Abstract
Owing to the increasing epidemiological and therapeutic challenges associated with infections due to ESBL producers, ESBL prevalence rate among some bacteria isolates from healthy and non-healthy human population in a metropolitan Nigerian setting was evaluated.
A total of one hundred and forty-five (145) bacteria strains were isolated from a total of four hundred and sixty (460) samples collected from urine, wound, throat and anal swabs of 220 healthy volunteers in the community and from 240 patients in 2 secondary and 2 tertiary hospitals (altogether, 4) in Enugu metropolis. The presumptive confirmatory test used for ESBL detection was the Double Disc Synergy Test (DDST) method. Conjugation and plasmid curing studies were also done for resistance factor determination.
Of the 145 isolates, 20 were ESBL producers with 35% of these ESBL producers being of community origin and 65% from hospitals. This translates to 4.8% and 9% incidences (comparably higher than established prevalence of 4.4% and 7.5 respectively) for community and hospital infections respectively. The ESBL isolates showed high resistance to tetracycline, gentamicin, pefloxacin, ceftriaxone, cefuroxime, ciprofloxacin and Augmentin® (Amoxicilin and clavulanic acid combination). Conjugation studies for Resistance plasmid transfer showed non-transference of resistance determinants between the ESBL transconjugants and recipient strains. Correspondingly, the plasmid curing studies revealed that the acridine orange could not effect a cure on the isolates as they still retained high resistance to the antibiotics after the treatment.
This study confirms the growing incidences/pool of ESBL strains in Nigeria and call for widespread and continuous monitoring towards an effective management of the potential therapeutic hurdle posed by this trend.
KEY WORDS: Enterobacteriaceae, Pseudomonas aeruginosa, extended spectrum beta lactamase (ESBL), antibiotics, resistance plasmid, conjugation
INTRODUCTION
The introduction of third- and fourth-generation cephalosporins as therapeutic agents has been followed by the dissemination of different extended-spectrum b-lactamases (ESBLs) that hydrolyse those b-lactam drugs and also monobactam antibiotics. ESBL are bacterial enzymes that hydrolyse and confer resistance to modern cephalosporin antibiotics. They constitute the major mechanism of resistance to second, third and fourth generation cephalosporins (for example: ce- furoxime, cefotaxime, ceftriaxone and ceftazidime) [1-3]. Additionally unfortunate is the discovery that ESBL-producing organisms often also possess resistance determinants to other important antibiotic groups, such as aminoglycosides and fluoroquinolones, leaving an extremely limited range of effective agents [4]. ESBLs have been found in a great number of different bacterial species, but more frequently in Escherichia coli and Klebsiella pneumoniae [3, 5]. There have also been reports of the growing concern of the Enterobacteriaceae and Pseudomonas spp producing extended-spectrum b-lactamases (ESBLs) among nosocomial and also community-acquired infections [6-11]. Clinicians, microbiologists, infection control practitioners, and hospital epidemiologists are concerned about ESBL-producing bacteria because of the increasing incidence of such infections, the limitations of effective antimicrobial drug therapy, and adverse patient outcomes [8, 12-15]. In Nigeria, there have been reports of the reoccurring cases of antimicrobial resistance by most pathogenic organisms against many antibiotics [16]. Moreover, fractional isolated studies establishing the presence of ESBL producing bacteria clinical isolates from specific localities within the Western and Eastern part of the country have also been reported [16-18]. These initial preliminary reports have further heightened the necessity of extending similar studies to cover more unsampled localities, to further harmonize the epidemiological data base, and the need for a continuous epidemiological monitoring of the prevalence rate of these ESBL bacteria isolates. Data generated from such foregoing exercises would not only define the existent bacteria resistance indices but also clearly serve as a useful baseline in determining the rational and effective chemotherapeutic options for the management of infectious disease due to the ESBL bacteria organisms. It is therefore based on these established premises that this present study was carried out to determine the prevalence rate of these pathogenic ESBL-producing enterobacteriaceae organisms in the sampled community.
MATERIALS AND METHODS
Microorganisms
Samples were collected over a five months period between October 2006 and February 2007. Informed consent and ethical approval was obtained. A total of four hundred and sixty (460) samples (urine (338), vaginal (24), anal (26), wound (59), and throat swabs (13)) were collected. Two hundred and twenty (220) samples from 70, 55, and 95 healthy human volunteers in the community aged 2-10, 11-19, and 20-60 years respectively, while two hundred and forty samples (240) from 185 and 55 patients aged 28-40, and 41-60 respectively from four (4) hospitals comprising University of Nigeria Teaching Hospital (UNTH); Enugu, National Orthopaedic Hospital, Enugu (NOHE), Ntasiobi Ndinona Afufu (NONA), and Reego Laboratories, Enugu. Sixty eight (68) and one hundred and seventy-two (172) samples from hospital patients were collected below and over 48 hours post-admission respectively. Characterization of isolates was according to recommended standard technique by the National Committee for Clinical Laboratory Standard (NCCLS).
Culture Media and Reagents
They include Nutrient broth (Oxoid, England), mannitol salt agar (Oxoid, England) nutrient agar (Fluka Spain) and peptone water. Sucrose, mannitol, Gram Staining reagents, buffer solution, Tris-ethylenediamine tetra- acetic acid sodium sulfate (TENS), sodium acetate, peptone, Ethidium bromide and Bromo – phenol blue were all analar grade reagents.
Antibiotic Discs
Antibiotic discs used were obtained from Oxoid (England) and they include ceftriaxone (30μg), Clindamycin (30μg), Ciprofloxacin (30μg), Cefuroxime (30μg), Augmentin® (Amoxicillin and clavulanic acid) (30μg), Gentamicin (30μg), Pefloxacin (30μg), Imipenem (30μg), Cefotaxime (30μg), Ceftazidime (30μg), Tetracycline (30μg).
Antibiotic Sensitivity test
Antibiotic sensitivity of the isolates was determined using previously established procedure [19]. Briefly, the isolates were cultured in nutrient broth at 37°C for 24 h. Two (2) loopfuls of the suspension of each isolate were inoculated into 20ml of sterile molten agar in 10 cm diameter Petri dishes and mixed. The plates were allowed to set and the antibiotic Sensitivity discs were aseptically placed on their surfaces. The plates were incubated at 37°C for 24 h and the resultant inhibition zone diameters (IZDs) were measured and recorded.
Test for ESBL Production
The isolates were tested for ESBL production using the double disc synergy test (DDST) method. A combination disc (Amoxicillin 20 μg and Clavulanic acid 10 μg) was placed at the centre of the Petri dish and antibiotics (Ciprofloxacin 30 μg and Cefuroxime 30 μg) were placed 15 mm apart on both sides of the plates. It was incubated at 37°C for 24 hours after which the various inhibition zone diameters were measured.
Resistance Transfer
Resistance transfer to ascertain if the resistance determinants were plasmid mediated was determined by conjugation test. ESBL positive isolates of Klebsiella pneumoniae, Klebsiella oxytoca, Escherichia coli and Pseudomonas aeruginosa were mated with an Escherichia coli recipient strain that is rifampicin resistant and susceptible to Antibiotic Resistant Markers (ARM) that will include gentamicin, ampicillin, tetracycline, chloramphenicol and trimethoprim/sulphamethoxazole. Recipient and donor strains were grown in Mueller Hinton broth at 37° C for 18-24 hr. The donor and recipient strains were mixed in a ratio of (1:10) and incubated for 3 hours at 37° C. Samples of 200 ml and 300 ml of donor to recipient mixture were spread-plated on the surface of MacConkey agar plates supplemented with 25 mg/ml of rifampicin and 100 mg/ml of ampicillin (oxoid U.K) and incubated at 37 ° C for 18 – 24 hr. Samples from the donor and recipient were used as control.
Plasmid Curing
Further confirmatory plasmid curing studies were carried out to further confirm the location of the resistance determinants. Briefly, the organisms were grown at 37 ° C for 18 – 24 hr in Mueller Hinton broth. The organisms were diluted to 1×105 cfu/ml in Mueller Hinton broth and 0.1 mg/ml of acridine orange was added to the 24 hr broth and incubated at 37 ° C for 24 hr. The cells were tested for plasmid curing by subjecting them to further antibiogram studies.
RESULTS
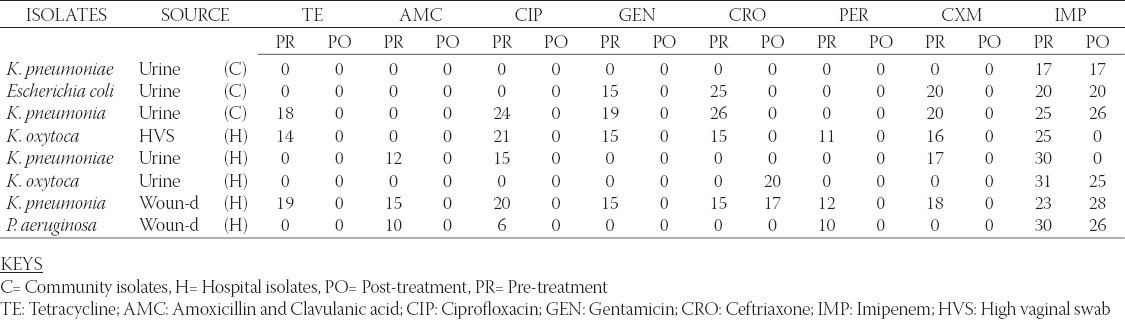
Table 1 shows the antibiogram of the hospital and community isolates used in the study. From the table, high resistance by the isolates to most of the antibiotics is reflected. Over 90% of the community isolates were resistant to the antibiotics- Tetracycline, Augmentin® (Amoxicillin and Clavulanic acid combination), ceftriaxone, ciprofloxacin, pefloxacin, gentamicin, and cefuroxime while only about 30% of the hospital isolates were resistant to the same antibiotics. All the isolates however showed high susceptibility to imipenem. Also, Table 1 records that the identified MDR-bacteria strains to be distributed largely between the Enterobacteriaceae and Pseudomonas groups. Their incidence rates among the prospective ESBL producing compartment are 85 % and 15 % respectively. Additionally, 13 out of 20 (65 %) bacteria strains sampled were MDR-hospital-isolates with 7 out of 20 (35 %) being MDR-community-isolates. Table 2 shows the Resistant, Intermediate and Susceptible percentage rates of the isolates to the different antibiotics with the community derived isolates having higher percentage resistance to the antibiotics. Result of the phenotypic confirmatory double disc synergy test for ESBL production is shown in Table 3. The test showed positive for the twenty (20) suspicious isolates of hospital and community origin. Of the 20 bacteria strains identified as ESBL producers, 14 (70 %) were Klebsiella spp, 3 (15 %) were E. coli while the remaining 3 (15 %) were Pseudomonas spp. Conjugation studies showed no visible growth inhibition on any of the plates suggesting that none of the transconjugated organism could transfer its plasmid DNA to the recipient strain. The subsequent plasmid curing studies (Table 4) reveal that the isolates still retained high resistance to the antibiotics after the treatment with acridine orange, the implication being that the resistance determinant might not be extrachromosomal.
TABLE 1.
Antibiogram of hospital and community isolates from ENUGU Inhibition Zone Diameter of antibiotics (mm)
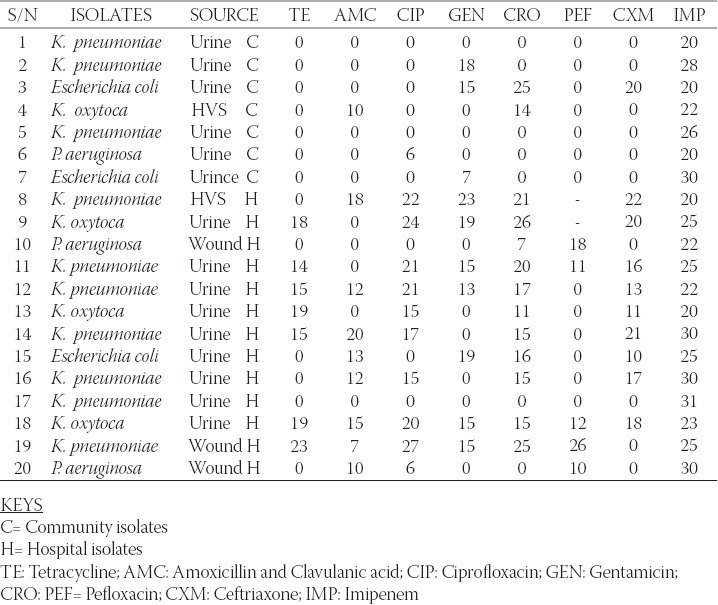
TABLE 2.
Percentage resistance and sensitivity pattern of the isolates
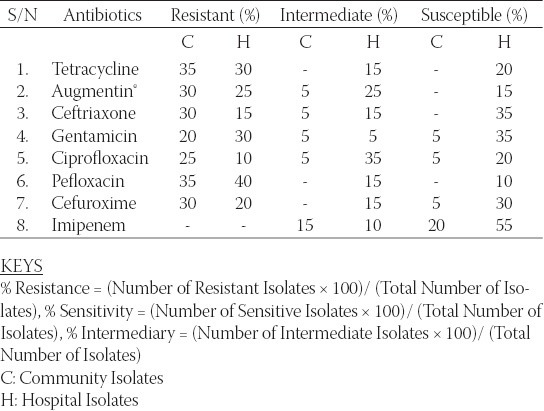
TABLE 3.
Phenotypic confirmation of esbl production using the Double Disc Synergy Test (DDST) Inhibition Zone Diametre (IZD)
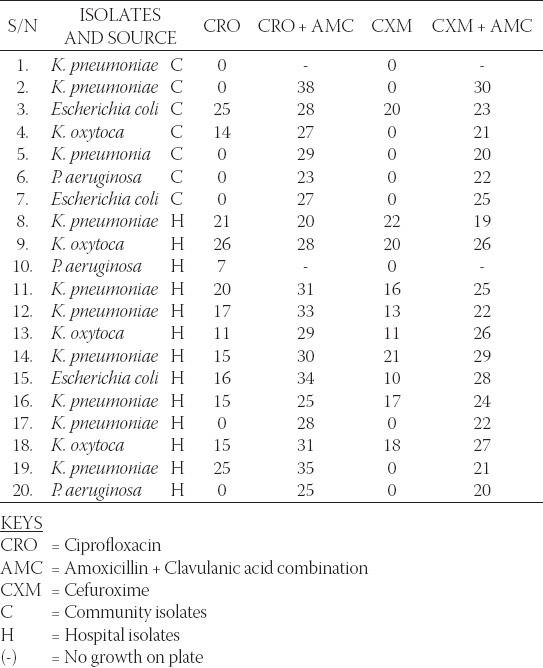
TABLE 4.
Pre- and post-plasmid curing sensitivity test results Inhibition Zone Diameter (mm)
DISCUSSION
Antibiotic resistant bacteria have been a source of ever-increasing therapeutic problem [20]. Continued mismanaged selective pressure has contributed towards the emergence of multiple-drug-resistant bacteria and that has been regarded as an inevitable genetic response to misappropriated exposures of microbial populations to antimicrobial therapy [21-22]. These misappropriated exposures to a wide and varied antimicrobial cocktails no doubt has contributed to the troublesome bacteria resistance pattern recorded worldwide especially in the developing countries. In tropical areas, early-onset infections may be caused by multi-drug resistant (MDR) bacteria strains. These organisms are usually resistant genera of the family Enterobacteriaceae, Pseudomonas spp., and Staphylococci [23]. In the present area under study also, we have equally identified the MDR bacteria strains to belong principally to the Enterobacteriaceae family and Pseudomonas genera with a greater percentage skewing towards the Enterobacteriaceae category. Another closer observation of the results obtained seem to suggest a pressurized influence of mismanaged chemotherapy of nosocomial infections given the greater proportional position occupied by the hospital isolates where 65 % of bacteria strains sampled were MDR-hospital-isolates and only 35 % being MDR-community-isolates (Table 1). However, further observation of the data presented shows that the community isolates displayed more extensive resistances coverage to a wider cocktail of antibiotics utilized in the study. This finding does not appear to be fully understood since the community origin of the isolates does not normally seem to point to any previously known heavy antibiotic exposure. This consideration nevertheless may suffer credibility given the relatively poor and porous antibiotics distribution and use control existent in Nigeria that keep ensuring that individuals within the community have easy, abusive and uncontrolled access to antibiotics [24]. And in certain cases, these misguided individuals employ the use of high-action-profile antibiotics in abusive proportions thus compromising the future positioning of this special category of antibiotics as treatment options of last resort in highly resistant bacteria infections. Therefore, adequate drug and antibiotics control if pursued as an indispensable project by all stakeholders involved in healthcare delivery is expected to immensely contribute towards controlling this ugly trend. Moreover, given that the isolates were sensitive to imipenem and not to the other majority of the employed antibiotics is quite suggestive that they are Extended Spectrum Beta- Lactamase (ESBL) producing organisms. This assertion was further confirmed by the double disc synergy test (DDST), thus leading to selection of 20 ESBL producing bacteria isolates among the sampled bacteria population were identified (Table 2). DDST for the detection and confirmation of ESBL production involves the recording of increased susceptibility to at least one of the extended spectrum cephalosporins in the presence of a combination disc (amoxicillin/clavulanic acid combination) when placed 15 mm apart centre to centre [1]. This means of detection of ESBL has been widely employed by many workers [25-26]. Meanwhile, from our findings, given that 70 % of the confirmed ESBL producers were identified as Klebsiella spp (Table 3) places this group in a luminous position suggesting its possibly major contributory role as a principal repository and transmitter of the putative extrachromosomal ESBL conferring determinants in the sampled area. Earlier reports of ESBL producing strains have showcased Klebsiella spp. as possessing a traditional role in the overall definition and expression of ESBL [27]. We have therefore the emerging problem of antibacterial chemotherapy that would involve an increasing plethora of ESBL-producing bacteria population strains in the tropics, and this poses a huge medical and economic challenge. It is equally important to recall that previous investigators working in Nigeria have reported related findings from neighbouring localities [16, 18]. And recently, the ESBL hospital and community prevalence in Southwest and Southeastern Nigeria were placed at 7.5% and 4.4% respectively [28, 29] falling slightly short of the values obtained in our study. This increasing emergence and development of ESBL producing bacteria strains which remains a decimating therapeutic impediment clearly point to a present and troublesome problem that could constitutes a great deal of menace to futuristic infectious diseases control exercises. Hence, a great deal of attention is required to conduct studies to both identify and fully understand this problem - its cause and scope - so as to create an enabling and useful baseline for effective handling of the ESBL threat. Conjugation studies for Resistance plasmid transfer showed non-transference of resistance determinants between the ESBL strains and the transconjugants. This therefore suggests a possible chromosomal location of all recorded ESBL traits since chromosomally located genetic determinants cannot be transferred through non-replicating bacteria conjugation. In a corresponding similar outcome, the plasmid curing studies (Table 4) also revealed that the acridine orange could not affect a cure on the isolates as they still retained relatively high resistances to the antibiotics after the treatment. This again would further corroborate the chromosomal location of the ESBL traits. However, an exception recorded was the singular isolated case of the Pseudomonas aeruginosa that swung from resistance status to sensitivity upon exposure to ceftriaxone (CRO) before and after the gene curing exercise. This nonetheless, the findings from the twin tests of plasmid gene curing exercise and conjugation studies strongly point to the chromosomal loci of the resistance gene markers. When this is considered in relation to other previous reports of ESBL occurrence coming from Nigeria there seem to be a palpable variability among the ESBL-producing strains with respect to the chromosomal and extra-chromosomal location of the resistance determinants. Enabulele et al. [30] reported that the observed resistance by gram negative wound isolates from the University of Benin were plasmid-mediated. However, in a later study Yah et al. [31] working with ESBL-producing bacteria strains from Ahmadu Bello University Teaching Hospital (ABUTH) Zaria, reported their findings that the bacteria population showed a distribution into sensitive and resistant post-plasmid-curing groups representing both chromosomal and extrachromosomal located resistance markers. Later, Iroha et al. [16, 18] accorded chromosomal location to the resistance determinants of the ESBL-producing bacteria strains they encountered. When these are considered in relation to our present findings they seem to underscore the established variability in the cellular location of the resistance markers. This scenario, apart from determining the ease of passage and acquisition of the resistance traits, also, underscores the need for an extensive and constant demographic coverage of the country for an antimicrobial surveillance studies especially the specialized ESBL-producing bacteria strains. These data would be useful for present and future intervention exercises.
CONCLUSION
The outcome of this study has demonstrated the palpable presence of ESBL producing bacteria strains from community and hospital subjects within Enugu metropolis. This kind of study remains relevant towards providing adequate baseline for the future projection and effective management of infectious diseases caused by the ESBL producing bacteria strains.
DECLARATION OF INTEREST
The authors declare that they have no competing interests.
ACKNOWLEDGEMENTS
The authors wish to appreciate the technical support of personnel of the Medical Laboratory and Microbiology Laboratory Units of the following institutions: Division of Pharmaceutical Microbiology Department of Pharmaceutics, University of Nigeria; National Orthopaedic Hospital Enugu; University of Nigeria Teaching Hospital, Enugu (UNTH); Nona Afulu Hospital, Enugu.
REFERENCES
- [1].Brun-Buisson C, Legrand P, Rauss A, Richard C, Montravers F, Besbes M, et al. Intestinal decontamination for control of nosocomial multiresistant gram-negative bacilli: Study of an outbreak in an intensive care unit. Ann Intern Med. 1989;110:873–81. doi: 10.7326/0003-4819-110-11-873. [DOI] [PubMed] [Google Scholar]
- [2].Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39(Suppl 6):1211–33. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lavilla S, González-López JJ, Miró E, Domínguez A, Llagostera M, Bartolomé RM, et al. Dissemination of extended-spectrum b-lactamase producing bacteria: the food-borne outbreak lesson. J Antimicrob Chemother. 2008;61:1244–51. doi: 10.1093/jac/dkn093. [DOI] [PubMed] [Google Scholar]
- [4].Weinbren MJ, Borthwick MA. Rapid detection of extended-spectrum b-lactamase (ESBL)-producing organisms in blood culture. J Antimicrob Chemother. 2005;55:131–132. doi: 10.1093/jac/dkh502. [DOI] [PubMed] [Google Scholar]
- [5].Nordmann P, Naas T. Sequence analysis of PER-1 extended-spectrum beta-lactamase from Pseudomonas aeruginosa and comparison with class A beta-lactamases. Antimicrob Agents Chemother. 1994;38:104–114. doi: 10.1128/aac.38.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Borer A, Gilad J, Menashe G, Peled N, Riesenberg K, Schlaeffer F. Extended-spectrum beta-lactamase-producing Enterobacteriaceae strains in community-acquired bacteremia in southern Israel. Med Sci Monit. 2002;8:44–47. [PubMed] [Google Scholar]
- [7].Canton R, Coque TM. The CTX-M b-lactamase pandemic. Curr Opin Microbiol. 2006;9:466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- [8].Paterson DL. Resistance in Gram-negative bacteria: Enterobacteriaceae. Am J Med. 2006;119:S20–S28. doi: 10.1016/j.amjmed.2006.03.013. [DOI] [PubMed] [Google Scholar]
- [9].Pena C, Gudiol C, Tubau F, Saballs M, Pujol M, Dominguez MA, et al. Risk factors for faecal carriage of Klebsiella pneumoniae producing extended spectrum blactamase (ESBL-KP) in the intensive care unit. J Hosp Infect. 1997;35:9–16. doi: 10.1016/s0195-6701(97)90163-8. [DOI] [PubMed] [Google Scholar]
- [10].Pitout JD, Nordmann P, Laupland KB, Poirel L. Emergence of Enterobacteriaceae producing extended-spectrum blactamases (ES-BLs) in the community. J Antimicrob Chemother. 2005;56:52–59. doi: 10.1093/jac/dki166. [DOI] [PubMed] [Google Scholar]
- [11].Ramphal R, Ambrose PG. Extended-spectrum blactamases and clinical outcomes: current data. Clin Infect Dis. 2006;42:S164–S172. doi: 10.1086/500663. [DOI] [PubMed] [Google Scholar]
- [12].Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clin Infect Dis. 2001;32:1162–71. doi: 10.1086/319757. [DOI] [PubMed] [Google Scholar]
- [13].Paterson DL, Ko WC, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, et al. Antibiotic therapy for Klebsiella pneumoniae bacteremia: implications of production of extended-spectrum betalactamases. Clin Infect Dis. 2004;39:31–37. doi: 10.1086/420816. [DOI] [PubMed] [Google Scholar]
- [14].Rossi F, Baquero F, Hsueh PR, Paterson DL, Bochicchio GV, Snyder TA, et al. In vitro susceptibilities of aerobic and facultatively anaerobic Gram-negative bacilli isolated from patients with intraabdominal infections worldwide: 2004 results from SMART (Study for Monitoring Antimicrobial Resistance Trends) J Antimicrob Chemother. 2006;58:205–210. doi: 10.1093/jac/dkl199. [DOI] [PubMed] [Google Scholar]
- [15].National Nosocomial Infections Surveillance (NNIS) System Report. Data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004;32:470–485. doi: 10.1016/S0196655304005425. [DOI] [PubMed] [Google Scholar]
- [16].Iroha IR, Amadi ES, Adikwu MU, Esimone CO. Detection of plasmid borne Extended Spectrum Beta Lactamase enzymes in clinical isolates of Escherichia coli from a community general hospital. Int J Med Adv. in press. [Google Scholar]
- [17].Albinu I, Odugbemi P, Brian JM. Extended spectrum beta lactamases in isolates of Klebsiella species and Escherichia coli from Lagos, Nigeria. Nig J Health and Biomed Science. 2003;2(Suppl 2):53–60. [Google Scholar]
- [18].Iroha IR, Oji AE, Esimone CO. Antimicrobial resistance pattern of plasmid mediated extended spectrum beta-lactamase producing strain of Escherichia coli. Sci Res Essay. 2008;3(Suppl 6):215–218. [Google Scholar]
- [19].Okore VC. Evaluation of chemical antimicrobial agents. In: Okore VC, editor. Pharmaceutical Microbiology: Principles of Pharmaceutical Applications of Antimicrobial Agents. 1st edition. Enugu: Demak Publishers; 2005. pp. 61–64. [Google Scholar]
- [20].Gutmann L, Williamson R, Moreau R, Kitzis MD, Collatz E, Acar JF, et al. Cross resistance to nalidixic acid, trimethoprim and chloramphenicol associated with alterations in the outer membrane proteins of Klebsiella, Enterobacter and Serratia. J Infect Dis. 1985;151:501–507. doi: 10.1093/infdis/151.3.501. [DOI] [PubMed] [Google Scholar]
- [21].Cohen ML, Auxe RV. Drug resistant Salmonella in the United States: an epidemiologic perspective. Science. 1992;234:964–970. doi: 10.1126/science.3535069. [DOI] [PubMed] [Google Scholar]
- [22].Sheikh AR, Afsheen A, Sadia K, Abdul W. Plasmid borne antibiotic resistance factors among indigenous Klebsiella. Pak J Bot. 2003;35(Suppl 2):243–248. [Google Scholar]
- [23].Begue P. Current orientation of antibiotic treatment in neonatal bacterial infection. Bull Soc Pathol Exot. 1991;84:712–720. [PubMed] [Google Scholar]
- [24].Hansotia JB, Agarwal V, Pathak AA, Saoji AM. Extended spectrum b-lactamase mediated resistance to third generation cephalosporins in Klebsiella pneumoniae in Nagpur, central India. Indian J Med Res. 1997;105:158–161. [PubMed] [Google Scholar]
- [25].Harris AD, McGregor JC, Johnson JA, Strauss SM, Moore AC, Standiford HC, et al. Risk Factors for Colonization with Extended-Spectrum β-Lactamase–Producing Bacteria and Intensive Care Unit Admission. Emerg Infect Dis. 2007;13(Suppl 8):1144–1149. doi: 10.3201/eid1308.070071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Amita J, Indranil R, Mahendra KG, Mala K, Agarwal SK. Prevalence of extended-spectrum-lactamaseproducing Gram-negative bacteria in septicaemic neonates in a tertiary care hospital. J Med Microbiol. 2003;52:421–425. doi: 10.1099/jmm.0.04966-0. [DOI] [PubMed] [Google Scholar]
- [27].Esimone CO, Nworu CS, Udeogaranya OP. Utilization of antimicrobial agents with and without prescription by outpatients in selected pharmacies in South-Eastern Nigeria. Pharm Sci Journal. 2007;11096:6. doi: 10.1007/s11096-007-9124-0. [DOI] [PubMed] [Google Scholar]
- [28].Olowe OA, Aboderin BW. Detection of extended spectrum β-lactamase producing strains of Escherichia coli and Klebsiella sp in a tertiary Health centre in Ogun State. Int J of Trop Med. 2010;5(3):62–64. [Google Scholar]
- [29].Afiukwa F.N, Iroha I.R, Afiukwa C.A, Ayogu T.E, Oji A.E, Onwa N.C. Presence of coliform producing extended spectrum beta lactamase in sachet-water manufactured and sold in Abakaliki, Ebonyi State, Nigeria, 2010. Int Res J Microb. 2010;1(2):32–36. [Google Scholar]
- [30].Enabulele OI, Aluyi HAS, Omokao O. Incidence of bacteraemia following teeth extraction at the dental clinic of the University of Benin Teaching Hospital, Benin City, Nigeria. Afr J Biotechnol. 2008;7(Suppl 10):1390–93. [Google Scholar]
- [31].Yah SC, Eghaforia NO, Oranusi S, Abono AM. Widespread plasmid resistance genes among Proteus species in diabetic wounds of patients in the Ahmadu Bello University Teaching Hospital (ABUTH) Zaria. Afr J Biotechnol. 2007;6(Suppl 15):1757–62. [Google Scholar]


