), high apoptosis and low proliferation (n = 16) and high apoptosis and high proliferation (n = 31). Kaplan–Meier curves were used to visualize differences between the patients groups.
Results
Enzyme digestion assay
Using methylation-sensitive restriction enzymes, we investigated the methylation status of key apoptosis genes Apaf1, Bcl2, p53, Fas and TRAILR2 in DNA extracted from frozen rectal cancer tissues. For quality control purposes, we verified performance of the enzyme assays using control genes B2m and MYOD1. Based on optimization of the assays using active housekeeping gene B2m and silenced gene MYOD1, incubation of enzyme reactions was set to 16 h (overnight) at 37 °C, followed by 20 min heat inactivation at 65 °C and subsequently cooling the samples at 4 °C for one hour. The mean methylation percentages were 86 % (78–100 %) for MYOD1 and 38 % (18–68 %) for B2m. Standard deviations for the controls included in every digestion and on every PCR plate ranged from 7.5 to 8.2 % for UMC DNA, and from 0.14 to 1.71 % for PBL DNA. The methylation percentages of these controls ranged from 65 to 100 % for UMC DNA, and from 1 to 6.5 % for PBL DNA. For every digestion procedure, we verified that >75 % of the DNA was actually digested in the digestion reactions, measured as Ct ≥2 between the MspI- and mock-treated samples, and the PCR product after digestion with HpaI had a lower Ct value than the product in the mock-treated samples. Correct PCR product sizes were confirmed by running the products on a 1 % gel. Based on these results, we continued with statistical analyses of the patient data.
Pilot study results for individual markers
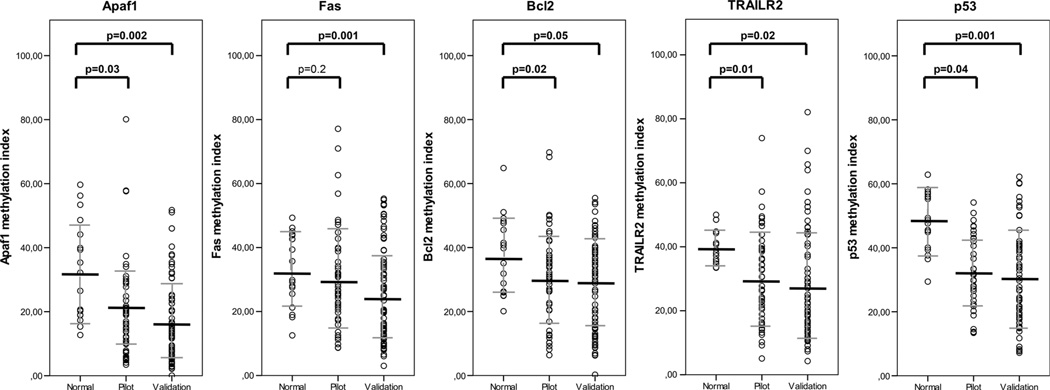
The patient cohort of the pilot study was representative of the complete non-irradiated patient cohort of the Dutch TME trial with respect to the main clinicopathological parameters (Table 1). Methylation percentages in the tumor samples were significantly different from the normal samples analyzed for Apaf1, Bcl2, TrailR2 and p53 (Fig. 1). No significant differences were observed for Fas. χ2 tests showed significant correlation of a high methylation of two of the markers with a low level of apoptosis based on M30 staining, with Apaf1 (p = 0.03) and p53 (p = 0.04). No significant correlation with apoptosis status was found for the markers Fas, Bcl2 or TrailR2 using χ2 analyses. However, correlation analyses did show a negative correlation for these markers between protein expression and methylation index, indicating a decreasing amount of methylation with a higher number of apoptotic cells, with values between −0.2 and −0.3. No significant difference in methylation of the individual markers, apoptosis (M30) or proliferation (Ki-67) was observed between the tumor stages (data not shown). Therefore, we continued with stage III patients for the validation study.
Fig. 1.
Methylation values and means of individual markers in normal, pilot and validation study tissues. Shown are methylation percentages for normal and tumor tissues (in both pilot and validation studies) for each of the apoptosis markers separately. Mean methylation percentages are indicated with horizontal bars. Methylation percentages were calculated as follows: methylation percent-age=2–(Ct HpaII treated samples–Ct mock treated samples) *100%. p-values comparing the pilot and validation tumor samples with their normal counterparts were calculated using paired samples t tests
Validation study results for individual markers
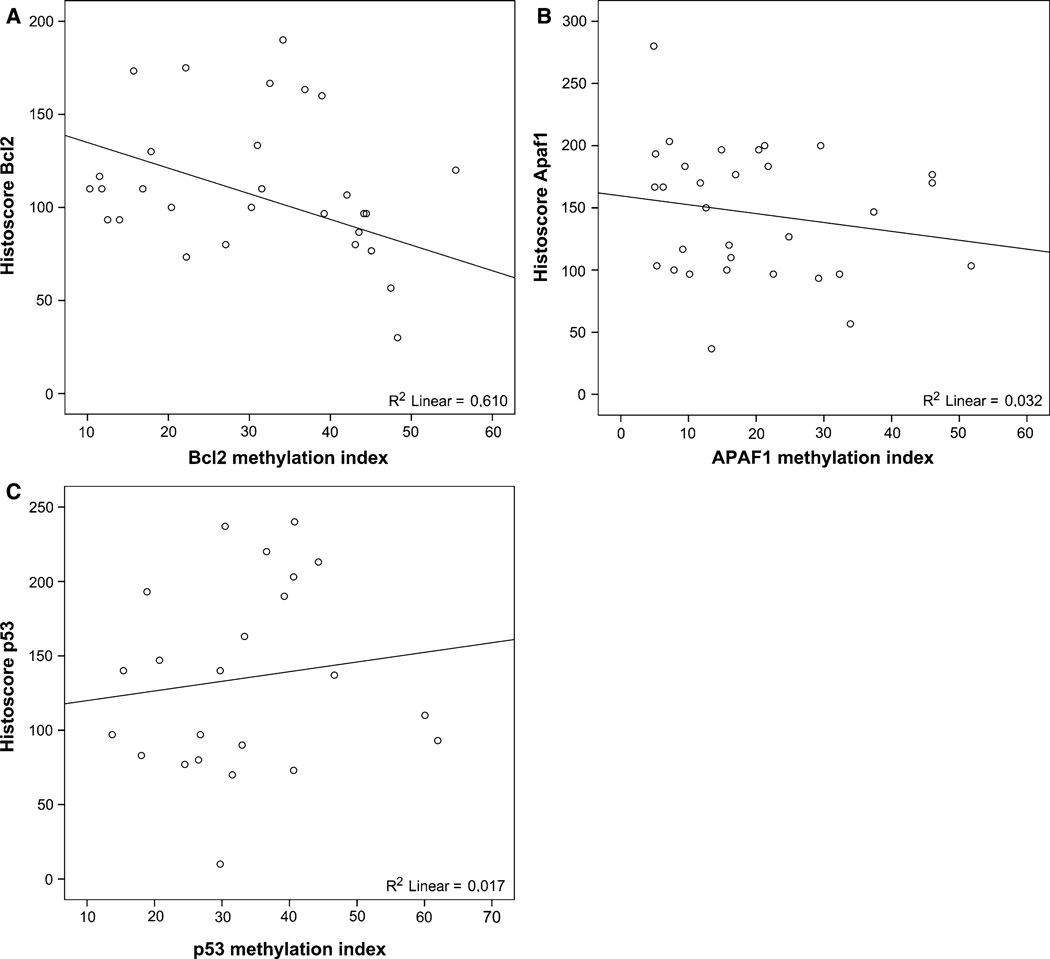
In the validation study, we included only stage III rectal cancer patients, as large differences in patient survival and tumor recurrence are observed within this specific patient group [37, 38], and these patients will likely benefit the most from finding new biomarkers that could complement the current TNM staging system. Patients included in the validation study were representative of the non-irradiated patient cohort of the Dutch TME trial with respect to the main clinicopathological parameters (Table 1). Mean methylation percentages in the tumor tissues in the validation study were similar to those found in the pilot study for all markers. The mean methylation percentages in the tumor samples were significantly different from the methylation percentages in the normal samples included in the validation study (Fig. 1). DNA methylation percentages of the individual markers were not normally distributed. Therefore, the median methylation percentage for each individual marker was used as a cut-off value to divide patients into low and high methylation groups. To verify that a lower methylation status indeed corresponded with higher apoptosis levels in the tumor, we compared the methylation percentage of each of the different markers to the known apoptotic status (based on M30 IHC data) in each of the tumors. A representative example of IHC staining results of M30 staining is shown in Supplementary Fig. 1. χ2 tests showed significant correlation of a high methylation of Apaf1 (p = 0.05) and Fas (p = 0.01) with a low level of apoptosis (Supplementary Fig. 2A). Methylation of the other markers (Bcl2, p53 and TrailR2) did show a similar correlation with M30 apoptosis levels, although not statistically significant. Linear regression using the number of M30-positive (apoptotic) cells per mm2 in each of the tumors and the methylation percentages of each of the individual markers as continuous variables showed a significant correlation between the methylation status and apoptosis status for Apaf1 (p = 0.03), Bcl2 (p = 0.01) and p53 (p = 0.04) (Supplementary Fig. 2B). No significant correlation was found for markers Fas and TrailR2. Subsequently, we analyzed if methylation of the three markers Apaf1, Bcl2 and p53, correlated to protein expression (representative IHC staining results are shown in Supplementary Fig. 1). As can be observed in the interpolation plots in Fig. 2, Bcl2 and Apaf1 methylation correlated well with protein expression, with R2 values of 0.610 (Bcl2, Fig. 2a) and 0.320 (Apaf1, Fig. 2b). For p53, methylation did not directly correlate with protein expression (Fig. 2c), which might be explained by differences in p53 mutation status. Unfortunately, p53 mutation status was not known for these patients. Survival analyses did not yield any significant difference between samples with high or low methylation for any of the individual markers, based on the median methylation percentage. Using Kaplan–Meier curves, we did observe that for Apaf1, Bcl2 and p53, high methylation in the tumor tissues was associated with shorter patient survival and higher probability of tumor recurrence, although not statistically significant. We hypothesized that combining the intrinsic apoptotic pathway markers might result in better classification of rectal cancer patients in our study cohort.
Fig. 2.
Methylation compared to protein expression for Apaf1, Bcl2 and p53. Interpolation plots showing methylation versus protein expression for Apaf1, Bcl2 and p53. Protein expression was scored as percentage of tumor cells showing negative, weak, moderate or strong IHC staining. The histoscore was calculated as follows: histoscore = (0 * percentage negative) + (1 * percentage weak) + (2 * percentage moderate) + (3 * percentage strong). R2 values indicate the degree of correlation between methylation and protein expression for each of the markers
Combined marker analyses
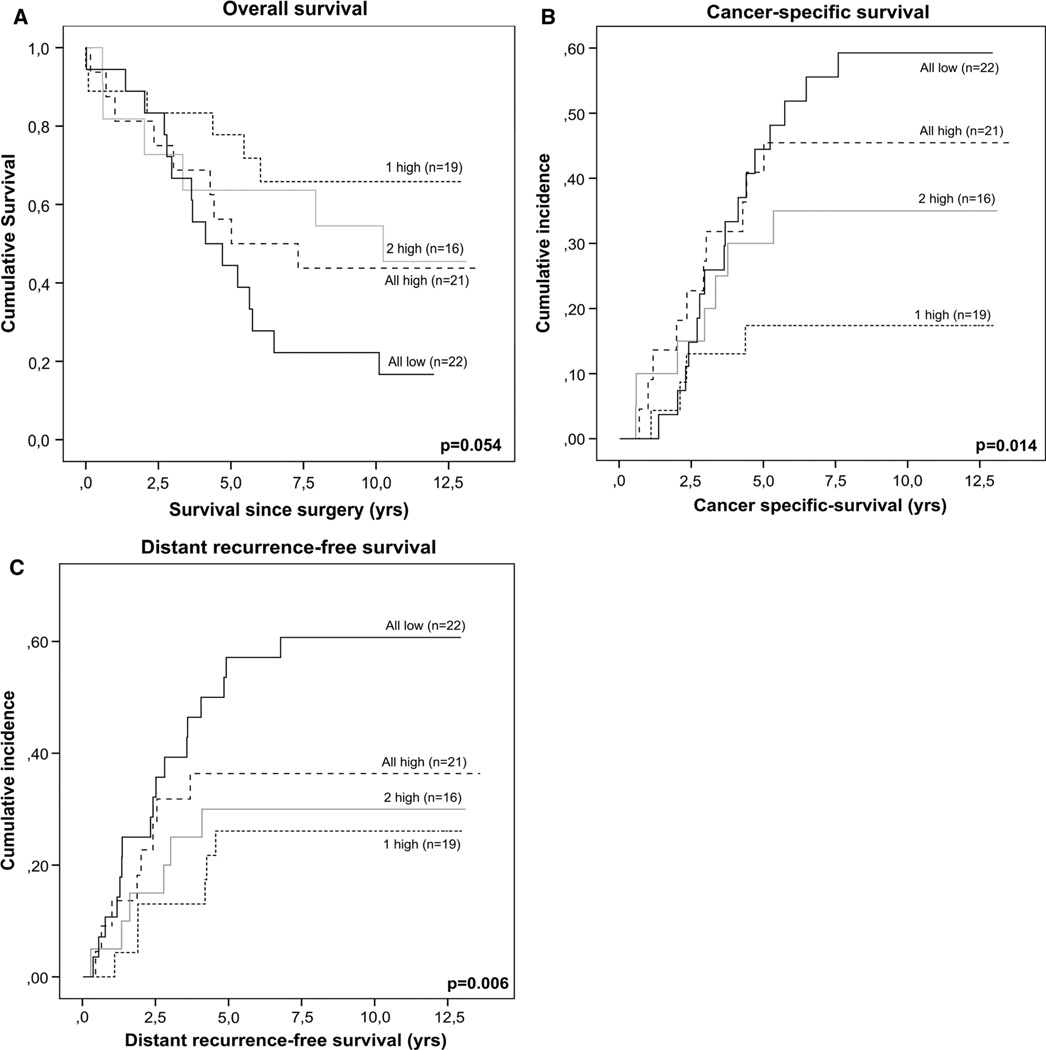
First, we studied combinations of two markers within the intrinsic or the extrinsic apoptosis pathway. A trend towards statistically significant differences between the patient groups was observed for the combination of Apaf1 and Bcl2 for LRFS, with p = 0.09 (HR 0.59, 95 % CI 0.33–1.09) in univariate and p = 0.1 (HR 0.58; 95 % CI 0.28–1.17) in multivariate analyses. Also for the combination of Bcl2 and p53 a trend was observed in multivariate analyses for DRFS, with p = 0.07 (HR 1.51; 95 % CI 0.96–2.38). For the other combinations, no significant differences were observed. To obtain more power for statistical analyses, all three intrinsic pathway markers (Apaf1, Bcl-2 and p53) were combined into a new variable. Methylation of markers Apaf1, Bcl2 and p53 was combined into one variable, based on the number of markers showing high methylation (all low, 1 high, 2 high, or all high). Methylation of the three combined markers showed a correlation to apoptosis status as measured by M30 (p = 0.07). Cox proportional hazard models were used to assess the differences in patient survival and tumor recurrence between the groups in both univariate and multivariate analyses (Table 2). In univariate analyses, significant differences were observed for OS (p = 0.05), CSS (p = 0.01) and DRFS (p = 0.006). Multivariate analyses included the covariates gender, age at the time of surgery, circumferential margin and distance to the anal verge. Significant differences were observed for OS (p = 0.01), CSS (p = 0.004), and DRFS (p = 0.001). No significant differences were observed for LRFS, in either univariate or multivariate analyses. The differences in patient survival and tumor recurrence between the combined marker groups were visualized using Kaplan–Meier survival curves (for OS) and cumulative incidence curves (for CSS and DRFS) (Fig. 3). The curves indicated that the more markers show high methylation, the shorter the survival and recurrence-free periods. The patient group with high methylation for only one out of the three markers (“1 high” group) showed the best survival, directly followed by the patient group with two out of three markers showing high methylation (“2 high” group). The patient group with high methylation of all three markers (“all high” group) showed even shorter survival and recurrence-free periods, but the shortest survival and disease free periods were observed for the patient group with low methylation on all three markers (“all low” group). In addition to low methylation of Apaf1, Bcl2 and p53 in the “all low” combined marker patient group, low methylation of Fas and TRAILR2 was observed for 96 and 81 % of the patients in this group, respectively. No difference in patient characteristics was observed between the combined marker groups that could explain these results, suggesting that other tumor-intrinsic factors might be responsible for the shorter patient survival and recurrence-free periods.
Table 2.
Univariate and multivariate analyses of combined markers Apaf1, Bcl2 and p53 in the validation series
| Variable | Overall survival | Cancer specific survival | Distant recurrence- free survival |
Local recurrence- free survival |
||||
|---|---|---|---|---|---|---|---|---|
| HR (95 % CI) | p value | HR (95 % CI) | p value | HR (95 % CI) | p value | HR (95 % CI) | p value | |
| Univariate analyses | ||||||||
| Combined marker group | 0.44 (0.24–0.89) |
0.05 | 0.19 (0.04–0.68) |
0.01 | 0.28 (0.16–0.87) |
0.006 | 0.14 (0.04–1.04) |
0.21 |
| Multivariate analysis | ||||||||
| Age | 1.94 | 0.5 | 1.46 | 0.85 | 1.89 | 0.43 | 1.69 | 0.68 |
| Gender | 0.68 | 0.32 | 0.72 | 0.47 | 0.84 | 0.69 | 1.29 | 0.75 |
| Circumferential margin | 2.1 | 0.1 | 3.04 | 0.03 | 4.62 | 0.005 | 6.29 | 0.03 |
| Tumor location | 2.1 | 0.11 | 2.93 | 0.07 | 2.78 | 0.12 | 5.37 | 0.12 |
| Combined marker group | 0.28 (0.09–0.83) |
0.01 | 0.13 (0.03–0.67) |
0.004 | 0.22 (0.05–0.84) |
0.001 | 0.07 (0.01–1.06) |
0.17 |
Bold values indicate statistically significant (p < 0.05)
Univariate and multivariate analyses are shown for combined markers Apaf1, Bcl2 and p53. The “all high” group (with high methylation of all three markers) was used as the reference group. Combined marker groups were defined as follows: all high (high methylation in all three markers), 2 high (2 out of 3 markers high methylation), 1 high (1 out of 3 markers high methylation) and all low (low methylation in all three markers). Hazard ratios are shown for both the univarite and the multivariate analyses, with a 95 % confidence interval for the combined marker group
Fig. 3.
Survival analyses of combined markers Apaf1, Bcl2 and p53. Intrinsic apoptotic pathway markers Apaf1, Bcl2 and p53 were combined based on the number of markers showing high methylation. Only patient with data for all three markers available were included in the survival analyses (n = 78). The resulting combined marker groups were: all low (low methylation in all three markers), 1 high (1 out of 3 markers high methylation), 2 high (2 out of 3 markers high methylation) and all high (high methylation in all three markers). Kaplan–Meier curves were made to visualize differences in patient survival and tumor recurrence between the different methylation groups for OS (a). Survival times were calculated as the time from surgery till an event (death or recurrence, resp.). Cumulative incidence curves were calculated for CSS (b) and DRFS (c)
Proliferation and apoptosis analyses
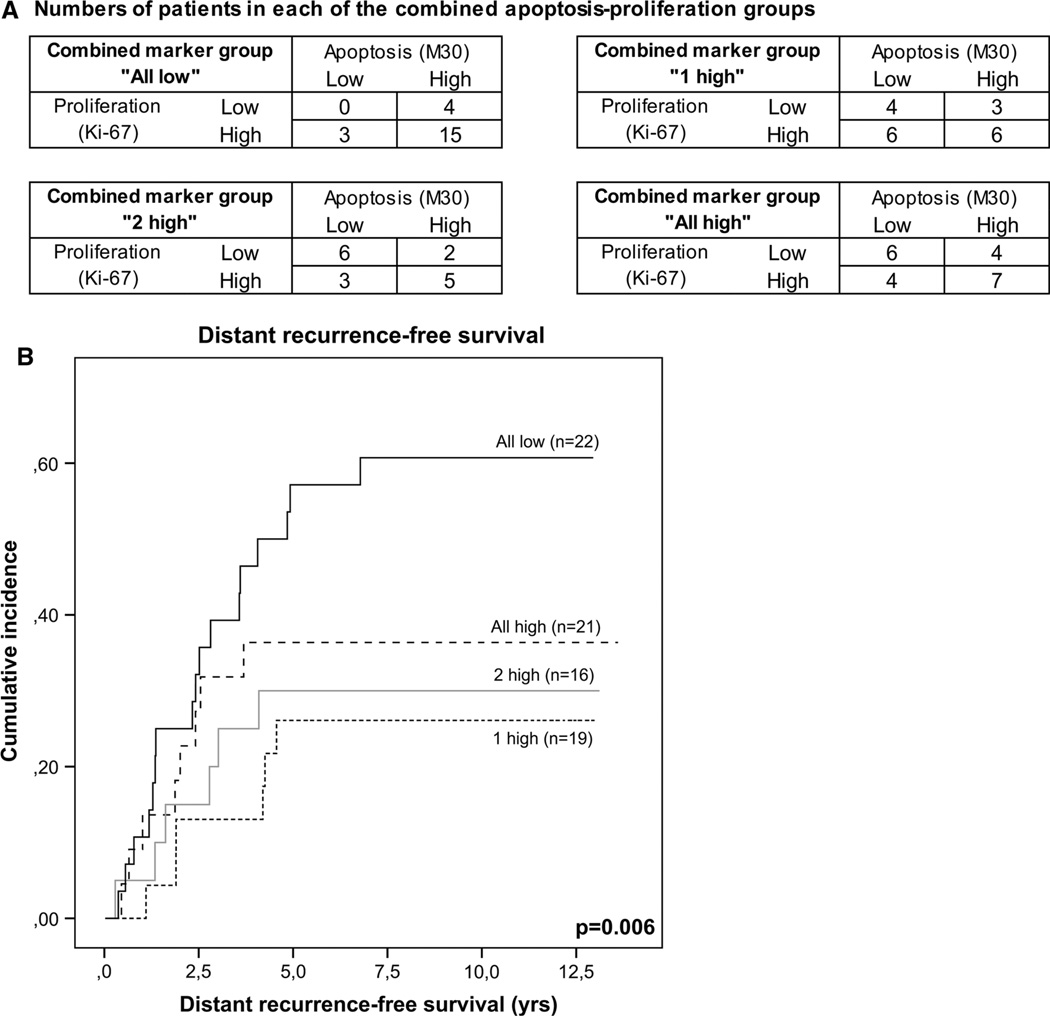
In order to explain the survival and recurrence data observed for the “all low” combined marker patient group, we also assessed tumor proliferation (as measured by Ki-67 IHC staining, see Supplementary Fig. 1) for patients in the validation study. Only patients with both M30 and Ki-67 data available (n = 76) were included in the analyses. Missing data were due to unavailability of tumor specimens or missing punches on the TMA sections. In the “all low” group showing low methylation of Apaf1, Bcl2 and p53, 15 out of 22 patients (68 %) showed both high apoptosis and high proliferation (Fig. 4a). For the other combined marker groups (1 high, 2 high and all high), the same numbers of patients were observed in each of the combined apoptosis-proliferation groups. As shown in Fig. 4b, this also translated into differences in survival for each of the combined apoptosis-proliferation patient groups, although not statistically significant (p = 0.19). The patient group with both high proliferation and high apoptosis indeed showed the shortest survival, which corresponds to the poor survival observed for patients with low methylation of all three apoptosis markers.
Fig. 4.
Apoptosis and proliferation for combined markers Apaf1, Bcl2 and p53. Apoptosis (M30, IHC) and proliferation (Ki-67, IHC) were combined and compared to the methylation percentages of the markers Apaf1, Bcl2 and p53. Patients with data for all markers available (n = 78) were classified into high or low apoptosis or proliferation groups based on the median number or percentage of positive tumor cells, respectively. For all combined marker groups, the number of patients in each of the combined apoptosis-proliferation categories was determined (a). Kaplan–Meier curves were made to visualize survival differences for the combined apoptosis-proliferation categories (b)
Discussion
Changes in the regulation of the apoptotic process, one of the hallmarks of cancer [1, 2], provides tumor cells with a survival advantage and could hence promote tumorigenesis. Epigenetic aberrations have been shown to contribute to the process of tumorigenesis in many ways [16]. Since the outcome of the apoptotic pathway has been proven to correlate with patient outcome parameters in rectal cancer [6], we hypothesized that studying the epigenetic regulation of the apoptotic process might provide more insight in this crucial but complicated cellular process. Furthermore, it may bring us one step closer to the discovery of new clinically relevant prognostic biomarkers in rectal cancer. In this study, methylation of key apoptosis genes using a restriction enzyme-based protocol was correlated to patient survival and tumor recurrence. We showed that combining multiple markers (intrinsic pathway markers Apaf1, Bcl2 and p53) resulted in better patient stratification and therefore better prognostication as compared to the individual markers or combinations of only two markers. With little modifications, including varying amounts of enzyme and DNA added to the digestion reactions, the enzyme-based protocol presented in this paper can be used to analyze small amounts of DNA without any loss of DNA due to prior processing steps, which is a major advantage compared to the current bisulfite modification-based methods used to detect DNA methylation. In a clinical setting this approach will be useful, as the amount of DNA available for analyses, usually derived from tumor biopsies, is limited and should be used with great care.
Current literature indicates that methylation of apoptosis proteins Apaf1, Bcl2 and p53 have prognostic value in various cancers [26, 39–42]. In colorectal cancer, reduced Apaf1 expression was found to be associated with tumor progression and adverse prognosis [25, 43]. High co-expression of Bcl2 and p53 proteins was found to be associated with poor prognosis in colorectal cancer [44], gastric MALT lymphoma [45] and B cell lymphoma [45, 46]. Piris et al. suggested aberrant expression of both p53 and Bcl2 to be part of a multistep process of dysregulation of the apoptotic machinery critical for progression of tumours [46]. Abnormal expression of p53 itself has been related to a poor patient prognosis in colorectal cancer [47]. Methylation of the investigated apoptosis markers Apaf1, Bcl2 and p53 has been reported in various cancers, including lung cancer (Bcl2; [39] ), renal cell carcinoma(Apaf1; [40] ), melanoma (Apaf1; [41] ) and ovarian cancer (p53; [42] ).
To our best knowledge, the study presented here is the first study combining methylation data of several apoptosis genes at the same time, based on their functions in the intrinsic apoptotic process [48]. The methylation percentages of the combined markers Apaf1, Bcl2 and p53 correlated significantly with the apoptotic status of the tumors (M30 IHC staining) and disease outcome (patient survival and disease recurrence). We therefore conclude that gene promoter methylation status can be a useful surrogate marker for the apoptotic status of individual tumors, but also provides additional information about specific apoptosis-related genes as compared to a ‘general’ apoptotic status as determined by M30 staining.
Previous research of our group has shown that the risk of a local recurrence is lower when tumor intrinsic levels of apoptosis are high [6]. In this study we showed that low methylation is correlated with higher levels of apoptosis, indeed correlating with better survival and longer recurrence-free periods. This finding is supported in literature, where high expression of Apaf1 and Bcl2 was reported to be significantly correlated with better OS [49]. Although mean methylation levels were significantly lower in tumor tissues as compared to normal tissues for all of the markers suggesting higher apoptotic activity, in individual tumors we found that higher levels of methylation of the combined markers Apaf1, Bcl2 and p53 correlated to shorter patient survival and recurrence-free periods as compared to the patients showing low expression of one or two of the markers. The patient group showing low methylation of all three markers, however, did not comply with our expectations, since this patient group showed the shortest survival and recurrence-free periods. To explain this phenomenon, we studied cell proliferation in addition to apoptosis. We observed that patients showing both high apoptosis and high proliferation, of which 68 % of the patients corresponded to the “all low” combined marker group, showed the shortest survival. This finding is supported by literature, in which both increased apoptosis and increased proliferation were reported in rectal tumors with lymph node metastases as compared to non-metastatic tumors [50]. The distorted balance between the apoptosis pathways and cellular proliferation in the “all low” combined marker group hence might explain the observed survival data. Dysfunctioning of the apoptotic pathway could not be demonstrated in this patient group based on the results presented for the studied apoptosis genes, as methylation was reported to be low for all five markers. Of course, there might be other apoptotic factors involved that could compromise proper functioning of the apoptotic pathway in this patient group, which have not been investigated in this study. Epigenetic mechanisms other than DNA methylation at gene promoters could also be involved in the regulation of apoptosis gene expression in these tumors, as was suggested by the work of Hinoue et al. [51] Using a genome-wide approach, cancer-specific methylation of multiple gene regions has been described in CpG island methylator phenotype (CIMP)-high colorectal cancers, with characteristic genetic and clinical features. In addition, 48 of 112 genes were also found to be transcriptionally downregulated in non-CIMP tumors, but this could not be correlated to higher DNA methylation at these specific regions, suggesting involvement of other (epigenetic) mechanisms. In contrast to these genome-wide studies, a more pathway-focused approach was used in our study that could facilitate the discovery of new biomarkers in rectal cancer prognosis.
In conclusion, in this study we found that methylation of apoptotic genes is correlated with the overall apoptotic status of a tumor, and that this status can be used to assess clinical outcome in terms of patient survival and tumor recurrence. The methylation analysis presented using methylation-sensitive restriction enzyme digestion provides a biological explanation for the differences in apoptotic status in individual tumors. High methylation of combined intrinsic apoptosis pathway markers Apaf1, Bcl2 and p53, suggesting deregulation of the apoptotic pathway, was associated with poor prognosis in our study cohort of colorectal cancer patients. High proliferation and high apoptosis were observed when methylation of the intrinsic apoptosis pathway markers was low. The study of epigenetic regulation of apoptosis genes will provide more insight in the tumorigenic process in rectal cancer and might be helpful in further refining treatment regimens for individual patients.
Supplementary Material
Acknowledgments
This study was funded by the Sacha Swarttouw-Hijmans Fund (The Netherlands), KWF (Dutch Cancer Society, The Netherlands), Collegium Chirurgicum Neerlandicum (The Netherlands) and the Leiden University Fund (LUF, The Netherlands). This work was also supported by Grant Number 1R01CA171767-01A1 from the National Institute of Health, National Cancer Institute (USA, DSBH) and the Ruth and Martin H Weil Fund (Los Angeles, CA; DSBH).
Abbreviations
- TME
Total mesorectal excision
- TNM
Tumor node metastasis
- IHC
Immunohistochemistry
- OS
Overall survival
- CSS
Cancer-specific survival
- LRFS
Local recurrence-free survival
- DRFS
Distant recurrence-free survival
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10495-014-1022-z) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that that they have no competing interests to disclose.
Contributor Information
Anne Benard, Department of Surgery, K6-R, Leiden University Medical Center, P.O. Box 9600, 2300 RC Leiden, The Netherlands; Department of Molecular Oncology, John Wayne Cancer, Institute, Santa Monica, CA, USA.
Eliane C. M. Zeestraten, Department of Surgery, K6-R, Leiden University Medical Center, P.O. Box 9600, 2300 RC Leiden, The Netherlands
Inès J. Goossens-Beumer, Department of Surgery, K6-R, Leiden University Medical Center, P.O. Box 9600, 2300 RC Leiden, The Netherlands
Hein Putter, Department of Medical Statistics and Bioinformatics, Leiden, University Medical Center, Leiden, The Netherlands.
Cornelis J. H. van de Velde, Department of Surgery, K6-R, Leiden University Medical Center, P.O. Box 9600, 2300 RC Leiden, The Netherlands
Dave S. B. Hoon, Department of Molecular Oncology, John Wayne Cancer, Institute, Santa Monica, CA, USA
Peter J. K. Kuppen, Email: p.j.k.kuppen@lumc.nl, Department of Surgery, K6-R, Leiden University Medical Center, P.O. Box 9600, 2300 RC Leiden, The Netherlands.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Zhang L. Apoptosis in human cancer cells. Curr Opin Oncol. 2004;16:19–24. doi: 10.1097/00001622-200401000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bruin EC, van de Velde CJ, van de Pas S, Nagtegaal ID, van Krieken JH, Gosens MJ, Peltenburg LT, Medema JP, Marijnen CA. Prognostic value of apoptosis in rectal cancer patients of the dutch total mesorectal excision trial: radiotherapy is redundant in intrinsically high-apoptotic tumors. Clin Cancer Res. 2006;12:6432–6436. doi: 10.1158/1078-0432.CCR-06-0231. [DOI] [PubMed] [Google Scholar]
- 6.de Heer P, de Bruin EC, Klein-Kranenbarg E, Aalbers RI, Marijnen CA, Putter H, de Bont HJ, Nagelkerke JF, van Krieken JH, Verspaget HW, van de Velde CJ, Kuppen PJ. Caspase-3 activity predicts local recurrence in rectal cancer. Clin Cancer Res. 2007;13:5810–5815. doi: 10.1158/1078-0432.CCR-07-0343. [DOI] [PubMed] [Google Scholar]
- 7.Marijnen CA, Nagtegaal ID, Mulder-Stapel AA, Schrier PI, van de Velde CJ, van Krieken JH, Peltenburg LT. High intrinsic apoptosis, but not radiation-induced apoptosis, predicts better survival in rectal carcinoma patients. Int J Radiat Oncol Biol Phys. 2003;57:434–443. doi: 10.1016/s0360-3016(03)00580-7. [DOI] [PubMed] [Google Scholar]
- 8.Valentini V, Aristei C, Glimelius B, Minsky BD, Beets-Tan R, Borras JM, Haustermans K, Maingon P, Overgaard J, Pahlman L, Quirke P, Schmoll HJ, Sebag-Montefiore D, Taylor I, Van CE, Van d V, Cellini N, Latini P. Multidisciplinary rectal cancer management: 2nd European rectal cancer consensus conference (EURECA-CC2) Radiother Oncol. 2009;92:148–163. doi: 10.1016/j.radonc.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 9.Dahlberg M, Glimelius B, Pahlman L. Improved survival and reduction in local failure rates after preoperative radiotherapy: evidence for the generalizability of the results of Swedish rectal cancer trial. Ann Surg. 1999;229:493–497. doi: 10.1097/00000658-199904000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapiteijn E, Marijnen CA, Nagtegaal ID, Putter H, Steup WH, Wiggers T, Rutten HJ, Pahlman L, Glimelius B, van Krieken JH, Leer JW, van de Velde CJ. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer. N.Engl J Med. 2001;345:638–646. doi: 10.1056/NEJMoa010580. [DOI] [PubMed] [Google Scholar]
- 11.Sebag-Montefiore D, Stephens RJ, Steele R, Monson J, Grieve R, Khanna S, Quirke P, Couture J, de MC, Myint AS, Bessell E, Griffiths G, Thompson LC, Parmar M. Preoperative radiotherapy versus selective postoperative chemoradiotherapy in patients with rectal cancer (MRC CR07 and NCIC-CTG C016): a multicentre, randomised trial. Lancet. 2009;373:811–820. doi: 10.1016/S0140-6736(09)60484-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birgisson H, Pahlman L, Gunnarsson U, Glimelius B. Late adverse effects of radiation therapy for rectal cancer—a systematic overview. Acta Oncol. 2007;46:504–516. doi: 10.1080/02841860701348670. [DOI] [PubMed] [Google Scholar]
- 13.Lange MM, van de Velde CJ. Faecal and urinary incontinence after multimodality treatment of rectal cancer. PLoS Med. 2008;5:e202. doi: 10.1371/journal.pmed.0050202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange MM, Marijnen CA, Maas CP, Putter H, Rutten HJ, Stig-gelbout AM, Meershoek-Klein KE, van de Velde CJ. Risk factors for sexual dysfunction after rectal cancer treatment. Eur J Cancer. 2009;45:1578–1588. doi: 10.1016/j.ejca.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 15.Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8:1409–1420. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 16.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 17.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16(1):R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 18.Jones PA. DNA methylation and cancer. Oncogene. 2002;21:5358–5360. doi: 10.1038/sj.onc.1205597. [DOI] [PubMed] [Google Scholar]
- 19.Hector S, Conlon S, Schmid J, Dicker P, Cummins RJ, Con-cannon CG, Johnston PG, Kay EW, Prehn JH. Apopto-some-dependent caspase activation proteins as prognostic markers in Stage II and III colorectal cancer. Br J Cancer. 2012;106(9):1499–1505. doi: 10.1038/bjc.2012.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pancione M, Forte N, Fucci A, Sabatino L, Febbraro A, Di BA, Daniele B, Parente D, Colantuoni V. Prognostic role of beta-catenin and p53 expression in the metastatic progression of sporadic colorectal cancer. Hum Pathol. 2010;41:867–876. doi: 10.1016/j.humpath.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 21.Perraud A, Akil H, Nouaille M, Petit D, Labrousse F, Jauberteau MO, Mathonnet M. Expression of p53 and DR5 in normal and malignant tissues of colorectal cancer: correlation with advanced stages. Oncol Rep. 2011;26:1091–1097. doi: 10.3892/or.2011.1404. [DOI] [PubMed] [Google Scholar]
- 22.Poincloux L, Durando X, Seitz JF, Thivat E, Bardou VJ, Giovannini MH, Parriaux D, Barriere N, Giovannini M, Delpero JR, Monges G. Loss of Bcl-2 expression in colon cancer: a prognostic factor for recurrence in stage II colon cancer. Surg Oncol. 2009;18:357–365. doi: 10.1016/j.suronc.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Strater J, Hinz U, Hasel C, Bhanot U, Mechtersheimer G, Lehnert T, Moller P. Impaired CD95 expression predisposes for recurrence in curatively resected colon carcinoma: clinical evidence for immunoselection and CD95L mediated control of minimal residual disease. Gut. 2005;54:661–665. doi: 10.1136/gut.2004.052696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uddin S, Hussain AR, Ahmed M, Al-Sanea N, Abduljabbar A, Ashari LH, Alhomoud S, Al-Dayel F, Bavi P, Al-Kuraya KS. Coexpression of activated c-Met and death receptor 5 predicts better survival in colorectal carcinoma. Am J Pathol. 2011;179:3032–3044. doi: 10.1016/j.ajpath.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujimoto A, Takeuchi H, Taback B, Hsueh EC, Elashoff D, Morton DL, Hoon DS. Allelic imbalance of 12q22–23 associated with APAF-1 locus correlates with poor disease outcome in cutaneous melanoma. Cancer Res. 2004;64:2245–2250. doi: 10.1158/0008-5472.can-03-2932. [DOI] [PubMed] [Google Scholar]
- 26.Haq R, Yokoyama S, Hawryluk EB, Jonsson GB, Frederick DT, McHenry K, Porter D, Tran TN, Love KT, Langer R, Anderson DG, Garraway LA, Duncan LM, Morton DL, Hoon DS, Wargo JA, Song JS, Fisher DE. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc Natl Acad Sci USA. 2013;110:4321–4326. doi: 10.1073/pnas.1205575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umetani N, Fujimoto A, Takeuchi H, Shinozaki M, Bilchik AJ, Hoon DS. Allelic imbalance of APAF-1 locus at 12q23 is related to progression of colorectal carcinoma. Oncogene. 2004;23:8292–8300. doi: 10.1038/sj.onc.1208022. [DOI] [PubMed] [Google Scholar]
- 28.Kapiteijn E, Kranenbarg EK, Steup WH, Taat CW, Rutten HJ, Wiggers T, van Krieken JH, Hermans J, Leer JW, van de Velde CJ. Total mesorectal excision (TME) with or without preoperative radiotherapy in the treatment of primary rectal cancer. Prospective randomised trial with standard operative and histopathological techniques. Dutch ColoRectal Cancer Group. Eur J Surg. 1999;165:410–420. doi: 10.1080/110241599750006613. [DOI] [PubMed] [Google Scholar]
- 29.van Gijn W, Marijnen CA, Nagtegaal ID, Kranenbarg EM, Putter H, Wiggers T, Rutten HJ, Pahlman L, Glimelius B, van de Velde CJ. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer: 12-year follow-up of the multicentre, randomised controlled TME trial. Lancet Oncol. 2011;12:575–582. doi: 10.1016/S1470-2045(11)70097-3. [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto K, Kokubun S, Itoi E, Roach HI. Improved quantification of DNA methylation using methylation-sensitive restriction enzymes and real-time PCR. Epigenetics. 2007;2:86–91. doi: 10.4161/epi.2.2.4203. [DOI] [PubMed] [Google Scholar]
- 31.Peeters KC, Marijnen CA, Nagtegaal ID, Kranenbarg EK, Putter H, Wiggers T, Rutten H, Pahlman L, Glimelius B, Leer JW, van de Velde CJ. The TME trial after a median follow-up of 6 years: increased local control but no survival benefit in irradiated patients with resectable rectal carcinoma. Ann Surg. 2007;246:693–701. doi: 10.1097/01.sla.0000257358.56863.ce. [DOI] [PubMed] [Google Scholar]
- 32.Kapiteijn E, van de Velde CJ. Developments and quality assurance in rectal cancer surgery. Eur J Cancer. 2002;38:919–936. doi: 10.1016/s0959-8049(02)00049-7. [DOI] [PubMed] [Google Scholar]
- 33.Peeters KC, Kapiteijn E, van de Velde CJ. Managing rectal cancer: the Dutch experience. Colorectal Dis. 2003;5:423–426. doi: 10.1046/j.1463-1318.2003.00513.x. [DOI] [PubMed] [Google Scholar]
- 34.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 35.Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples) Biometrika. 1965;52:591–611. [Google Scholar]
- 36.Putter H, Fiocco M, Geskus RB. Tutorial in biostatistics: competing risks and multi-state models. Stat Med. 2007;26:2389–2430. doi: 10.1002/sim.2712. [DOI] [PubMed] [Google Scholar]
- 37.Hoeben KW, van Steenbergen LN, van de Wouw AJ, Rutten HJ, van Spronsen DJ, Janssen-Heijnen ML. Treatment and complications in elderly stage III colon cancer patients in the Netherlands. Ann Oncol. 2013;24:974–979. doi: 10.1093/annonc/mds576. [DOI] [PubMed] [Google Scholar]
- 38.van den Broek CB, Bastiaannet E, Dekker JW, Portielje JE, de Craen AJ, Elferink MA, van de Velde CJ, Liefers GJ, Kapiteijn E. Time trends in chemotherapy (administration and costs) and relative survival in stage III colon cancer patients—a large population-based study from 1990 to 2008. Acta Oncol. 2013;52:941–949. doi: 10.3109/0284186X.2012.739730. [DOI] [PubMed] [Google Scholar]
- 39.Nagatake M, Osada H, Kondo M, Uchida K, Nishio M, Shimokata K, Takahashi T, Takahashi T. Aberrant hypermethylation at the bcl-2 locus at 18q21 in human lung cancers. Cancer Res. 1996;56:1886–1891. [PubMed] [Google Scholar]
- 40.Ahmad ST, Arjumand W, Seth A, Saini AK, Sultana S. Methylation of the APAF-1 and DAPK-1 promoter region correlates with progression of renal cell carcinoma in North Indian population. Tumour Biol. 2012;33:395–402. doi: 10.1007/s13277-011-0235-9. [DOI] [PubMed] [Google Scholar]
- 41.Soengas MS, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, McCombie R, Herman JG, Gerald WL, Lazebnik YA, Cordon-Cardo C, Lowe SW. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- 42.Chmelarova M, Krepinska E, Spacek J, Laco J, Beranek M, Palicka V. Methylation in the p53 promoter in epithelial ovarian cancer. Clin Transl Oncol. 2013;15:160–163. doi: 10.1007/s12094-012-0894-z. [DOI] [PubMed] [Google Scholar]
- 43.Zlobec I, Minoo P, Baker K, Haegert D, Khetani K, Tornillo L, Terracciano L, Jass JR, Lugli A. Loss of APAF-1 expression is associated with tumour progression and adverse prognosis in colorectal cancer. Eur J Cancer. 2007;43:1101–1107. doi: 10.1016/j.ejca.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 44.Papagiorgis PC, Zizi AE, Tseleni S, Oikonomakis IN, Sofras L, Patsouris E, Nikiteas NI. Disparate clinicopathological correlations of p53 and Bcl-2 in colorectal cancer. Mol Med Rep. 2012;5:377–382. doi: 10.3892/mmr.2011.687. [DOI] [PubMed] [Google Scholar]
- 45.He M, Gao L, Zhang S, Tao L, Wang J, Yang J, Zhu M. Prognostic significance of miR-34a and its target proteins of FOXP1, p53, and BCL2 in gastric MALT lymphoma and DLBCL. Gastric Cancer. 2013 doi: 10.1007/s10120-013-0313-3. [DOI] [PubMed] [Google Scholar]
- 46.Piris MA, Pezzella F, Martinez-Montero JC, Orradre JL, Villuendas R, Sanchez-Beato M, Cuena R, Cruz MA, Martinez B, Pezella F. p53 and bcl-2 expression in high-grade B-cell lymphomas: correlation with survival time. Br J Cancer. 1994;69:337–341. doi: 10.1038/bjc.1994.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munro AJ, Lain S, Lane DP. P53 abnormalities and outcomes in colorectal cancer: a systematic review. Br J Cancer. 2005;92:434–444. doi: 10.1038/sj.bjc.6602358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeestraten EC, Benard A, Reimers MS, Schouten PC, Liefers GJ, van de Velde CJ, Kuppen PJ. The prognostic value of the apoptosis pathway in colorectal cancer: a review of the literature on biomarkers identified by immunohistochemistry. Biomark Cancer. 2013;5:13–29. doi: 10.4137/BIC.S11475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krajewska M, Kim H, Kim C, Kang H, Welsh K, Matsuzawa S, Tsukamoto M, Thomas RG, Assa-Munt N, Piao Z, Suzuki K, Perucho M, Krajewski S, Reed JC. Analysis of apoptosis protein expression in early-stage colorectal cancer suggests opportunities for new prognostic biomarkers. Clin Cancer Res. 2005;11:5451–5461. doi: 10.1158/1078-0432.CCR-05-0094. [DOI] [PubMed] [Google Scholar]
- 50.Kim YH, Lee JH, Chun H, Nam SJ, Lee WY, Song SY, Kwon OJ, Hyun JG, Sung IK, Son HJ, Rhee PL, Kim JJ, Paik SW, Rhee JC, Choi KW. Apoptosis and its correlation with proliferative activity in rectal cancer. J Surg Oncol. 2002;79:236–242. doi: 10.1002/jso.10063. [DOI] [PubMed] [Google Scholar]
- 51.Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk CM, Toll-enaar RA, Laird PW. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.