Abstract
Aberrant protein folding and assembly contribute to a number of diseases, and efforts to rationalize how pathogenic mutations cause this phenomenon represent an important imperative in biochemical research. However, for α-helical membrane proteins, this task is complicated by the fact that membrane proteins require intricate machinery to achieve structural and functional maturity under cellular conditions. In this work, we utilized the ΔG predictor algorithm (www.dgpred.cbr.su.se) to survey 470 known pathogenic mutations occurring in five misfolding-prone α-helical membrane proteins for their predicted effects on the translocon-mediated membrane integration of transmembrane helices, a critical step in biosynthesis and folding of nascent membrane proteins. The results suggest that about 10 % of these mutations are likely to have adverse effects on the topogenesis of nascent membrane proteins. These results suggest that the misfolding of a modest but nonetheless significant subset of pathogenic variants may begin at the translocon. Potential implications for therapeutic design and personalized medicine are discussed.
Keywords: Misfolding disease, Rhodopsin, Peripheral myelin protein 22, Cystic fibrosis transmembrane regulator, Vasopressin V2 receptor, KCNQ1
Introduction
Improper folding and assembly of nascent proteins have been implicated in the molecular basis of a growing number of diseases. Efforts to elucidate the various mechanisms by which pathogenic mutations alter protein conformation and function or prompt the formation of cytotoxic protein aggregates address one of the central questions in biomedical research. However, a precise and comprehensive understanding of the effects of pathogenic mutations often remains elusive (Kelly and Balch 2006), and the delineation of the effects of these mutations on protein biosynthesis, folding, assembly, and function represents a daunting challenge. This is especially true for pathogenic mutations occurring within α-helical membrane proteins, which require elaborate cellular machinery to achieve and maintain their functional conformations within chemically diverse cellular membranes (Sanders and Myers 2004).
There are numerous ways by which pathogenic mutations can compromise the ability of α-helical membrane proteins to acquire their native, functional structure. The difficulty in assessing such mechanisms is highlighted by recent work showing that a single pathogenic variant of a sodium channel that misfolds under certain conditions is capable of folding into a constitutively active channel under other conditions (Cestèle et al. 2013). Furthermore, considering the genotype-specific efficacy exhibited by many therapeutic compounds (Okiyoneda et al. 2013; Rowe and Verkman 2013; Van Goor et al. 2009), efforts to differentiate the specific effects of each mutation may be of great importance for personalized medicine. In this work, we explore one potential mechanism by which pathogenic mutations may compromise membrane protein bioassembly, namely, the potential disruption of the cotranslational membrane integration of nascent membrane proteins.
Nascent membrane proteins must traverse a perilous pathway to achieve functional maturity within the cell. For the majority of eukaryotic membrane proteins, this pathway begins with cotranslational membrane integration of the nascent transmembrane (TM) helices into the endoplasmic reticulum (ER) membrane by the Sec 61 translocon, a protein-conducting channel in the ER membrane that extends the ribosomal exit tunnel into the lumen (Denks et al. 2014; Egea and Stroud 2010; Van den Berg et al. 2004). During translation, the nascent chain transiently samples the ER membrane through the lateral gate of the translocon (Egea and Stroud 2010; Heinrich et al. 2000; Hessa et al. 2005), which allows TM helices to partition into the membrane and to establish their initial topology (topogenesis). This topogenic process, which requires the recognition of tens of thousands of different TM helices with diverse sequences, presents an unenviable challenge to the translocon. The amino acid sequences of many TM helices exhibit favorable membrane partitioning through the burial of hydrophobic residues within the membrane core and/or interaction of positively charged residues with negatively charged phospholipid head groups (Hessa et al. 2005, 2007; Öjemalm et al. 2011; von Heijne 1992; White and von Heijne 2008). However, many TM helices contain functionally essential charged or polar residues buried within the membrane core (Adamian and Liang 2002; Illergard et al. 2011; Popot and Engelman 2000), which pose a challenge to this topogenic process. In some cases, the energetic penalties associated with the partitioning of these residues into the membrane may be offset by the formation of tertiary contacts with neighboring residues or TM helices (Meindl-Beinker et al. 2006; White and von Heijne 2008). In other cases, these polar helices are not integrated into the membrane in the nascent form of the protein (Gafvelin and von Heijne 1994; Kanki et al. 2002; Lu et al. 2000), and instead require subsequent topological rearrangement to achieve the native fold (Kauko et al. 2010; Virkki et al. 2014). However, these topological decisions are not always cut-and-dried; the nascent forms of some multi-pass α-helical membrane proteins have heterogeneous topologies (Moss et al. 1998). Thus, only a fraction of some nascent membrane proteins may achieve the correct topology during topogenesis (von Heijne 2006). While aberrant topomers appear to be capable of acquiring the correct topology in some cases (Kanki et al. 2002; Kanner et al. 2002; Lu et al. 2000), this process appears to be quite slow (Lu et al. 2000) and likely competes with the recognition and degradation of topological intermediates by cellular quality control (Buck and Skach 2005). The nature of this process suggests that pathogenic mutations may be capable of tipping the energetic balances involved in topogenesis. However, given that there are a multitude of potential mechanisms by which membrane protein biosynthesis may be disrupted, it is currently unclear just how common the disruption of topogenesis is as a pathogenic mechanism.
To assess whether pathogenic mutations are likely to commonly influence topogenesis, we carried out a limited bioinformatic analysis of known pathogenic mutations in α-helical membrane proteins implicated in misfolding diseases using the ΔG predictor algorithm (www.dgpred.cbr.su.se) (Hessa et al. 2007). From amino acid sequence, this knowledge-based algorithm can be used to predict the topological decisions of the translocon as well as the degree of membrane partitioning of isolated TM segments with considerable reliability (Hessa et al. 2007; Kauko et al. 2010; Virkki et al. 2014). We utilized this algorithm to survey the effects of 470 known pathogenic missense mutations occurring within or near TM helices on the predicted membrane partitioning of nascent TM helices. By comparing predictions for wild-type and mutant sequences, we find that about 10 % of these mutations are predicted to significantly disfavor integration of their host TM helices into the membrane by the translocon. In more rare cases, the results suggest that certain pathogenic mutations may also influence the position of the initially inserted segment with respect to the membrane, which could potentially influence the kinetics and thermodynamics associated with the post-translocon formation of tertiary and quaternary structure. These results may inform efforts to interpret the effects of pathogenic mutations in α-helical membrane proteins, and suggest that the ΔG prediction server (www.dgpred.cbr.su.se) may help to differentiate pathogenic mutations that primarily influence early folding events in membrane protein biogenesis from those that may instead influence post-translational folding events or the function of the mature protein. Potential implications of such findings for therapeutic approaches are also discussed.
Methods
Topological Predictions of Pathogenic Variants of α-Helical Membrane Proteins
Pathogenic mutations in rhodopsin, vasopressin 2 receptor (V2R), the cystic fibrosis transmembrane conductance regulator (CFTR), the voltage-gated potassium channel KCNQ1, and peripheral myelin protein 22 (PMP22) were gathered from the Human Gene Mutation Database (HGMD, www.hgmd.cf.ac.uk). Predictions of the energetics of translocon-mediated membrane integration of wild-type and mutant TM helices were acquired from full amino acid sequence scans with the ΔG prediction server v1.0 (www.dgpred.cbr.su.se). In two instances (TM7 of rhodopsin and the S4 helix of the voltage sensor domain of KCNQ1), the TM helices were not identified in full-sequence scans. In these cases, the identity of the TM helices was inferred from available structural data for these proteins. Based on the crystal structure of bovine rhodopsin (PDB code 1U19), TM7 was judged to correspond to residues 285–309. Based on solution NMR characterization of the voltage sensor domain of KCNQ1 (Peng et al. 2014), the S4 helix of KCNQ1 was judged to correspond to residues 221–243. ΔGapp,pred and ΔΔGapp,pred values for the wild-type and mutant variants of these helices were calculated using single-helix ΔG predictions with the ΔG prediction server v1.0. The fraction of integrated TM helices was determined from ΔGapp,pred values at a temperature of 30 °C, which is the temperature at which the data used to train the ΔG prediction server were acquired (Hessa et al. 2005, 2007).
Results and Discussion
Selection of Pathogenic Mutations in Misfolding-Prone α-Helical Membrane Proteins
To survey the influence of pathogenic mutations on the energetics of translocon-mediated membrane integration of TM helices, we first selected five multi-pass α-helical membrane proteins for which certain pathogenic mutations are known to prompt protein misfolding and misassembly. We chose to analyze pathogenic mutations in the G protein-coupled receptors (GPCRs) rhodopsin and the vasopressin 2 receptor (V2R), as well as the cystic fibrosis transmembrane conductance regulator (CFTR) chloride channel, the voltage-gated potassium channel KCNQ1, and the tetraspan protein known as peripheral myelin protein 22 (PMP22). As logged in the Human Gene Mutation Database (HGMD, www.hgmd.cf.ac.uk), pathogenic missense mutations in these proteins cause a variety of diseases including retinitis pigmentosa (RP, 118 mutations in rhodopsin), nephrogenic diabetes insipidus (NDI, 129 mutations in V2R), cystic fibrosis (CF, 864 mutations in CFTR), long QT syndrome (296 mutations in KCNQ1), and Charcot–Marie–Tooth disease (CMT, 44 mutations in PMP22). In some cases, the pathogenesis arising from these mutations may be strictly due to the effects of the mutations on the native function of the mature protein. However, biochemical characterizations have suggested that some, possibly most, pathogenic mutations within each of these proteins result in improper folding and assembly as their fundamental defect (Hwa et al. 1997; Kaushal and Khorana 1994; Morello et al. 2000; Myers et al. 2008; Sanders et al. 2001; Sung et al. 1991; Welsh and Smith, 1993; Wilson et al. 2005). The structural and functional diversity of these proteins and the widely distinct pathologies that arise from these mutations together suggest that the selected set of mutations represents reasonable cross section of pathogenic missense mutations occurring within the TM helices α-helical membrane proteins.
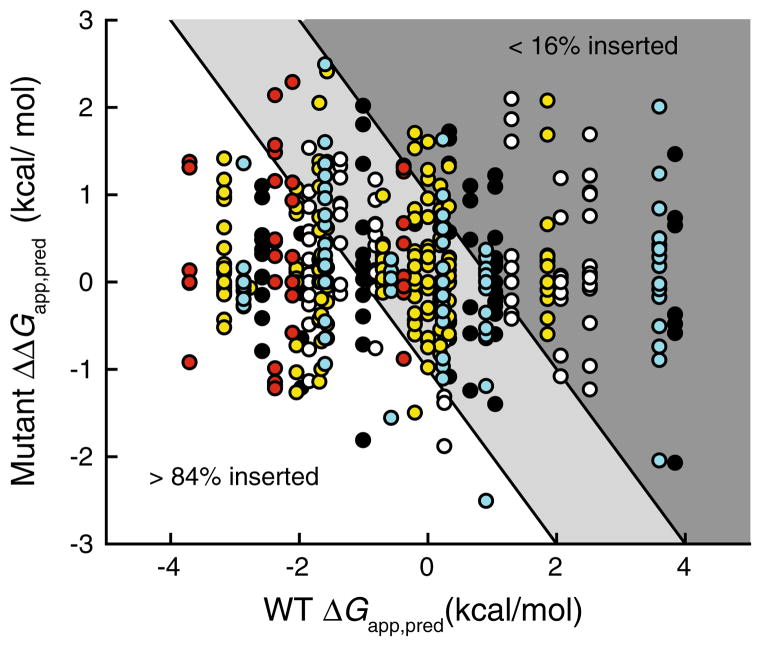
Collectively, rhodopsin (7), V2R (7), CFTR (12), KCNQ1 (6), and PMP22 (4) contain 36 TM helices. To interpret topogenic predictions for wild-type and mutant proteins, we make the simplifying assumption that most transmembrane helices partition into the membrane independently of neighboring helices, which is reasonable considering the energetic effects of interhelical hydrogen bonds typically appear to be modest (Meindl-Beinker et al. 2006). The degree to which the sequences of these helices are suitable for membrane partitioning is predicted to vary widely (Table 1); the predicted apparent free energy for partitioning of the TM helix into the ER membrane (ΔGapp,pred) varies from −3.71 (TM1 of PMP22) to 3.84 (TM7 of rhodopsin) kcal/mol (negative values indicate favorable partitioning into the membrane phase). For simplicity, we arbitrarily binned the predicted ΔGapp,pred values of both wild-type and mutant TM helices reported in this work into three regimes. For TM segments that are projected to exhibit favorable partitioning energetics, we chose an upper limit of −1 kcal/mol in the predicted ΔGapp,pred values, which would correspond to at least 84 % integration of the nascent helices. TM segments with predicted ΔGapp,pred values ranging from −1 to +1 kcal/mol, which correspond to a range of membrane integration from 84 to 16 % of the nascent helices, were considered to have intermediate partitioning energetics. Finally, those with predicted ΔGapp,pred values greater than +1 kcal/mol, which would correspond to less than 16 % integration of the nascent helices, were considered likely to exhibit unfavorable partitioning energetics. TM segments with favorable predicted partitioning energetics are likely to spontaneously partition into the membrane regardless of the sequence context, while the membrane integration of TM segments that fall within the other regimes may sometimes hinge on the formation of tertiary or quaternary contacts. Of the 36 wild-type TM helices in these proteins, 16 were predicted to have favorable partitioning energetics, 13 were predicted to have intermediate partitioning energetics, and 7 were predicted to have unfavorable partitioning energetics. These variations are consistent with the distribution of predicted ΔGapp,pred values in genomic surveys of the TM helices in multi-pass α-helical membrane proteins (Hessa et al. 2007; White and von Heijne 2008), which again suggests the chosen proteins may serve as a representative cross section of multi-pass α-helical membrane proteins. The predicted partitioning energetics of these helices is generally insensitive to mutations occurring distant from the TM segments (data not shown). For this reason, we narrowed our analysis to pathogenic mutations occurring within or near segments predicted to partition into the membrane, which included 70 mutations within rhodopsin, 87 mutations within V2R, 171 mutations within CFTR, 105 mutations within KCNQ1, and 37 mutations within PMP22 (470 missense mutations total).
Table 1.
Predicted partitioning energetics of wild-type TM helices
| Protein | TM helix | TM segment start | TM segment end | ΔGapp,pred (kcal/ mol) | TM Helix partitioning |
|---|---|---|---|---|---|
| Rhodopsin | 1 | 39 | 61 | −2.58 | Favorable |
| 2 | 74 | 96 | 0.66 | Intermediate | |
| 3 | 115 | 137 | 0.33 | Intermediate | |
| 4 | 157 | 179 | 1.04 | Unfavorable | |
| 5 | 203 | 224 | −1.01 | Favorable | |
| 6 | 250 | 269 | −1.97 | Favorable | |
| 7 | 285a | 309a | 3.84a | Unfavorable | |
| V2R | 1 | 41 | 63 | −1.37 | Favorable |
| 2 | 71 | 91 | 1.30 | Unfavorable | |
| 3 | 117 | 137 | 2.52 | Unfavorable | |
| 4 | 160 | 179 | 0.26 | Intermediate | |
| 5 | 202 | 224 | −0.82 | Intermediate | |
| 6 | 269 | 290 | −1.85 | Favorable | |
| 7 | 302 | 320 | 2.06 | Unfavorable | |
| CFTR | 1 | 73 | 94 | 0.19 | Intermediate |
| 2 | 117 | 138 | −3.17 | Favorable | |
| 3 | 195 | 216 | −0.20 | Intermediate | |
| 4 | 218 | 240 | −1.57 | Favorable | |
| 5 | 308 | 328 | −0.70 | Intermediate | |
| 6 | 331 | 352 | 0.00 | Intermediate | |
| 7 | 862 | 884 | −2.76 | Favorable | |
| 8 | 913 | 935 | 0.32 | Intermediate | |
| 9 | 989 | 1,011 | −2.04 | Favorable | |
| 10 | 1,014 | 1,034 | −1.69 | Favorable | |
| 11 | 1,101 | 1,122 | −1.69 | Favorable | |
| 12 | 1,128 | 1,147 | 1.86 | Unfavorable | |
| KCNQ1 | S1 | 123 | 145 | −2.87 | Favorable |
| S2 | 153 | 175 | −0.58 | Intermediate | |
| S3 | 193 | 215 | 0.903 | Intermediate | |
| S4 | 221a | 243a | 3.60a | Unfavorable | |
| S5 | 262 | 283 | −1.41 | Favorable | |
| S6 | 331 | 353 | 0.82 | Intermediate | |
| PMP22 | 1 | 2 | 24 | −3.71 | Favorable |
| 2 | 68 | 90 | −2.38 | Favorable | |
| 3 | 96 | 118 | −0.38 | Intermediate | |
| 4 | 134 | 156 | −2.11 | Favorable |
These helices were not identified as TM segments in full-sequences scans using the ΔG predictor algorithm. The identities of these helices were from available experimental data as described in the “Methods” section. The ΔGapp,pred values for these segments were determined using single-helix prediction with the ΔG predictor algorithm
Energetic Effects of Pathogenic Mutations on the Membrane Partitioning of TM Helices
Full-sequence scans of the 470 chosen pathogenic variants were performed using the ΔG predictor algorithm, and the predicted ΔGapp,pred value for each mutant TM helix was recorded. The effects of these mutations on the predicted partitioning energetics varied greatly; certain mutations were predicted to disfavor while others were predicted to enhance membrane integration. To visualize the range of predicted effects of these mutations, we plotted the change in the ΔGapp,pred values of the mutant helix (ΔΔGapp,pred) against the corresponding ΔGapp,pred value of the WT helix (Fig. 1). In this plot, the variants form vertical columns, each of which reflects all of the pathogenic variants of a single TM helix. Many of these columns span the indicated boundaries of the favorable (white), intermediate (light gray), and unfavorable (dark gray) partitioning regimes, which suggest the magnitude of the energetic effects of some mutations may be sufficient to change the partitioning behavior of certain TM helices. 63 of the 470 surveyed mutations (13 %) have a predicted ΔΔGapp,pred value greater than 1 kcal/mol (Table 2), which is typically a sufficient energetic effect to render a change in the partitioning regime. This suggests that topological defects likely constitute the pathogenic mechanism for a modest but nontrivial fraction of pathogenic mutations in transmembrane helices. Of course, it should be recognized that due to the formation of tertiary contacts between neighboring helices at or near the translocon (Meindl-Beinker et al. 2006; White and von Heijne 2008), the topogenic behavior for some of these helices may deviate from these predictions in the context of the full-length protein. Thus, the relevance of these predictions to the topogenic process and, ultimately, to the efficiency of the biogenesis of these variants will require experimental validation. Nevertheless, these results highlight the utility of the ΔG prediction algorithm in generating testable hypothesis involving the molecular basis of disease.
Fig. 1.
Effects of pathogenic mutations on the predicted energetics of translocon-mediated membrane integration of transmembrane helices. Full-sequence scans of wild-type and pathogenic variants of rhodopsin (black), V2R (white), CFTR (yellow), KCNQ1 (cyan), and PMP22 (red) were performed using the ΔG predictor algorithm, and the predicted perturbation of the free energy of translocon-mediated membrane integration of the mutated TM helix (ΔΔGapp,pred) caused by the mutation is plotted against the predicted free energy of translocon-mediated membrane integration (ΔGapp,pred) of the wild-type TM helix. Negative ΔGapp,pred and ΔΔGapp,pred values indicate favorable membrane integration (Color figure online)
Table 2.
Pathogenic mutations predicted to significantly disfavor membrane partitioning of TM helices
| Protein | Variant | TM helix | WT TM Helix Partitioning | ΔΔGapp,pred (kcal/mol) | Mut TM Helix Partitioning |
|---|---|---|---|---|---|
| Rhodopsin | G51R | 1 | Favorable | 1.10 | Favorable |
| Rhodopsin | L88P | 2 | Intermediate | 1.10 | Unfavorable |
| Rhodopsin | L125R | 3 | Intermediate | 1.73 | Unfavorable |
| Rhodopsin | L131P | 3 | Intermediate | 1.64 | Unfavorable |
| Rhodopsin | A164E | 4 | Unfavorable | 1.22 | Unfavorable |
| Rhodopsin | C167R | 4 | Unfavorable | 1.09 | Unfavorable |
| Rhodopsin | I214N | 5 | Favorable | 2.02 | Unfavorable |
| Rhodopsin | M216K | 5 | Favorable | 1.81 | Intermediate |
| Rhodopsin | M216R | 5 | Favorable | 1.35 | Intermediate |
| Rhodopsin | A292E | 7 | Unfavorable | 1.47 | Unfavorable |
| V2R | L44P | 1 | Favorable | 1.33 | Intermediate |
| V2R | I46K | 1 | Favorable | 1.41 | Intermediate |
| V2R | L53R | 1 | Favorable | 1.20 | Intermediate |
| V2R | L83P | 2 | Unfavorable | 1.61 | Unfavorable |
| V2R | L83Q | 2 | Unfavorable | 2.10 | Unfavorable |
| V2R | A84D | 2 | Unfavorable | 1.87 | Unfavorable |
| V2R | G122D | 3 | Unfavorable | 1.03 | Unfavorable |
| V2R | M123K | 3 | Unfavorable | 1.69 | Unfavorable |
| V2R | Y128D | 3 | Unfavorable | 1.01 | Unfavorable |
| V2R | A132D | 3 | Unfavorable | 1.22 | Unfavorable |
| V2R | F214S | 5 | Intermediate | 1.17 | Intermediate |
| V2R | L274P | 6 | Favorable | 1.04 | Intermediate |
| V2R | L282P | 6 | Favorable | 1.54 | Intermediate |
| V2R | L309P | 7 | Unfavorable | 1.19 | Unfavorable |
| CFTR | L88S | 1 | Intermediate | 1.11 | Unfavorable |
| CFTR | G126D | 2 | Favorable | 1.42 | Favorable |
| CFTR | L137H | 2 | Favorable | 1.18 | Favorable |
| CFTR | L137P | 2 | Favorable | 1.03 | Favorable |
| CFTR | L210P | 3 | Intermediate | 1.53 | Unfavorable |
| CFTR | E217G | 3 | Intermediate | 1.71 | Unfavorable |
| CFTR | L227R | 4 | Favorable | 1.38 | Intermediate |
| CFTR | V232D | 4 | Favorable | 2.41 | Intermediate |
| CFTR | I336K | 6 | Intermediate | 1.18 | Unfavorable |
| CFTR | I340N | 6 | Intermediate | 1.28 | Unfavorable |
| CFTR | L346P | 6 | Intermediate | 1.61 | Unfavorable |
| CFTR | Y917D | 8 | Intermediate | 1.36 | Unfavorable |
| CFTR | L927P | 8 | Intermediate | 1.33 | Unfavorable |
| CFTR | A1006E | 9 | Favorable | 1.08 | Intermediate |
| CFTR | V1020E | 10 | Favorable | 1.39 | Intermediate |
| CFTR | A1025D | 10 | Favorable | 2.05 | Intermediate |
| CFTR | M1028R | 10 | Favorable | 1.09 | Intermediate |
| CFTR | Y1032N | 10 | Favorable | 1.30 | Intermediate |
| CFTR | M1137R | 12 | Unfavorable | 2.08 | Unfavorable |
| CFTR | M1140K | 12 | Unfavorable | 1.69 | Unfavorable |
| KCNQ1 | L134P | S1 | Favorable | 1.36 | Favorable |
| KCNQ1 | I235N | S4 | Unfavorable | 2.01 | Unfavorable |
| KCNQ1 | L239P | S4 | Unfavorable | 1.24 | Unfavorable |
| KCNQ1 | L266P | S5 | Favorable | 1.35 | Intermediate |
| KCNQ1 | I268S | S5 | Favorable | 1.07 | Intermediate |
| KCNQ1 | G269D | S5 | Favorable | 1.36 | Intermediate |
| KCNQ1 | G272D | S5 | Favorable | 1.60 | Intermediate |
| KCNQ1 | L273R | S5 | Favorable | 2.50 | Intermediate |
| KCNQ1 | V280E | S5 | Favorable | 1.22 | Intermediate |
| KCNQ1 | A341E | S6 | Intermediate | 1.63 | Unfavorable |
| PMP22 | L16Pa | 1 | Favorable | 1.38 | Favorable |
| PMP22 | L19Pa | 1 | Favorable | 1.31 | Favorable |
| PMP22 | L71Pa | 2 | Favorable | 1.16 | Favorable |
| PMP22 | L78P | 2 | Favorable | 1.49 | Intermediate |
| PMP22 | L80P | 2 | Favorable | 1.57 | Intermediate |
| PMP22 | L105R | 3 | Intermediate | 1.27 | Intermediate |
| PMP22 | L108P | 3 | Intermediate | 1.33 | Intermediate |
| PMP22 | C109R | 3 | Intermediate | 1.31 | Intermediate |
| PMP22 | L147R | 4 | Favorable | 2.29 | Intermediate |
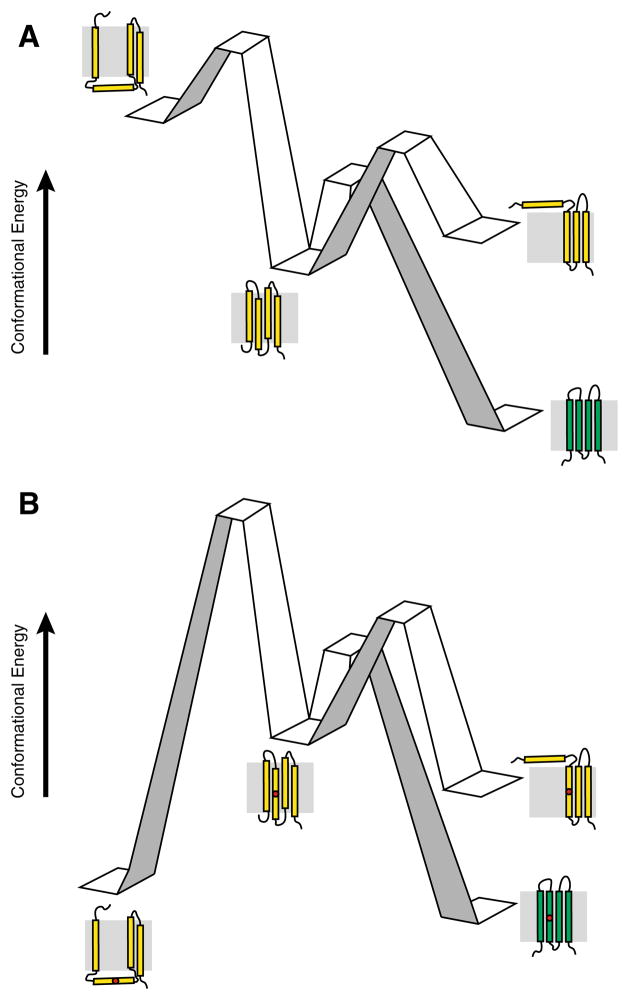
Topological decisions are governed by the principles of physical chemistry (White and von Heijne 2008). For this reason, the proposed effects of mutations that disrupt this process can be rationalized with the concept of a reaction coordinate. In Fig. 2, we have illustrated this concept with a hypothetical reaction coordinate encompassing the biosynthesis of a tetraspan membrane protein. A topological ensemble featuring the native topology (center) and two alternative non-native topomers (left and right), is established for the nascent protein (yellow) by the translocon based on the energetics of translocon-membrane partitioning. For most wild-type proteins, the native topology will be the most energetically favorable and abundant topomer, which can readily fold into the native conformation (green). Folding of the minor populations with non-native topologies may be inefficient due to the fact that they must traverse energy barriers associated with topological rearrangement in order to fold productively. The introduction of mutations that disfavor the membrane integration of a TM helix may selectively stabilize aberrant topomers relative to the native topology (Fig. 2b), which may increase the fraction of non-native topomers and decrease the fraction of nascent protein capable of achieving the native conformation prior to recognition by cellular quality control. This is, of course, only one set of examples among many for how mutations may impact topogenesis and specifically perturb the conformational energetics of a given membrane protein.
Fig. 2.
Examples of potential influence of pathogenic mutations on the topogenesis of multi-pass α-helical membrane proteins. Cartoons depict hypothetical reaction coordinates for nascent multi-pass α-helical membrane proteins in membranes. a Wild-type. The native topology (center) predominates in the topological ensemble of the nascent wild-type membrane protein (yellow). Nascent membrane proteins with incorrect topologies (left and right) must overcome the energy barriers associated with reorientation of TM helices across the membrane in order to achieve the native topology prior to acquiring the native fold (green). b Pathogenic variant. The incorporation of a pathogenic mutation within a TM helix (red circle) disfavors the native topology in the nascent ensemble. A diminished fraction of the nascent proteins are synthesized in the correct topology, and prohibitive energy barriers may prevent defective nascent proteins from achieving the correct topology prior to recognition by cellular quality control machinery associated with degradation pathways (Color figure online)
Influence of Pathogenic Mutations on the Position of Nascent TM Helices
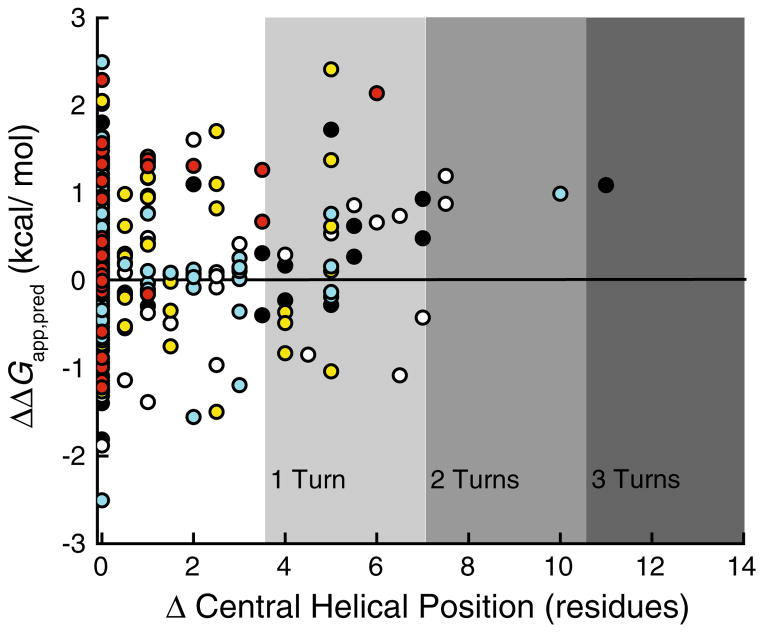
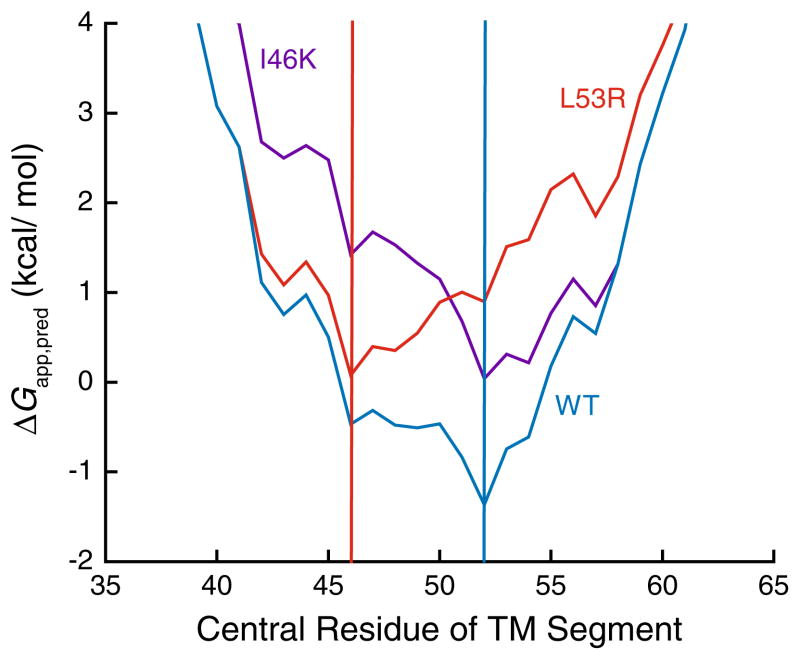
In addition to the energetics associated with the integration of isolated TM helices into the membrane, the ΔG predictor algorithm is also capable of identifying the portions of the proteins that are most likely to partition into the membrane. These segments do not always precisely correspond to the TM segments in the native structure, as previously demonstrated (Kauko et al. 2010; Virkki et al. 2014). While a change in the predicted partitioning energetics for a given TM segment was the most commonly observed effect of these pathogenic mutations, in several cases we noticed predicted changes in the N and C-terminal residues for the TM segment. In the most dramatic example (C167R Rhodopsin), the mutation was predicted to shift the center of the inserted segment by 11 residues. To visualize these effects, we plotted the ΔΔGapp,pred of these mutations against the number of resides by which the center of the helix is predicted to shift (Fig. 3). Most of these mutations surveyed in this work are predicted to have little or no effect on the position of the inserted segment. Nevertheless, several are predicted to shift the segment by the equivalent of two or more turns of a α-helix. Paradoxically, several mutations predicted to shift the sequence of the inserted segments are predicted to have minimal influence on the partitioning energetics. This suggests that, rather than shunting the mutated helix into the aqueous phase, the translocon may instead facilitate the membrane integration of an alternative sequence segment, which usually will overlap on one end or the other with the original wild-type TM segment. To illustrate such a possibility, we compared 23-residue window scans of the sequence near TM1 of wild-type, I46 K, and L53R V2R (Fig. 4). The energetic minimum in the wild-type protein (blue) corresponds to an inserted segment that begins at residue 41 and ends at residue 63. The I46 K mutation (purple) introduces a positively charged residue near the N-terminal edge of this nascent TM helix, which simply increases (disfavors) the predicted free energy of membrane partitioning, while maintaining the position of the free energy minimum. On the other hand, the L53R mutation (red) introduces a positively charged residue near the center of the wild-type TM helix. In this variant, a new energetic minimum is established 7 residues away, which would correspond to the integration of residues 35–54, placing this non-native arginine among the negatively charged phospholipid head groups in the cytoplasmic membrane interface rather than within the membrane core. This outcome may arise for TM helices that are not flanked by highly polar loop sequences. Together, these findings highlight two potentially distinct ways in which pathogenic mutations may influence topogenesis: (i) alteration of the efficiency of TM helix integration or (ii) a shift in the position of the nascent helix. Either alteration could affect the rate and/or efficiency with which nascent proteins achieve their native fold.
Fig. 3.
Effects of pathogenic mutations on the predicted position of the nascent TM helix. Full-sequence scans of wild-type and pathogenic variants of rhodopsin (black), V2R (white), CFTR (yellow), KCNQ1 (cyan), and PMP22 (red) were performed using the ΔG predictor algorithm, and the predicted perturbation of the free energy of translocon-mediated membrane integration of the mutated TM helix (ΔΔGapp,pred) caused by the mutation is plotted against the shift in the predicted position of the center of the nascent TM helix. Negative ΔGapp,pred and ΔΔGapp,pred values indicate favorable membrane integration (Color figure online)
Fig. 4.
Effects of pathogenic mutations on the predicted free energy minimum corresponding to the translocon-mediated integration of the first transmembrane helix of V2R into the ER membrane. The predicted free energy of translocon-mediated membrane integration (ΔGapp,pred) was determined for sequential 23-residue segments of wild-type (blue), I46K (purple), and L53R (red) V2R using the ΔG predictor algorithm, and the ΔGapp,pred values were plotted against the central residue of the 23-residue segment. Vertical lines indicate the predicted central position of the first transmembrane helix of wild-type and I46K (blue), as well as for L53R (red) V2R. Negative ΔGapp,pred values indicate favorable membrane integration (Color figure online)
Reconciling Predictions with Biochemical Properties of Pathogenic Rhodopsin Variants
Pathogenic variants that exhibit serious topological defects are likely to be recognized by cellular quality control, retained within the ER, and degraded prior to maturation (Buck and Skach 2005). Furthermore, such proteins are likely to be incapable of forming a native ligand-binding pocket. We sought to assess whether the modest fraction of disease mutations predicted to adversely influence topogenesis do indeed exhibit enhanced cellular turnover, intracellular retention, and diminished ligand binding when expressed in mammalian cells. Of the surveyed proteins, the biochemical properties of pathogenically misfolded rhodopsin variants seem to have been studied the most extensively, at least in terms of the number of mutant forms subjected to study (Hwa et al. 1997; Kaushal and Khorana 1994; Sung et al. 1991). We, therefore, compiled qualitative observations of the steady-state expression level, cellular localization, and retinal binding propensity for 22 pathogenic rhodopsin variants in mammalian cells and compared them to the predicted effects of these mutations on the energetics of membrane partitioning (Table 3). 15 of these mutations are predicted to have minimal effects on membrane integration or to enhance integration, which suggests that the conformational defects caused by these mutations are manifested after translocon-mediated membrane integration of the nascent TM helices. Alternatively, three of these mutations are predicted to have a ΔΔGapp,pred > 1 kcal/mol (disfavoring correct membrane partitioning), and four additional mutations are predicted to significantly decrease the efficiency with which their TM helix partitions into the membrane. Of these seven variants, six exhibit both significantly decreased steady-state expression levels and a significantly reduced capacity to bind its native retinal ligand. The available observations also show that these variants are retained within the ER, suggesting that they are largely incapable of passing quality control in the early secretory pathway. While these observations are consistent with the expected manifestations of topological defects, there was one notable outlier. The L125R mutation, which is known to distort the native tertiary structure of rhodopsin (Garriga et al. 1996), is predicted to significantly disfavor the partitioning of TM3 into the membrane (ΔΔGapp,pred = 1.7 kcal/mol). However, this variant exhibits normal steady-state expression levels and cellular trafficking, and is capable of binding its native retinal ligand (Kaushal and Khorana 1994). These observations suggest that the L125R mutation does not prevent the protein from acquiring its native topology, but more likely results in loss of rhodopsin function through some other, more subtle mechanism.
Table 3.
Biochemical properties of pathogenic rhodopsin variants
| Variant | WT TM helix insertion | ΔΔGapp,pred (kcal/mol) | Mutant TM helix insertion | Expression | Localization | Retinal binding |
|---|---|---|---|---|---|---|
| L125R | Intermediate | 1.73 | Unfavorable | Normala | PMa | Normala |
| G51R | Favorable | 1.10 | Favorable | Poora | ERa | Poora |
| C167R | Unfavorable | 1.09 | Unfavorable | Poora | ERa | Poora |
| V87D | Intermediate | 0.94 | Unfavorable | Poorb | ERb,a | Poorb,a |
| S297R | Unfavorable | 0.74 | Unfavorable | Poorc | – | Poorc |
| G89D | Intermediate | 0.48 | Unfavorable | Poorb | ERb,a | Poorb,c,a |
| G90D | Intermediate | 0.48 | Unfavorable | Poora | ERa | Poora |
| R135G | Intermediate | 0.28 | Intermediate | Normala | ||
| Y178C | Unfavorable | 0.17 | Unfavorable | Poorb | ERa | |
| P53R | Favorable | 0.06 | Favorable | Poor | ERa | Diminisheda |
| T58R | Favorable | 0.05 | Favorable | Poorb | PM/ERb | Poorb |
| F45L | Favorable | −0.15 | Favorable | Normalb,a | PMb | Diminishedb,c |
| H211P | Favorable | −0.15 | Favorable | Diminishedc,a | ERa | Poorc,a |
| A164V | Unfavorable | −0.22 | Intermediate | Diminishedc | ||
| G51A | Favorable | −0.41 | Favorable | Diminishedc | ||
| R135L | Intermediate | −0.44 | Intermediate | Poorb | PM/ERb | Poorb |
| R135W | Intermediate | −0.46 | Intermediate | Poorb | PM/ERb | Poorb |
| K296E | Unfavorable | −0.48 | Unfavorable | Diminisheda | ERa | Poora |
| P267R | Favorable | −0.64 | Favorable | Poorc | Poorc | |
| G51V | Favorable | −0.79 | Favorable | Normala | ERa | Diminishedc,a |
| P267L | Favorable | −1.21 | Favorable | Diminishedc | Diminishedc | |
| P171L | Unfavorable | −1.40 | Intermediate | Poora | ERa | Poora |
Biochemical data were gathered from the following references:
To illuminate how the L125R variant may be capable of compensating for the introduction of this non-native charge in TM3 during topogenesis, we inspected the structure of bovine rhodopsin. There appear to be two negatively charged residues at comparable depth within the membrane in the crystal structure of bovine rhodopsin. One is a conserved glutamate residue within the same helix (TM3) that resides one helical turn away from the mutated residue, which may be capable of forming interhelical contacts that could facilitate the membrane integration of the arginine side chain. The other residue is a conserved aspartate located on the adjacent TM2 at a similar membrane depth, which may enable the formation of an interhelical salt-bridge to help enable integration of the mutated helix into the membrane. While experimental evidence will be needed to determine whether these non-native contacts are indeed formed at or near the translocon during biosynthesis of this variant, these observations offer a reasonable explanation for how this outlier may achieve its native topology despite the hazardous introduction of this seemingly poorly placed, non-native side chain. Together, these findings confirm the utility of the ΔG predictor algorithm to generate testable hypothesis regarding the effects of certain pathogenic mutations on the early stages of membrane protein folding.
Conclusions and Outlook
Heterogeneity in the mechanisms by which pathogenic mutations are capable of prompting the cellular misfolding of integral membrane proteins has undermined attempts to identify therapeutic compounds with broad efficacy (Rowe and Verkman 2013). Thus, an understanding for the prevalence of certain misfolding mechanisms and how these relate to common pathogenic mutations will be of increasing importance in rational drug design efforts. In this work, we sought to assess whether pathogenic mutations in transmembrane helices commonly influence topogenesis. Using the freely available ΔG predictor server, we surveyed the effects of 470 pathogenic mutations implicated in diseases of membrane protein misfolding on the energetics of translocon-mediated membrane integration of transmembrane helices. The results suggest that roughly 10 % of the mutations that fall within or near TM helices may adversely influence the efficiency with which these variants acquire their correct membrane topology. This is as opposed to other mutations that primarily influence post-translational folding and/or function. The intrinsic differences between the structural properties of topologically defective disease variants relative to other disease variants may have implications for therapeutic design. For instance, variants with incorrect membrane topology may be incapable of binding ligands, which may limit the utility of ligand-based pharmacological chaperones. However, considering that correction of aberrant topologies by cellular quality control machinery may compete with degradation pathways (Buck and Skach 2005), these variants may instead be more amenable to pharmacological rescue mediated by the so-called proteostasis regulators (Balch et al. 2008; Mu et al. 2008; Powers et al. 2009), which alter the activity of cellular quality control networks. Such possibilities provide the impetus to elucidate the linkage between the structural and biochemical defects caused by these mutations and the corresponding efficacy of various therapeutics.
Acknowledgments
This work was supported by US NIH Grants RO1 DC007416, RO1 HL122010, and U54 GM094608. JPS was supported by US NIH F32 GM110929.
References
- Adamian L, Liang J. Interhelical hydrogen bonds and spatial motifs in membrane proteins: polar clamps and serine zippers. Proteins. 2002;47:209–218. doi: 10.1002/prot.10071. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Buck TM, Skach WR. Differential stability of biogenesis intermediates reveals a common pathway for aquaporin-1 topological maturation. J Biol Chem. 2005;280:261–269. doi: 10.1074/jbc.M409920200. [DOI] [PubMed] [Google Scholar]
- Cestèle S, Schiavon E, Rusconi R, Franceschetti S, Mantegazza M. Nonfunctional NaV1.1 familial hemiplegic migraine mutant transformed into gain of function by partial rescue of folding defects. Proc Natl Acad Sci USA. 2013;110:17546–17551. doi: 10.1073/pnas.1309827110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denks K, Vogt A, Sachelaru I, Petriman NA, Kudva R, Koch HG. The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol Membr Biol. 2014;31:58–84. doi: 10.3109/09687688.2014.907455. [DOI] [PubMed] [Google Scholar]
- Egea PF, Stroud RM. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc Natl Acad Sci USA. 2010;107:17182–17187. doi: 10.1073/pnas.1012556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafvelin G, von Heijne G. Topological “frustration” in multispanning E. coli inner membrane proteins. Cell. 1994;77:401–412. doi: 10.1016/0092-8674(94)90155-4. [DOI] [PubMed] [Google Scholar]
- Garriga P, Liu X, Khorana HG. Structure and function in rhodopsin: correct folding and misfolding in point mutants at and in proximity to the site of the retinitis pigmentosa mutation Leu-125–>Arg in the transmembrane helix C. Proc Natl Acad Sci USA. 1996;93:4560–4564. doi: 10.1073/pnas.93.10.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich SU, Mothes W, Brunner J, Rapoport TA. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- Hwa J, Garriga P, Liu X, Khorana HG. Structure and function in rhodopsin: packing of the helices in the transmembrane domain and folding to a tertiary structure in the intradiscal domain are coupled. Proc Natl Acad Sci USA. 1997;94:10571–10576. doi: 10.1073/pnas.94.20.10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illergard K, Kauko A, Elofsson A. Why are polar residues within the membrane core evolutionary conserved? Proteins. 2011;79:79–91. doi: 10.1002/prot.22859. [DOI] [PubMed] [Google Scholar]
- Kanki T, Sakaguchi M, Kitamura A, Sato T, Mihara K, Hamasaki N. The tenth membrane region of band 3 is initially exposed to the luminal side of the endoplasmic reticulum and then integrated into a partially folded band 3 intermediate. Biochemistry. 2002;41:13973–13981. doi: 10.1021/bi026619q. [DOI] [PubMed] [Google Scholar]
- Kanner EM, Klein IK, Friedlander M, Simon SM. The amino terminus of opsin translocates “posttranslationally” as efficiently as cotranslationally. Biochemistry. 2002;41:7707–7715. doi: 10.1021/bi0256882. [DOI] [PubMed] [Google Scholar]
- Kauko A, Hedin LE, Thebaud E, Cristobal S, Elofsson A, von Heijne G. Repositioning of transmembrane alpha-helices during membrane protein folding. J Mol Biol. 2010;397:190–201. doi: 10.1016/j.jmb.2010.01.042. [DOI] [PubMed] [Google Scholar]
- Kaushal S, Khorana HG. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry. 1994;33:6121–6128. doi: 10.1021/bi00186a011. [DOI] [PubMed] [Google Scholar]
- Kelly JW, Balch WE. The integration of cell and chemical biology in protein folding. Nat Chem Biol. 2006;2:224–227. doi: 10.1038/nchembio0506-224. [DOI] [PubMed] [Google Scholar]
- Lu Y, Turnbull IR, Bragin A, Carveth K, Verkman AS, Skach WR. Reorientation of aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol Biol Cell. 2000;11:2973–2985. doi: 10.1091/mbc.11.9.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl-Beinker NM, Lundin C, Nilsson I, White SH, von Heijne G. Asn- and Asp-mediated interactions between transmembrane helices during translocon-mediated membrane protein assembly. EMBO Rep. 2006;7:1111–1116. doi: 10.1038/sj.embor.7400818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello JP, Salahpour A, Laperriere A, Bernier V, Arthus MF, Lonergan M, Petaja-Repo U, Angers S, Morin D, Bichet DG, Bouvier M. Pharmacological chaperones rescue cell-surface expression and function of misfolded V2 vasopressin receptor mutants. J Clin Invest. 2000;105:887–895. doi: 10.1172/JCI8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss K, Helm A, Lu Y, Bragin A, Skach WR. Coupled translocation events generate topological heterogeneity at the endoplasmic reticulum membrane. Mol Biol Cell. 1998;9:2681–2697. doi: 10.1091/mbc.9.9.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu TW, Ong DS, Wang YJ, Balch WE, Yates JR, 3rd, Segatori L, Kelly JW. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134:769–781. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JK, Mobley CK, Sanders CR. The peripheral neuropathy-linked Trembler and Trembler-J mutant forms of peripheral myelin protein 22 are folding-destabilized. Biochemistry. 2008;47:10620–10629. doi: 10.1021/bi801157p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öjemalm K, Higuchi T, Jiang Y, Langel Ü, Nilsson I, White SH, Suga H, von Heijne G. Apolar surface area determines the efficiency of translocon-mediated membrane-protein integration into the endoplasmic reticulum. Proc Natl Acad Sci USA. 2011;108:E359–E364. doi: 10.1073/pnas.1100120108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T, Veit G, Dekkers JF, Bagdany M, Soya N, Xu H, Roldan A, Verkman AS, Kurth M, Simon A, Hegedus T, Beekman JM, Lukacs GL. Mechanism-based corrector combination restores DeltaF508-CFTR folding and function. Nat Chem Biol. 2013;9:444–454. doi: 10.1038/nchembio.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D, Kim JH, Kroncke BM, Law CL, Xia Y, Droege KD, Van Horn WD, Vanoye CG, Sanders CR. Purification and structural study of the voltage-sensor domain of the human KCNQ1 potassium ion channel. Biochemistry. 2014;53:2032–2042. doi: 10.1021/bi500102w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popot JL, Engelman DM. Helical membrane protein folding, stability, and evolution. Annu Rev Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Rowe SM, Verkman AS. Cystic fibrosis transmembrane regulator correctors and potentiators. Cold Spring Harb Perspect Med. 2013;3:a009761. doi: 10.1101/cshperspect.a009761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C, Myers J. Disease-related misassembly of membrane proteins. Annu Rev Biophys Biomol Struct. 2004;33:25–51. doi: 10.1146/annurev.biophys.33.110502.140348. [DOI] [PubMed] [Google Scholar]
- Sanders CR, Ismail-Beigi F, McEnery MW. Mutations of peripheral myelin protein 22 result in defective trafficking through mechanisms which may be common to diseases involving tetraspan membrane proteins. Biochemistry. 2001;40:9453–9459. doi: 10.1021/bi010894f. [DOI] [PubMed] [Google Scholar]
- Sung CH, Schneider BG, Agarwal N, Papermaster DS, Nathans J. Functional heterogeneity of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci USA. 1991;88:8840–8844. doi: 10.1073/pnas.88.19.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B, Clemons WM, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- Van Goor F, Hadida S, Grootenhuis PD, Burton B, Cao D, Neuberger T, Turnbull A, Singh A, Joubran J, Hazlewood A, Zhou J, McCartney J, Arumugam V, Decker C, Yang J, Young C, Olson ER, Wine JJ, Frizzell RA, Ashlock M, Negulescu P. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc Natl Acad Sci USA. 2009;106:18825–18830. doi: 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virkki MT, Agrawal N, Edsbacker E, Cristobal S, Elofsson A, Kauko A. Folding of aquaporin 1: multiple evidence that helix 3 can shift out of the membrane core. Protein Sci. 2014 doi: 10.1002/pro.2483. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- Welsh MJ, Smith AE. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-r. [DOI] [PubMed] [Google Scholar]
- White SH, von Heijne G. How translocons select transmembrane helices. Annu Rev Biophys. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- Wilson AJ, Quinn KV, Graves FM, Bitner-Glindzicz M, Tinker A. Abnormal KCNQ1 trafficking influences disease pathogenesis in hereditary long QT syndromes (LQT1) Cardiovasc Res. 2005;67:476–486. doi: 10.1016/j.cardiores.2005.04.036. [DOI] [PubMed] [Google Scholar]