Abstract
Purpose
Genomic profiling studies suggest triple-negative breast cancer (TNBC) is a heterogeneous disease. In this study we sought to define TNBC subtypes and identify subtype-specific markers and targets.
Patients and Methods
RNA and DNA profiling analyses were conducted on 198 TNBC tumors (ER-negativity defined as Allred Scale value ≤2) with >50% cellularity (discovery set: n=84; validation set: n=114) collected at Baylor College of Medicine. An external data set of 7 publically-accessible TNBC studies was used to confirm results. DNA copy number, disease-free survival (DFS) and disease-specific survival (DSS) were analyzed independently using these datasets.
Results
We identified and confirmed four distinct TNBC subtypes: (1) Luminal-AR (LAR); 2) Mesenchymal (MES); 3) Basal-Like Immune-Suppressed (BLIS), and 4) Basal-Like Immune-Activated (BLIA). Of these, prognosis is worst for BLIS tumors and best for BLIA tumors for both DFS (logrank test p=0.042 and 0.041, respectively) and DSS (logrank test p=0.039 and 0.029, respectively). DNA copy number analysis produced two major groups (LAR and MES/BLIS/BLIA), and suggested gene amplification drives gene expression in some cases (FGFR2 (BLIS)). Putative subtype-specific targets were identified: 1) LAR: androgen receptor and the cell surface mucin MUC1; 2) MES: growth factor receptors (PDGF receptor A; c-Kit); 3) BLIS: an immune suppressing molecule (VTCN1); and 4) BLIA: Stat signal transduction molecules and cytokines.
Conclusion
There are four stable TNBC subtypes characterized by the expression of distinct molecular profiles that have distinct prognoses. These studies identify novel subtype-specific targets that can be targeted in the future for effective treatment of TNBCs.
Keywords: breast cancer, estrogen receptor-negative, “triple-negative” breast cancer, genomic profiling, personalized medicine
INTRODUCTION
Recent studies have demonstrated that breast cancer heterogeneity extends beyond the classic immunohistochemistry (IHC)-based divisions of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (Her2)1. 10-20% of primary breast cancers are triple-negative breast cancers (TNBCs)2, which lack expression of ER, PR and Her2, present with higher grade, often contain mutations in TP533, and have a poor prognosis4. Molecularly-targeted therapy has shown limited benefit so far in TNBCs, and although PARP inhibitors in the BRCA-mutant setting are promising5,6, new strategies for classifying and treating women affected by this aggressive disease are urgently needed.
The intrinsic subtyping of breast cancer by gene expression analyses7 was recently supported by The Cancer Genome Atlas (TCGA) Program through mRNA, miRNA, DNA, and epigenetic analyses8. The basal-like subtype, traditionally defined by RNA profiling or cytokeratin expression9, account for 10-25% of all invasive breast cancers10. In addition, basal-like breast cancers account for 47-88% of all TNBCs8,11,12. Tumors of the “claudin-low” (CL) subtype13,14 have particularly poor prognoses compared to hormone-sensitive tumors15. The results from an aggregate analysis of publically available expression data sets performed by Lehmann et al.12 suggested that TNBCs are more heterogeneous than previously described, and identified 6 subtypes: 1) androgen receptor positive; 2) claudin-low-enriched mesenchymal; 3) mesenchymal stem-like; 4) immune response; and two cell cycle-disrupted basal subtypes 5) BL-1 and 6) BL-2. However, immunohistochemical (IHC) detection of ER, PR, and HER2 protein is the clinical standard used to define TNBC. In the study by Lehmann et al., when tumors with IHC-confirmed ER, PR, and HER2 protein expression were analyzed, only 5 of the 6 described subtypes were observed (see supplemental figures 4 and 5 in Lehmann et al.12). Therefore, while previous genomic studies have advanced our understanding of TNBCs, stable subtypes, as well as subtype-specific molecular targets still need to be identified.
In this study, we investigated 198 previously uncharacterized TNBCs using mRNA expression and DNA profiling, and identified 4 stable TNBC subtypes: 1) Luminal/Androgen Receptor (LAR), 2) Mesenchymal (MES), 3) Basal-Like Immune Suppressed (BLIS), and 4) Basal-Like Immune Activated (BLIA). Using independent TNBC datasets, we show that BLIS and BLIA tumors have the worst and best prognoses, respectively (independently of other known prognostic factors), compared to the other subtypes. Our DNA studies demonstrate unique subtype-specific gene amplification, with CCND1, EGFR, FGFR2, and CDK1 amplified in the LAR, MES, BLIS and BLIA subtypes, respectively. Collectively, our RNA and DNA genomic results identify stable, reproducible TNBC subtypes characterized by specific RNA and DNA markers, and identify potential targets for more effective treatment of TNBCs.
MATERIALS AND METHODS
Patients and study recruitment
278 anonymized tissues collected from multiple U.S. and European sites were obtained from the Lester and Sue Smith Breast Cancer Tumor Bank at Baylor College of Medicine (BCM), diagnosis-confirmed and flash frozen. BCM purchased these tumors (with clinical information, including: age, menopausal status, histology, AJCC stage, tumor grade) from Asterand USA. No treatment or outcome data was available for these tumors. Tissues were managed by the BCM Breast Center's Human Tissue Acquisition and Pathology (HTAP) shared resource. Cellularity, histology, and IHC ER, PR, and HER2 receptor status in discovery and validation samples were assessed by Breast Center pathologists. Only tumors exhibiting >50% tumor cellularity were used. ER-receptor negativity is defined as Allred Scale ≤2.
RNA/DNA extraction and array experiments
For extraction and quality control details, see supplemental material. Briefly, tumors were profiled using the Affymetrix U133 Plus 2.0 gene expression array and affy16 package in R17. Discovery and validation set SNP experiments were performed on Illumina 610K and 660K platforms, respectively. Common SNPs were analyzed after independent processing in Illumina Genome Studio v2011 Genotyping Module 1.9.4.
PAM50, TNBCType, and ERSig
TNBCs were assigned to previously described subtypes using the TNBCType tool18. Intrinsic subtypes were established with the PAM50 Breast Cancer Intrinsic Classifier19, and compared to 67 non-TNBC randomly sampled tumors representing 80% of the assigned sample (confirmed by Pearson Correlation). This comparison was used to create a 32-gene centroid signature (derived from Williams et al.'s estrogen receptor 1 (ESR1) downstream targets gene list20, accessed via the Molecular Signatures Database (MSigDB)21) in order to correlate TNBCs with ER activation (“ERSig”).
Gene selection, NMF clustering, differential expression, and centroid signatures
Genes were sorted by aggregate rank of median absolute deviations (MADs) across all samples and the MAD across each of the two most predominant clusters (approximating basal-like versus the remaining intrinsic subtypes) for the discovery set using R package Differential Expression via Distance Summary (DEDS)22. The top 1000 median-centered genes were utilized for clustering and split into 2000 positive input features23. The ideal rank basis and factorization algorithm was determined using the R package Non-negative Matrix Factorization (NMF)24 before taking the 1000-iteration consensus for a final clustering basis of 4.
Genes were sorted by DEDS using: 1) Goeman's Global Test (GGT)25 applied to each set individually for all 18,209 genes, using a Benjamini-Hochberg (BH) False Discovery Rate (FDR) multi-test correction; and 2) computed log2(Fold Change) (“FC”) values. The top 20 unique genes by p-value and log2(FC) became a classifier comprising 80 genes and representing the median quantiles of all 80 genes for each discover set cluster, with cases assigned by minimum average Euclidean distances of quantile gene expression data. Non-significant p-values (p>0.05 by 10,000 permutations) or deviations from any centroid >0.25 were left unclassified.
Preprocessing and assignment of expression data for publically-accessible cases
Normalization and quality control procedures identical to the primary study sets (but using the Partek Genomics Suite program26 to perform ANOVA-based batch correction across the 221 arrays prior to summarization of probe set data) were performed on 7 publically-accessible studies in Gene Expression Omnibus (GEO) with TNBCs (by IHC) profiled on the Affymetrix U133 Plus 2.0 array (“external set”). Series GEO matrices and accompanying TNBC tumor clinical data from the Sabatier27 (also included in external set) and Curtis11 studies were assigned using gene-centric representation of array data.
Ingenuity pathways analysis
Significant genes (BH correction p-value<0.001 from GGT) for each dataset group were uploaded independently into Ingenuity Systems’ Interactive Pathway Analysis (IPA) software (www.ingenuity.com). A 0.05 significance threshold was used for pathway enrichment. Molecules, chemicals, or groups with regulatory function(s) were analyzed by IPA to produce final gene lists.
Copy number segmentation and analysis
Allele-Specific Piecewise Constant Fitting (ASPCF) analysis and Allele-Specific Copy Number (CN) Analysis of Tumors (ASCAT, default values)28 of 84 discovery and 58 validation set tumors yielded 62 and 46 samples, respectively, with assigned reliable DNA ploidy- and tumor percentage-corrected integer CNs. These segments were uploaded collectively and individually by assigned expression-based subtypes to Genomic Identification of Significant Targets in Cancer (GISTIC) 2.029 (default settings, with a 0.5 linear margin for gains and losses).
Survival analyses
Survival curves were constructed using the Kaplan-Meier product limit method and compared between subtypes with the log-rank test using publically available datasets for which disease-free survival and disease-specific survival results are available; however, no treatment information was available for these datasets. Cox proportional hazard regression model adjusted for available prognostic clinical covariates was performed to calculate subtype-specific hazard ratios, 95% confidence intervals, and disease-free and overall survival (DFS and OS, respectively). Survival analyses were performed using the R package survival.
RESULTS
Patient population
198 TNBCs were assigned to discovery (n=84) or validation (n=114) sets based on chronological acquisition of tissue. Subjects were predominantly postmenopausal, Caucasian, and of mean and median age of 53 (Table 1). 95% of TNBCs were invasive ductal carcinomas, predominantly Stages I-III (1% were metastatic breast cancers), and >75% of tumors were >2cm at diagnosis.
Table 1.
Clinical characteristics of the patients and tumor samples used in study.
| Characteristic | Both Sets | Discovery Set | Validation Set | p-value | |||
|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | ||
| Number of Tumors | 198 | 84 | 42 | 114 | 58 | ||
| Age | 192 | 84 | 108 | 0.02 | |||
| <50 yrs | 81 | 42 | 43 | 51 | 38 | 35 | |
| ≥50 yrs | 111 | 58 | 41 | 49 | 70 | 65 | |
| Missing | 6 | 0 | 6 | ||||
| Race | 194 | 80 | 114 | 0.10 | |||
| Caucasian | 187 | 96 | 75 | 94 | 112 | 98 | |
| Asian/Pacific Islander | 7 | 4 | 5 | 6 | 2 | 2 | |
| Missing | 4 | 4 | 0 | ||||
| Menopausal status | 167 | 71 | 96 | 0.24 | |||
| Premenopausal | 62 | 37 | 31 | 44 | 31 | 32 | |
| Menopausal | 11 | 7 | 3 | 4 | 8 | 8 | |
| Postmenopausal | 94 | 56 | 37 | 52 | 57 | 59 | |
| Missing | 31 | 13 | 18 | ||||
| Body mass index | 166 | 65 | 101 | 0.65 | |||
| Underweight (< 18.5) | 3 | 2 | 2 | 3 | 1 | 1 | |
| Normal (18.5-24.9) | 46 | 28 | 17 | 26 | 29 | 29 | |
| Overweight (25-29.9) | 61 | 37 | 26 | 40 | 35 | 35 | |
| Obese (≥30) | 56 | 33 | 20 | 31 | 36 | 35 | |
| Missing | 32 | 19 | 13 | ||||
| Tumor size | 195 | 83 | 112 | 0.01 | |||
| <2 cm | 35 | 18 | 10 | 12 | 25 | 22 | |
| 2-5 cm | 139 | 71 | 60 | 72 | 79 | 71 | |
| >5 cm | 12 | 6 | 10 | 12 | 2 | 2 | |
| Any size with direct extension | 9 | 5 | 3 | 4 | 6 | 5 | |
| Cannot be assessed | 3 | 1 | 2 | ||||
| Positive lymph nodes | 150 | 66 | 84 | 0.14 | |||
| 0 | 74 | 49 | 29 | 44 | 45 | 54 | |
| 1-3 | 49 | 33 | 28 | 42 | 21 | 25 | |
| 4-9 | 17 | 11 | 6 | 9 | 11 | 13 | |
| >10 | 10 | 7 | 3 | 5 | 7 | 8 | |
| Unknown | 48 | 18 | 30 | ||||
| Metastases | 146 | 64 | 82 | 0.86 | |||
| No metastases | 144 | 99 | 63 | 98 | 81 | 99 | |
| Metastases found | 2 | 1 | 1 | 2 | 1 | 1 | |
| Unknown | 52 | 20 | 32 | ||||
| Histology | 198 | 84 | 114 | ||||
| Infiltrating ductal carcinoma (IDC) | 188 | 95 | 82 | 98 | 106 | 93 | 0.41 |
| Infiltrative lobular carcinoma (ILC) | 1 | 0.5 | 0 | 0 | 1 | 1 | |
| Adenocarcinoma/carcinoma, not otherwise specified | 7 | 3.5 | 2 | 2 | 5 | 4 | |
| Medullary carcinoma | 2 | 1 | 0 | 0 | 2 | 2 | |
mRNA profiling of TNBCs reveals four stable molecular phenotypes
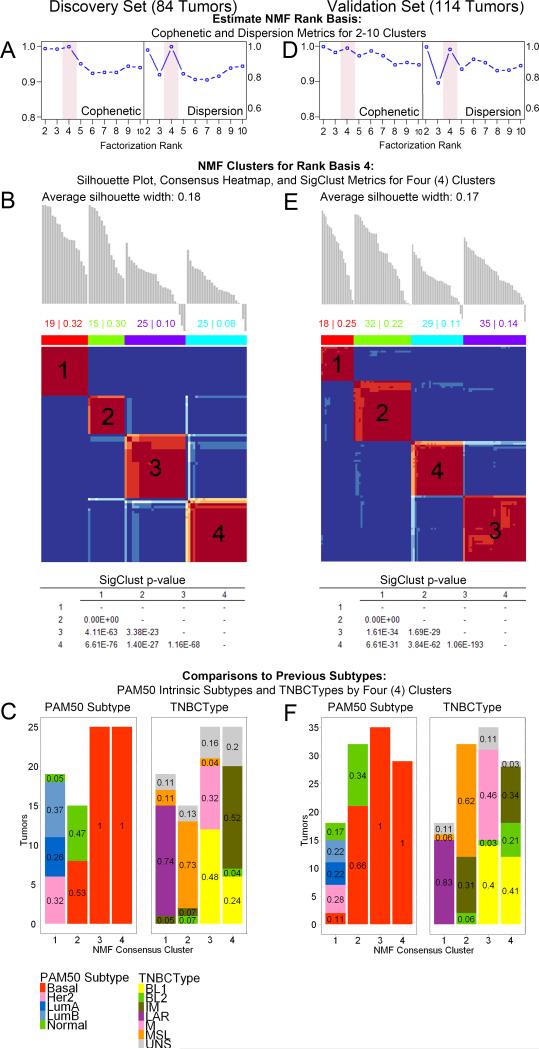
Using RNA gene expression profiling, we explored TNBC molecular phenotypes. NMF was performed on 1000 discovery set genes selected to maximize separation across and within conventional intrinsic subtypes. These tumors were most stably divided into 4 clusters by cophenetic, dispersion, silhouette, and Statistical Significance of Clustering (SigClust)30 metrics, in addition to visual inspection of the consensus heat map (Figures 1A-B, S1). This quadrilateral division of data was also observed in the validation set tumors using the same input features (Figures 1D-E, S2). ER-, PR- and Her2-negativity was IHC-confirmed by our participating pathologist, Dr. Contreras (Figures S3). Differentially-expressed genes (BH-adjusted p-value<0.001 from GGT) were significantly enriched only within corresponding discovery and validation set clusters (Fisher Exact test p=4.01E-30, 3.47E-17, 2.88E-46, and 3.61E-10, respectively, Tables S1-5), independently confirming the 4 molecular phenotypes observed. Additionally, significant enrichment of discovery set IPA results in the validation set also support the four cluster separation (Tables S6-10).
Figure 1. Classification of TNBCs by mRNA profiling reveals four stable molecular phenotypes.
84 (discovery set) and 114 TN breast tumors (validation set) both demonstrate 4 stable clusters by NMF of mRNA expression across the top 1000 genes (IQR summarized) selected by DEDS aggregate rank of median absolute deviations (see complete methods) of the discovery set. A & D. Cophenetic and dispersion metrics for NMF across 2-10 clusters with 50 runs suggest 4 stable clusters. Full metrics are available for each set in Supplemental Figures 1 and 2. B & E. Silhouette analyses and consensus plots for rank basis 4 NMF clusters (1000 runs, nsNMF factorization). Average silhouette widths worsened with increasing clusters beyond the 4 shown. SigClust was significant for all pairwise comparisons with this feature set. C & F. PAM50 intrinsic subtypes and TNBCType distributions by 4 NMF clusters.
Comparison of our NMF results to Perou's “PAM50” TNBC molecular classification (luminal A, luminal B, HER-2-positive, basal-like and normal-like subtypes)9 shows clusters 3 and 4 to be entirely basal-like, containing 86% and 74% of all PAM50 basal-like tumors in the discovery and validation sets, respectively (Figure 1C). Conversely, cluster 1 contains all luminal A, luminal B, and Her2-positive PAM50 tumors, and cluster 2 contains basal-like and normal-like PAM50 tumors.
We then compared our NMF results with the Lehmann/Pietenpol “TNBC Type” molecular classification (basal-like-1, basal-like-2, immunomodulatory, luminal androgen receptor (LAR), mesenchymal, and mesenchymal stem-like subtypes)12, in which “claudin-low” tumors are split between the mesenchymal and mesenchymal stem-like subtypes. Our results show cluster 1 contains all of Lehmann's LAR tumors, and cluster 2 contains most of Lehmann's mesenchymal stem-like and some claudin-low mesenchymal tumors (Figures 1F, S4B, S5). Conversely, our TNBC clustering did not separate Lehmann's12 “basal-like 1” and “basal-like 2” types even when utilizing all six subtype signatures described in Lehmann et al.12 in a semi-supervised NMF (2188 genes; Figure S4). Instead, Lehmann's basal-like-1 and basal-like-2 tumors are split between clusters 3 and 4 (Figure S4). Finally, Lehmann's remaining claudin-low mesenchymal tumors reside in cluster 3, while the immunomodulatory tumors are distributed across clusters 2 and 4, which express common signaling pathways (Figures S4-5).
Gene signatures define four prognostically-distinct TNBC subtypes
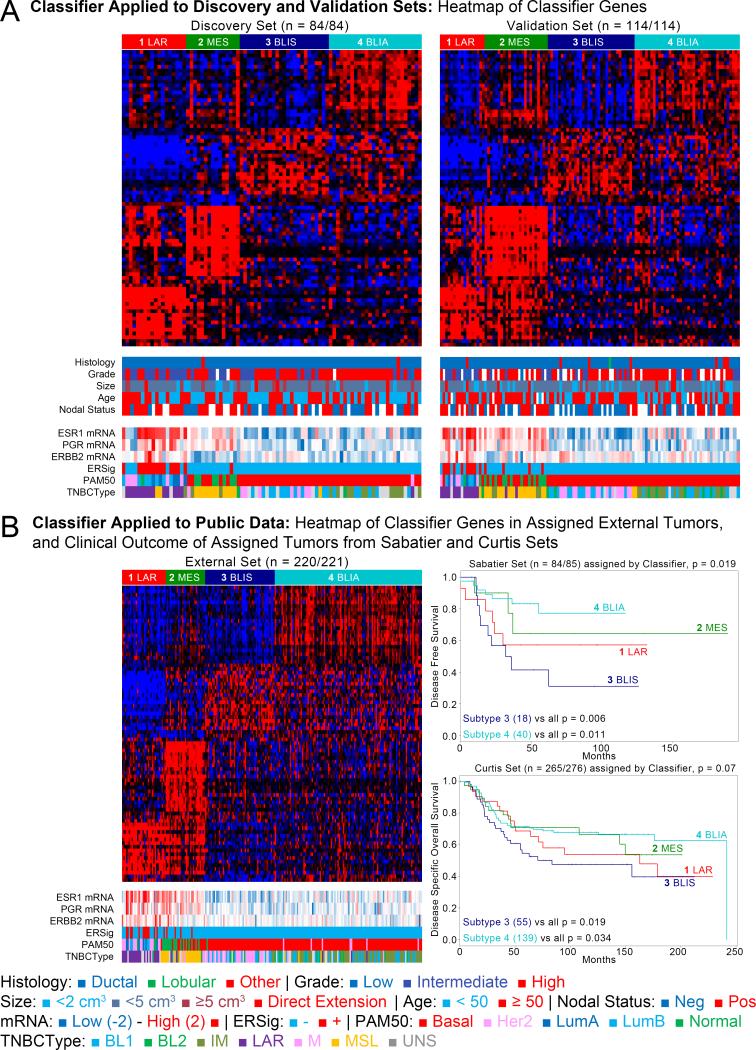
Using the discovery and validation sets we developed and confirmed an 80-gene signature for these clusters (Figure 2A, Tables S11-16). This analysis was repeated using an independent set of 221 publically accessible TNBCs with IHC data (“external set”, Tables S17-18, Figure 2B), and other publically accessible datasets with available clinical data (Tables S19-20). Comparisons of group assignment against existing NMF clusters demonstrated strong reproducibility, with Rand indices of 0.94 (p<0.0001) and 0.82 (p<0.0001), respectively (Tables S21-22).
Figure 2. Gene signature defines four subtypes of TNBC with prognostic differences.
Discovery, validation, and external sets tumors with intermediate grade, high ESR1, PGR, and ERBB2 expression, activated ER downstream targets, and luminal A/B subtypes are enriched in subtype 1. A. The four assigned subtypes in both the discovery (84/84) and validation sets (114/114). B. Gene signature applied successfully to 220 of 221 external set TNBCs. C. Clinical outcomes from independent sets classified by the discovery set-based signature. Subtype 4 has a better prognosis for both disease-free and disease-specific survival.
Clinical outcome data was available for this publically available “external set” of TNBCs. However, treatment information for the “external set” data is not available. Analysis of disease-free survival (DFS) and disease-specific survival (DSS) showed that subtype 3 has the worst prognosis of all 4 subtypes, while subtype 4 has a relatively good prognosis for DFS (logrank test p=0.042 and 0.041, respectively) and disease-specific survival (DSS; logrank test p=0.039 and 0.029, respectively) (Tables S23-24, Figure 2C). The associations between subtypes 3 and 4 and DFS and DSS remained significant in multivariate models adjusted for available prognostic clinical covariates..
TNBC subtype-specific enrichment of molecular pathways
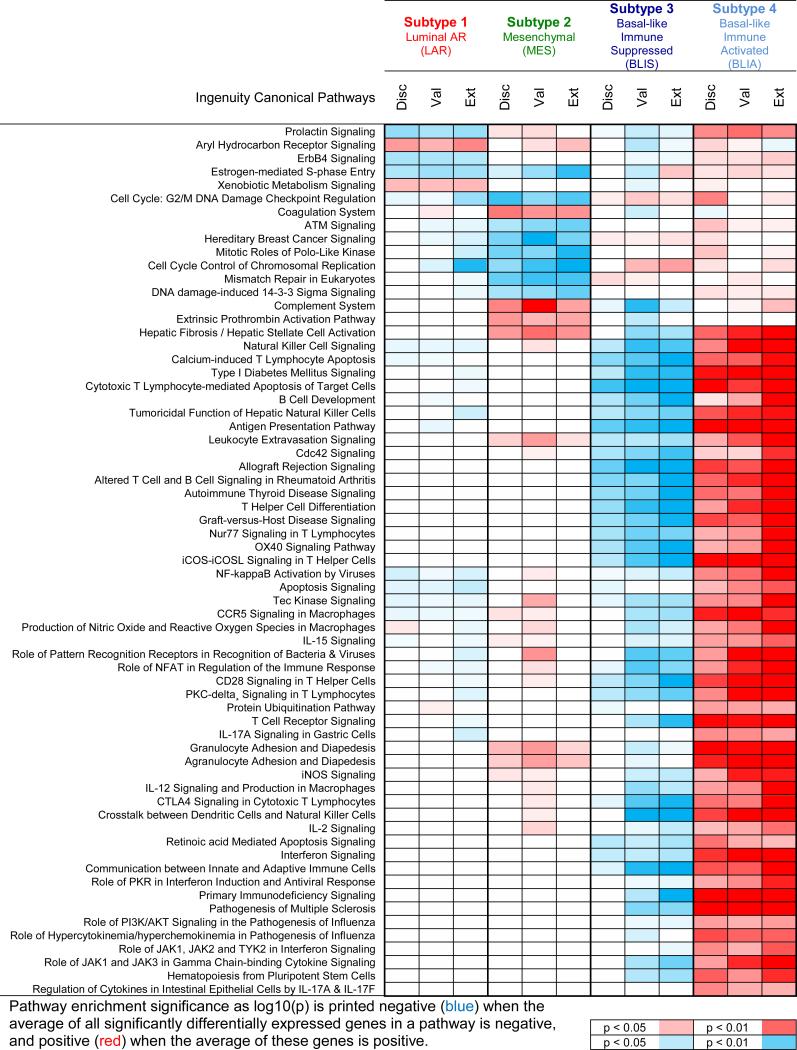
Differentially expressed genes from each subtype (BH-adjusted p-value<0.001 from GGT) were analyzed for pathway enrichment. Results from the validation and external sets significantly overlapped the discovery set, with predicted regulator activation and inhibition patterns stable across the three datasets but distinct between subtypes (Tables S25-29, Figure 3).
Figure 3. Molecular pathways enriched in the four identified subtypes of TNBCs.
Significant pathways from the discovery set also found in validation and external sets are listed for the LAR, MES, BLIS and BLIA subtypes.
Subtype 1 tumors exhibit androgen receptor, ER, prolactin, and ErbB4 signaling (Figure 3), but ER-alpha-negative IHC staining. Gene expression profiling demonstrates expression of ESR1 (the gene encoding ERα; Figure S6), and other estrogen-regulated genes (PGR, FOXA, XBP1, GATA3). Thus, these “ER-negative” tumors demonstrate molecular evidence of ER activation. This may be because 1% of these tumor cells express low levels of ER protein, defining them as “ER-negative” by IHC analysis. These observations suggest subtype 1 tumors may respond to traditional anti-estrogen therapies as well as to anti-androgens, as previously suggested12. To be consistent with previous studies12, we termed Subtype 1 the Luminal/Androgen Receptor (LAR) subtype.
Subtype 2 is characterized by pathways known to be regulated in breast cancer, including cell cycle, mismatch repair and DNA damage networks, and hereditary breast cancer signaling pathways (Figure 3). Additionally, genes normally exclusive to osteocytes (OGN) and adipocytes (ADIPOQ, PLIN1), and important growth factors (IGF-1) are highly expressed in this subtype, previously described as “mesenchymal stem-like” or “claudin-low” (Figure S7). Therefore, we named Subtype 2 the Mesenchymal (MES) subtype.
Subtype 3 is one of two basal-like clusters, and exhibits downregulation of B cell, T cell, and natural killer cell immune-regulating pathways and cytokine pathways (Figure 3). This subtype has the worst DFS and DSS, and low expression of molecules controlling antigen presentation, immune cell differentiation, and innate and adaptive immune cell communication. However, this cluster uniquely expresses multiple SOX family transcription factors. We termed Subtype 3 the Basal-Like Immune Suppressed (BLIS) subtype.
Immune regulation pathways are upregulated in Subtype 4, the other basal-like cluster (Figure 3). Contrary to BLIS, Subtype 4 tumors display upregulation of genes controlling B cell, T cell, and natural killer cell functions. This subtype has the best prognosis, exhibits activation of STAT transcription factor-mediated pathways, and has high expression of STAT genes. To contrast BLIS tumors, we termed Subtype 4 the Basal-Like Immune Activated (BLIA) subtype.
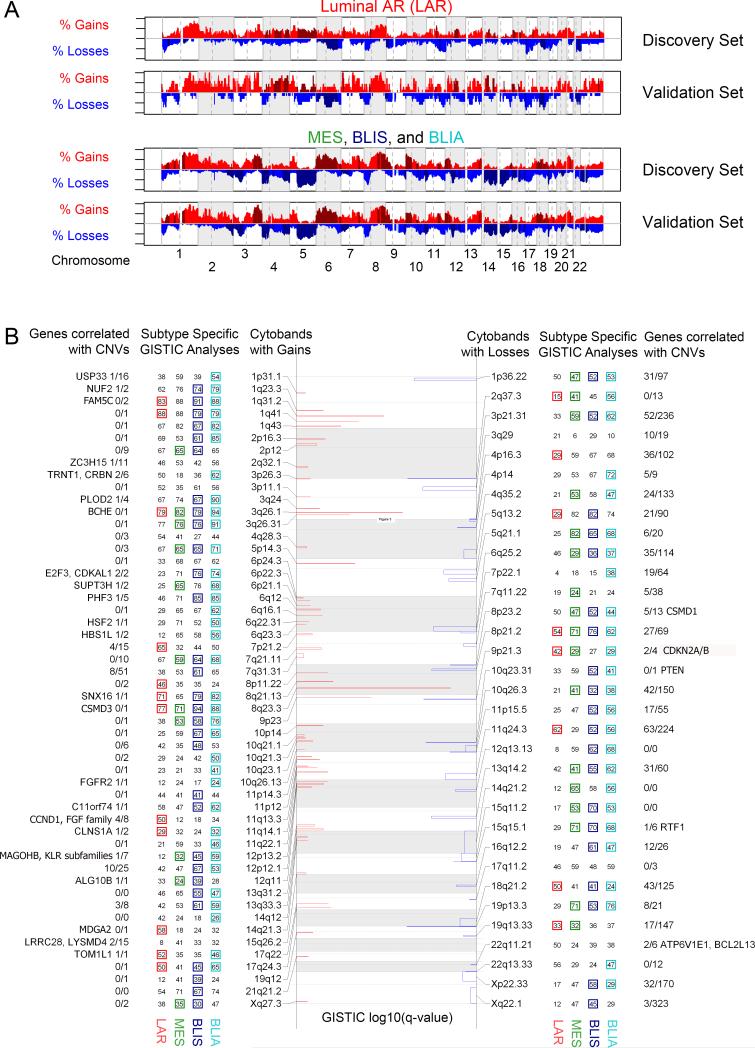
DNA copy number analysis identifies TNBC subtype-specific focal changes
We next investigated TNBC subtype-defined CN variation (CNV) by ploidy- and tumor percentage-correcting 62 discovery and 46 validation set TNBCs, before analyzing them together in GISTIC 2.0. Overall, genomes were very unstable and exhibited common TNBC chromosomal arm gains and deletions (Tables S30-35, Figure 4A, S7-8). Focal variations present in all 4 TNBC subtypes include: 1) focal gains on 8q23.3 (CSMD3), 3q26.1 (BCHE), and 1q31.2 (FAM5C), which are the greatest gains and characterize >84% of all tumors; and 2) focal losses on 9p21.3 (CDKN2A/B), 10q23.31 (PTEN), and 8p23.2 (CSMD1) (Figure 4B).
Figure 4. DNA Copy Number Analysis identifies focal changes in TNBC subtypes.
DNA copy number changes observed in each subtype are listed. A. Focal gains (red) and losses (blue) detected by GISTIC 2.0 are plotted by log10(q-value) and reported by cytoband. Adjacent numbers are percentages of subtype specific cases (n = 24, 17, 33, 34, respectively) with this focal aberration. Presence of a colored square demonstrates this region was detected by subtype-specific GISTIC 2.0 analysis as well. All structural events for each subtype and set are available in the supplemental. B. Broad copy number events distinguish the LAR subtype from all others. Gains (red) and losses (blue) are plotted along the genome, with darker colors representing a region enriched to the displayed subtype by Fisher Exact Test.
Subtype-specific variation is greatest between LAR and the remaining 3 subtypes (Figure 4). LAR tumors have focal gains twice as frequently on 11q13.3 (CCND1, FGF family) and 14q21.3 (MDGA2), but 1/3 as frequently on 12p13.2 (MAGOHB, KLR subfamilies) and 6p22.3 (E2F3, CDKAL1) compared to MES, BLIS, and BLIA tumors (Figure 4). The LAR subtype also has more frequent deletions of 6q, lacks arm-wide deletions across 5q, 14q, and 15q, and has significantly fewer focal deletions on 5q13.2 (RAD17, ERBB2IP), 12q13.13 (CCNT1, ERBB3), 14q21.2 (FOXA1), and 15q11.2 (HERC2) (Figures 4, S8). MES and BLIA tumors, which exhibit increased normal (diploid) immune cell infiltration, are characterized by lower aberrant cell fractions than LAR and BLIS tumors (Figure S9). Additional subtype-specific gene overexpression includes: 1) LAR: AR, MUC1; 2) MES: IGF-1, ADRB2, EDNRB, PTGER3/4, PTGFR, PTGFRA; 3) BLIS: VTCN1; 4) BLIA: CTLA4 (Tables 2, S36-39).
Table 2.
Selected genes from pathway analysis with significant relative overexpression (>2-fold, BH p≤0.05) in discovery and validation sets.
| TNBC Subtype | Symbol | Description | Discovery Fold-Change | Druggable | CNV Seen |
|---|---|---|---|---|---|
| 1: Luminal AR (LAR) | DHRS2 | dehydrogenase/reductase (SDR family) member 2 | 68.6 | ||
| PIP | prolactin-induced protein | 21.1 | |||
| AGR2 | anterior gradient 2 homolog (Xenopus laevis) | 17.1 | Yes | ||
| FOXA1 | forkhead box A1 | 17.1 | Yes | ||
| ESR1 | estrogen receptor 1 | 13.9 | Yes | ||
| ERBB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | 11.3 | Yes | ||
| CA12 | carbonic anhydrase XII | 11.3 | Yes | ||
| AR | androgen receptor | 9.8 | Yes | ||
| TOX3 | TOX high mobility group box family member 3 | 7.5 | Yes | ||
| KRT18 | keratin 18 | 4.3 | Yes | ||
| MUC1 | mucin 1, cell surface associated | 4.3 | Yes | ||
| PGR | progesterone receptor | 3.5 | Yes | ||
| ERBB3 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | 3 | Yes | ||
| RET | ret proto-oncogene | 2.5 | Yes | ||
| ITGB5 | integrin, beta 5 | 2.1 | Yes | ||
| 2: Mesenchymal (MES) | ADH1B | alcohol dehydrogenase 1B (class I), beta polypeptide | 42.2 | Yes | |
| ADIPOQ | adiponectin, C1Q and collagen domain containing | 32 | |||
| OGN | osteoglycin | 16 | |||
| FABP4 | fatty acid binding protein 4, adipocyte | 14.9 | |||
| CD36 | CD36 molecule (thrombospondin receptor) | 14.9 | |||
| NTRK2 | neurotrophic tyrosine kinase, receptor, type 2 | 6.1 | Yes | ||
| EDNRB | endothelin receptor type B | 5.7 | Yes | ||
| GHR | growth hormone receptor | 4.9 | Yes | ||
| ADRA2A | adrenoceptor alpha 2A | 4.6 | Yes | ||
| PLA2G2A | phospholipase A2, group IIA (platelets, synovial fluid) | 4.6 | Yes | ||
| PPARG | peroxisome proliferator-activated receptor gamma | 4 | Yes | ||
| ADRB2 | adrenoceptor beta 2, surface | 3.5 | Yes | ||
| PTGER3 | prostaglandin E receptor 3 (subtype EP3) | 3.2 | Yes | ||
| IL1R1 | interleukin 1 receptor, type I | 3 | Yes | ||
| TEK | TEK tyrosine kinase, endothelial | 2.8 | Yes | ||
| 3: Basal-like Immune Suppressed (BLIS) | ELF5 | E74-like factor 5 (ets domain transcription factor) | 7 | ||
| HORMAD1 | HORMA domain containing 1 | 5.7 | Yes | ||
| SOX10 | SRY (sex determining region Y)-box 10 | 4.9 | Yes | ||
| SERPINB5 serpin peptidase inhibitor, clade B (ovalbumin), member 5 | 4.6 | ||||
| FOXC1 | forkhead box C1 | 4.6 | |||
| SOX8 | SRY (sex determining region Y)-box 8 | 4.3 | |||
| TUBB2B | tubulin, beta 2B class IIb | 3.2 | Yes | ||
| VTCN1 | V-set domain containing T cell activation inhibitor 1 | 3 | |||
| SOX6 | SRY (sex determining region Y)-box 6 | 3 | |||
| KIT | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | 2.5 | Yes | ||
| FGFR2 | fibroblast growth factor receptor 2 | 2 | Yes | Yes | |
| 4: Basal Immune Activated (BLIA) | CXCL9 | chemokine (C-X-C motif) ligand 9 | 5.3 | ||
| IDO1 | indoleamine 2,3-dioxygenase 1 | 4.9 | |||
| CXCL11 | chemokine (C-X-C motif) ligand 11 | 4.9 | |||
| RARRES1 | retinoic acid receptor responder (tazarotene induced) 1 | 4 | Yes | ||
| GBP5 | guanylate binding protein 5 | 4.3 | Yes | ||
| CXCL10 | chemokine (C-X-C motif) ligand 10 | 4.3 | Yes | ||
| CXCL13 | chemokine (C-X-C motif) ligand 13 | 4.3 | |||
| LAMP3 | lysosomal-associated membrane protein 3 | 3.7 | Yes | ||
| STAT1 | signal transducer and activator of transcription 1, 91kDa | 3 | |||
| PSMB9 | proteasome (prosome, macropain) subunit, beta type, 9 | 2.8 | Yes | ||
| CD2 | CD2 molecule | 2.5 | Yes | ||
| CTLA4 | cytotoxic T-lymphocyte-associated protein 4 | 2.5 | Yes | ||
| TOP2A | topoisomerase (DNA) II alpha 170kDa | 2.1 | Yes | Yes | |
| LCK | lymphocyte-specific protein tyrosine kinase | 2.1 | Yes | ||
DISCUSSION
Using RNA and DNA profiling, we identified four stable, molecularly-defined TNBC subtypes, LAR, MES, BLIS, and BLIA, characterized by distinct clinical prognoses, with BLIS tumors having the worst and BLIA tumors having the best outcome. DNA analysis demonstrated subtype-specific gene amplifications, suggesting the possibility of using in situ hybridization techniques to identify these TNBC subsets. Our results also demonstrate subtype-specific molecular expression, thereby enabling TNBC subtype classification based on molecules they do express as opposed to molecules they do not express.
Many highly expressed molecules in specific TNBC subtypes can be targeted using available drugs (Tables 2, S36-39). Our results suggest that AR antagonists12 and MUC1 vaccines may prove effective for the treatment of AR- and MUC1-overexpressing LAR tumors, while beta-blockers, IGF inhibitors, or PDGFR inhibitors may be useful therapies for MES tumors. Conversely, immune-based strategies (e.g., PD1 or VTCN1 antibodies) may be useful treatments for BLIS tumors, whereas STAT inhibitors, cytokine or cytokine receptor antibodies, or the recently FDA-approved CTLA4 inhibitor, ipilumimab31 may be effective treatments for BLIA tumors. Thus, these studies have identified novel TNBC subtype-specific markers that distinguish prognostically distinct TNBC subtypes and may be targeted for more effective treatment of TNBCs.
Lehmann's TNBC-subtyping study identified six TNBC subtypes through the combined analysis of 14 RNA profiling datasets (“discovery dataset”)12. Assignment to these subtypes was confirmed using a second dataset comprised of 7 other publically-available datasets, however all six subtypes were not detected when subtyping was limited to only those tumors with ER, PR, and HER2 IHC data. In addition, basal-like-1 and basal-like-2 tumors are not readily distinguishable by hierarchical clustering of public TNBC data sets using Lehmann's gene signatures32, despite demonstration of molecular heterogeneity beyond the classic intrinsic subtypes. In Lehmann's study, TNBCs strongly segregated into stromal, immune, and basal gene modules, partially supporting our model. Additional studies have also demonstrated that an immune signature is an important clinical predictor for ER-negative tumors33,27,34. The large set of ER-, PR-, and HER2-characterized tumors used in our study enabled us to further separate TNBCs into LAR, MES (including “claudin-low”), BLIS, and BLIA subtypes, and define the clinical outcome of each subtype.
Previous genomic profiling studies have not demonstrated this degree of heterogeneity in basal-like breast tumors. Profiling of TCGA data across miRNA, DNA, and methylation data supported the intrinsic subtypes of breast cancer and grouped all basal-like tumors8. In the Curtis dataset11, unsupervised clustering by CNV-driven gene expression did not identify multiple basal-like subtypes, confirming that CNV alone does not distinguish these tumor subtypes. However, our integrated DNA and mRNA data demonstrate that gene amplification drives several subtype-specific genes. The CCND1 and FGFR2 genes are amplified in LAR tumors, while MAGOHB is more commonly amplified in MES, BLIS and BLIA tumors. Conversely, CDK1 is amplified in all 4 TNBC subtypes (most highly in BLIA tumors) and thus represents a potential target. While broad and focal CNs differentiate LAR tumors from the remaining subtypes, they cannot dissociate BLIS and BLIA tumors.
All LAR and most mesenchymal stem-like tumors identified by the Pietenpol group12 fall within our LAR and MES subtypes. However, our study splits the remaining proposed subtypes, including Lehmann's basal-like-1 and basal-like-2 tumors into distinct BLIS and BLIA subtypes based on immune signaling. Furthermore, stratification of our subtypes is based on a few broad biological functions. LAR and MES tumors downregulate cell cycle regulators and DNA repair genes, while MES and BLIA tumors upregulate immune signaling and immune-related death pathways (Table S36-39). Conversely, our BLIS and BLIA subtypes show a relative lack of P53-dependent gene activation (P53 mutations characterize most TNBC tumors), and BLIA tumors highly express and activate STAT genes. Both our current study and the study by Lehmann et al. used RNA-based gene profiling to subtype TNBCs. Until more TNBC datasets are analyzed, it will not be clear which specific subgrouping will ultimately be most clinically useful. The study by Lehmann et al. subdivided TNBCs into 6 subtypes while this manuscript describes subgrouping of TNBCs into 4 distinct subtypes, 2 of which overlap with Lehmann et al. (LAR & MES), while our other 2 subtypes (BLIS & BLIA) contain mixtures of the other 4 Lehmann subgroups (see Figure 1 C&F). Our attempt at reproducing the 6 Lehmann et al. subgroups by clustering our data using their gene signatures was unsuccessful (n = 198, Figure S5). The exact subdivision of these TNBC subtypes, while important, is less important than the clinical prognosis defined by each subtype, and most importantly, the specific molecular targets identified within the subtypes. To this point, the identification of specific targets that modulate the immune system in the BLIA and BLIS subtypes is one of the most important and unique findings in this study.
In summary, using RNA profiling we have defined 4 stable, clinically-relevant subtypes of TNBC characterized by distinct molecular signatures. Our results uniquely define TNBCs by the molecules that are expressed in each subtype as opposed to molecules that are not expressed. Furthermore, these newly defined subtypes are biologically diverse, activate distinct molecular pathways, have unique DNA CNVs, and exhibit distinct clinical outcomes. By identifying molecules highly expressed in each TNBC subtype, this study provides the foundation for future TNBC subtype-specific molecularly-targeted and/or immune-based strategies for more effective treatment of these aggressive tumors.
Supplementary Material
Statement of Translational Relevance.
This study describes the results of RNA and DNA genomic profiling of a large set of triple-negative breast cancers. We identified four stable triple-negative breast cancer subgroups with distinct clinical outcomes defined by specific over-expressed or amplified genes. The four subgroups have been named the “Luminal / Androgen Receptor (LAR)”, “mesenchymal (MES)”, “basal-like / immune-suppressed (BLIS)”, and “basal-like / immune activated (BLIA)” groups. We also identified specific molecules that define each subgroup, serving as subgroup-specific biomarkers, as well as potential targets for the treatment of these aggressive breast cancers. Specific biomarkers and targets include the androgen receptor, MUC-1, and several estrogen-regulated genes for the LAR subgroup; IGF-1, prostaglandin F receptor for the MES subgroup; SOX transcription factors and the immune regulatory molecule VTCN1 for the BLIS subgroup; and STAT transcription factors for the BLIA group. Thus, these studies form the basis to develop molecularly targeted therapy for triple-negative breast cancers.
Acknowledgements
The authors acknowledge important contributions from Mr. Aaron Richter for administering and coordinating the Komen Promise Grant, Ms. Samantha Short for her administrative assistance, Lester and Sue Smith for support of the Baylor College of Medicine tumor bank, Ms. Carol Chenault and Mr. Bryant L. McCue for their management of this tumor bank, and the significant contribution from the women who provided tumor samples for this study.
Financial Support: This work was funded by the MD Anderson Cancer Center Support Grant (CCSG) (1CA16672), the Dan L. Duncan Cancer Center Support Grant, Baylor College of Medicine, and a Susan G. Komen Promise Grant (KG081694, P.H.B., G.B.M).
Footnotes
Author Contributions:
Conception and design: Matthew D. Burstein, Ching Lau, Jenny Chang, C. Kent Osborne, Susan Hilsenbeck, Gordon Mills, and Powel H. Brown
Development of methodology: Ching Lau and Powel Brown
Acquisition of data: Anna Tsimelzon, Susan Hilsenbeck, Alejandro Contreras, Suzanne Fuqua, Jenny Chang
Analysis and interpretation of data: Matthew D. Burstein, Anna Tsimelzon, Susan Hilsenbeck, Graham Poage, Kyle Covington, Ching Lau, Gordon Mills, Powel Brown
Writing, review and/or revision of the manuscript: Matthew Burstein, Michelle Savage, Ching Lau, Gordon Mills, and Powel Brown, with input from remaining authors
Administrative, technical or material support: Michelle Savage
Study supervision: Ching Lau and Powel Brown
Financial support: Powel H. Brown and Gordon B. Mills (co-PIs of the Susan G. Komen for the Cure Promise Grant), C. Kent Osborne (for support of the Lester and Sue Smith Baylor College of Medicine Breast Tumor Bank)
No prior or subsequent publication: The authors confirm that this manuscript, nor any similar manuscript, in whole in or part (aside from an abstract), is under consideration, in press, or published elsewhere.
Conflict of interest: PH Brown is on the Scientific Advisory Board of Susan G. Komen for the Cure. All remaining authors declare no actual, potential, or perceived conflict of interest that would prejudice the impartiality of this article.
REFERENCES
- 1.Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol. 2005 Oct 10;23(29):7350–60. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
- 2.Morris GJ, Naidu S, Topham AK, Guiles F, Xu Y, McCue P, et al. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: a single-institution compilation compared with the National Cancer Institute's Surveillance, Epidemiology, and End Results database. Cancer. 2007 Aug 15;110(4):876–84. doi: 10.1002/cncr.22836. [DOI] [PubMed] [Google Scholar]
- 3.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnson H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001 Sep 11;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malorni L, Shetty PB, De Angelis C, Hilsenbeck S, Rimawi MF, Elledge R, et al. Clinical and biologic features of triple-negative breast cancers in a large cohort of patients with long-term follow-up. Breast Cancer Res Treat. 2012 Dec;136(3):795–804. doi: 10.1007/s10549-012-2315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shastry M, Yardley DA. Updates in the treatment of basal/triple-negative breast cancer. Curr Opin Obstet Gynecol. 2013 Feb;25(1):40–8. doi: 10.1097/GCO.0b013e32835c1633. [DOI] [PubMed] [Google Scholar]
- 6.Lee JM, Ledermann JA, Kohn EC. PARP Inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Ann Oncol. 2014 Jan;25(1):32–40. doi: 10.1093/annonc/mdt384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000 Aug 17;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012 Oct 4;490(7428):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2011;16(Suppl 1):61–70. doi: 10.1634/theoncologist.2011-S1-61. [DOI] [PubMed] [Google Scholar]
- 10.Bertucci F, Finetti P, Birnbaum D. Basal breast cancer: a complex and deadly molecular subtype. Curr Mol Med. 2012 Jan;12(1):96–110. doi: 10.2174/156652412798376134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012 Apr 18;486(7403):346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011 Jul;121(7):2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011 Feb;5(1):5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004 Feb 12;20(3):307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 17.R Core Team R: A Language and Environment for Statistical Computing, (Ver.2.12.2). R Foundation for Statistical Computing. 2012 [Google Scholar]
- 18.Chen X, Li J, Gray WH, Lehmann BD, Bauer JA, Shyr Y, et al. TNBCtype: A Subtyping Tool for Triple-Negative Breast Cancer. Cancer Inform. 2012;11:147–56. doi: 10.4137/CIN.S9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009 Mar 10;27(8):1160–7. doi: 10.1200/JCO.2008.18.1370. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams C, Edvardsson K, Lewandowski SA, Ström A, Gustafsson JA. A genome-wide study of the repressive effects of estrogen receptor beta on estrogen receptor alpha signaling in breast cancer cells. Oncogene. 2008 Feb 7;27(7):1019–32. doi: 10.1038/sj.onc.1210712. [DOI] [PubMed] [Google Scholar]
- 21.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011 Jun 15;27(12):1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang YH, Xiao Y, Segal MR. Identifying differentially expressed genes from microarray experiments via statistic synthesis. Bioinformatics. 2005 Apr 1;21(7):1084–93. doi: 10.1093/bioinformatics/bti108. [DOI] [PubMed] [Google Scholar]
- 23.Kim PM, Tidor B. Subsystem identification through dimensionality reduction of large-scale gene expression data. Genome Res. 2003 Jul;13(7):1706–18. doi: 10.1101/gr.903503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010 Jul 2;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004 Jan 1;20(1):93–9. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 26.Partek Inc. Partek® Discovery SuiteTM, (Ver. 6.3) Partek Inc.; St. Louis: 2008. [Google Scholar]
- 27.Sabatier R, Finetti P, Mamessier E, Raynaud S, Cervera N, Lambaudie E, et al. Kinome expression profiling and prognosis of basal breast cancers. Mol Cancer. 2011 Jul 21;10:86. doi: 10.1186/1476-4598-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Loo P, Nordgard SH, Lingjærde OC, Russnes HG, Rye IH, Sun W, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010 Sep 28;107(39):16910–5. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Hayes DN, Nobel A, Marron J. Statistical Significance of Clustering for High-Dimension, Low–Sample Size Data. Journal of the American Statistical Association. 2008;103:1281–1293. [Google Scholar]
- 31.Stagg J, Allard B. Immunotherapeutic approaches in triple-negative breast cancer: latest research and clinical prospects. Ther Adv Med Oncol. 2013 May;5(3):169–81. doi: 10.1177/1758834012475152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18(2):123–33. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8(8):R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rody A, Karn T, Liedtke C, Pusztai L, Ruckhaeberle E, Hanker L, et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2201. 2011 Oct 6;13(5):R97. doi: 10.1186/bcr3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.