Abstract
Biometric sensors and portable devices are being increasingly embedded into our everyday life, creating the need for robust physiological models that efficiently represent, analyze, and interpret the acquired signals. We propose a knowledge-driven method to represent electrodermal activity (EDA), a psychophysiological signal linked to stress, affect, and cognitive processing. We build EDA-specific dictionaries that accurately model both the slow varying tonic part and the signal fluctuations, called skin conductance responses (SCR), and use greedy sparse representation techniques to decompose the signal into a small number of atoms from the dictionary. Quantitative evaluation of our method considers signal reconstruction, compression rate, and information retrieval measures, that capture the ability of the model to incorporate the main signal characteristics, such as SCR occurrences. Compared to previous studies fitting a predetermined structure to the signal, results indicate that our approach provides benefits across all aforementioned criteria. This paper demonstrates the ability of appropriate dictionaries along with sparse decomposition methods to reliably represent EDA signals and provides a foundation for automatic measurement of SCR characteristics and the extraction of meaningful EDA features.
Index Terms: Dictionary design, electrodermal activity, (orthogonal) matching pursuit, skin conductance response, sparse representation
I. Introduction
Recent technological achievements are enabling the increasing use of wearable devices that allows the sensing of physiological signals for health [1], [2], medical [3], [4] and other [5], [6] purposes. These applications stem from the need to monitor individuals over long periods of time overcoming the limits imposed by traditional nonambulatory technology and providing new insights into diagnostic and therapeutic means [3]. They typically rely on small sensors for capturing data, portable devices for temporal storage, and the use of wireless networks for (periodic) data transfer to a database [7]. The large volume of recordings from these sensors, their use in everyday life and beyond specialized places like clinics, the potential variability in the data quality, and the limited resources of appropriately trained people for the corresponding signal analysis underscore the need of automatic ways to process them. A central goal is to derive meaningful quantitative constructs from the measured signal data. Robust representations of physiological signals should consider their characteristic structure, efficiently encode the underlying information, and provide reliable interpretation.
Electrodermal activity (EDA) is one of the most widely used psychophysiological signals. It is related to the sympathetic nerve activity through the changes in the levels of sweat in the ducts [8], [9], that result in the observed skin conductance response (SCR) fluctuations. EDA has been used in a variety of laboratory settings examining attention, memory, decision making, emotion, as well as a predictor of normal and abnormal behavior and other psychological constructs [8], [9].
Recently the focus has shifted to collecting EDA signals outside the laboratory “in the wild” [10], [11] with wearable devices that can measure continuously, privately logging and/or wirelessly streaming data [12]–[14]. This unobtrusive long-term recording of EDA results in large amounts of data that require deriving appropriate signal representation and interpretation with two separate goals. EDA models should first target efficient signal compression yielding reduced memory allocation and fast network transfer, accompanied with low reconstruction error. Second, since the availability (and analysis ability) of human experts is not always guaranteed, in the light of human–machine applications, automated interpretation of the acquired physiological patterns is essential. The reliable detection of SCRs, linked to various indexes of psychophysiological interest [8], can serve as a step toward this goal. Eventually, inference of internal states based on EDA representations could afford us more insights by comparing the parameters of the corresponding models across various conditions of emotion, arousal, and attention [15], [16].
Previous research has introduced parameterized [17] and generative causal [15], [18], [19] models that consider the characteristic structure of the signal or mimic the physiological processes of the sweat ducts. These methods tend to explain the cause and not the observed signal. They are mostly evaluated through empirical findings about hidden variables for inferring internal states [20], although several efforts have taken into account signal reconstruction criteria [15], [16].
Sparse representation techniques model a signal as a linear combination of a small number of atoms chosen from an overcomplete dictionary aiming to reveal certain structures of a signal and represent them in a compact way [21]. Since psychophysiological signals, such as EDA, show typical patterns over time, their sparse decomposition can yield accurate representations of scientific and translational value and contribute to scalable implementations (e.g., on mobile devices). Noting that the desired information contained in EDA signals is low dimensional, we introduce the use of sparsity to robustly represent them. The innovation of our approach lies in that we model the EDA signal directly by taking into account its variability through the use of overcomplete knowledge-driven dictionaries and evaluate our method explicitly through both signal reconstruction and information retrieval measures.
We propose EDA-specific dictionaries that take into account the characteristic signal structure in time. Dictionaries include tonic and phasic atoms capturing the signal levels and SCR fluctuations, respectively. We represent the characteristic SCR shape with previously used sigmoid-exponential [17] and Bateman functions [15], as well as a chi-square function, introduced in this study, and decompose EDA into a small number of tonic and phasic atoms with matching pursuit [22] and orthogonal matching pursuit [23]. Through postprocessing of the selected phasic atoms, we automatically detect the SCRs occurring in the signal. We evaluate the usefulness of our approach through goodness of fit criteria and SCR detection measures with respect to human annotated groundtruth. Due to potential portable device applications for signal storage and transmission, we further analyze the compression rate of our algorithm. Our results indicate that the proposed approach provides benefits compared to the least-squares fit method that estimates predetermined model parameters [17], with respect to reconstruction, discrimination, and efficiency metrics.
In the following, we briefly survey previous EDA models (see Section II). In Section III, we present our sparse EDA representation approach with knowledge-driven dictionaries. We next describe the data for this study (see Section IV) and the corresponding results (see Section V). Finally, we discuss our results and conclusions in Sections VI and VII, respectively.
II. Background and Related Work
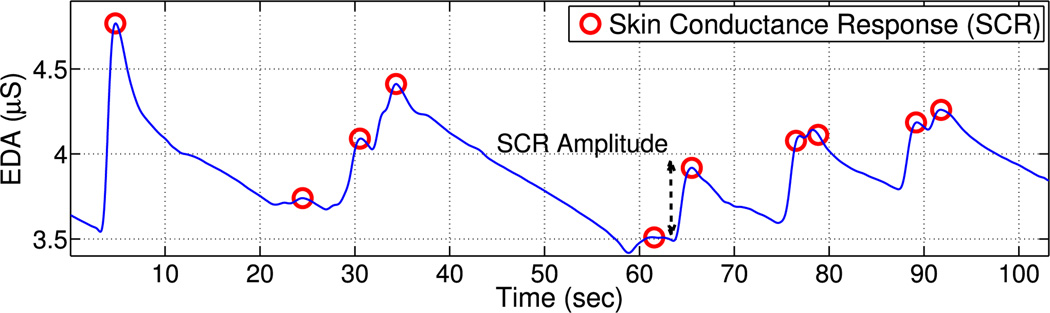
The EDA signal is decomposed into tonic and phasic components. The slow moving tonic part, called skin conductance level (SCL), depicts the general trend, whereas the fast fluctuations superimposed onto the tonic signal are the skin conductance responses (SCR) (see Fig. 1) [8]. The shape of SCRs is characterized by a steep increase in the signal and a slow recovery [24]. Amplitude is the most commonly reported SCR measure, quantified as the amount of increase in conductance from the onset of the response to its peak (see Fig. 1) with typical values ranging between 0.1 and 1.0 μS [8]. SCRs are caused by the burst of the sympathetic sudomotor nerves controlling the sweat glands linked to emotion, arousal, and attention [8]. Previous work has modeled explicitly the shape of the resulting signal with appropriate functions [17], [25] or implicitly the causal relation between the underlying activity of sudomotor nerves and the observable SCRs [15], [18], [19]. A review on EDA models can be found in [19].
Fig. 1.
Example of an electrodermal activity (EDA) signal of skin conductance responses (SCR), marked with red “o,” and an indicative notation of SCR amplitude measure.
Several studies have mathematically expressed the SCR shape. Lim et al. [17] developed a parameterized sigmoid-exponential model of EDA fitted into signal segments. Results were found to be correlated with previously established automatic scoring methods [24]. Storm et al. [25] used a quadratic polynomial fit to sequential groups of datapoints to detect SCRs, whose total number was compared to manual counting.
In the context of causal EDA modeling, Alexander et al. [18] represented the SCR shape as a biexponential function and used deterministic inverse filtering to estimate the driver of nerve bursts. Evaluation of the method was performed by visual inspection and by finding significant correlations of the resulting SCR measures with variables of gender and age. Benedek et al. [15], [26] assumed EDA to be the convolution of a driver function, reflecting the activity of sweat glands, with an impulse response depicting the states of neuron activity. This method was evaluated through the reconstruction error. It was also compared to standard peak detection for a set of noise burst events and was found to give results more likely to confound with these environmental conditions. Finally, Bach et al. [16], [19], [27], [28] have described a dynamic causal model (DCM) using Bayesian inversion to infer the underlying activity of sweat glands. Each sudomotor activity burst is modeled as a Gaussian function, which serves as an input to a double convolution operation yielding the EDA signal. The correlation of the estimated bursts with the number of SCRs from semivisual analysis was reported. This model was found to be a good predictor of anxiety in public speaking.
EDA decomposition was cast as a convex optimization problem in [29]. The minimization of a quadratic cost function was used to estimate the tonic and phasic signal components, represented with a cubic spline and a biexponential Bateman function, respectively. The EDA features extracted from the model parameters yielded statistically significant differences between neutral and high-arousal stimuli.
Despite their encouraging results, some of these research efforts [17], [25] tend to impose restrictions on the signal structure. Also, studies modeling EDA through its relationship with sympathetic arousal [15], [18], [19] assume a linear-time invariant system, which is not always justified by empirical evidence [20].While several studies take into account signal reconstruction measures [15], [16], evaluation is mostly performed by visual inspection or implicitly through expected empirical assumptions correlating the systems’ SCR measures with physical, mental, and behavioral states. The novelty of this study lies in the fact that it directly models the EDA signal with sparsity constraints and takes into account the SCR shape variability. We evaluate our approach through both signal reconstruction criteria and measures comparing automatically detected SCRs to human-annotated SCRs.
III. Proposed Approach
Many psychophysiological signals carry distinctive structures in time making the use of sparse decomposition techniques appealing. Their small number of nonzero components contain important information about the signal characteristics, which can potentially be related to various medical conditions and psychological constructs. For this reason, dictionaries have to be carefully designed so that they capture the signal variability and their underlying information. We propose the use of parameterized EDA-specific dictionaries that are able to represent the tonic and phasic parts of the signal (see Section III-A). The benefit of introducing these predetermined atoms is that we know their properties in advance, such as amplitude and rise and recovery time, which are of interest in a variety of psychophysiological studies [30]. We further decompose EDA into a small number of atoms (see Section III-B) which are used to retrieve information about its specific constructs, such as SCRs (see Section III-C). We compare the proposed parametric EDA representation to the least-means-squares parametric fit proposed by Lim et al. [17] (see Section III-D) with respect to signal reconstruction, information retrieval, and compression rate criteria (see Section III-E).
In the following, we will use x = [x(1) … x(L)]T ∈ ℜL to denote a vector of length L, x(t), t = 1, …, L, its corresponding value at the tth sample in time and its p-order norm. The inner product between two vectors x1 and x2 will be referred as x1T x2. A matrix with L1 raws and L2 columns will be written in bold capital letters as A ∈ ℜL1 × L2.
A. Dictionary Design
Since our approach involves a knowledge-based representation of signals, the choice of dictionary atoms is critical. Specifically for EDA, we introduce two kinds of atoms that model the tonic and phasic components of the signal (see Table I).
TABLE I.
Equations and Parameter Values of Dictionary Atoms Representing Tonic and Phasic EDA Components
| Atom | EDA part | Equation | Parameter Values | ||
|---|---|---|---|---|---|
| Straight Line | Tonic | gβ (t) = Δ0 + Δ · t | Δ0 ∈ {−20, −10, 1} Δ ∈ {−0.010, −0.009, …, −0.001, 0, 0.01, 0.02, …, 0.10} |
||
| Sigmoid-exponential | Phasic |
Trise ∈ {0.5, 0.8, …, 2.9} Tdecay ∈ {0.3, 0.6, …, 3} |
s ∈ {0.06, 0.07, …, 0.14} | ||
| Bateman | Phasic |
gγ(2) (t) = (e−a (st −t0) − e−b (st −t0)) u(t − t0) a < b |
a ∈ {0.2, 0.3, …, 2} b ∈ {0.4, 0.6, …, 2} |
t0 ∈ {0, 10, 20, …, 310} | |
| Chi-Square | Phasic | gγ(3) (t) = χ2 (st − t0, k)u(t − t0) | k ∈ {2.70, 2.73, …, 5.37} | ||
u(t) = 1, t ≥ 0, and u(t) = 0 otherwise, χ2 (t, k): the value of k-degrees chi-square distribution at point t.
The first group takes into account the tonic part, i.e., the slow varying signal level. It is quantified with straight lines having an offset Δ0 and a slope Δ. Tonic atoms will be referred as gβ ∈ℜL, where β = (Δ0, Δ) is an element of the set 𝔹 = ℜ2. Negative values of Δ indicate decreasing lines, while positive values represent lines with increasing slope.
The second group of atoms captures the phasic component of EDA, meaning the rapid fluctuations superimposed on the signal, also known as SCRs. We use three types of functions that can potentially represent the characteristic steep rise and slow decay of SCRs: the sigmoid-exponential, the Bateman, and the chi-square functions. The sigmoid-exponential was introduced in [17] for representing SCRs with high values of Trise and Tdecay resulting in long rise and decay time. The Bateman function [15], inspired by the laws of diffusion at the skin pores, represents the shape of an impulse response, which when convolved with a driver function results in the EDA signal. Parameters a and b control the steepness of recovery and onset, respectively; the higher they are, the less time it takes for the SCR to transition to and from a peak. Finally, the probability density function of the chi-squared distribution, referred here as chi-square function, is introduced by the authors of this paper as an alternative way to represent the SCR shape. It involves only one parameter k, with large values resulting in wider SCRs. Although no apparent physiological explanation could be found for the chi-square, each dictionary atom is encoded with one fewer parameter. Taking into account the large size of (overcomplete) dictionaries in sparse representation techniques, this function benefits computational cost and memory allocation, an important factor for mobile device applications.
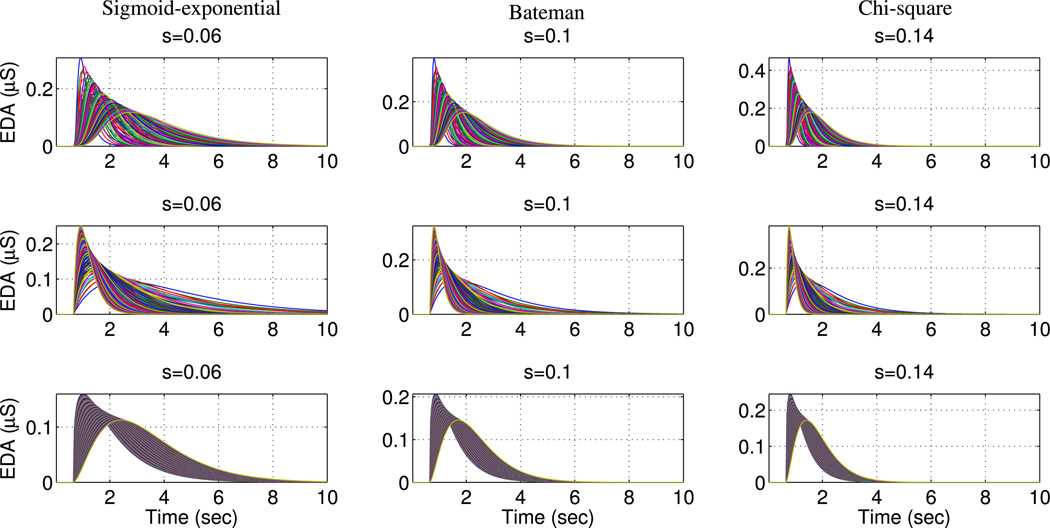
In order to ensure large variety in the dictionary atoms, we considered two additional parameters common for all phasic atom types. The time scale s is responsible for the compression and dilation of the atom in time with lower values resulting in wider SCRs and vice-versa (see Fig. 2). Since there is no a priori information about the location of SCRs within an analysis frame, the dictionary contains shifted versions of all atoms over a range of time offsets t0 spanning one signal frame.
Fig. 2.
Examples of normalized phasic atoms represented with sigmoid-exponential, Bateman and chi-square functions for different time scale parameters s = 0.06, 0.1, 0.14 and time offset t0 = 20. For each time-scale value, plots were created using all combinations of corresponding atom-specific parameters.
We will refer to the three types of sigmoid-exponential, Bateman, and chi-square SCR atoms as gγ(1) ∈ ℜL, gγ(2) ∈ ℜL, and gγ(3) ∈ ℜL, respectively, with corresponding parameters γ(1) = (Trise, Tdecay, s, T0) ∈ Γ(1), γ(2) = (a, b, s, T0) ∈ Γ(2) and γ(3) = (k, s, T0) ∈ Γ(3), where Γ(1) = ℜ+4, Γ(2) = ℜ+4, and Γ(3) = ℜ+3. By combining the possible parameters, we created all tonic and phasic atoms which are merged into three different dictionaries D(1) = {gβ(t), gγ(1) (t)}, D(2) = {gβ(t), gγ(2) (t)}, and D(3) = {gβ(t), gγ(3) (t)}, containing sigmoid-exponential, Bateman, and chi-square functions, respectively. In the case of sigmoid-exponential dictionary (see Table I), for example, there are nine possible values for scale s, 32 for time offset t0, 9 for Trise, and 10 for Tdecay, resulting in 25 920 phasic atoms. Combining the 3 and 21 different values for Δ0 and Δ, respectively, we get additionally 63 tonic atoms, that result in a dictionary size of 25 983 atoms in total. Similarly, Bateman and chi-square dictionaries contain eventually the same number of phasic and tonic atoms with the sigmoid-exponential one. All dictionary atoms are normalized for unit standard deviation. We introduced more phasic than tonic atoms in order to capture the large variety of SCR shapes compared to the signal level, which remains fairly constant through an analysis frame. The parameter values (see Table I) were empirically set so that the resulting atom shapes are similar to the observed SCRs. In the future, we plan to automatically learn the dictionary parameters for more accurate representation.
B. Sparse Decomposition
Sparse representation of a signal f ∈ ℜL can described by the following equation providing a nonconvex problem [21]:
| (1) |
where D ∈ ℜL × Q is an overcomplete dictionary matrix with Q prototype signal-atoms and c ∈ ℜQ are the atom coefficients with N ≪ Q nonzero elements.
One way to approach this NP-hard problem is to use greedy strategies, that abandon exhaustive search in favor of locally optimal updates. The most well known are MP [22] and OMP [23], [31] reaching suboptimal solutions. Another way of solving Eq. (1) is through relaxation of the discontinuous l0-norm leading to basis pursuit [32] and focal underdetermined system solver [33], [34]. Despite the fact that these relaxation approaches can obtain global solutions, they are far more complicated and less efficient in terms of running time [21], rendering MP algorithms compelling because of their simplicity and low computational cost. Theoretical guarantees of correctness [35] along with empirical experiments [36]–[38], led us to the use of MP and OMP in this paper. (O)MP will denote cases referring to both these algorithms.
Let D be a dictionary described in Section III-A containing a total of Q tonic and phasic atoms from either the sigmoid-exponential, Bateman, or chi-square functions, i.e., D = D1, or D = D2, or D = D3, and gζn an atom from the dictionary, i.e., gζn ∈ D: ζn ∈ 𝔹 ∪ Γ(1), or ζn ∈ 𝔹 ∪ Γ(2), or ζn ∈ 𝔹 ∪ Γ(3). According to MP [22], the initial signal f can be written as a linear combination of infinite atoms as follows:
| (2) |
where cn are the atom coefficients. MP selects a set of atoms from D that best matches the signal structure. If gζ0 ∈ D is the atom at the first iteration, the signal f can be written as follows:
| (3) |
where Rf is the signal residual after approximating f in the direction gζ0. The selection of gζ0 is greedily performed in order to maximize the similarity of the atom to the original signal, i.e., such that fT gζ0 is maximum. Note that in the selection criterion, we did not use the absolute value, as in the original MP algorithm. If we allowed negative inner product values, i.e., negative atom coefficients, the shape of the predefined atoms would change, resulting in a totally different signal interpretation. Negative coefficients of the SCR atoms, for example, would not have any physical meaning and would disturb the structure of the original EDA signal.
After the first iteration, residual Rf is similarly decomposed by selecting an atom from dictionary D that best matches its structure. Iteratively, the residual Rn+1 f can be written as follows:
| (4) |
where gζn ∈ D is selected to maximize (Rn f)T gζn. After N iterations, the original signal f is approximated by fN:
| (5) |
where δn ∈ {0, 1} selects either a tonic or a phasic atom (see Table I), βn ∈ 𝔹 and γn ∈ Γ(1), or γn ∈ Γ(2), or γn ∈ Γ(3) are the parameters of the nth atom.
Since processing of long duration signals is involved, we perform segmentation and frame-by-frame analysis. Let the superscript (k) denote the kth time frame of length L, where k = 1, …, K with K being the total number of frames. Therefore, the kth frame of the original signal in terms of the atoms selected by MP can be approximated by
| (6) |
We use a step of samples in order to avoid discontinuities introduced by the start and end of the signal frame. As will be described in Section III-C, SCR detection is based on the middle part of each signal frame for the same reason. A similar approach has been followed with other biosignals [39].
OMP [23], [31], [35], [40] is a refinement of MP which adds a least-squares minimization to obtain the best approximation over the atoms that have already been chosen at each iteration. Let DSN = [gζ0, …, gζN−1] ∈ ℜL × N be the matrix containing the selected atoms after N iterations. The least-squares approximation fN of signal f is
| (7) |
where the term in brackets corresponds to the N nonzero coefficients used to scale the selected atoms. OMP updates the residual by projecting the original signal onto the linear subspace spanned by the selected atoms; therefore, it never selects the same atom twice. Similar expressions hold for the signal of the kth time frame.
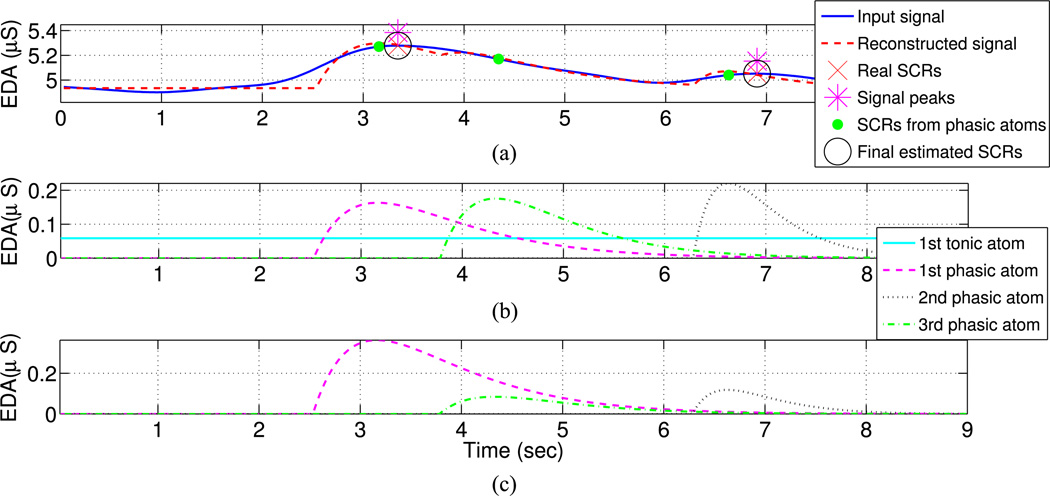
A schematic representation of the way OMP sparsely decomposes a signal frame is presented in Fig. 3. The original and reconstructed signals are shown with solid and dashed lines [see Fig. 3(a)]. OMP first selects a straight line atom in order to represent the signal level and then two phasic atoms centered around the 100th and 220th sample to capture the two SCRs. The original normalized atoms as well as the resulting phasic atoms when scaled with the coefficients computed by OMP are shown in Fig. 3(b) and (c). We also notice that the phasic atom centered around sample 100 has the highest energy [see Fig. 3(c)], since it represents the wider SCR of the frame. Finally, the third selected phasic atom is centered toward the end of the first annotated SCR around sample 140. This happens because the first phasic atom did not capture the entire energy of the corresponding SCR; therefore, another atom with much smaller coefficient was introduced. In Section III-C, we will describe how we combine these two phasic atoms located at samples 100 and 140 in order to reliably represent an SCR.
Fig. 3.
EDA representation scheme and SCR detection for an example signal frame. (a) Input and reconstructed signal with solid blue and dashed red lines. The location of expert hand-annotated SCRs is marked (red “×”), along with the signal peaks (magenta “*”) and the SCRs estimated based on the phasic atoms of the sparse decomposition algorithm (green “•”). The final SCRs (black “○”) are located by combining the SCRs from the phasic atoms according to their location and mapping them to the signal peaks. (b) The normalized dictionary atoms selected by the first four iterations of OMP. The first tonic atom (solid cyan line) captures the signal level, the first and third phasic atoms (dashed magenta and dash-dotted green lines) the first SCR, while the second phasic atom (black dotted line) the second SCR. (c) The normalized phasic atoms multiplied by the corresponding OMP coefficients (dashed magenta, dotted black, and dash-dotted green lines). The energy of each atom is indicative of the order they have been selected by OMP with higher energy atoms selected first.
C. SCR Detection
Let be the phasic atoms selected after N (O)MP iterations at the kth signal frame with M < N, as at least one tonic atom will compensate for the signal level. Since the dictionary is predetermined, we know exactly the location of the maximum amplitudes of the phasic atoms, denoted as . As shown in Fig. 3 for atoms centered around samples 100 and 140, more than one phasic atoms might be selected to represent a single SCR because the fit between dictionary atoms and signal is not always perfect. Taking advantage of their time proximity, we can group phasic atoms according to their location with a histogram of the values with Nb bins. Each nonempty histogram bin h will contain all phasic atoms capturing one SCR, which is represented by the linear combination of the grouped phasic atoms:
| (8) |
The weights correspond to the atom coefficients computed by (O)MP. Because of the simple shape of the above SCR signal, we are able to find its amplitude and time of peak by simply locating its maximum point. SCRs with amplitude less than a predetermined threshold athr were omitted [8], in accordance to the manual annotation (see Section IV-B).
To estimate the final SCR locations for the whole signal, we merged the SCRs from all analysis frames. Since the starting and ending samples of each frame are more prone to errors due to discontinuity issues, we only take into account SCRs located in the middle samples of each frame. The value was chosen to coincide with the length of the analysis step in order to ensure that each SCR is taken into account once. For the first and last frame we also consider SCRs from the first and last samples, respectively. A signal frame of 10 s (320 samples), for example, will result in analysis step of approximately 3.3 s (107 samples), in which the shape of an SCR is easily identifiable, as shown in Fig. 3.
Since our approach involves local decisions made on each signal frame, a postprocessing step that takes into account the global EDA shape is necessary. Simple peak detection was performed and the located SCRs were then mapped to the nearest detected peak within a distance threshold dthr. SCRs not paired with a neighboring peak were omitted and those matched to the same peak were only counted once. In Fig. 3, for example, the phasic atoms centered around samples 100 and 140 were grouped together based on the proximity of their location and were further mapped to the peak of sample 110 to represent the first SCR. In order to capture the second SCR, the phasic atom centered around sample 220 was mapped to the peak from sample 230.
D. Description of the Least-Squares Fit Model for EDA
We compare our approach against the least-squares fit EDA representation model by Lim et al. [17], since it is conceptually the closest to our method involving a parameterization of the signal. The authors proposed a eight-parameter model that incorporates the signal tonic level, at most two SCRs and a recovery phase. SCL is represented by a straight line with offset c. SCRs are captured by sigmoid-exponential functions with common rise time tr and decay time td. They are located at time offsets Tos1, Tos2, and have amplitudes g1, g2, respectively. Finally, the recovery phase is assumed to follow the exponential decay of SCRs with the same time constant decay td and an initial amplitude a0. Therefore, the eight-parameter model representing an EDA signal frame can be written as follows:
| (9) |
| (10) |
The nonlinear least-squares fit [41] was used to estimate the model parameters from the data. Time offset and amplitude of SCRs were initialized from peak detection. If less than two peaks were detected in a signal frame, only the corresponding parameters were estimated and the remaining were set to zero.
E. Evaluation
Evaluation of the proposed model is performed with respect to the quality of signal reconstruction, the ability to capture EDA-specific characteristics and the compression rate.
1) Signal Reconstruction
Signal reconstruction is evaluated with the root mean square (RMS) error, defined as follows:
| (11) |
where f(k) (l) denotes the value of the lth sample at the kth signal frame with k = 1, …, K and l = 1, …, L; similarly for the reconstructed signal with N (O)MP iterations. Low RMS error values reflect high-quality signal representation.
2) Information Retrieval
Evaluation with RMS error provides a limited view of the representation quality, since an overly complex model may result in perfect representation of the existing data, but might perform poorly on unseen patterns [42]. One technique to avoid overfitting with greedy algorithms, such as (O)MP, is early stopping, that might result in a better estimate of the original signal without introducing too many erroneous or noisy entries [43], [44]. Indeed, our results (see Section VA2) suggest that 4–7 (O)MP iterations provide the most reliable EDA representation. An alternative way is the use of quantitative and qualitative criteria, depending on the application that assess the amount of meaningful information captured by the proposed models [45], [46]. In this study, we measure the ability of our approach to detect SCRs, an important EDA component [8], [9]. SCR detection is assessed in two ways, each containing complementary accuracy metrics.
We first quantify the quality of SCR representation based on the amount of detected SCRs and their distance from the human annotated reference ones. Let Sr = {pri} and Sd = {pdi} be the sets of locations of real and detected SCRs, respectively. The absolute relative difference between the number of real and detected SCRs captures the portion of hand annotated SCRs that is correctly located by our system:
| (12) |
where | · | denotes the cardinality of a set. In order to see how many of the detected SCRs are correct, we compute the mean distance of each detected SCR from the closest real one
| (13) |
We further calculate precision and recall measures. If Srd = {pdi : ∃j s.t. minprj |pdi − prj| < dmax} are the estimated SCRs whose distance to their closest real SCR is less than a maximum threshold dmax, precision equals to the portion of detected SCRs that are close to a real one within the distance boundary dmax: Precision = |Srd|/|Sd|. Recall captures the fraction of hand-annotated SCRs that are detected from our system within the same boundary: Recall = |Srd|/|Sr|.
F-score is the harmonic mean of precision and recall, providing a single measure from these complementary ones:
| (14) |
These goodness of detection measures improve as the distance boundary dmax increases, since evaluation becomes more forgiving to errors. Note that the distance boundary dmax used for evaluation of SCR detection is different from the distance threshold dthr for mapping SCRs detected from the phasic atoms to the signal peaks (see Section III-C).
One can observe that precision is related to the MinDistmetric quantifying how close an estimated SCR is to a real one. On the other hand, recall is closer to RDiff that captures the algorithm’s ability to reliably detect the hand-annotated SCRs.
SCR detection results (see Section V-A2) are reported with a tenfold cross validation by randomly splitting the data from the 37 subjects. At each fold, the training set was used to find the optimal combination of dthr and Nb (see Section III-C) from a pool of parameters dthr ∈ {32, 64, … 320 samples} and N ∈ {2, 3, 4, 5 bins}, selected to maximize the Fscore of the corresponding data, since this yields a combined measure of the amount of missed and erroneously detected SCRs. The average value of each metric on the test set is reported based on the optimal parameters of each fold, as found from the training set. This reduces the risk of overfitting and is more indicative of the performance of our algorithm on novel data.
3) Compression Rate
Compression rate is evaluated as the number of bits used for representing the EDA signal over 1 s with low values indicating better compression ability.
IV. Data Description
A. Data Collection
Our data contain 10-min recordings of 37 children watching a preferred television show in order to sustain attention and keep the subject stationary. Two silver-silver chloride electrodes were placed on the index and middle finger of the child’s nondominant hand. These were connected to the BIOPACMP150 system that recorded EDA at a sampling rate of 32 Hz. Details about the experiment can be found in [47].
B. Preprocessing and Annotation
High-frequency noise artifacts were removed with a low-pass Blackman filter [48] of 32 samples (1 s).
Annotation of SCRs was performed by an expert using the visualization capabilities of BIOPAC’s AcqKnowledge software. SCR scoring involved visual inspection of the denoised EDA signal within a 30-s time frame and detection of signal “peaks” with the typical steep rise and slow decay. Each SCR amplitude was visually determined (see Fig. 1) and a minimum threshold of 0.05 μS was set for an SCR to be eligible for annotation [8]. In order to ensure reliable annotations, 25% of the data were double coded, yielding interannotator agreement of 96%.
V. Experiments
We evaluate the performance of our method based on the collected and annotated data (see Section IV) and compare it against the least-squares fit approach (see Section V-A). Results concern the mean of each evaluation metric over all subjects. We further discuss how the analysis frame length and the SCR detection parameters affect our results (see Sections V-B and V-C).
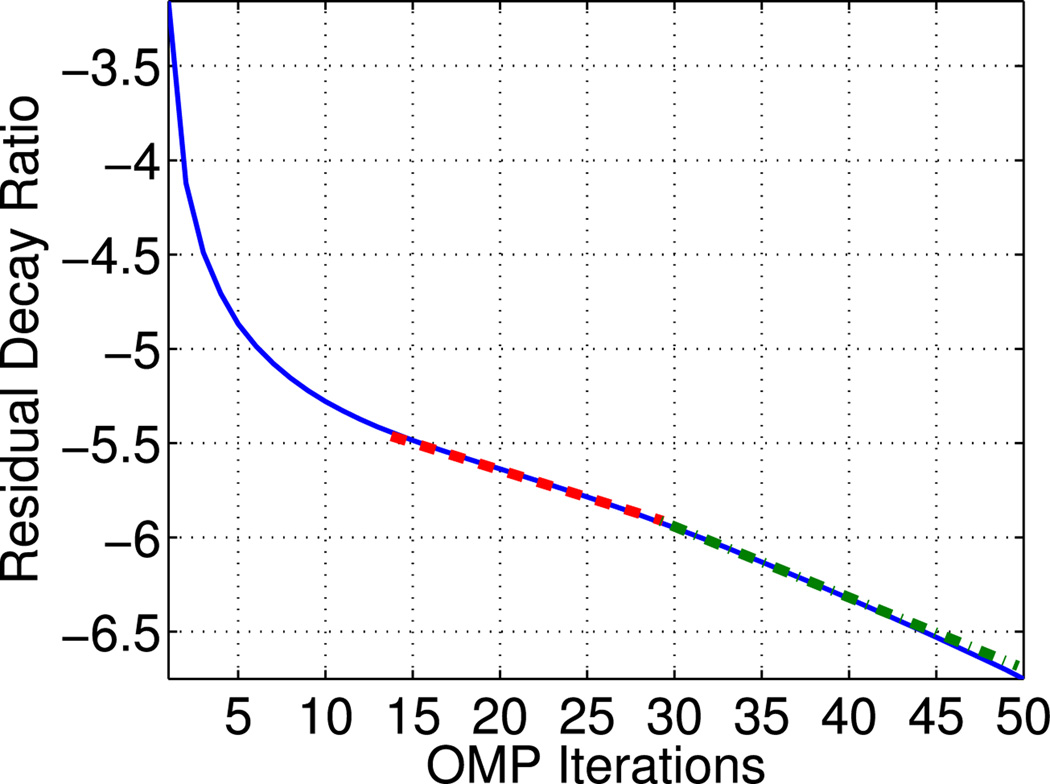
In the following experiments, the maximum number of (O)MP iterations was determined so that the coherent signal structures, i.e., the nonzero signal components, are likely to be recovered. As explained in [22], this occurs when the residual decay ratio r = 2 · log10(‖Rn f‖2/‖f‖2) decays at a constant rate. Our data suggest that this occurs after 15 OMP iterations with a dictionary of Bateman phasic atoms, while after 30 iterations decay happens at an even steeper constant slope (see Fig. 4). Similar results are obtained from other types of dictionaries, omitted for the sake of simplicity. For this reason, we examine 1–15 (O)MP iterations and in fact, our experimental results indicate that even a smaller number can achieve reliable representation (see Section V-A).
Fig. 4.
Logarithmic ratio between the second-order norms of the residual Rn f and original signal f, r = 2 · log10 (‖Rn f ‖2/‖f‖2), computed for 1–50 orthogonal matching pursuit (OMP) iterations based on a dictionary of tonic and Bateman phasic atoms. Red-dashed line denotes a decay of approximately constant rate between 15–30 iterations. Green dashed-dotted line denotes a steeper constant decay between 30–50 OMP iterations.
A. EDA Representation Results
For consistency with [17], in which 10-s isolated EDA segments were evaluated, results in this section are reported and compared to [17] with the same analysis frame.
1) Signal Reconstruction
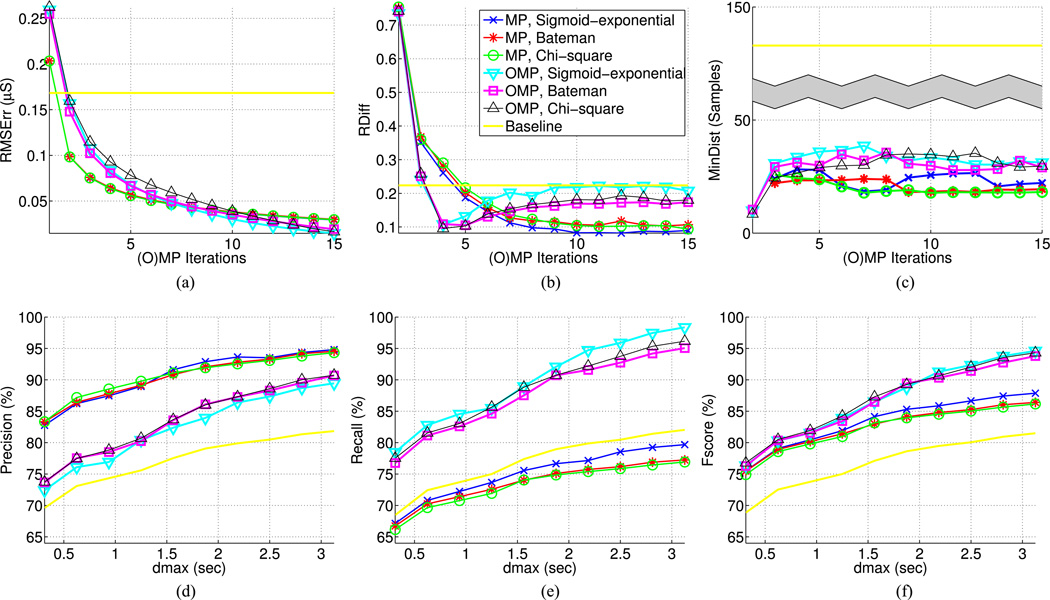
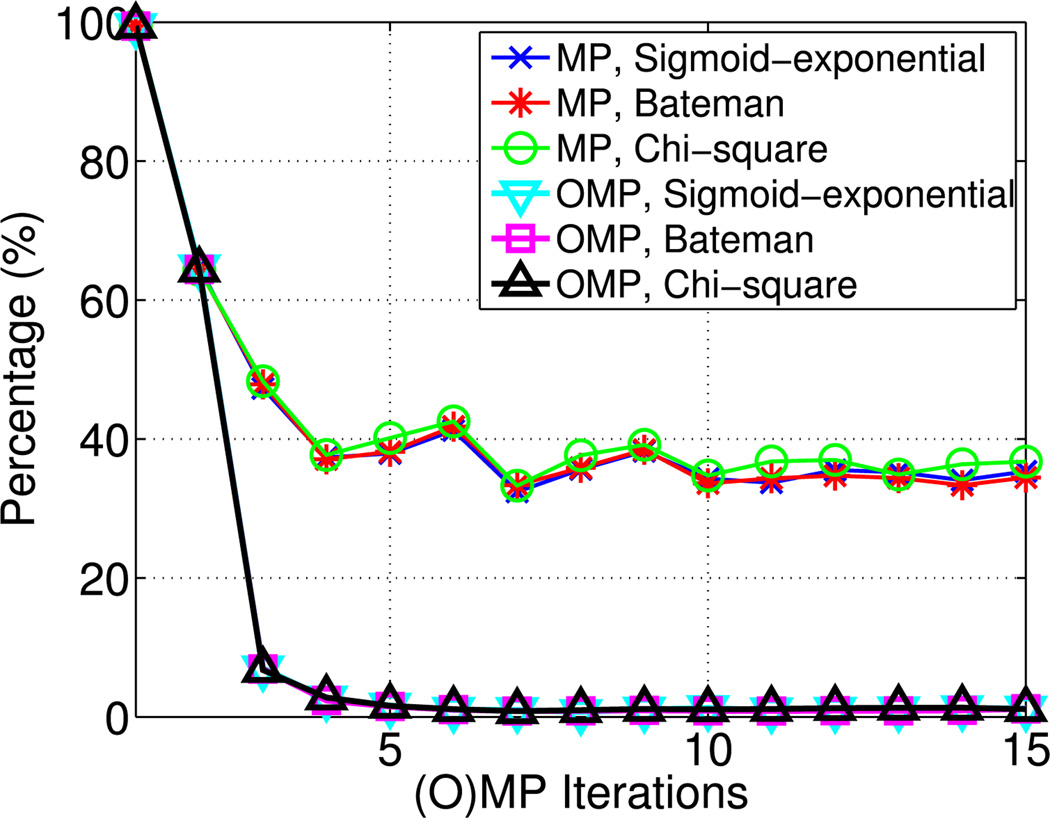
During the first iteration, MP yields better signal reconstruction than OMP [see Fig. 5(a)]. Since MP does not reestimate the atom coefficients, it selects more tonic atoms in order to adjust signal levels after the addition of new phasic ones. On the contrary, OMP readjusts the levels of the already selected tonic atoms. It is also apparent from Fig. 6 that tonic atoms are mostly selected by OMP during the first two iterations, whereas this occurs very often in MP. Therefore, during the first iteration, MP tends to capture part of the signal energy that is more attributed to the signal level and less to the signal fluctuations, resulting in lower RMS values. As more iterations occur, OMP achieves lower RMS indicating its ability to more reliably capture the signal variability.
Fig. 5.
Signal reconstruction and SCR detection results. (a) Root mean square (RMS) error between original and reconstructed signal with respect to (w.r.t.) the number of (orthogonal) matching pursuit ((O)MP) iterations. (b) Absolute number of relative difference between real and estimated SCRs w.r.t. the number of (O)MP iterations. (c) Mean distance of estimated SCRs from their closest real SCR w.r.t. (O)MP iterations. (d), (e), (f) Precision, recall, and Fscore of SCR detection with 6 (O)MP iterations w.r.t maximum distance threshold dmax between real and detected SCRs, the latter ranging between 10–100 samples, or 0.3125–3.125 s. Results on Fig. (b)– (f) are reported based on a tenfold cross validation, during which SCR detection on the test set is performed using the parameter combination (dthr, Nb) that gave the best results on the training data, where dthr is the distance threshold for mapping estimated SCRs to the nearest signal peaks and Nb is the number of histograms bins for grouping the selected phasic atoms of each analysis frame. Same legend applies to all plots.
Fig. 6.
Percentage of selected tonic atoms with respect to the number of iterations for (orthogonal) matching pursuit ((O)MP) and the various dictionaries (with sigmoid-exponential, Bateman and chi-square phasic atoms).
2) Information Retrieval
Since OMP introduces more phasic atoms than MP for a given iteration, the absolute relative difference between real and estimated SCRs RDiff is lower and becomes optimal around 4–7 iterations [see Fig. 5(b)]. This can be also justified by the fact that in a 10 s frame, we have on average 1–3 tonic and 2–4 phasic atoms, the latter capturing the 1–3 expected SCRs. To achieve similar RDiff, MP needs more iterations due to the tonic atoms it keeps selecting.
Because MP selects less phasic atoms than OMP, it tends to capture the most distinct SCRs, resulting in smaller distances MinDist from the estimated to the real SCRs [see Fig. 5(c)] and higher precision [see Fig. 5(d)]. For the same reason MP shows poorer recall [see Fig. 5(e)], as it misses many SCRs. Compared to the least-squares fit approach, Fscore is higher for both MP and OMP [see Fig. 5(f)], indicating the ability of our system to reliably detect SCRs.
3) Compression Rate
In a 10 s segment, we compute the number of encoding bits for the raw signal, the least-squares fit and the proposed models. Long integers and single-precision binary floating-point numbers of 32 bits are assumed.
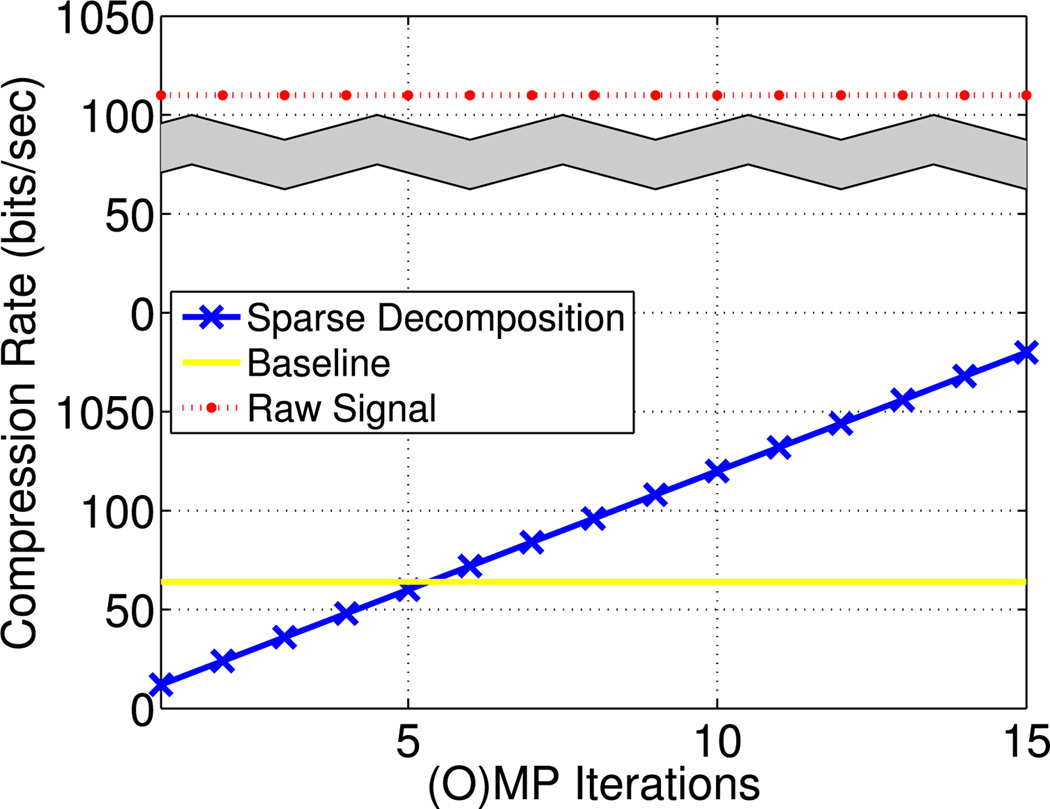
Since the original signal was recorded with 32 Hz, a 10 s signal segment is represented with 320 samples (1280 Bytes); therefore, its compression rate is CRraw = 1024 bits/s.
Lim et al. [17] use a 8-parameter model to represent a 10 s signal. Assuming that the time offset parameters Tos1, Tos2 are long integers and the remaining parameters are floating points, we need 32 Bytes, resulting in CRb = 25.6 bits/s.
In our approach, given a known dictionary, each atom is represented by its indexed location in the dictionary and the corresponding coefficient. A 16 bit integer can encode the location of the 25 983 atoms used in our experiments. Assuming that each atom coefficient is a single-precision floating point, N selected atoms can be encoded by (16·N + 32·N) bits, which is equivalent to compression rate CRp = 4.8N bits/s.
The proposed model achieves compression rate between 14.2 to 213 times upon the original signal depending on the number of (O)MP iterations (see Fig. 7). Compared to the least-squares approach, signal decomposition with 2–5 atoms also yields lower compression rate (see Fig. 7) and lower RMS error [see Fig. 5(a)].
Fig. 7.
Compression rate of the original EDA signal and the EDA representation with the proposed sparse decomposition and the least-squares fit methods. (Y-axis break between 100 and 900 bits/s).
B. Effect of Analysis Frame Length
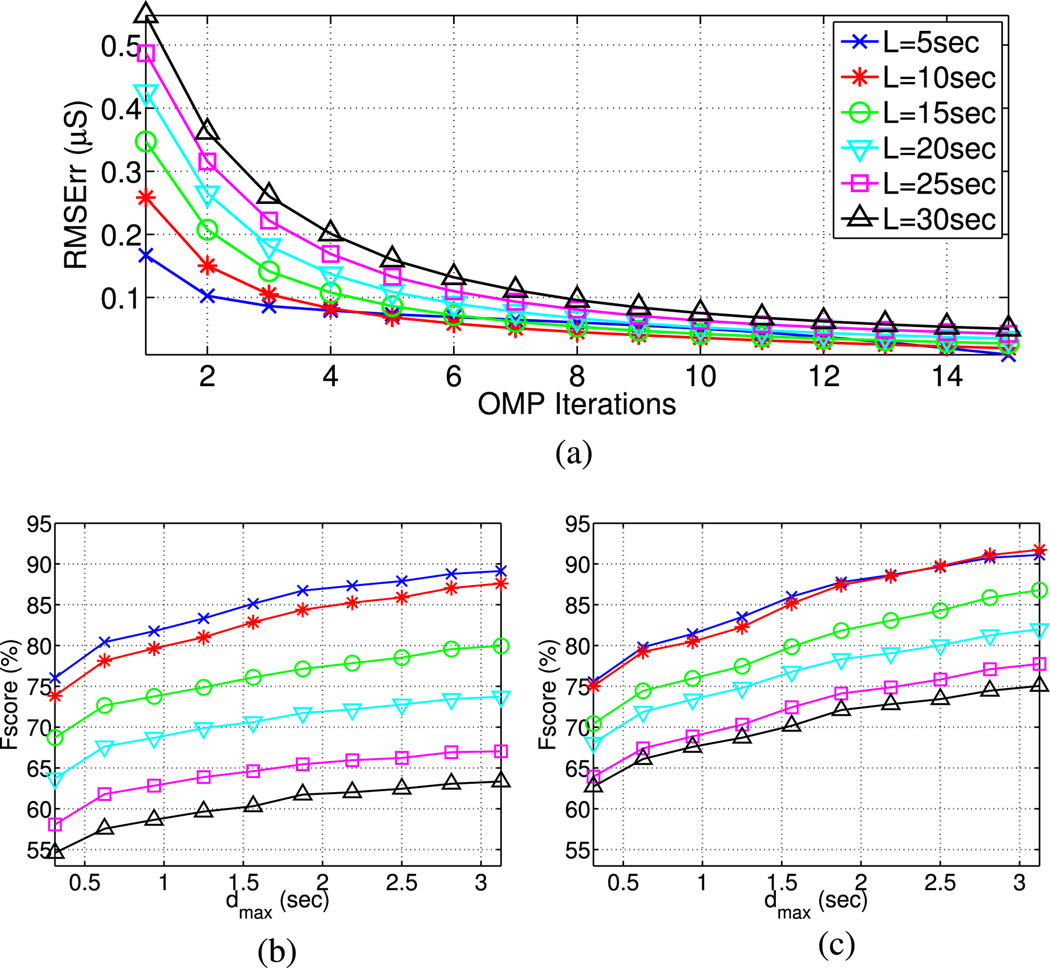
The different time scales over which tonic and phasic EDA components coevolve, the first being much slower, create the need of analyzing multiple window lengths. Since the tonic component of the signal lies in the frequency range between 0–0.05 Hz [49], the upper limit corresponding to 20 s, we examined EDA representation metrics of variable frame lengths between 5–30 s. For the sake of simplicity, our experiments for this section are performed with OMP based on Bateman phasic atoms, but no significant differences occur from other combinations.
Representation results for the various analysis windows are greatly influenced by the number of (O)MP iterations. Signal reconstruction based on a few selected atoms is better for shorter than longer analysis frames, while this difference converges as the number of OMP iterations increases [see Fig. 8(a)]. Similar patterns occur with SCR detection Fscore, although the short-term nature of the phasic part makes short analysis frames more compelling with respect to that [see Fig. 8(b) and (c)].
Fig. 8.
Effect of the analysis frame length L (in seconds) to signal reconstruction and SCR detection. (a) Root mean square (RMS) error between original and reconstructed signal for 1–15 orthogonal matching pursuit (OMP) iterations. (b) and (c) Fscore of SCR detection with 4 and 10 OMP iterations. Same legend applies to both plots.
C. Effect of SCR Detection Parameters
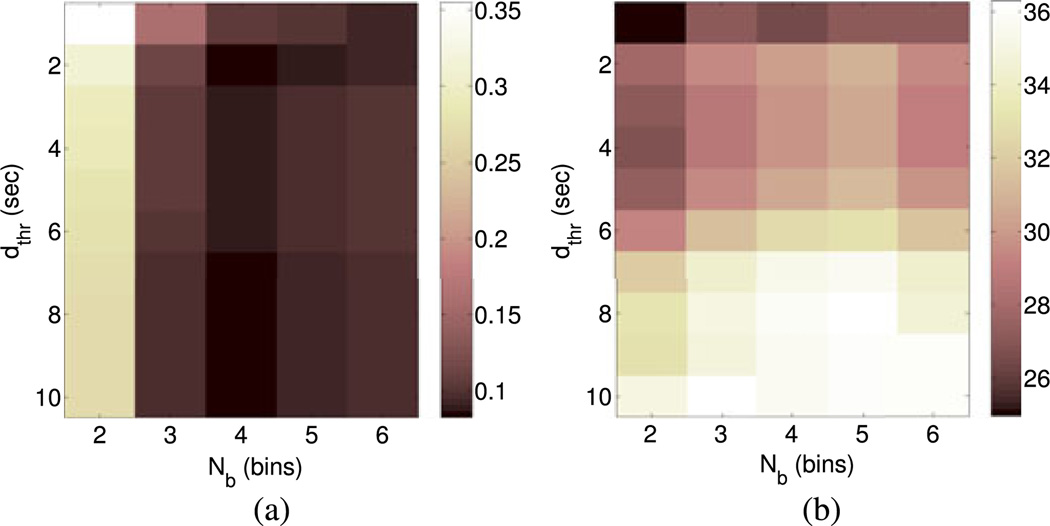
We analyze SCR detection with respect to the distance threshold dthr used for mapping the estimated SCRs to the nearest signal peaks and with respect to the number of histograms bins Nb for grouping the selected phasic atoms of each analysis frame. As given in Section V-B, we report results using OMP with Bateman atoms.
When changing dthr, the absolute relative difference RDiff between the number of real and detected SCRs remains almost unaltered [see Fig. 9(a)]. This is not the case for their minimum average distance MinDist, which decreases as dthr becomes larger [see Fig. 9(b)], suggesting that more information about signal peaks benefits SCR detection.
Fig. 9.
SCR detection metrics with respect to the distance threshold dthr for mapping estimated SCRs to signal peaks and the number of histogram bins Nb for grouping the selected phasic atoms from sparse decomposition. (a) Absolute relative difference RDiff of real and detected SCRs. (b) Mean distance MinDist of detected SCRs to the closest real one.
The number of histogram binsNb strongly affects the absolute relative difference RDiff between real and estimated SCRs, since the latter directly depends on the number of groups in which the phasic atoms are combined. In contrast, metric MinDist is not largely affected by this parameter.
VI. Discussion
Our method involves the design of EDA-specific dictionaries that take into account the tonic and phasic parts of the signal and the use of sparse decomposition techniques to represent the EDA. Compared to previous EDA models, our approach incorporates the idea of sparsity and is quantitatively evaluated directly on the signal as well as its human annotated characteristics. We examined three different types of phasic atoms of similar nature. These provided equivalent results in terms of signal reconstruction and information retrieval metrics (see Fig. 5); therefore, no definite conclusions can be drawn with respect to the most suitable dictionary for representing EDA among these variants. A larger amount of data from variable sources could potentially address this further in the future.
Sparsity is important for representing biomedical signals, especially for efficient storage and transmission in ambulatory (mobile device based) applications. Parameterization of EDA in light of compression was considered by Lim et al. [17], who used a eight-parameter sigmoid exponential model. This effort has several constraints, as it assumes the existence of at most two SCRs within 10 s and is tested on predefined signal segments. In real-world applications, however, the shape of the signal is more unpredictable and can be better captured by the variability of the proposed EDA-specific dictionaries. Sparsity has also been examined by the DCM approach [16], that computes the onset and amplitude of spontaneous fluctuations every 2 s, resulting in 10 parameters for every 10 s. In our sparse EDA model, each atom is represented by its location in the dictionary and the coefficient to which it is multiplied. This results in 2N parameters for a 10 s segment, where N is the number of (O)MP iterations. Taking into account that 5 (O)MP iterations give adequately reliable results (see Fig. 5), the two algorithms are fairly equivalent with respect to compression rate. Sparse representations can be further used to represent other psychophysiological signals with characteristic structure in time, such as electroencephalogram (ECG), electrocardiogram (ECG), and photoplethysmograph (PPG).
Our approach offers also advantages in terms of quantitative evaluation criteria. Most previous studies were implicitly validated with statistical analysis of the resulting parameters. Lim et al. [17] visualized an example residual error and reported residuals below 5% of the signal amplitude, while Bach et al. [19] plotted an original example and reconstructed signal with no further quantitative results. Bach et al. [16] reported the negative log-likelihood of the mean sum of squares of the residual, referred as Log Bayes Factor (LBF), with DCM reaching LBF values around −3 × 106. In their nonnegative deconvolution approach, Benedek et al. [15] examined signal reconstruction with average RMS error of 0.019 μS. In our study, OMP running for 15 iterations achieves mean RMS error of 0.015, 0.019, 0.017 μS for dictionaries containing sigmoid-exponential, Bateman, and chi-square phasic atoms, respectively. Although 15 iterations are not indicative of the representation quality of our algorithm, a smaller number of iterations appears more suitable for the inherent low-dimensionality of EDA (see Fig. 5), they give the least compressed representation in our setup and are more comparable with [15]. Despite the fact that comparisons are performed on different data and in the context of slightly different approaches, these results indicate a promising framework for our method. Since dictionaries were empirically designed, our future work will examine dictionary learning techniques for data-specific dictionaries that can improve signal reconstruction measures.
The high correlation between SCRs and sympathetic nerve activity bursts and their extensive use in various psychophysiological studies [8] suggest that reliable SCR detection mechanisms are necessary. It is therefore essential to validate automatic scoring systems with human-annotated ground truth, since the increasing use of longitudinal data renders manual annotation techniques laborious and time consuming. To the best of our knowledge, only Storm et al. [25] compared the number of automatically detected and manually annotated SCRs with linear regression analysis. For 0.05 μS minimum SCR amplitude, the magnitude of linear regression ranged between 0.75 and 0.98 depending on the model parameters and the data. Although it is useful to compare the total number of real and estimated SCRs, information about their relative location is also important. One of the novelties of this paper lies in the use of SCR detection criteria assessing both the sensitivity and specificity of automatic SCR scoring. These include two complementary pair measures: 1) the relative absolute difference between the number of real and detected SCRs, the average distance of detected SCRs to their closest real one and 2) the precision and recall of SCR detection, yielding the Fscore. Fscore is 83.43% using six OMP iterations and a tolerance threshold of 1.25 s between real and estimated SCRs [see Fig. 5(f)]. This improves upon [17], which achieves 75.1% Fscore for the same conditions, and suggests that our approach appears promising for an automatic system.
Continuous advances in wearable technology suggest that physiological models should go beyond traditional signal reconstruction and pattern detection to signal interpretation in the context of internal human state tracking that could lead to individualized assessment and intervention [3], [7]. Previous efforts have provided meaningful interpretations of EDA models in terms empirical expectations (e.g., public versus nonpublic speaking [19], baseline versus anticipation [19], different noise stimuli [15], neutral, negative and positive arousing images [16]). Other studies have analyzed EDA patterns in the context of human–computer interaction relatively to individual’s affective states [6], [50]. The parameterized nature of the dictionary atoms in our approach can capture SCR characteristics, such as width, rise, and recovery time, which could further lead to more intuitive interpretations of EDA signals in terms of cognitive, social, and emotional stimuli. Data that allow these types of comparisons, including in the context of clinical translation such as therapy for Autism [51], will be explored as part of our future work.
A common problem when measuring changes in EDA is overlapping SCRs. It is typically agreed that when a second response occurs before completion of the first, one would count two overlapping responses [30]. Such an example is shown in Fig. 3(a) with two overlapping SCRs at samples 110 and 230, which we notice that our sparse decomposition approach is capable of discriminating. Previous studies have also assessed this by considering the underlying processes that contribute to SCRs as a linear time invariant system and measuring the event onsets of the system generator [27], [28].
VII. Conclusion
We propose a knowledge-driven EDA model that sparsely decomposes the signal into a small number of atoms from a dictionary. We focus on the dictionary design, which contains tonic and phasic atoms relevant to the signal levels and SCR fluctuations, respectively. With appropriate postprocessing, we automatically retrieve the SCRs from the EDA. Evaluation of our approach includes signal reconstruction, SCR detection, and compression rate analysis. These measures are improved upon the least-square fit model, that uses a parameterized template, providing benefits in terms of all criteria.
Acknowledgments
This work was supported by National Science Foundation, National Institute of Health; National Institute of Dental and Craniofacial Research (1R34DE022263-01, ClinicalTrials.gov identifier: NCT02077985), the Eunice Kennedy Shriver Institute of Child Health and Human Development of the National Institutes of Health (T32HD064578), and a California Foundation of Occupational Therapy Research Grant.
Biographies

Theodora Chaspari (S’12) received the Diploma degree in electrical and computer engineering from the National Technical University of Athens, Athina, Greece, in 2010 and the M.S. degree from the University of Southern California, Los Angeles, CA, USA, 2012, where she is currently working toward the Ph.D. degree.
Since 2010, she has been a Member of the Signal Analysis and Interpretation Laboratory. Her research interests include the area of biomedical signal processing, speech analysis, and behavioral signal processing.
Ms. Chaspari received the USC Annenberg Graduate Fellowship in 2010– 2012 and the IEEE Signal Processing Society Travel Grant in 2014.

Andreas Tsiartas (S’10–M’14) was born in Nicosia, Cyprus, in 1981. He received the B.Sc. degree in electronics and computer engineering from the Technical University of Crete, Crete, Greece, in 2006, and the M.Sc. and Ph.D. degrees from the Department of Electrical Engineering, University of Southern California, Los Angeles, CA, USA, in 2012 and 2014, respectively.
He is currently a Postdoctoral Fellow at the Stanford Research Institute, Menlo Park, CA, USA. His research interests include speech-to-speech translation, acoustic and language modeling for automatic speech recognition, voice activity detection and cognitive state representation through multimodal signals.
Dr. Tsiartas received the USC Viterbi School Deans Doctoral Fellowship in 2006 and the USC Department of Electrical Engineering best teaching assistant awards for the years 2009 and 2010.

Leah I. Stein received the B.S. degree in neuroscience and behavioral biology from Emory University, Atlanta, GA, USA, in 2004, and the M.A. and Ph.D. degrees in occupational science and occupational therapy from the University of Southern California, Los Angeles, CA, USA, in 2006 and 2013, respectively.
She is currently a Postdoctoral Fellow at the Mrs. T.H. Chan Division of Occupational Science and Occupational Therapy at the University of Southern California. Her research interests include autism, pediatric oncology, the use of wearable sensors to collect physiological data, and the impact of environmental modifications to reduce distress in children during healthcare procedures.

Sharon A. Cermak received the B.S. degree in occupational therapy from Ohio State University, Columbus, OH, USA, and the M.S. degree and Ed.D. degrees from Boston University, Boston, MA, USA, in occupational therapy and special education, respectively.
She is currently a Professor of occupational science and occupational therapy at the Mrs. TH Chan Division of Occupational Science and Occupational Therapy, the University of Southern California (USC), Los Angeles, CA, USA, and is a Professor of Pediatrics at the Keck School of Medicine at USC. Her research focuses on autism spectrum disorders, dyspraxia, and sensory processing. He is PI on a grant from the National Institute of Dental and Craniofacial Research to adapt sensory environments in the dental clinic to enhance oral care for children with ASD.
Dr. Cermak is a Charter Member of the AOTF Academy of Research Academy and a Fulbright Scholar. She is an internationally renowned Scholar, Researcher, and Clinician with more than 170 publications.

Shrikanth S. Narayanan (S’88–M’95–SM’02–F’09) received his M.S., Engineer, and Ph.D., degrees in electrical engineering, from UCLA in 1990, 1992, and 1995, respectively. From 1995–2000 he was with AT&T Labs-Research, Florham Park and AT&T Bell Labs, Murray Hill–first as a Senior Member and later as a Principal member of its Technical Staff. He is currently a Professor of engineering at the University of Southern California, Los Angeles, CA, USA, where he directs the Signal Analysis and Interpretation Laboratory. He holds appointments as a Professor of Electrical Engineering, Computer Science, Linguistics, and Psychology, and as the Founding Director of the Ming Hsieh Institute, Ming Hsieh Department of Electrical Engineering, USC, Los Angeles, CA, USA. He has published more than 600 papers and has been granted 16 U.S. patents. His research interests include human-centered signal and information processing and systems modeling with an interdisciplinary emphasis on speech, audio, language, multimodal and biomedical problems, and applications with direct societal relevance.
Dr. Narayanan is a Fellow of the Acoustical Society of America and the American Association for the Advancement of Science and a member of Tau Beta Pi, Phi Kappa Phi, and Eta Kappa Nu. He is also an Editor for the Computer Speech and Language Journal and an Associate Editor for the IEEE TRANSACTIONS ON AFFECTIVE COMPUTING, APSIPA Transactions on Signal and Information Processing, and the Journal of the Acoustical Society of America. He was also previously an Associate Editor of the IEEE TRANSACTIONS OF SPEECH AND AUDIO PROCESSING (2000–2004), IEEESIGNAL PROCESSINGMAGAZINE (2005– 2008) and the IEEE TRANSACTIONS ONMULTIMEDIA (2008–2011). He received of a number of honors, including Best Transactions Paper awards from the IEEE Signal Processing Society in 2005 (with A. Potamianos) and 2009 (with C. M. Lee) and selection as an IEEE Signal Processing Society Distinguished Lecturer for 2010–2011. Papers coauthored with his students have won awards at Interspeech 2014 Cognitive and Physical Load Challenge, Interspeech 2013 Social Signal Challenge, Interspeech 2012 Speaker Trait Challenge, Interspeech 2011 Speaker State Challenge, Interspeech 2013 and 2010, Interspeech 2009 Emotion Challenge, IEEE DCOSS 2009, IEEE MMSP 2007, IEEE MMSP 2006, ICASSP 2005, and ICSLP 2002.
Footnotes
Color versions of one or more of the figures in this paper are available online at http://ieeexplore.ieee.org.
Contributor Information
Theodora Chaspari, Email: chaspari@usc.edu, Signal Analysis and Interpretation Laboratory, Ming Hsieh Department of Electrical Engineering, University of Southern California, Los Angeles, CA 90089 USA.
Andreas Tsiartas, Stanford Research Institute.
Leah I. Stein, Division of Occupational Science and Occupational Therapy, Herman Ostrow School of Dentistry, University of Southern California
Sharon A. Cermak, Division of Occupational Science and Occupational Therapy, Herman Ostrow School of Dentistry, University of Southern California
Shrikanth S. Narayanan, Signal Analysis and Interpretation Laboratory, Ming Hsieh Department of Electrical Engineering, University of southern California.
References
- 1.Milenković A, et al. Wireless sensor networks for personal health monitoring: Issues and an implementation. Comput. Commun. 2006;29(13):2521–2533. [Google Scholar]
- 2.Mitra U, et al. KNOWME: A case study in wireless body area sensor network design. IEEE Commun. Mag. 2012 May;50(5):116–125. [Google Scholar]
- 3.Binkley P. Predicting the potential of wearable technology. IEEE Eng. Med. Biol. Mag. 2003 May-Jun;22(3):23–27. [Google Scholar]
- 4.Vuorela T, et al. Design and implementation of a portable long-term physiological signal recorder. IEEE Trans. Inform. Technol. Biomed. 2010 May;14(3):718–725. doi: 10.1109/TITB.2010.2042606. [DOI] [PubMed] [Google Scholar]
- 5.Coyne J, et al. Applying real time physiological measures of cognitive load to improve training. Proc. 5th Int. Conf. Foundations Augmented Cognition. Neuroergonomics Operational Neurosci. 2009:469–478. [Google Scholar]
- 6.Leite I, et al. Sensors in the wild: exploring electrodermal activity in child-robot interaction. Proc. ACM/IEEE Int. Conf. Human-Robot Interaction. 2013:41–48. [Google Scholar]
- 7.Bonato P. Wearable sensors/systems and their impact on biomedical engineering: An overview from the guest editor. IEEE Eng. Med. Biol. Mag. 2003;22(3):18–20. doi: 10.1109/memb.2003.1213622. [DOI] [PubMed] [Google Scholar]
- 8.Dawson M, et al. The electrodermal system. In: Cacioppo J, Tassinary L, Berntson G, editors. Handbook of Psychophysiology. 3rd ed. New York, NY, USA: Cambridge Univ. Press; 2007. pp. 159–181. [Google Scholar]
- 9.Boucsein W. Electrodermal Activity. New York, NY, USA: Springer; 2012. [Google Scholar]
- 10.Picard R. Future affective technology for autism and emotion communication. Phil. Trans. R. Soc. 2009;364:3575–3584. doi: 10.1098/rstb.2009.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Picard R. Measuring affect in the wild. Proc. 4th Int. Conf. Affective Comput. Intelligent Interaction. 2011:3. [Google Scholar]
- 12.Poh M, et al. A wearable sensor for unobtrusive, long-term assessment of electrodermal activity. IEEE Trans. Biomed. Eng. 2010 May;57(5):1243–1252. doi: 10.1109/TBME.2009.2038487. [DOI] [PubMed] [Google Scholar]
- 13.Burns A, et al. SHIMMERTM —Awireless sensor platform for noninvasive biomedical research. IEEE Sensors J. 2010 Sep.10(9):1527–1534. [Google Scholar]
- 14.Lee Y, et al. Wearable EDA sensor gloves using conducting fabric and embedded system. Proc. World Congr. Med. Phys. Biomed. Eng. 2006:883–888. doi: 10.1109/IEMBS.2006.260947. [DOI] [PubMed] [Google Scholar]
- 15.Benedek M, Kaernbach C. Decomposition of skin conductance data by means of nonnegative deconvolution. Psychophysiol. 2010;47(4):647–658. doi: 10.1111/j.1469-8986.2009.00972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bach D, et al. An improved algorithm for model-based analysis of evoked skin conductance responses. Biol. Psychol. 2013;94(3):490–497. doi: 10.1016/j.biopsycho.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim C, et al. Decomposing skin conductance into tonic and phasic components. Int. J. Psychophysiol. 1997;25(2):97–109. doi: 10.1016/s0167-8760(96)00713-1. [DOI] [PubMed] [Google Scholar]
- 18.Alexander D, et al. Separating individual skin conductance responses in a short interstimulus-interval paradigm. J. Neurosci. Methods. 2005;146(1):116–123. doi: 10.1016/j.jneumeth.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Bach D, et al. Dynamic causal modeling of spontaneous fluctuations in skin conductance. Psychophysiology. 2011;48(2):252–257. doi: 10.1111/j.1469-8986.2010.01052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bach D, Friston K. Model–based analysis of skin conductance responses: Towards causal models in psychophysiology. Psychophysiology. 2013;50(1):15–22. doi: 10.1111/j.1469-8986.2012.01483.x. [DOI] [PubMed] [Google Scholar]
- 21.Elad M. Sparse and Redundant Representations: From Theory to Applications in Signal and Image Processing. New York, NY, USA: Springer; 2010. [Google Scholar]
- 22.Mallat S, Zhang Z. Matching pursuits with time-frequency dictionaries. IEEE Trans. Signal Process. 1993 Dec.41(12):3397–3415. [Google Scholar]
- 23.Pati Y, et al. Orthogonal matching pursuit: Recursive function approximation with applications to wavelet decomposition. Proc. 27th Asilomar Conf. Signals, Syst. Comput. 1993:40–44. [Google Scholar]
- 24.Boucsein W. Electrodermal Activity. New York, NY, USA: Plenum Univ. Press; 1992. [Google Scholar]
- 25.Storm H, et al. The development of a software program for analyzing spontaneous and externally elicited skin conductance changes in infants and adults. Clin. Neurophysiol. 2000;111(10):1889–1898. doi: 10.1016/s1388-2457(00)00421-1. [DOI] [PubMed] [Google Scholar]
- 26.Benedek M, Kaernbach C. A continuous measure of phasic electrodermal activity. J. Neurosci. Methods. 2010;190(1):80–91. doi: 10.1016/j.jneumeth.2010.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bach D, et al. Dynamic causal modelling of anticipatory skin conductance responses. Biol. Psychol. 2010;85:163–170. doi: 10.1016/j.biopsycho.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bach D, et al. Modelling event-related skin conductance responses. Int. J. Psychophysiol. 2010;75:349–356. doi: 10.1016/j.ijpsycho.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greco A, et al. Electrodermal activity processing: A convex optimization approach. Proc. Annu. Int. Conf. IEEE Med. Biol. Soc. 2014:2290–2293. doi: 10.1109/EMBC.2014.6944077. [DOI] [PubMed] [Google Scholar]
- 30.Boucsein W, et al. Committee report: Publication recommendations for electrodermal measurements. Psychophysiology. 2012;49:1017–1034. doi: 10.1111/j.1469-8986.2012.01384.x. [DOI] [PubMed] [Google Scholar]
- 31.Davis G, et al. Adaptive greedy approximations. Constructive Approximation. 1997;13(1):57–98. [Google Scholar]
- 32.Chen S, et al. Atomic decomposition by basis pursuit. SIAM J. Sci. Comput. 1998;20(1):33–61. [Google Scholar]
- 33.Rao B, Kreutz-Delgado K. An affine scaling methodology for best basis selection. IEEE Trans. Signal Process. 1999 Jan.47(1):187–200. [Google Scholar]
- 34.Rao B, et al. Subset selection in noise based on diversity measure minimization. IEEE Trans. Signal Process. 2003 Mar.51(3):760–770. [Google Scholar]
- 35.Tropp J. Greed is good: Algorithmic results for sparse approximation. IEEE Trans. Inform. Theory. 2004 Oct.50(10):2231–2242. [Google Scholar]
- 36.Tan Q, et al. Improved simultaneous matching pursuit for multi-lead ECG data compression. Proc. Int. Conf. Measuring Technol. Mechatronics Autom. 2010;2:438–441. [Google Scholar]
- 37.Polania L, et al. Compressed sensing based method for ECG compression. Proc. IEEE Int. Conf. Acoust., Speech Signal Process. 2011:761–764. [Google Scholar]
- 38.Polania L, et al. Exploiting prior knowledge in compressed sensing wireless ECG systems. IEEE J. Biomed. Health Informat. 2014 May; doi: 10.1109/JBHI.2014.2325017. [DOI] [PubMed] [Google Scholar]
- 39.Zhu X, et al. Real-time signal estimation from modified short-time Fourier transform magnitude spectra. IEEE Trans. Audio, Speech, Language Process. 2007 Jul.15(5):1645–1653. [Google Scholar]
- 40.Cai T, Wang L. Orthogonal matching pursuit for sparse signal recovery with noise. IEEE Trans. Inform. Theory. 2011 Jul.57(7):4680–4688. [Google Scholar]
- 41.Marquardt D. An algorithm for least-squares estimation of nonlinear parameters. J. Soc. Ind. Appl. Math. 1963;11(2):431–441. [Google Scholar]
- 42.Duda R, et al. Pattern Classification. 2nd ed. New York, NY, USA: Wiley; 2000. Multiple discriminant analysis. [Google Scholar]
- 43.Fletcher A, Rangan S. Orthogonal matching pursuit from noisy random measurements: A new analysis. Adv. Neural Inform. Process. Syst. 2009:540–548. [Google Scholar]
- 44.Fletcher A, Rangan S. Orthogonal matching pursuit: A Brownian motion analysis. IEEE Trans. Signal Process. 2012 Mar.60(3):1010–1021. [Google Scholar]
- 45.Lustig M, et al. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magnetic Resonance Med. 2007;58(6):1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 46.Pichevar R, et al. Auditory-inspired sparse representation of audio signals. Speech Commun. 2011;53(5):643–657. [Google Scholar]
- 47.Stein L. Ph.D. dissertation. Los Angeles, CA, USA: Univ. of Southern California; 2013. Oral care and sensory sensitivities in children with autism spectrum disorders. [Google Scholar]
- 48.Oppenheim A, et al. Discrete-Time Signal Processing. Upper Saddle River, NJ, USA: Prentice-Hall; 1999. [Google Scholar]
- 49.Ishchenko A, Shev’ev P. Automated complex for multiparameter analysis of the galvanic skin response signal. Biomed. Eng. 1989;23(3):113–117. [Google Scholar]
- 50.Leijdekkers R, et al. CaptureMyEmotion: A mobile app to improve emotion learning for autistic children using sensors; Proc. Int. Symp. Comput.-Based Med. Syst; 2013. pp. 381–384. [Google Scholar]
- 51.Chaspari T, et al. A non-homogeneous Poisson process model of skin conductance responses integrated with observed regulatory behaviors for Autism intervention; Proc. IEEE Int. Conf. Acoust., Speech Signal Process; 2014. pp. 1611–1615. [Google Scholar]