Abstract
Background
Brain metastases are one of the most malignant complications of lung cancer and constitute a significant cause of cancer related morbidity and mortality worldwide. Recent years of investigation suggested a role of LKB1 in NSCLC development and progression, in synergy with KRAS alteration. In this study, we systematically analyzed how LKB1 and KRAS alteration, measured by mutation, gene expression (GE) and copy number (CN), are associated with brain metastasis in NSCLC.
Materials and Methods
Patients treated at University of North Carolina Hospital from 1990 to 2009 with NSCLC provided frozen, surgically extracted tumors for analysis. GE was measured using Agilent 44,000 custom-designed arrays, CN was assessed by Affymetrix GeneChip Human Mapping 250K Sty Array or the Genome-Wide Human SNP Array 6.0 and gene mutation was detected using ABI sequencing. Integrated analysis was conducted to assess the relationship between these genetic markers and brain metastasis. A model was proposed for brain metastasis prediction using these genetic measurements.
Results
17 of the 174 patients developed brain metastasis. LKB1 wild type tumors had significantly higher LKB1 CN (p < 0.001) and GE (p = 0.002) than the LKB1 mutant group. KRAS wild type tumors had significantly lower KRAS GE (p < 0.001) and lower CN, although the latter failed to be significant (p = 0.295). Lower LKB1 CN (p = 0.039) and KRAS mutation (p = 0.007) were significantly associated with more brain metastasis. The predictive model based on nodal (N) stage, patient age, LKB1 CN and KRAS mutation had a good prediction accuracy, with area under the ROC curve of 0.832 (p < 0.001).
Conclusion
LKB1 CN in combination with KRAS mutation predicted brain metastasis in NSCLC.
Keywords: NSCLC, Brain Metastasis, Prognostic model, LKB1, KRAS
Introduction
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related deaths in the United States with brain metastasis as one of the most dreaded complications [1]. Historically, the prognosis of NSCLC with brain metastasis has been poor, with a median overall survival of 4.5 months for patients treated with standard whole brain radiation therapy (WBRT) and 4–11 weeks in untreated patients [2, 3]. The prevalence of brain metastasis in NSCLC is reported to be increasing, possibly due to improved diagnosis in brain imaging and prolonged survival with new systemic treatment options [4]. Therefore, identification of biomarkers that have critical roles in cell growth, metabolism, and tumor recurrence would provide valuable information in disease prognosis and better treatment choices.
In the past few years, several lines of evidence implicate the importance of liver kinase B1 (LKB1, aka, serine-threonine kinase or STK11) as a tumor suppressor gene in lung cancer development and progression in both human and model organisms [5, 6]. LKB1 was first identified in 1997 as the causative mutation in the autosomal-dominant inherited Peutz–Jeghers Syndrome (PJS) [7]. LKB1 loss is one of the most frequent genetic alterations in NSCLC [8], the inactivation of which has also been proposed to be associated with tumor metastasis in lung cancer and other tumor types [5, 6, 9]. Specifically, LKB1 mutation or loss of heterozygosity (LOH) of 19p13.2 which harbors the LKB1 gene was observed in a much higher proportion in brain metastases of lung cancer patients than in the primary tumors [5, 10].
As with many tumor suppressor genes, identifying patients with LKB1 inactivation remains a challenge, with potential mechanisms including homozygous deletion, point mutations and epigenetic silencing [5, 6]. The discrepancy between the high frequency of LOH (often over 50%) of 19p13.3 [11] and the reported rate of LKB1 mutation [5, 8] suggests that many “second hits” to the gene may go undetected by current sequencing techniques or that epigenetic silencing or other inactivating events may be more prevalent than previously recognized. In any case, for the purposes of clinical assessment, investigators are challenged to assess the gene through multiple mechanisms to gain confidence in characterizing the gene as intact or altered. In addition, multiple investigators have now reported coordination between losses of LKB1 and the oncogene, KRAS, particularly in smokers suggesting that coordinated assessment may be clinically relevant.
In this study, we seek to identify how LKB1 alteration, assessed by gene mutation, gene expression (GE) and copy number (CN) change, can predict brain metastasis in a group of NSCLC patients in conjunction with KRAS aberration, which has been shown to have a synergistic effect with LKB1 inactivation in lung cancer development and metastasis [6].
Material and Method
Tumor collection and clinical data abstraction
Frozen tumors were collected from patients who received curative surgery at the University of North Carolina (UNC) hospital with NSCLC diagnosis from December 1990 to September 2009. Tissues were flash-frozen and stored at −80 °C until time of analysis. Tumor histology includes adenocarcinoma [12], adenosquamous carcinoma, bronchioloalveolar carcinoma, large cell carcinoma and squamous cell carcinoma [13]. Patient outcomes were assessed by retrospective chart review for vital status and tumor recurrence, including brain metastasis through the end of the study, January 2011. For any patients whose follow-up was not at the UNC, records were requested from outside treating facilities. Assessment of brain metastasis was made by review of all radiology reports of brain imaging or pathology in cases of brain tissue resection. Patients were only assessed as having a metastasis if a radiology report concluded definitively that a brain metastasis was present in the summary conclusion of the official report. In cases where no brain imaging was performed, a patient was assessed as negative for brain metastasis. In cases where a patient had both imaging and tissue confirmation of brain metastasis, the time to recurrence was estimated based of the first positive report. The study was approved by Institutional Review Board (IRB) under protocols 90-0573 and 07-0120.
Gene expression microarray
GE was measured by Agilent 44 K microarrays (human tumor). Total RNA from tumor tissues was isolated using the RNeasy kit following the manufacturer’s protocols (Qiagen, Valencia, CA, USA). Total RNA-1ug was converted to labeled cRNA with nucleotides coupled to a fluorescent dye (Cy3) using the Quick Amp Kit (Agilent Technologies, Palo Alto, CA). Universal RNA from Invitrogen was labeled with Cy5 as a reference. Samples were purified using an RNeasy kit (Qiagen) and quantified for dye integration using a Nanodrop-8000 (Thermo Scientific). Following quantification, samples were hybridized overnight in a rotating hybridization oven and washed/scanned using an Agilent scanner. Microarrays were processed by normexp background correction and loess normalization [13, 14].
LKB1 and KRAS mutations
Genomic DNA was extracted from tumor tissues using Qiagen QiaAmp DNA kit and sent to Polymorphic DNA Technologies, Inc. (Almeda CA) for direct exon sequencing on ABI 3730XL DNA sequencers to detect LKB1 and KRAS mutations. Regions of LKB1 and KRAS sequencing were described elsewhere [12], with all nine exons of LKB1 and exon 2 of KRAS, which harbors more than 95% of KRAS mutation [15] sequenced. Non-synonymous or splice site differences compared to reference sequence were considered as mutations [16].
LKB1 and KRAS CN assessment
CN microarray of tumor DNA was performed using the Affymetrix GeneChip Human Mapping 250K Sty Array or the Genome-Wide Human SNP Array 6.0 (Affymetrix, Inc., Santa Clara, CA) according to the manufacturer’s instructions. CN for each marker was calculated using CRMA_v2 [17], which performs log2 transformation on preprocessed signal intensity. CN for each marker was taken to be log2(tumor sample/normal estimate), where the normal estimate was calculated using the mean intensity from all normal specimens. CN for LKB1 and KRAS in each sample was taken as the mean values of estimated copy numbers across all markers that are within the 100 kb region upstream or downstream of the genes.
Statistical analysis
All statistical analysis was performed using R 2.10.1 software (http://cran.r-project.org) unless otherwise stated. Patients follow up time was calculated using “reverse” Kaplan-Meier analysis in which the outcomes ‘dead’ and ‘censored’ are exchanged [18]. This method distinguishes the observation time between patients who were lost to follow up and patients who died during the study. The unobservable follow up time for a deceased patient is considered as the potential follow up time that would have been obtained had that patient not died [19]. Pairwise association between patients’ baseline characteristics, including gender, race, stage, tumor histology and smoking status, and genetic biomarkers, including LKB1 and KRAS mutation, GE and CN, were tested using Fisher’s exact test for categorical variables and two sample t-test for continuous variables. Logistic regression was used to test the association between each of the variables and brain metastasis. Variables showed significant association with brain metastasis at α = 0.05 level in univariate analysis were included in multivariate analysis. For all the analyses, a complete case approach was used to handle missing data. All statistical tests were two sided tests and all reported confidence intervals were constructed at a two sided 95% confidence level.
Result
Patient characteristics with respect to genetic biomarkers
174 of the patients provided sufficient tissue for at least one measurement of LKB1 alteration and were included in subsequent analysis, in which 172 had GE measurement, 162 had CN and 172 had mutation data. Diagnosis age ranges from 39 to 90 with a median of 66 years; approximately half of these patients (88) are males and most of them (161) had smoking history. The majority of these patients (153) were diagnosed when the tumor was still small (T1 or T2). Half of the patients (87) had adenocarcinoma, and most of the others had squamous cell carcinoma (57) or adenosquamous carcinoma (10). The median follow up time calculated from the reverse KM method was 91 months. Only 11 patients were lost to follow up before 60 months, with a median follow up time of 51 months. The median survival time of all 174 patients was 42 months (95% CI: 33–58 months). Seventeen of these patients had brain recurrence with a median survival time after brain metastasis of 6.8 months (95% CI: 2.67 –49.9 months). 3 of 17 patients developed brain metastases within 6 months of cancer diagnosis. An additional 13 patients developed recurrence within 5 years at a median and mean of 12 and 17 months respectively. One patient developed an unusual late brain only recurrence at 86 months which was nonetheless clinically determined to be originating from the remote lung cancer. Brain only recurrence was seen in 13 of 17 patients as the first sight of recurrence at a median of 8 months after initial diagnosis. The remaining 4 patients developed brain metastasis at later stages of the disease or in conjunction with multiple sites of disease at a median of 19 months after initial diagnosis.
Table 1 summarized how patient characteristics associated with genetic biomarkers LKB1 and KRAS. Overall, 21 samples (12.2%) sequenced for LKB1 had non-synonymous or splice site mutation and 22 (12.9%) had canonical mutations in KRAS. Consistent with previous research [8, 20], LKB1 mutations were more common in adenocarcinoma (13/85) than in non-adenocarcinoma (8/87), although the difference failed to be significant (p = 0.25). Similarly, KRAS mutations were more frequent in adenocarcinoma (20/85) than other tumor histology (2/86, p < 0.001). Accordingly, adenocarcinomas had significantly lower LKB1 expression (p < 0.001), lower LKB1 CN (p = 0.003) and higher KRAS expression (p = 0.035) compared to the non-adenocarcinoma group. Also consistent with previous reports, smoking was associated with LKB1 and KRAS mutation [20]: all samples that were mutant for LKB1 were smokers and only one KRAS mutant was a non-smoker, although the association was not significant. Both LKB1 and KRAS mutation were associated with earlier T stage. Gender and race were not associated with LKB1 or KRAS measurement.
Table 1.
Patient characteristics by genetic biomarker
| LKB1 mutation status | KRAS mutation status | LKB1 GE mean | LKB1 CN mean | |||
|---|---|---|---|---|---|---|
| WT | MT | WT | MT | |||
| # of Samples | 151 | 21 | 149 | 22 | 0.002 | |
|
| ||||||
| Gender | ||||||
| Female | 79 | 7 | 76 | 10 | 0.531 | 0.103 |
| Male | 72 | 14 | 73 | 12 | 0.585 | 0.107 |
|
| ||||||
| Race | ||||||
| White ζ | 125 | 15 | 120 | 20 | 0.564 | 0.112 |
| Black | 23 | 6 | 26 | 2 | 0.522 | 0.099 |
|
| ||||||
| Smoking Status | ||||||
| Current/Former Smoker ζ | 138 | 21 | 137 | 21 | 0.553 | 0.110 |
| Never/Light Smoker | 12 | 0 | 11 | 1 | 0.649 | 0.043 |
|
| ||||||
| Histology | ||||||
| Adenocarcinoma ζ | 72 | 13 | 65** | 20** | 0.430 ** | 0.036* |
| Non-adenocarcinoma | 79 | 8 | 84** | 2** | 0.683** | 0.172* |
|
| ||||||
| T stage ζ | ||||||
| T1-T2 | 134 | 18 | 130 | 21 | 0.558 | 0.099 |
| T3-T4 | 15 | 3 | 17 | 1 | 0.549 | 0.144 |
|
| ||||||
| N stage ζ | ||||||
| N0 | 100 | 15 | 104 | 11 | 0.584 | 0.096 |
| N1 or above | 45 | 6 | 41 | 9 | 0.511 | 0.120 |
p < 0.001
p < 0.05
number doesn’t sum to total because of missing values
Associations between genetic markers
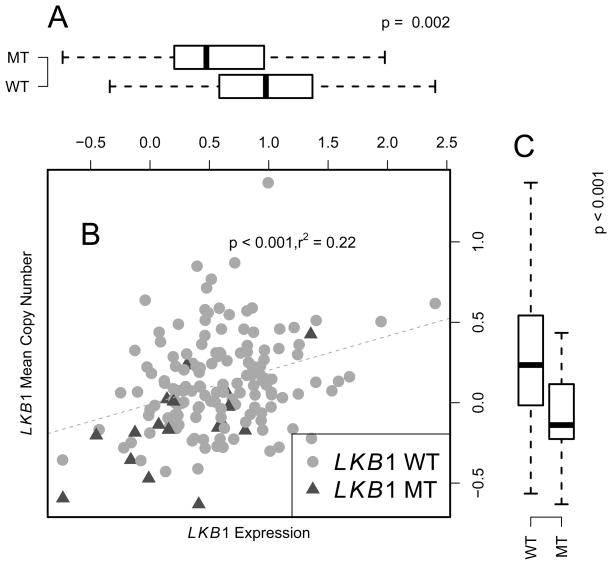
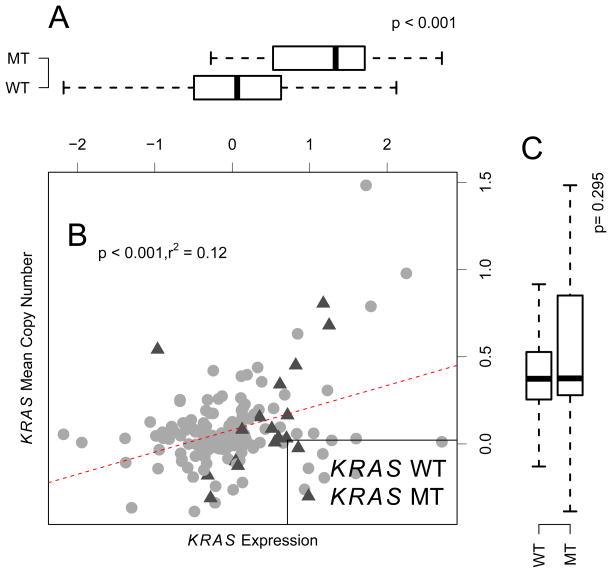
Alterations of LKB1 and KRAS were further interrogated as a function of GE and CN (Figure 1–2). LKB1 mutation was significantly associated with lower GE (Figure 1A, p = 0.002) and lower CN (Figure 1C, p < 0.001). On the contrary, KRAS mutation was associated with higher expression (Figure 2A, p < 0.001). There is no significant association between KRAS mutation and KRAS CN (Figure 2C). CN and GE are positively correlated in both LKB1 and KRAS (Figure 1B, 2B, p < 0.001).
Figure 1.
Correlation between LKB1 gene expression and copy number measurement. Panel A: LKB1 wild type group had significantly higher gene expression. Panel B: LKB1 GE and CN were positively correlated. Panel C: Wild type group had significantly higher CN. CN for each marker was calculated as log2 intensity ratio between tumor samples and normal samples using CRMA_v2 [17]. GE microarray was preprocessed by Loess normalization and GE values are unit-less values.
Figure 2.
Correlation between KRAS gene expression and copy number measurement.. Panel A: KRAS wild type samples had a significantly lower gene expression. Panel B: KRAS expression and copy number were positively correlated. Panel C: Wild type group had significantly lower copy number. CN for each marker was calculated as log2 intensity ratio between tumor samples and normal samples using CRMA_v2 [17]. GE microarray was preprocessed by Loess normalization and GE values are unit-less values.
Univariate association of patient characteristics and genetic markers with brain recurrence
Seventeen of the patients had brain metastasis during the follow up. Patients’ characteristics with respect to brain metastasis were summarized in Table 2. Neither gender nor race was associated with brain recurrence. All but one patient with brain metastasis were current/former smokers. Of all patients with brain recurrence, only one had late T (T3 or T4) stage at diagnosis. However, the association failed to be significant because of the small number of brain recurrence. Clinical N stage was significantly associated with brain metastasis (OR = 4.87, CI: 1.74–14.9). Brain recurrence in adenocarcinoma (11/87) was more frequent than in non-adenocarcinoma (6/87), although the association failed to be significant (p = 0.21). Of the genetic markers, KRAS mutation and LKB1 CN were significantly associated with brain metastasis (p = 0.007 and 0.039 respectively). Higher LKB1 expression and LKB1 wild type were also associated with fewer brain metastases, although the result did not achieve statistical significance.
Table 2.
Patient characteristics and genetic marker by disease outcome
| brain recurrence (Column %) | no brain recurrence (Column %) | Odds Ratio ϕ (95% CI) | P values | |
|---|---|---|---|---|
| # of patients | 17 | 157 | ||
|
| ||||
| Age at Diagnosis(mean) | 57.4 | 65.7 | ||
|
| ||||
| Gender | ||||
| Female | 9(52.9) | 77(49.0) | 1.17 | 0.76 |
| Male | 8(47.1) | 80(51.0) | (0.43–3.26) | |
|
| ||||
| Race | ||||
| Whiteζ | 12(70.6) | 129(82.1) | 0.47 | 0.183 |
| Black | 5(29.4) | 25(15.9) | (0.157–1.56) | |
|
| ||||
| Smoking status ζ | ||||
| Current/Former Smoker ζ | 16(94.1) | 145 (92.9) | 1.214 | 0.857 |
| Never/Light Smoker | 1(5.88) | 11(7.05) | (0.21–22.9) | |
|
| ||||
| Tumor histology ζ | ||||
| Adenocarcinoma | 11(64.7) | 76(48.4) | 1.95 | 0.21 |
| Non-adenocarcinoma | 6(35.3) | 81(51.6) | (0.707–5.91) | |
|
| ||||
| T stages ζ | ||||
| T3-T4 | 1(5.88) | 18(11.6) | 0.476 | 0.48 |
| T0-T1-T2 | 16(94.1) | 137(88.4) | (0.026–2.56) | |
|
| ||||
| N stages ζ | ||||
| N1 or above | 11(64.7) | 41(27.3) | 4.87 | 0.003 |
| N0 | 6(35.3) | 109(72.7) | (1.74–14.9) | |
|
| ||||
| LKB1 mutation ζ | ||||
| Mutant | 3(17.6) | 18(11.6) | 1.63 | 0.47 |
| Wild type | 14(82.4) | 137(88.4) | (0.35–5.62) | |
|
| ||||
| LKB1 expression ζ | 0.574 | 0.32 | ||
| mean | 0.447 | 0.570 | (0.187–1.65) | |
|
| ||||
| LKB1 copy number ζ | 0.103 | 0.039 | ||
| (mean) | −0.051 | 0.120 | (0.01–0.81) | |
|
| ||||
| KRAS mutation ζ | ||||
| Mutant | 6(35.3) | 16(10.4) | 4.71 | 0.007 |
| Wild type | 11(64.7) | 138(89.6) | (1.46–14.2) | |
|
| ||||
| KRAS expression ζ | 0.83 | 0.63 | ||
| mean | −0.127 | −0.041 | (0.388–1.69) | |
|
| ||||
| KRAS copy number ζ | 0.552 | 0.67 | ||
| mean | 0.0506 | 0.0776 | (0.024–5.52) | |
number doesn’t sum to total because of missing values
For each variable, the reported value was the odds ratio of brain metastasis in patients with characteristics in the first row compared to the patients with characteristics in the second row. For example, for gender, the odds of having brain recurrence in females are 1.429 times the odds of brain metastasis in males. Odds ratios, confidence intervals and p values were generated by fitting logistic models using each of the variables with brain recurrence as response variable.
Multivariate association of patient characteristics and genetic markers with brain recurrence
Variables that were significantly associated with brain recurrence in univariate models were considered for inclusion in the multivariate model (Table 3). 154 patients had complete data for all the gene measurements and were included in the multivariate models. LKB1 CN and KRAS mutation were significantly associated with brain recurrence after adjusting for patient age and nodal status. Patients with higher LKB1 CN or wild type KRAS had lower risk of developing brain recurrence. The odds of brain metastasis in mutant KRAS patients were estimated to be 5.52 (CI: 1.31–22.6) times higher than the odds of brain recurrence in patients with wild type KRAS, after adjusting for age, LKB1 CN and N stage. The odds ratio of brain metastasis was ~20 times higher in patients with one decrease in LKB1 CN values, after controlling for age, KRAS mutation and N stage. Considering the fact that variables may confound or interact with each other in multivariate model, another multivariate model which includes all variables that had p value < 0.20 was fitted and the result was similar to the result in table 3 (not shown).
Table 3.
Multivariate association between genetic biomarkers and disease outcome
| Odds Ratio | 95% CI | P values | |
|---|---|---|---|
| Age at diagnosis | 0.95 | 0.89–1.00 | 0.056 |
| LKB1 copy number loss | 19.04 | 1.59–307 | 0.026 |
| KRAS mutation | 5.52 | 1.31–22.6 | 0.016 |
| N stage | 4.46 | 1.26–17.5 | 0.023 |
Model predictions of brain metastasis
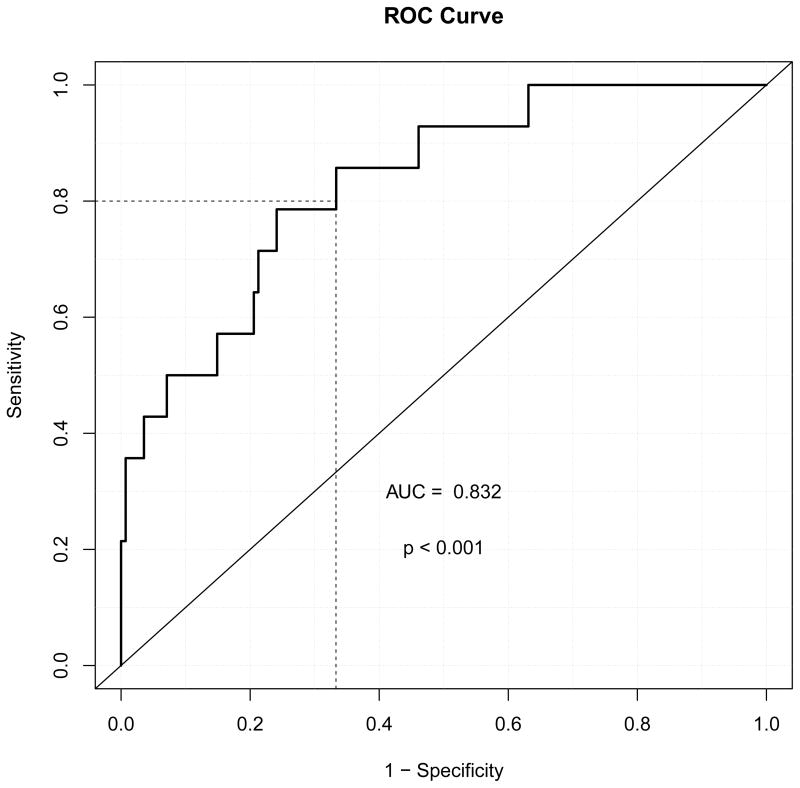
A linear predictor based on LKB1 CN, KRAS mutation, N stage and diagnosis age was constructed from the previously described multivariate model by using the model estimates. ROC curve was used to evaluate the prediction of this multivariate model (Figure 3). Area under the curve (AUC) was estimated to be 0.832 and significantly different from 0.5 (p < 0.001, CI: 76.6%–93.5%). The ROC curve suggests the test performance over a range of potential cut points on this linear predictor. The optimal cut points cannot be clearly defined because this will depend on user preference for defining false positives and false negatives. For example, with a false positive rate of approximately 30%, the model successfully captured 80% of the true brain metastasis patients.
Figure 3.
ROC curve for the multivariant predictive model. Predictors in this model include LKB1 CN, KRAS mutation, patients’ age at diagnosis and nodal stage. P values were generated by testing the hypothesis that area under the cuver (AUC) is 0.5.
Discussion
The poor outcomes of patients with lung cancer have been widely reported, including the frequent occurrence of brain metastases in patients who have otherwise controlled their disease through primary therapy. In small cell lung cancer progress has been made towards preventing brain metastases through prophylactic cranial radiation which has a proven survival benefit [21]. Attempts to extend this benefit to patients with NSCLC have similarly documented a small benefit in terms of prevention of brain metastasis but without impact on survival [22, 23]. Such data suggest that biomarkers to enrich for patients at highest risk for brain metastasis may be necessary to make progress in NSCLC.
LKB1 is one of the most important tumor suppressor genes and is observed to be inactivated in approximately 30% of all NSCLCs [8]. LKB1 encodes a widely expressed serine/threonine protein kinase whose primary action is through 5′-AMP-activated protein kinase (AMPK) to regulate metabolism and ensure efficient energy production in times of stress [24]. Decreased expression of AMPK pathway genes have also been shown to be related to metastasis in NSCLC [25]. AMPK controls cell proliferation through the mammalian target of rapamycin (mTOR) kinase, which regulates numerous downstream targets [26]. LKB1 loss impairs downstream AMPK signaling, leading to unsuppressed cell proliferation. LKB1 deficiency can be associated with increased expression of genes believed to control angiogenesis and metastatic potential [9].
LKB1 can be inactivated through a variety of mechanisms, including gene mutation, deletion and epigenetic events, like promoter methylation. Somatic mutations, mainly nonsense or frameshift mutation, can result in truncated and dysfunctional proteins [27]. As has been shown in this study as well as previous research [5, 28], somatic mutation can account for only a small fraction of tumors and cannot be the sole reason of LKB1 inactivation. Promoter methylation, resulting in reduced expression, was shown to account for a small percentage of depressed LKB1 expression as well [29]. Gene deletion is a frequent mechanism of LKB1 loss, which can be assessed by CN [30]. The fact that the LKB1 mutant group also had lower CN is consistent with the common two-hit model for tumorigenesis which requires an individual to be heterozygous for a mutant tumor suppressor gene to lose the normal allele in order for tumor development to occur, which is frequently achieved through deletion of the normal allele. Based on clear evidence in animal models that LKB1 haploinsufficiency accelerates KRAS driven lung cancer in mice [6], even a single copy inactivation of LKB1 might be oncogenic. A striking result from murine melanocyte models showed that somatically LKB1 inactivation and KRAS activation can induce highly metastatic melanoma with 100% penetrance, suggesting that LKB1 inactivation can greatly facilitate recurrence, especially in the context of RAS activation [31].
Studies on effects of KRAS alteration on metastasis in NSCLC is less conclusive than of LKB1. A recent report [32] on a specific stage IV NSCLC patient population indicated that KRAS was not associated with increase brain metastases; however, the result cannot be extrapolated directly to the surgically treated NSCLC patient population such as in the current study where the goal is prediction of future brain metastasis. The current study assessed the effect of LKB1 and KRAS in the same model, and may clarify that brain metastasis is part of the adverse outcomes of the combined LKB1/KRAS abnormality. CN might be a good proxy for LKB1 mutation, supported by our result that the mutated group is associated with reduced CN. It is also possible that a combination of these events is at work in inducing cancers and tumor invasion. Finding the best measurement that can adequately predict brain metastasis and is relatively straightforward to estimate in the clinical setting is very helpful in patient management.
The current study has limitations inherent to retrospective genomic analyses of clinical outcomes. The overall number of brain metastases was limited and the sample size was modest. It is therefore important to put the current study in the context of prior case reports of brain relapses in lung cancer. It is well established that the rate of brain metastasis in lung cancer is associated with both increasing tumor stage and adenocarcinoma histology. While autopsy series have reported incidence as high as 54% in lung adenocarcinoma, surgical case series of mixed histologic types have generally documented lower rates of brain recurrence in a stage specific manner. A large study by Figlin reported rates as low as 7% in a population of surgically treated patients of all histologic subtypes which is comparable to the rate of 9.7% seen in the current analysis [34]. As expected, both in the report by Figlin and in the current study, increasing tumor stage and adenocarcinoma subtype either trended towards or were significantly associated with brain recurrence. In both reports, relapse in the brain alone, without other systemic disease, was the most common pattern. We conclude that although the current study is small in size, the patterns seen are reflective of those seen in larger surgical case series.
From a statistical modeling standpoint, since the overall number of brain metastases was limited, validation techniques such as split sample cross validation were excluded. Therefore, the estimated odds ratio should be used as an indication of association direction, rather than being a concrete measurement of genetic effect. On the other hand, a significant p value with a modest sample size usually entails a potentially large effect size. The aim of this study is to find clinical relevant markers which can help with patient management, instead of evaluating the mechanism by which LKB1 is involved in NSCLC brain metastasis. On the other hand, the hypothesis of this study was driven by previous reports that KRAS and LKB1 predominant subtypes identified by unbiased expression profiling were associated with adverse events, including a preliminary report of increased brain metastasis [12] as well as profiling of metastatic lesions noted to have LOH for LKB1. Additionally, while SNP chips similar to those used in the current study are available for clinical use, in general their clinic use is the assessment of inherited chromosomal abnormalities rather than somatic alterations in tumors [35]. As such any conclusions must be validated through additional larger patient cohorts and using reagents appropriate for the assessment of somatic alterations in tumors.
In conclusion, we present a predictive model for the occurrence of brain metastases in lung cancer based on common coordinated alterations in NSCLC. If validated these findings could be the basis on which future therapies and diagnostics could be developed for the treatment of brain metastases in this disease.
We analyzed how LKB1 and KRAS alteration, measured by mutation, gene expression and copy number (CN), are associated with brain metastasis in NSCLC.
LKB1 CN and KRAS mutation were significantly associated with brain metastasis after adjusting for other variables.
We constructed a predictive model based on LKB1 CN and KRAS mutation for brain metastasis in NSCLC with good prediction accuracy.
If validated, the prediction model may serve as the basis on which further diagnosis and therapies for the treatment of brain metastasis in NSCLC.
Acknowledgments
Funding: This study was supported by the Thomas G. Labreque Foundation, through Joan’s Legacy Foundation and by a Clinical/Translational Award from the UNC Lineberger Comprehensive Cancer Center.
Footnotes
Conflict of Interest Statement: D. Neil Hayes and N. Zhao hold a provisional patent on the predictive model of brain metastasis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Laskin JJSA, Johnson DH. Philadelphia, PA, editor. Non-Small Cell Lung Cancer. Philadelphia, PA: 2005. [Google Scholar]
- 2.Gaspar L, Scott C, Rotman M, Asbell S, Phillips T, Wasserman T, McKenna WG, Byhardt R. Recursive partitioning analysis (RPA) of prognostic factors in three Radiation Therapy Oncology Group (RTOG) brain metastases trials. Int J Radiat Oncol Biol Phys. 1997;37:745–751. doi: 10.1016/s0360-3016(96)00619-0. [DOI] [PubMed] [Google Scholar]
- 3.Ampil F, Caldito G, Milligan S, Mills G, Nanda A. The elderly with synchronous non-small cell lung cancer and solitary brain metastasis: does palliative thoracic radiotherapy have a useful role? Lung Cancer. 2007;57:60–65. doi: 10.1016/j.lungcan.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Eichler AF, Chung E, Kodack DP, Loeffler JS, Fukumura D, Jain RK. The biology of brain metastases-translation to new therapies. Nat Rev Clin Oncol. 2011;8:344–356. doi: 10.1038/nrclinonc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsumoto S, Iwakawa R, Takahashi K, Kohno T, Nakanishi Y, Matsuno Y, Suzuki K, Nakamoto M, Shimizu E, Minna JD, Yokota J. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene. 2007;26:5911–5918. doi: 10.1038/sj.onc.1210418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C, Wu MC, Shimamura T, Perera SA, Liang MC, Cai D, Naumov GN, Bao L, Contreras CM, Li D, Chen L, Krishnamurthy J, Koivunen J, Chirieac LR, Padera RF, Bronson RT, Lindeman NI, Christiani DC, Lin X, Shapiro GI, Janne PA, Johnson BE, Meyerson M, Kwiatkowski DJ, Castrillon DH, Bardeesy N, Sharpless NE, Wong KK. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 7.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Hoglund P, Jarvinen H, Kristo P, Pelin K, Ridanpaa M, Salovaara R, Toro T, Bodmer W, Olschwang S, Olsen AS, Stratton MR, de la Chapelle A, Aaltonen LA. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, Westra WH, Herman JG, Sidransky D. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62:3659–3662. [PubMed] [Google Scholar]
- 9.Zhuang ZG, Di GH, Shen ZZ, Ding J, Shao ZM. Enhanced expression of LKB1 in breast cancer cells attenuates angiogenesis, invasion, and metastatic potential. Mol Cancer Res. 2006;4:843–849. doi: 10.1158/1541-7786.MCR-06-0118. [DOI] [PubMed] [Google Scholar]
- 10.Tiainen M, Ylikorkala A, Makela TP. Growth suppression by Lkb1 is mediated by a G(1) cell cycle arrest. Proc Natl Acad Sci U S A. 1999;96:9248–9251. doi: 10.1073/pnas.96.16.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobottka SB, Haase M, Fitze G, Hahn M, Schackert HK, Schackert G. Frequent loss of heterozygosity at the 19p13. 3 locus without LKB1/STK11 mutations in human carcinoma metastases to the brain. J Neurooncol. 2000;49:187–195. doi: 10.1023/a:1006442024874. [DOI] [PubMed] [Google Scholar]
- 12.Wilkerson MD, Yin X, Walter V, Zhao N, Cabanski CR, Hayward MC, Miller CR, Socinski MA, Parsons AM, Thorne LB, Haithcock BE, Veeramachaneni NK, Funkhouser WK, Randell SH, Bernard PS, Perou CM, Hayes DN. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLoS One. 2012;7:e36530. doi: 10.1371/journal.pone.0036530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkerson MD, Yin X, Hoadley KA, Liu Y, Hayward MC, Cabanski CR, Muldrew K, Miller CR, Randell SH, Socinski MA, Parsons AM, Funkhouser WK, Lee CB, Roberts PJ, Thorne L, Bernard PS, Perou CM, Hayes DN. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res. 2010;16:4864–4875. doi: 10.1158/1078-0432.CCR-10-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 15.Aviel-Ronen S, Blackhall FH, Shepherd FA, Tsao MS. K-ras mutations in non-small-cell lung carcinoma: a review. Clin Lung Cancer. 2006;8:30–38. doi: 10.3816/CLC.2006.n.030. [DOI] [PubMed] [Google Scholar]
- 16.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bengtsson H, Ray A, Spellman P, Speed TP. A single-sample method for normalizing and combining full-resolution copy numbers from multiple platforms, labs and analysis methods. Bioinformatics. 2009;25:861–867. doi: 10.1093/bioinformatics/btp074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shuster JJ. Median follow-up in clinical trials. J Clin Oncol. 1991;9:191–192. doi: 10.1200/JCO.1991.9.1.191. [DOI] [PubMed] [Google Scholar]
- 19.Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Control Clin Trials. 1996;17:343–346. doi: 10.1016/0197-2456(96)00075-x. [DOI] [PubMed] [Google Scholar]
- 20.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba, Ozenberger B, Good PJ, Chang AC, Beer DG, Watson MA, Ladanyi M, Broderick S, Yoshizawa A, Travis WD, Pao W, Province MA, Weinstock GM, Varmus HE, Gabriel SB, Lander ES, Gibbs RA, Meyerson M, Wilson RK. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slotman B, Faivre-Finn C, Kramer G, Rankin E, Snee M, Hatton M, Postmus P, Collette L, Musat E, Senan S. Prophylactic cranial irradiation in extensive small-cell lung cancer. N Engl J Med. 2007;357:664–672. doi: 10.1056/NEJMoa071780. [DOI] [PubMed] [Google Scholar]
- 22.Stuschke M, Eberhardt W, Pottgen C, Stamatis G, Wilke H, Stuben G, Stoblen F, Wilhelm HH, Menker H, Teschler H, Muller RD, Budach V, Seeber S, Sack H. Prophylactic cranial irradiation in locally advanced non-small-cell lung cancer after multimodality treatment: long-term follow-up and investigations of late neuropsychologic effects. J Clin Oncol. 1999;17:2700–2709. doi: 10.1200/JCO.1999.17.9.2700. [DOI] [PubMed] [Google Scholar]
- 23.Gore EM, Bae K, Wong SJ, Sun A, Bonner JA, Schild SE, Gaspar LE, Bogart JA, Werner-Wasik M, Choy H. Phase III comparison of prophylactic cranial irradiation versus observation in patients with locally advanced non-small-cell lung cancer: primary analysis of radiation therapy oncology group study RTOG 0214. J Clin Oncol. 2010;29:272–278. doi: 10.1200/JCO.2010.29.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.William WN, Kim JS, Liu DD, Solis L, Behrens C, Lee JJ, Lippman SM, Kim ES, Hong WK, Wistuba, Lee HY. The impact of phosphorylated AMP-activated protein kinase expression on lung cancer survival. Ann Oncol. 2011;23:78–85. doi: 10.1093/annonc/mdr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nanjundan M, Byers LA, Carey MS, Siwak DR, Raso MG, Diao L, Wang J, Coombes KR, Roth JA, Mills GB, Wistuba, Minna JD, Heymach JV. Proteomic profiling identifies pathways dysregulated in non-small cell lung cancer and an inverse association of AMPK and adhesion pathways with recurrence. J Thorac Oncol. 5:1894–1904. doi: 10.1097/JTO.0b013e3181f2a266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makowski L, Hayes DN. Role of LKB1 in lung cancer development. Br J Cancer. 2008;99:683–688. doi: 10.1038/sj.bjc.6604515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Launonen V. Mutations in the human LKB1/STK11 gene. Hum Mutat. 2005;26:291–297. doi: 10.1002/humu.20222. [DOI] [PubMed] [Google Scholar]
- 28.Bignell GR, Barfoot R, Seal S, Collins N, Warren W, Stratton MR. Low frequency of somatic mutations in the LKB1/Peutz-Jeghers syndrome gene in sporadic breast cancer. Cancer Res. 1998;58:1384–1386. [PubMed] [Google Scholar]
- 29.Esteller M, Avizienyte E, Corn PG, Lothe RA, Baylin SB, Aaltonen LA, Herman JG. Epigenetic inactivation of LKB1 in primary tumors associated with the Peutz-Jeghers syndrome. Oncogene. 2000;19:164–168. doi: 10.1038/sj.onc.1203227. [DOI] [PubMed] [Google Scholar]
- 30.Gill RK, Yang SH, Meerzaman D, Mechanic LE, Bowman ED, Jeon HS, Roy Chowdhuri S, Shakoori A, Dracheva T, Hong KM, Fukuoka J, Zhang JH, Harris CC, Jen J. Frequent homozygous deletion of the LKB1/STK11 gene in non-small cell lung cancer. Oncogene. 2011;30:3784–3791. doi: 10.1038/onc.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu W, Monahan KB, Pfefferle AD, Shimamura T, Sorrentino J, Chan KT, Roadcap DW, Ollila DW, Thomas NE, Castrillon DH, Miller CR, Perou CM, Wong KK, Bear JE, Sharpless NE. LKB1/STK11 inactivation leads to expansion of a prometastatic tumor subpopulation in melanoma. Cancer Cell. 2012;21:751–764. doi: 10.1016/j.ccr.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doebele RC, Lu X, Sumey C, Maxson DA, Weickhardt AJ, Oton AB, Bunn PA, Jr, Baron AE, Franklin WA, Aisner DL, Varella-Garcia M, Camidge DR. Oncogene status predicts patterns of metastatic spread in treatment-naive nonsmall cell lung cancer. Cancer. 118:4502–4511. doi: 10.1002/cncr.27409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox JD, Yesner RA. Adenocarcinoma of the lung: recent results from the Veterans Administration Lung Group. Am Rev Respir Dis. 1979;120:1025–1029. doi: 10.1164/arrd.1979.120.5.1025. [DOI] [PubMed] [Google Scholar]
- 34.Figlin RA, Piantadosi S, Feld R. Intracranial recurrence of carcinoma after complete surgical resection of stage I, II, and III non-small-cell lung cancer. N Engl J Med. 1988;318:1300–1305. doi: 10.1056/NEJM198805193182004. [DOI] [PubMed] [Google Scholar]
- 35.Shaikh TH. Oligonucleotide arrays for high-resolution analysis of copy number alteration in mental retardation/multiple congenital anomalies. Genet Med. 2007;9:617–625. doi: 10.1097/gim.0b013e318148bb81. [DOI] [PubMed] [Google Scholar]