Abstract
We developed a novel 3D immunomagnetic flow assay for the rapid detection of pathogenic bacteria in a large-volume food sample. Antibody-functionalized magnetic nanoparticle clusters (AbMNCs) were magnetically immobilized on the surfaces of a 3D-printed cylindrical microchannel. The injection of a Salmonella-spiked sample solution into the microchannel produced instant binding between the AbMNCs and the Salmonella bacteria due to their efficient collisions. Nearly perfect capture of the AbMNCs and AbMNCs-Salmonella complexes was achieved under a high flow rate by stacking permanent magnets with spacers inside the cylindrical separator to maximize the magnetic force. The concentration of the bacteria in solution was determined using ATP luminescence measurements. The detection limit was better than 10 cfu/mL, and the overall assay time, including the binding, rinsing, and detection steps for a 10 mL sample took less than 3 min. To our knowledge, the 3D immunomagnetic flow assay described here provides the fastest high-sensitivity, high-capacity method for the detection of pathogenic bacteria.
Harmful pathogenic bacteria cause a variety of diseases, including food poisoning, which poses a significant public health problem. Because microorganisms readily proliferate over time, the rapid detection of bacteria during the early stages is highly important for preventing food poisoning.1 Although cultivation techniques are widely used to detect pathogenic bacteria, they cannot provide on-site feedback because cultivation requires several days. Recently, novel detection methods based on functional magnetic nanoparticles have been used for the rapid detection of bacteria. Antibody-functionalized magnetic nanoparticles can capture, separate, and concentrate pathogenic bacteria. Captured bacteria may be detected without cultivation using a variety of analytical techniques, such as quartz crystal microbalance (QCM),2 surface plasmon resonance (SPR),3 electrochemical impedance spectroscopy (EIS),4 light absorbance5, surface-enhanced Raman scattering (SERS),6,7 and fluorescence spectroscopy.8
Previous immunoassays that have been developed to use magnetic nanoparticles have suffered from long assay times required for handling large-volume samples. Conventional immunomagnetic assays generally handle only small-volume samples (<1 mL) because the magnetic nanoparticle collection times can increase dramatically with the sample volume. In the real world, however, the size of a food sample needed for inspection should be sufficiently large to appropriately test for contamination because in most cases, only certain portions of a sample may be contaminated by pathogenic bacteria. The volume of a solution required to wash pathogenic bacteria from a sample specimen should be larger than a few tens of milliliters to suppress false negative errors. In addition to the long magnetic separation times needed for handling large volume samples, inefficient binding between the antibody-functionalized magnetic nanoparticles and the pathogenic bacteria can extend the time required for binding steps. The concentrations of magnetic nanoparticles and pathogenic bacteria in the sample solutions are generally quite low; therefore, binding between the particles and bacteria is not efficient in the bulk solution phase. Long incubation times are required to achieve conjugation. These problems have confined conventional immunomagnetic assays to laboratory experiment, and these assays have not yet been successfully implemented in on-site inspections.
In this study, we developed a novel 3D immunomagnetic flow assay for the rapid and sensitive detection of Salmonella bacteria in a large-volume sample solution. Binding between the antibody-functionalized magnetic nanoparticle clusters (AbMNCs) and Salmonella bacteria was facilitated by magnetically immobilizing the AbMNCs on the surface of a three-dimensional microchannel in a hollow cylinder. The strength of the magnetic field was maximized by stacking permanent magnets with spacers inside the cylinder, which allowed for the capture of the AbMNC-bacteria complexes and a postrinsing step under a high flow rate, and consequently reduced the assay time required to handle a large-volume sample. Combined with luminescence measurements, our method successfully detected the presence of pathogenic bacteria in 10 mL of a lettuce wash solution with a sensitivity of 10 cfu/mL (colony-forming units per mL) within 3 min (including conjugation, rinsing, and detection). To our knowledge, this is the fastest and the most sensitive rapid detection method yet reported.
EXPERIMENTAL SECTION
Materials
Iron(III) chloride hexahydrate, sodium citrate, polyacrylamide, urea, 3-amino-propyltriethoxysilane (APTES), glutaraldehyde, and Tween 20 were purchased from Aldrich (St. Louis, MO) and were used without further purification. Deionized water (18.3 MΩ. cm−1) was obtained using a reverse osmosis water purification system and was used to prepare the phosphate buffer (PB) solution. The monoclonal Salmonella antibody and goat antimouse IgG antibody were purchased from Abcam Inc. (Cambridge, MA). Disk-shaped permanent magnets were purchased from Seoul Magnetic (Seoul, Korea). The remanence of the magnets was 1.22 T, and the diameter and thickness were 3 and 0.5 cm, respectively.
Preparation of the Antibody-Immobilized Magnetic Nanoparticle Clusters (AbMNCs)
The Fe3O4 magnetic nanoparticle clusters were synthesized using a hydrothermal method9. In brief, 4 mmol of FeCl3, 12 mmol of urea, and 8 mmol of sodium citrate were sequentially added to 80 mL of DI water, followed by 0.6 mmol of polyacrylamide. After magnetic stirring for a few minutes to dissolve the polyacrylamide completely at room temperature, the solution was heated at 200 °C in a Teflon-lined autoclave for 10 h. After cooling to room temperature, the MNCs were separated and rinsed several times with water and absolute ethanol. MNCs were functionalized successively with 1% APTES in ethanol and 0.5% glutaraldehyde in water to produce amine-reactive aldehyde groups on the surfaces. A total of 10 μg of goat antimouse IgG antibody was immobilized onto the MNCs as a linker to produce oriented immobilization of the Salmonella antibody. After the addition of 1 wt % Tween 20 in PB, which prevented nonspecific binding, 10 μg of the Salmonella antibody was immobilized.
Fabrication of the HEMOS
The efficient conjugation and separation of the AbMNCs-bacteria complexes was implemented using a three-dimensional microchannel-containing hollow cylinder, the High-capacity Efficient Magnetic O-shaped Separator (HEMOS). The HEMOS device was fabricated using a stereolithography method.10 In brief, 3D CAD designs of the device were outlined using the Autodesk Inventor (San Rafael, CA), and the device was built by FineLine Prototyping, Inc. (Raleigh, North Carolina) using a 3D Systems Viper SL system (Rock Hill, SC) in high-resolution mode with a natural finish. DSM Somos Watershed XC 11122 (Heerlen, The Netherlands) was selected as the resin due to its transparency and tendency not to swell when in contact with water.
Bioluminescence Measurements Using a Portable ATP Luminometer
The concentrated AbMNCs-Salmonella complexes obtained from the HEMOS were added to 150 μL of the benzalkonium chloride solution to extract ATP from Salmonella bacteria. The ATP-extracted solution was added to lyophilized luciferin and luciferase powder, and the luminescence intensity was measured using a specially designed disposable test tube and a portable luminometer (Kikkoman PD-20). The mechanism underlying the photochemical reaction is described elsewhere.11,12 In brief, the adenylation of luciferin by ATP is catalyzed by luciferase, thereby generating an intermediate, luciferyl adenylate, which is immediately oxidized by dissolved oxygen in solution to produce excited oxyluciferin. Finally, light was emitted by the excited oxyluciferin, which then decayed to a stable state. The luminescence measurements, including the extraction and reactions, were completed within 1 min.
RESULTS AND DISCUSSION
Characterization of the MNCs and HEMOS
Figure 1a shows a scanning electron microscopy image of magnetic nanoparticle clusters (MNCs). The average size of the MNCs was approximately 150 nm, and each MNC consisted of a few hundred 15 nm Fe3O4 nanoparticles. Because the magnetic force experienced by the Fe3O4 nanoparticles was proportional to the particle volume, the MNCs were separated from the analyte solutions more effectively than were the small Fe3O4 nanoparticles. Figure 1b,c shows the 3D CAD design and photographic image of the HEMOS with stacked permanent magnets, respectively. The dimensions of the cylindrical microchannel were 500 μm in width, 3 cm in diameter, and 3.75 cm in height. Note that the female luer connectors were integrated into the design to form a tight seal and prevent leakage, which has been a common problem in conventional microfluidic devices. The luer connectors permitted convenient connections to a syringe pump or other laboratory equipment using barbed adapters.
Figure 1.
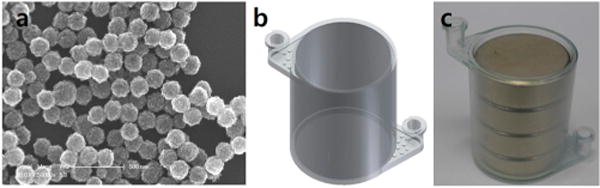
(a) SEM image of magnetic nanoparticles, (b) 3D CAD design of HEMOS, and (c) photographic image of the HEMOS containing stacked permanent magnets.
Decrease of the Linear Flow Velocity and Increase of the Magnetic Flux Density
The microchannel in the HEMOS possesses a unique cylindrical geometry that reduces the linear flow velocity, which is suitable for handling a large-volume sample solution under a high flow rate. Note that the flow rate used in the HEMOS assay was 25 mL/min and required only 24 s to handle a 10 mL sample solution. By contrast, typical flow rates in conventional microfluidic devices are <100 μL/min, and at least 100 min are required to handle the same amount of a sample solution.13,14 If the magnetic force applied to the AbMNCs or AbMNCs-Salmonella bacteria complexes was not sufficiently strong, they could be easily rinsed away from the microchannel under a high flow rate. The cylindrical shape of the microchannel surrounding the permanent magnets inside HEMOS allowed the sample solution to flow through the channel while maintaining a minimal distance from the magnet. This configuration immobilized the AbMNCs inside the microchannel under high flow rates. The magnetic force experienced by the AbMNCs is given by15
| (1) |
| (2) |
where B is magnetic flux density, μ0 is the magnetic permeability in space, H is the magnetic field strength, M is the magnetization of the material, and m is a point-like magnetic dipole moment. Because m decreases inversely with the cube of the distance from the magnet, the wall between the magnet and the microchannel was made to be as thin as possible in order to fully utilize the maximum magnetic force. The thickness of the inner wall was designed to be 500 μm, the minimum thickness for a wall to provide adequate rigidity. If the width of the microchannel were increased, the linear flow rate decreased but the reduced magnetic force due to the increased distance could not stably immobilize the AbMNCs. In addition, the increased microchannel volume degraded the binding efficiency due to the low probability of collisions, as described earlier.
This problem was addressed here by maximizing the magnetic field of the permanent magnets by stacking the magnets and inserting spacers between the magnets. Numerical simulations were performed using the finite element method (FEM) to examine the influence of the arrangement of magnets inside the HEMOS on the efficiency of magnetophoresis. The simulations were performed using the COMSOL Multiphysics 4.3 package (COMSOL Multiphysics GmbH, Berlin, Germany). The 2D plot in Figure 2 shows the calculated magnitude of the magnetic flux density in the y direction, which represented the outward radial direction in the 3D model. The MNCs were attracted along the magnetic flux density gradient, and the line indicating the region of concentrated magnetic field was called a “node”. Perfect disk magnets stacked directly on top of one another create nodes only at the bottom and top of the stack and produce only two nodes, as shown in Figure 2a. Typical cylindrical magnets with round edges in Figure 2b produce (n + 1) nodes due to the presence of tapered edges between two magnets, where n is the number of magnets included in the stack. The number of nodes increases to 2n when the round-edged magnets are stacked to include a certain gap between adjacent magnets (Figure 2c). The optimal thickness of the gap depends on the specifications of the magnet used and was calculated to be half the thickness of the magnet using FEM simulations. The gap between strong magnets may be maintained by inserting disk-shaped plastic spacers with half the thickness of the magnet between adjacent magnets. AbMNCs loaded into the HEMOS were distributed along the magnetic nodes, and a greater number of nodes enabled the AbMNCs-Salmonella bacteria complexes to adhere to the surface of the microchannel under a high flow rate. Additional simulation results for the magnetic flux density as a function of the gap distance between the magnets can be found in Figures S1 and S2 of the Supporting Information.
Figure 2.
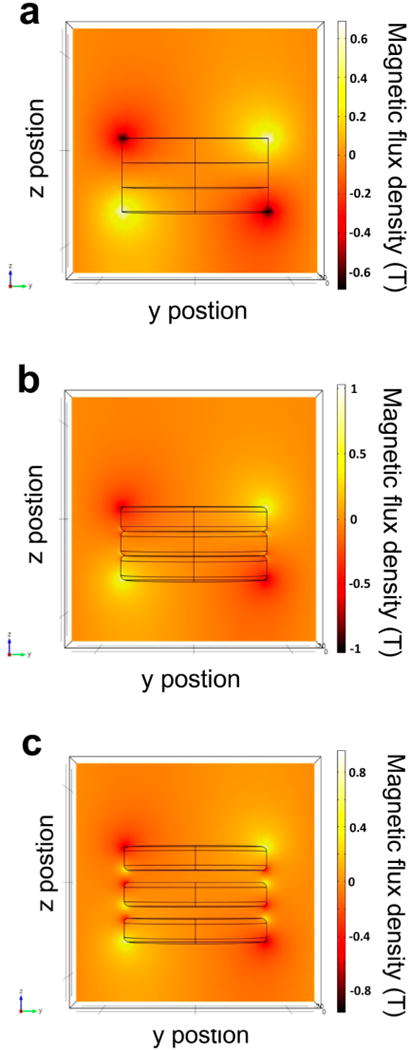
Magnitude of the magnetic flux density, depending on the stacked magnet configuration, obtained using numerical simulations. The color bar indicates the values of the y-component of the magnetic flux density. (a) Perfect disk-shaped magnets without spacers, (b) round-edged magnets without spacers, and (c) round-edged magnets separated by spacers.
MNC Trapping Performance of the HEMOS
The MNC trapping performance of the HEMOS with or without the spacers was characterized using UV–vis absorption measurements. A 100 μg/mL MNC suspension in a PB solution was prepared for this evaluation and was diluted to a concentration between 0% and 100% relative to the original concentration (i.e., the input solution). The absorbance spectra of the solutions were measured and were used as standards (black curves in Figure 3a) to examine the output sample concentration. The absorbance of the solution flowing out of the HEMOS (i.e., the output solution) without spacers was ~25% (the blue curve in Figure 3a) compared to the absorbance of the original input sample concentration, indicating that only ~75% of the MNCs were captured by the HEMOS prepared without spacers. By contrast, the absorbance of the output sample solution obtained from the HEMOS prepared with spacers was negligible (the red curve shown in Figure 3a), indicating that nearly perfect capture of MNCs was achieved in the device prepared with spacers between the magnets. Note that the magnetophoretic separation was conducted under a high flow rate (25 mL/min). Figure 3b shows optical microscopy images of the solutions before and after magnetophoretic separation using the HEMOS prepared with the magnet—spacer assembly as well as the captured solution. The color of the output solution was almost transparent compared to that of the input solution, indicating that most of the MNCs were separated. The color of the captured solution was much darker than that of the input solution, indicating that the MNCs had been concentrated.
Figure 3.
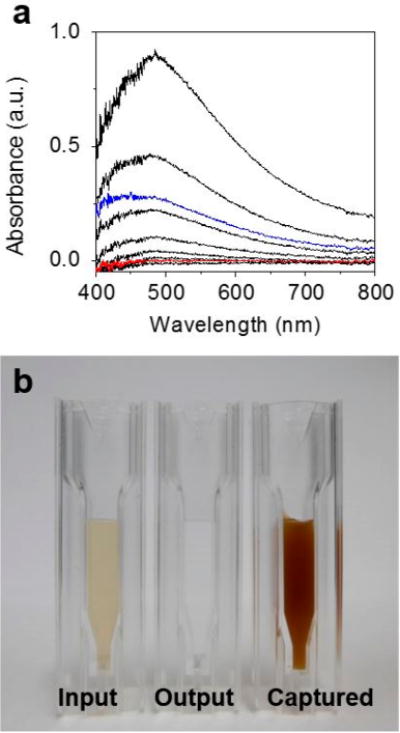
(a) UV–visible absorbance spectrum of the output MNC samples having a range of concentrations relative to the input sample concentration. The signals in black represent relative concentrations of 100%, 50%, 25%, 10%, 5%, 1%, and the baseline control, from top to bottom. The blue signal represents the output relative concentration obtained from a device prepared without spacers between the magnets, and the red signal represents the output relative concentration obtained from a device prepared with spacers. (b) Optical images of the MNC samples obtained from the input, output, and captured solutions in the HEMOS.
3D Immunomagnetic Flow Assay for the Separation of Salmonella Bacteria
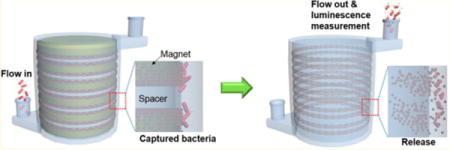
The procedure corresponding to the 3D immunomagnetic flow assay using HEMOS is illustrated in Figure 4. A volume of 2 mL of a 100 μg/mL AbMNC solution were loaded into the HEMOS prior to the bacteria capturing process, and the magnetic nanoparticles were trapped on the inner wall of the HEMOS microchannel. A sample solution spiked with Salmonella bacteria was injected into the AbMNCs-loaded HEMOS for 24 s at a flow rate of 25 mL/min. No incubation time was needed because nearly instant binding between the AbMNCs and Salmonella bacteria was achieved during injection into the HEMOS. The rapid binding was attributed to the magnetically concentrated and immobilized AbMNCs inside the narrow microchannels, which facilitated efficient collision between the Salmonella bacteria and the AbMNCs. By contrast, binding between similar concentrations of the Salmonella bacteria and AbMNCs did not occur efficiently in the bulk solution due to the low probability of collisions. Their conjugation in bulk solution generally required long incubation times over 30 min.
Figure 4.
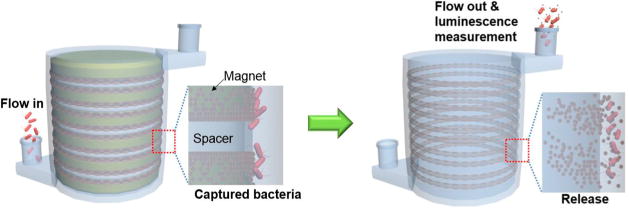
Schematic illustrations of 3D immunomagnetic flow assay. The magnet-spacer assembly was placed in the center opening of the HEMOS during the capture and rinsing of the AbMNCs-Salmonella complexes. After removing the magnet assembly from the HEMOS, the concentrated AbMNCs-Salmonella solution was collected using a disposable syringe.
After the binding between the AbMNCs and Salmonella bacteria, the microchannel was rinsed with a PB solution for 1 min. Whereas conventional assays require the collection of samples, followed by the rinsing of each bath multiple times to remove nonspecifically bound residues, the HEMOS rinsing process reached completion upon injection of a PB solution. This simple rinsing process was sufficient because the AbMNCs-Salmonella complexes were magnetically captured inside the microchannels of the HEMOS under a high flow rate. After removing the magnet assembly from the HEMOS, the solution inside the microchannel was removed using a disposable syringe with a magnet attached to its outer wall for the subsequent luminescence measurements.
Detection of Salmonella Bacteria Using a Portable ATP Luminometer
The performance of the 3D immunomagnetic flow assay was examined using various concentrations of Salmonella bacteria-spiked buffer solutions. After the separation of Salmonella bacteria from a sample solution using the HEMOS, the ATP luminescence was measured as a function of the concentration of Salmonella bacteria. Figure 5a shows that the intensity of the luminescence signal increased with the Salmonella concentration. The detection limit for the Salmonella bacteria-spiked PB solution was found to be better than 10 cfu/mL. Comparable experiments were conducted using a real food matrix (a Salmonella bacteria-spiked lettuce solution) to investigate the influence of lettuce-derived interferences on the detection sensitivity. A Salmonella bacteria-spiked lettuce solution was prepared by suspending 20 g of fresh lettuce in 50 mL of Salmonella-spiked DI water, followed by incubation at room temperature for 3 h with shaking. After incubation, 10 mL of the lettuce solution was sampled for the experiment. The color of the sample solution was light green, indicating that it contained both the Salmonella bacteria and substantial amounts of the interferences extracted from the lettuce. Although a variety of interferences were present in the lettuce solution, the luminescence intensity in Figure 5b was comparable to that obtained from buffer samples, and the detection limit remained better than 10 cfu/mL. The selectivity of the assay was examined using control experiments with a 10 mL buffer solution spiked with 105 cfu/mL Vibrio bacteria or Escherichia coli bacteria. Each solution was introduced into the HEMOS preloaded with Salmonella antibody-functionalized MNCs. The ATP luminescence intensity measured from the control experiments was lower than the noise level, as shown in Figure 5c, confirming that nonspecific binding was negligible.
Figure 5.
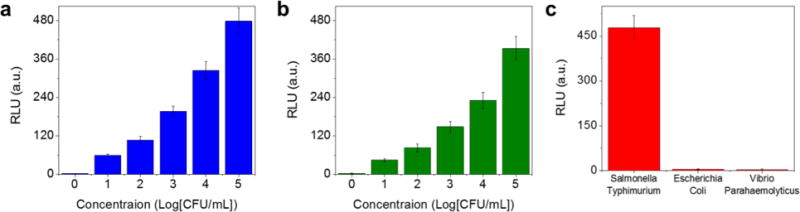
Intensities of the ATP luminescence, measured from (a) a buffer solution and (b) a lettuce solution spiked with different concentrations of Salmonella bacteria. (c) The intensities of the ATP luminescence, measured from the buffer solution spiked with different bacteria at concentrations of 105 cfu/mL. Concentrations of the control samples that are not spiked with bacteria (i.e., blank samples) were labeled “0” for convenience.
CONCLUSIONS
In summary, we developed a novel method for the rapid detection of pathogenic bacteria using an immunomagnetic assay based on a 3D-printed microfluidic device and a luminescent ATP detection kit. The detection limit of the assay was found to be better than 10 cfu/mL for a Salmonella bacteria-spiked lettuce solution. More importantly, the total assay time for the inspection of a 10 mL food sample was less than 3 min, including 24 s for the conjugation, 1 min for the rinsing, and 1 min for the detection steps. The short assay time was realized by magnetically concentrating and immobilizing AbMNCs inside the narrow microchannels, thereby facilitating efficient binding between the AbMNCs and the Salmonella bacteria and rinsing of the resulting complexes. The assay time required to inspect a larger volume of food sample solution (50 mL) was 4 min, including 2 min for the conjugation step. To our knowledge, the 3D immunomagnetic flow assay developed in this study is the fastest bacteria detection method yet reported without the loss of sensitivity and selectivity. Because the developed method can be easily extended to detect other pathogenic bacteria associated with a variety of diseases, it has great potential to contribute to ensuring public health.
Supplementary Material
Acknowledgments
This research was supported by the Public Welfare & Safety Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2012M3A2A1051679) and a grant (10162MFDS995) from the Ministry of Food and Drug Safety in 2014. W.L. acknowledges the grant from POSTECH Presidential Fellowship.
Footnotes
Supporting Information
Additional figure for simulations of the magnetic flux density as a function of the gap distance between the magnets, photographic image of the HEMOS with the magnet–spacer assembly, and experimental setup. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
W.L. and D.K. contributed equally.
Notes
The authors declare no competing financial interest.
References
- 1.Hobson NS, Tothill I, Turner APF. Biosens Bioelectron. 1996;11:455–477. doi: 10.1016/0956-5663(96)86783-2. [DOI] [PubMed] [Google Scholar]
- 2.Shen Z-Q, Wang J-F, Qiu Z-G, Jin M, Wang X-W, Chen Z-L, Li J-W, Cao F-H. Biosens Bioelectron. 2011;26:3376–3381. doi: 10.1016/j.bios.2010.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Torun Ö, Hakki Boyaci Ï, Temiir E, Tamer U. Biosens Bioelectron. 2012;37:53–60. doi: 10.1016/j.bios.2012.04.034. [DOI] [PubMed] [Google Scholar]
- 4.Liébana S, Spricigo DA, Cortés MP, Barbé J, Llagostera M, Alegret S, Pividori MI. Anal Chem. 2013;85:3079–3086. doi: 10.1021/ac3024944. [DOI] [PubMed] [Google Scholar]
- 5.Wang C, Irudayaraj J. Small. 2010;6:283–289. doi: 10.1002/smll.200901596. [DOI] [PubMed] [Google Scholar]
- 6.Guven B, Basaran-Akgul N, Temur E, Tamer U, Boyac IH. Analyst. 2011;136:740–748. doi: 10.1039/c0an00473a. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Ravindranath S, Irudayaraj J. Anal Bioanal Chem. 2011;399:1271–1278. doi: 10.1007/s00216-010-4453-6. [DOI] [PubMed] [Google Scholar]
- 8.Wen C-Y, Hu J, Zhang Z-L, Tian Z-Q, Ou G-P, Liao Y-L, Li Y, Xie M, Sun Z-Y, Pang D-W. Anal Chem. 2012;85:1223–1230. doi: 10.1021/ac303204q. [DOI] [PubMed] [Google Scholar]
- 9.Kwon D, Joo J, Lee J, Park K-H, Jeon S. Anal Chem. 2013;85:7594–7598. doi: 10.1021/ac401717f. [DOI] [PubMed] [Google Scholar]
- 10.Au AK, Lee W, Folch A. Lab Chip. 2014;14:1294–1301. doi: 10.1039/c3lc51360b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito K, Nishimura K, Murakami S, Arakawa H, Maeda M. Anal Chim Acta. 2000;421:113–120. [Google Scholar]
- 12.Qiu J, Zhou Y, Chen H, Lin J-M. Talanta. 2009;79:787–795. doi: 10.1016/j.talanta.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Li P, Kilinc D, Ran Y-F, Lee GU. Lab Chip. 2013;13:4400–4408. doi: 10.1039/c3lc50816a. [DOI] [PubMed] [Google Scholar]
- 14.Pamme N, Manz A. Anal Chem. 2004;76:72S0–72S6. doi: 10.1021/ac049183o. [DOI] [PubMed] [Google Scholar]
- 15.Pankhurst QA, Connolly J, Jones SK, Dobson JJ. Phys D: Appl Phys. 2003;36:R167. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


