Abstract
Natural products containing carbon-phosphorus bonds elicit important bioactivity in many organisms. L-phosphinothricin contains the only known naturally-occurring carbon-phosphorus-carbon bond linkage. In actinomycetes, the cobalamin-dependent radical S-adenosyl-L-methionine (SAM) methyltransferase PhpK catalyzes the formation of the second C-P bond to generate the complete C-P-C linkage in phosphinothricin. Here we use electron paramagnetic resonance and nuclear magnetic resonance spectroscopies to characterize and demonstrate the activity of a cobalamin-dependent radical SAM methyltransferase denoted SD_1168 from Shewanella denitrificans OS217, a marine bacterium that has not been reported to synthesize phosphinothricin. Recombinant, refolded, and reconstituted SD_1168 binds a four-iron, four-sulfur cluster that interacts with SAM and cobalamin. In the presence of SAM, a reductant, and methylcobalamin, SD_1168 surprisingly catalyzes the P-methylation of N-acetyl-demethylphosphinothricin and demethylphosphinothricin to produce N-acetyl-phosphinothricin and phosphinothricin, respectively. In addition, this enzyme is active in the absence of methylcobalamin if the strong reductant titanium (III) citrate and hydroxocobalamin are provided. When incubated with [methyl-13C] cobalamin and titanium citrate, both [methyl-13C] and unlabeled N-acetylphosphinothricin are produced. Our results suggest that SD_1168 catalyzes P-methylation using radical SAM-dependent chemistry with cobalamin as a coenzyme. In light of recent genomic information, the discovery of this P-methyltransferase suggests that S. denitrificans produces a phosphinate natural product.
Keywords: radical S-adenosyl-L-methionine, methyltransferase, cobalamin, phosphinate
1. Introduction
Phosphonate and phosphinate natural products contain carbon-phosphorus (C-P) bonds and have noteworthy bioactivities due to their structural similarities to phosphate esters, carboxylic acids, and various tetrahedral intermediates in enzymatic reactions [1]. These compounds are widely used as antimicrobial, antifungal, and herbicidal agents [2-4]. Such applications, as well as the extraordinary biochemical reactions thought to be involved in the biosynthesis of C-P compounds, have inspired a wealth of research. The majority of C-P compound biosynthetic pathways begin with the reaction catalyzed by phosphoenolpyruvate mutase (PepM), which forms the initial C-P bond via isomerization of phosphoenolpyruvate to generate phosphonopyruvate. This energetically unfavorable reaction is usually driven by decarboxylation catalyzed by phosphonopyruvate decarboxylase (Ppd) to generate phosphonoacetaldehyde [1].
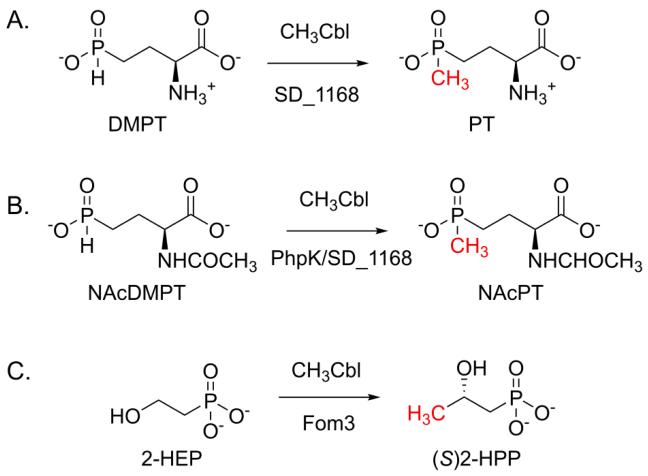
L-phosphinothricin (PT; L-2-amino-4-hydroxymethylphosphinylbutanoate) is an unusual amino acid that contains the only known naturally occurring C-P-C bond sequence (Figure 1A). PT is a L-glutamate analog and has herbicidal and antimicrobial activity through inhibiting plant and bacterial glutamine synthetases [5-7]. PT is produced as part of a tripeptide by Streptomyces hygroscopicus, Streptomyces viridochromogenes, and Kitasatospora phosalacinea [8-10]. In the two Streptomyces species, at least 24 genes are required for biosynthesis of the PT tripeptide, L-PT-Ala-L-Ala (PTT) [11, 12]. Although the biosynthetic pathway for phosalacine (L-PT-L-Ala-L-Leu) in K. phosalacinea has not been investigated, it is likely to be similar. These tripeptides are readily absorbed by target cells, where intracellular peptidases release the active PT antibiotic. In the latter stages of PT biosynthesis, the P-methyltransferase PhpK is thought to append a methyl group to the phosphinate precursor 2-acetylamino-4-hydroxyphosphinylbutanoate (N-acetyldemethylphosphinothricin or NAcDMPT) to produce 2-acetylamino-4-hydroxymethylphosphinylbutanoate (N-acetylphosphinothricin or NAcPT), which contains the final C-P-C bond sequence (Figure 1B) [11, 13-15]. In a randomly-generated S. hygroscopicus mutant that could not catalyze P-methylation, NAcDMPT and its tripeptide, N-acetyldemethylphosphinothricin tripeptide (NAcDMPTT) accumulated, suggesting these two N-acetylated metabolites were substrates for PhpK (Figure 1B) [14]. Furthermore, only the N-acetylated precursors were methylated by S. hygroscopicus cell lysates while the corresponding non-acetylated precursors, demethylphosphinothricin (DMPT; 2-amino-4-hydroxyphosphinylbutanoate) and DMPT tripeptide (DMPTT), were not methylated. Isotopic labeling studies demonstrated that methylcobalamin (CH3Cbl) was the methyl group donor for the P-methylation reaction [5,13].
Figure 1.
P-methylation and C-methylation reactions of interest.
In 2001, PhpK was identified as a radical S-adenosyl-L-methionine (SAM) superfamily member based on the presence of a highly conserved CxxxCxxC motif whose cysteine residues coordinate a four-iron, four-sulfur ([4Fe-4S]) cluster [16]. The reduced [4Fe-4S]+1 cluster and SAM are used to generate a 5′-deoxyadenosyl radical (Ado-CH2•) that abstracts a hydrogen atom from a substrate as the first step of catalysis (Figure 2) [17]. PhpK belongs to a subset of radical SAM enzymes that contain a cobalamin (Cbl)-binding domain [16, 18]. In vitro studies in our laboratory demonstrated that PhpK from K. phosalacinea catalyzes the P-methylation of NAcDMPT to produce NAcPT in a SAM-, sodium dithionite-, and CH3Cbl-dependent manner (Figure 1B) [15]. Three related Cbl-dependent radical SAM methyltransferases TsrM, GenK, and Fom3, have been reported upon since the initial PhpK work [19-21]. These enzymes are found in bacterial biosynthetic pathways for the antibiotics thiostrepton, gentamicin, and fosfomycin, respectively. Although some similarities exist between known members of the Cbl-dependent radical SAM family, a variety of differences have been reported, and many mechanistic details remain unresolved.
Figure 2.
Radical SAM cleavage.
Here, we describe the characterization of a Cbl-dependent radical SAM methyltransferase encoded by the sden_1168 gene from the denitrifying marine bacterium Shewanella denitrificans OS217 [22]. We will refer to the resulting protein as SD_1168. S. denitrificans OS217 has not been reported to biosynthesize known C-P compounds. However, its genome encodes phosphoenolpyruvate mutase (PepM; sden_1161) and phosphonopyruvate decarboxylase (Ppd; sden_1162, annotated as a thiamine pyrophosphate-dependent enzyme) (Figure 3) near the sden_1168 gene, suggesting that this organism has the capacity to produce C-P compounds [23].
Figure 3.
Comparison of putative S. denitrificans OS217 and Kitasatospora NRRL F-6133 C-P homologous genes [23]. Genes in color are homologous. Red indicates Cbl-dependent radical SAM methyltransferases. Genes in gray are not homologous. Not all genes from each putative pathway are shown. PepM: phosphoenolpyruvate mutase, Ppd: phosphonopyruvate decarboxylase.
Although the biological function of SD_1168 is currently unknown, the protein shares significant identity and similarity with both Fom3 and PhpK (34% and 53%, and 13% and 31%, respectively), methyltransferases involved in C-P compound biosynthesis [15, 21]. Fom3 is required for the penultimate step of fosfomycin biosynthesis, where it adds a methyl group to the sp3-hybridized C-2 carbon of 2-hydroxyethylphosphonate (2-HEP) to generate S-2-hydroxypropylphosphonate (S-2-HPP) (Figure 1C) [21,24,25]. The significant sequence similarity between SD_1168 and Fom3 led us to hypothesize that SD_1168 might catalyze a similar, or even the same, C-methylation as Fom3. To date, we have not observed SD_1168 C-methylation activity upon 2-HEP. Instead, we found that SD_1168 unexpectedly catalyzes P-methylation upon both NAcDMPT and DMPT to generate the final C-P-C bond sequence found in PT (Figures 1A and B). We show that SD_1168 is likely to perform radical SAM chemistry using a Cbl coenzyme for this difficult reaction. Since PT and its derivatives are the only naturally-occurring phosphinates discovered to date, our results suggest that S. denitrificans synthesizes an as-yet undiscovered phosphinate natural product.
2. Materials and Methods
2.1 Materials
Reagents were obtained from typical suppliers unless otherwise indicated. Titanium(III) (Ti) citrate and [methyl-13C]Cbl were synthesized as described elsewhere [26,27]. SAM was acquired from Safeway and was purified as described elsewhere [15]. 2-HEP was synthesized as described elsewhere [21].
2.2 Cloning of sden_1168 from S. denitrificans OS217 (GenBank Accession ABE54454.1)
S. denitrificans OS217 was obtained from the American Type Culture Collection (ATCC) (ATCC-BAA 1090), was reconstituted in liquid media according to ATCC recommendations, and was streaked onto marine broth 2216 (Difco, Sparks, MD) agar plates to obtain isolated colonies. A single colony was used to inoculate 5 mL of marine broth, and the culture was incubated with shaking at 30 °C overnight. Genomic DNA was isolated from the overnight culture using the Wizard Genomic DNA purification kit (Promega, Madison, WI). The sden_1168 gene was amplified from S. denitrificans genomic DNA using PCR. The forward primer was 5′-TATATACATATGCGACCAAATTTT TA-3′ and the reverse primer was 5′-TTTTGCGGCCGCTCATTCTTCATAAGC TT-3′. Restriction sites for NdeI and NotI, respectively, are underlined in each primer. Digestion and ligation of sden_1168 into pET-30a(+) (EMD Millipore/Merck KGaA) was performed using standard molecular biology techniques. Ligation mixtures were transformed into NovaBlue GigaSingles competent cells (EMD Millipore/Merck KGaA). Plasmids were isolated with the Spin Miniprep Kit (Qiagen, Valencia, CA) and sequenced to confirm the expected nucleotide sequence for sden_1168. The resulting construct was denoted sden_1168-pET-30a.
2.3 Overproduction, purification, and iron-sulfur cluster reconstitution of SD_1168
Overproduction and purification of SD_1168 was conducted as described elsewhere with minor modifications [15,21,28]. All purification and protein manipulations took place in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI). SD_1168 was allowed to refold overnight by itself (denoted “SD_1168a”), in the presence of SAM (denoted “SD_1168”), or in the presence of hydroxocobalamin (HOCbl; denoted “SD_1168b”). Protein concentrations were estimated using the method of Waddell [29]. Aliquots of SD_1168 were frozen anaerobically and stored in liquid nitrogen.
2.4 Iron and sulfide content and spectrophotometric analysis
The amount of iron and sulfide bound by SD_1168 was determined with an Agilent 8453 ultraviolet-visible (UV-Vis) spectrophotometer (Agilent Technologies, Santa Clara, CA) using methods described previously [21,30-32]. To determine iron content, samples contained 0.55-3.27 nmol of SD_1168, and a standard curve was developed using 1-25 nmol FeCl3. For sulfide determination, samples contained 0.22-0.87 nmol of SD_1168 and a standard curve was generated with 1-5 nmol Na2S. For spectrophotometric measurements, purified and reconstituted SD_1168 was first thawed in the anaerobic chamber. The protein was diluted with 20 mM potassium (K+) 4-(2-hydroxyethyl)piperazine-1-propanesulfonic acid (EPPS) and 1 mM magnesium sulfate (Buffer A) to a final concentration of ~30 μM, and 300 μL was transferred to an anaerobic cuvette for measurement.
2.5 DMPT synthesis
DMPT was synthesized according to a procedure developed by AsisChem Inc. (Watertown, MA) with some modifications [15]. To 14.8 g (0.04 mol) Z-L-glutamic acid α-benzyl ester (Chem-Impex, Wood Dale, IL) was added 50 mL benzene, 0.4 g copper acetate, and 224 μl pyridine. The mixture was stirred at 20 °C for 1 h, then 50 mL benzene and 8.8 g (0.027 mol) lead acetate were added. The reaction was refluxed under argon for 24 h, filtered through a celite pad, and rinsed with ethyl acetate (3 × 20 mL). The bright turquoise filtrate was washed with water (3 × 100 mL) and brine (3 × 100 mL), and the resulting yellow/brown organic layer was dried over 4 g anhydrous sodium sulfate. The product was purified by column chromatography over silica gel (ethyl acetate:hexanes, 1:9). Fractions containing the desired product were combined and concentrated by rotary evaporation. To the resulting yellow syrup was added 2 mL methanol, 8 mL 50% aqueous hypophosphorus acid, and 0.16 g 2,2′-azobis(2-methylpropionitrile). The reaction was stirred at 75 °C for 9 h. 30 mL water was added and the pH was adjusted to ~8 with saturated sodium carbonate. This mixture was washed with methyl tert-butyl ether (2 × 40 mL). The aqueous layer was adjusted to pH ~2 with 6 M hydrochloric acid. The desired product was extracted with dichloromethane (2 × 30 mL) and dried over anhydrous sodium sulfate. After concentration via rotary evaporation, 15 mL 6 M hydrochloric acid was added and the reaction was refluxed under argon for 3 h. The resulting DMPT product was concentrated using rotary evaporation, and the yellow gel was resuspended in ~3 mL water. DMPT was purified using cation exchange resin (Dowex AG-50, Acros Organics). The resin was washed stepwise with water followed by 10 mM sodium carbonate at pH 8, 9, and 10. DMPT was eluted with 10 mM sodium carbonate, pH 11. The eluant was concentrated via rotary evaporation, and the resulting yellow oil was transferred into the anaerobic chamber to deaerate. The concentration of DMPT was determined by 31P nuclear magnetic resonance (NMR) spectroscopy by comparison with a phosphonate standard of known concentration. DMPT was resuspended in 1 M K+ EPPS, pH 8 at a concentration of ~80-100 mM and stored in aliquots at −80 °C.
2.6 NAcDMPT synthesis
Crude DMPT was N-acetylated by adding 20 mL acetic acid, 28 μl triethylamine, and 4 mL acetic anhydride (in 500 μl aliquots over ~5 min). Acetylation was complete after ~24 h. The resulting NAcDMPT was concentrated and purified over silica gel in 90% methanol followed by cation exchange (Dowex AG-50, Acros Organics) eluted with water. Fractions containing NAcDMPT were combined and concentrated by rotary evaporation to leave a yellow oil. The concentration of NAcDMPT was determined by NMR spectroscopy by comparison with a phosphonate standard of known concentration. NAcDMPT was resuspended in 1 M K+ EPPS, pH 8 at a concentration of ~100 mM and stored at −80 °C.
2.7 Electron paramagnetic resonance (EPR) sample preparation and spectral collection
SD_1168 was thawed in the anaerobic chamber and diluted to a final concentration of ~100 μM in Buffer A. The protein was mixed with various combinations of the following components: sodium dithionite (4 mM), SAM (1 mM), NAcDMPT (1 mM), and CH3Cbl (0.5-1 mM). After ~1 h, the samples were transferred to 4 mm EPR tubes (Norell, Landisville, NJ), frozen in isopentane, and stored in liquid nitrogen. Low temperature X-band EPR spectra were obtained on a Bruker EMXplus spectrometer equipped with a Bruker EMXpremium microwave bridge, an Oxford Instruments ESR 900 continuous flow cryostat, and an Oxford Instruments ITC503 temperature controller. Bruker Xenon software (version 1.1b50) was used to acquire and manipulate spectra. Spectra were recorded under the following conditions: 9.379 GHz, 16 G modulation amplitude, 3400 G center field, 1000 G sweep width, 1 mW power, 0.3 s time constant, 10 K, 4 × 2 min scans.
2.8 Evaluation of enzymatic activities
SD_1168 was thawed in the anaerobic chamber and diluted to ~100 μM with Buffer A. Ti(III) citrate (~13.6 mM) or sodium dithionite (4 mM) was added to the protein and the mixture was incubated for 30-60 min. When dithionite was used as a reductant, the protein was exchanged into Buffer A to remove excess dithionite using a PD-10 desalting column (Sephadex G-25 M, GE Healthcare, Fairfield, CT). Then SAM (1 mM), NAcDMPT (1 mM) or DMPT (800 μM) or 2-HEP (1 mM), and CH3Cbl (1 mM), [methyl-13C]Cbl (1 mM), and/or HOCbl (1 mM) were added. Control reactions were performed in which one or more of the reaction components were omitted. Reactions were set up in triplicates (3 × 1 mL) except for the [methyl-13C]Cbl experiment, which was set up as 6 × 1 mL reactions. Reaction mixtures were incubated anaerobically for ~18 h at ~20 °C before removal from the anaerobic chamber. Centrifugation (10,000 rpm, 10 min) removed precipitated protein; any remaining protein was removed using polyethersulfone centrifugal filters (VWR, Radnor, PA). The resulting filtrate from reactions containing NAcDMPT was partially purified with water elution from cation exchange resin (AG-50, Acros Organics, Geel, Belgium) and concentrated via rotary evaporation. The resulting filtrate from reactions containing DMPT or 2-HEP were partially purified using a reverse-phase C4 column (Grace Vydac, Deerfield, IL, 10 mm × 250 mm) connected to a high performance liquid chromatography (HPLC) instrument (Beckman Coulter, System Gold, Brea, CA). The elution included a 20 min wash (0.1% formic acid in water, 1 mL/min) followed by a linear gradient from 0-100% acetonitrile. The phosphinates and phosphonates eluted during the initial wash and were collected and concentrated via rotary evaporation. The concentrated, partially purified reaction mixtures were resuspended in 550 μl deuterium oxide (Cambridge Isotope Labs, Tewksbury, MA) for NMR analysis. One-dimensional (1-D) 1H or 31P and two-dimensional (2-D) H-P gradient heteronuclear single quantum correlation (gHSQC) NMR spectra were collected at the Washington State University NMR Center using a Varian 600 MHz spectrometer at 22 °C [15,22]. gHSQC data were collected for 12-16 h for each enzymatic reaction sample. Peak positions varied between samples (up to 2 ppm along 1H and/or 10 ppm along 31P) due to differences in final pH/pD of the partially purified reactions.
2.9 Partial unfolding of SD_1168a to produce SAM-free protein (“SD_1168 -(SAM)”)
SD_1168a (~100 μM) that had originally been refolded without SAM and without HOCbl was thawed in the anaerobic chamber at room temperature (20 °C). Next, the enzyme (3 mL) was dialyzed stepwise into increasing concentrations of urea (1-3 M) in Buffer A (1 L) using a Slide-A-Lyzer cassette (Thermo Fisher Scientific, Rockford, IL). Three rounds of dialysis for 3 h each were performed at 20 °C in the anaerobic chamber. After the final round of dialysis (3 M urea), SD_1168 was light yellow in color, and the UV-Vis spectrum indicated that the protein no longer contained a [4Fe-4S] cluster. These results suggested that the enzyme had partially unfolded and had released any bound SAM. The enzyme was then transferred to a new container, 6-fold molar excess ferrous ammonium sulfate and sodium sulfide and 5 mM dithiothreitol (DTT) were added, and the mixture was diluted tenfold in Buffer A. This mixture was incubated overnight in the anaerobic chamber at ~20 °C. The protein was then concentrated using centrifugal filter units (Amicon Ultra-15, 30 kDa cut-off, EMD Millipore/Merck KGaA) spun at 5000 rpm and 4 °C to ~100 μM. The final protein, denoted “SD_1168 (−SAM)” was dialyzed into Buffer A and stored in liquid nitrogen.
3. Results and Discussion
3.1 Overexpression and refolding
The gene encoding SD_1168 from S. denitrificans was cloned into pET-30a(+) for protein overexpression. SD_1168 was found exclusively in inclusion bodies when overexpressed in either Escherichia coli or Streptomyces lividans. All efforts to express soluble recombinant protein were unsuccessful, as observed in our laboratory for related methyltransferases [15,21]. Therefore, we refolded SD_1168 using an established refolding procedure based upon a literature precedent [15,21,28]. In brief, the inclusion bodies were solubilized in urea. The soluble protein was subsequently refolded by dilution in urea-free buffer in the presence of iron, sulfide, and DTT. The protein was then concentrated, the [4Fe-4S] clusters were reconstituted, and the protein was dialyzed to remove excess iron, sulfide, and reductant. The final, typical yield was ~12 mg of SD_1168 protein per g of E. coli cell paste. SD_1168 was ~75% pure as assessed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE; Figure S1) and it migrated at its expected molecular mass of 64.8 kDa.
3.2 [4Fe-4S] cluster characterization
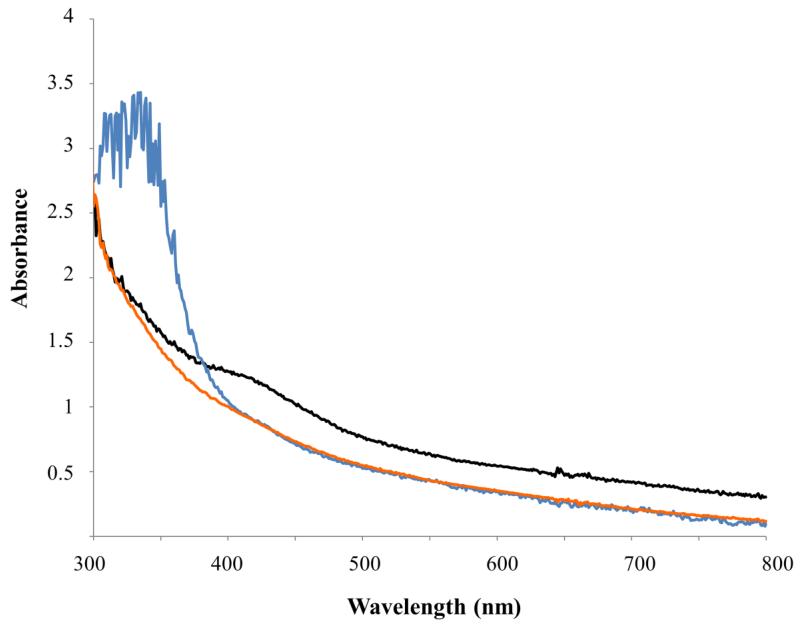
Reconstituted SD_1168 was dark brown in color and displayed a UV-Vis spectrum with a broad shoulder having an absorbance maximum at 420 nm (Figure 4, black line), characteristic of proteins containing [Fe-4S]+2 clusters [33,34]. This absorbance was quenched upon the addition of sodium dithionite or Ti citrate (Figure 4, orange and blue lines, respectively). Analysis of iron and sulfide content showed that SD_1168 bound 6.1 ± 0.5 mol of iron per mol protein and 4.0 ± 0.8 mol of sulfide per mol protein. The slightly higher than expected iron content (6.1 ± 0.5 mol iron per mol protein) is comparable to our data for the related Fom3 methyltransferase from S. wedmorensis [21] and suggests that adventitious iron may be bound elsewhere in the protein.
Figure 4.
UV-Vis spectra of ~30 μM SD_1168. Black: reconstituted SD_1168; orange: SD_1168 + 4 mM sodium dithionite; blue: SD_1168 + 10 mM Ti citrate.
To better characterize the [4Fe-4S] cluster, low temperature X-band EPR spectroscopy was employed since it is a valuable method for characterizing the oxidation state and chemical environment of paramagnetic centers in proteins [35]. The g-value and shape of the observed signal assist in the characterization of different types of unpaired electron spin in different chemical environments [36]. Refolded and reconstituted SD_1168 was EPR-silent (Figure 5A), which was consistent with the resting [4Fe-4S]+2 clusters we observed with UV-Vis spectrophotometry. Signals for an incomplete [3Fe-4S]+1 and/or oxidized [4Fe-4S]+3 cluster were not observed. Such catalytically-inactive forms of the cluster have been found in other radical SAM enzymes after purification and reconstitution [37-41], but our refolding and reconstitution procedure produced the complete [4Fe-4S]+2 cluster in SD_1168. Upon incubation with sodium dithionite, an EPR signal centered at g=1.93 was observed (Figure 5B). This feature is characteristic of the [4Fe-4S]+1 cluster in radical SAM enzymes.
Figure 5.
EPR spectra of SD_1168. Spectrum A: Refolded and reconstituted SD_1168, Spectrum B: SD_1168 + dithionite, Spectrum C: SD_1168 refolded with HO-Cbl + dithionite, Spectrum D: SD_1168 refolded with HO-Cbl + dithionite + SAM, Spectrum E: SD_1168 (−SAM) + dithionite.
When the protein was refolded in the presence of HOCbl (SD_1168b) and reduced with dithionite, a more intense signal for the +1 cluster was observed (Figure 5C) compared to the sample containing reduced SD_1168 refolded in the presence of added SAM (Figure 5B). This result suggests that Cbl binds near the [4Fe-4S] cluster and may raise its reduction potential, thus facilitating generation of the +1 cluster [42]. Addition of SAM and dithionite to SD_1168b produced new features in the EPR signal at g~2.04 (Figure 5D), providing evidence that SAM is a ligand for the cluster as observed for other radical SAM enzymes [43,44]. A concomitant decrease in the intensity of the EPR signal for the +1 cluster (compare Figures 5C and D) was also observed. This may be due to uncoupled cleavage of SAM that yields the EPR-silent +2 form of the cluster together with methionine and AdoCH3. When SAM is reductively cleaved to generate Ado-CH2• in the first step of radical SAM catalysis, the [4Fe-4S] cluster is oxidized to the EPR-silent +2 state (Figure 2). Many radical SAM enzymes catalyze uncoupled cleavage of SAM in the absence of substrate, especially when dithionite is used as a reducing agent [37,45-47]. However, we have not detected these cleavage products using other means (e.g., HPLC).
3.3 Enzymatic activities
After determining that SD_1168 demonstrated some of the expected characteristics of a radical SAM enzyme, we set out to determine its in vitro activity. Reaction mixtures were analyzed using 2-D gHSQC NMR spectroscopy [15]. Approximate levels of turnover were determined by integrating the resulting 2-D cross-peak volumes and comparing with the original amount of starting organophosphorus substrate. Activity results for SD_1168 are summarized in Table 1.
Table 1.
Methylation activity of SD_1168 upon various substrates. Unless otherwise noted, reactions were incubated anaerobically for ~18 h prior to NMR analysis. NR = No reaction.
| Enzyme preparation |
Reductant | Organophosphorus substrate |
Cobalamin substrate |
SAM present(+)/ absent(−) |
% Turnover |
[Product] (μM) |
Figure |
|---|---|---|---|---|---|---|---|
| − | Ti(III) citrate |
2-HEP | CH3Cbl | + | NR | NR | − |
| SD_1168 | Na2SO4 | 2-HEP | CH3Cbl | + | NR | NR | S2 |
| SD_1168 | Ti(III) citrate |
2-HEP | CH3Cbl | + | NR | NR | − |
| − | Ti(III) citrate |
NAcDMPT | CH3Cbl | + | NR | NR | S4A |
| SD_1168 | Na2SO4 | NAcDMPT | CH3Cbl | + | 0.3% | 18 | S3 |
| SD_1168 | Ti(III) citrate |
NAcDMPT | CH3Cbl | + | 3.7% (5 min) |
220 | Not shown |
| SD_1168 | Ti(III) citrate |
NAcDMPT | CH3Cbl | + | 4.8% (2 h) |
290 | Not shown |
| SD_1168 | Ti(III) citrate |
NAcDMPT | CH3Cbl | + | 7.4% | 400 | 5A |
| SD_1168 | − | NAcDMPT | CH3Cbl | + | NR | NR | S4B |
| SD_1168 | Ti(III) citrate |
NAcDMPT | − | + | NR | NR | S4C |
| SD_1168 (refolded without exogenous SAM) |
Ti(III) citrate |
NAcDMPT | CH3Cbl | − | 8.4% | 460 | S4D |
| SD_1168(− SAM) |
Ti(III) citrate |
NAcDMPT | CH3Cbl | − | NR | NR | S6 |
| SD_1168 | Ti(III) citrate |
NAcDMPT | HOCbl | + | 8.5% | 460 | 5C |
| SD_1168 | Ti(III) citrate |
NAcDMPT | [methyl- 13C]Cbl |
+ | 11.0% | 1100 | 5D |
| SD_1168 | Ti(III) citrate |
DMPT | CH3Cbl | + | 3.5% | 190 | 5B |
Given the sequence similarities between SD_1168, Fom3, and PhpK, we examined SD_1168 for both C-methyltransferase and P-methyltransferase activities. Due to the high sequence similarity between SD_1168 and Fom3, we initially hypothesized that SD_1168 would catalyze C-methylation. The Fom3 substrate, 2-HEP, and product, 2-HPP (Figure 1C), have distinguishable 1-D 31P and 2-D 1H-31P gHSQC NMR spectra [21]. Depending on the pH/pD of the final sample, the peaks for 2-HEP and 2-HPP are approximately 0.5-1 ppm apart along the phosphorus axis. The 2-D gHSQC experiment, which is two-to-fourfold more sensitive than 1-D 31P detection, was used to examine SD_1168 for C-methylation activity upon 2-HEP. However, when SD_1168 was incubated with dithionite or Ti citrate, SAM, 2-HEP, and CH3Cbl, C-methylation activity was not observed (Table 1; Figure S2). This result suggests that SD_1168 may not be a C-methyltransferase, although we have previously found that Fom3 is also quite difficult to assay for reproducible turnover [21].
Knowing that SD_1168 shared lower but still significant similarity with PhpK, the P-methyltransferase found in K. phosalacinea, S. hygroscopicus, and S. viridochromogenes Tü494, we decided to test SD_1168 for P-methylation activity. The potential substrates for SD_1168, NAcDMPT and DMPT, and the products of P-methylation, NAcPT and PT, respectively (Figures 1A and B), have easily distinguishable 1-D 31P and 2-D 1H-31P gHSQC NMR spectra [15]. We assessed SD_1168 for P-methylation activity using the more sensitive 2-D gHSQC method [15]. In these experiments, the signal positions depend upon final pH/pD and therefore vary as much as 10 ppm along the phosphorus axis and 2 ppm along the proton axis between samples. Standards at varying pH/pD and enzymatic reaction samples spiked with authentic product were used to confirm the identities of observed products.
Under our partial purification conditions, the phosphorus-coupled C-4 methylene protons of NAcDMPT typically produce a strong cross-peak centered at ~1.7 ppm (1H) and ~35 ppm (31P) (Figure 6A). When SD_1168 was incubated with dithionite, SAM, NAcDMPT, and CH3Cbl, a new cross-peak corresponding to the methyl phosphinate protons of NAcPT was observed (Table 1, Figure S3). This was an unexpected result since the genome of S. denitrificans does not encode most of the known enzymes required for PT biosynthesis. Turnover was poor under these conditions and resulted in only 0.3% product formation (Table 1). This is a significantly lower amount of turnover than PhpK from K. phosalacinea is capable of under similar conditions [15]. This result suggested that the reaction conditions used were suboptimal, perhaps due to differences in reduction potentials of the [4Fe-4S]+2/+1 couple or to as-yet unknown differences in the active sites of the two enzymes. The reduction potential of similar [4Fe-4S]+2/+1 interconversions in biotin synthase and clostridial lysine-2,3-aminomutase has been estimated to be ~-450 mV [42, 48]. In comparison, the reduction potential of sodium dithionite has been measured and or estimated to fall between −327 and −511 mV, depending upon concentration, pH, and purity [49-52].
Figure 6.
H-P gHSQC NMR spectra of partially purified, Ti citrate-reduced SD_1168-catalyzed reactions of interest. A. NAcDMPT + CH3Cbl reaction. The cross-peaks at (1.66, 35) ppm and (1.95, 35) ppm correspond to the C-4 methylene protons and the C-3 methylene protons, respectively, of NAcDMPT. The cross-peak at (1.35, 56) ppm corresponds to the methyl protons of NAcPT. B. DMPT + CH3Cbl reaction. The cross-peaks at (1.45, 26) ppm and (1.90, 26) ppm correspond to the C-4 methylene protons and the C-3 methylene protons, respectively, of DMPT. The cross-peak at (1.12, 41) ppm corresponds to the methyl protons of PT. C. NAcDMPT + [methyl-13C]Cbl reaction. The two cross-peaks indicated by the double daggers (‡) at (1.30, 56) ppm and (1.09, 56) ppm correspond to the methyl protons of [methyl-13C]NAcPT, which are split by the NMR-active 13C and 31P nuclei. The cross-peak indicated by the asterisk (*) at (1.20, 56) ppm corresponds to the methyl protons of unlabeled NAcPT. D. NAcDMPT + HOCbl reaction. The major cross-peak at (1.68, 35) ppm corresponds to the C-4 methylene protons of NAcDMPT, and the cross-peak at (1.29, 56.5) ppm corresponds to the methyl protons of NAcPT.
Our previous work with multiple Cbl-dependent radical SAM methyltransferases including PhpK and Fom3 has revealed that dithionite can inadvertently react with CH3Cbl to produce the presumably catalytically-incompetent Cbl(II). We do not observe dithionite-reduced SD_1168 P-methylation unless the enzyme is desalted prior to adding the other reaction components. In addition, dithionite binds to Cbl(II) [53], which may affect the ability of SD_1168 and related enzymes to perform more than one turnover. Finally, sodium dithionite is largely impure, contains a variety of breakdown products, and is unstable even after purification efforts [54]. Therefore, we sought an alternative cluster reductant.
Ti citrate is a one-electron reductant with a redox potential of −500 mV that has been successfully used to reduce [4Fe-4S] cofactors found in carbon monoxide dehydrogenase/acetyl-CoA synthase and the corrinoid iron-sulfur protein [55-57]. When Ti citrate was used in SD_1168 reactions instead of dithionite, significantly greater P-methylation activity (7.4% vs. 0.3%, Table 1) was observed as evidenced by the cross-peak at 1.35 ppm (1H) and 57.0 ppm (31P) (Figure 6A). In our hands, PhpK also appears to be more active in the presence of Ti citrate compared with dithionite [58]. For these reasons, Ti citrate was used for subsequent experiments with SD_1168.
Since titanium(III) is paramagnetic and interferes with the EPR signal for the +1 cluster, we did not obtain EPR evidence for Ti citrate reduction of the [4Fe-4S] cluster. Instead, we used UV-Vis spectrophotometry to monitor the cluster’s oxidation state. In the presence of Ti citrate, the broad shoulder characteristic of the [4Fe-4S]+2 cluster disappeared (Figure 4, blue line). This is evidence for successful reduction of the +2 cluster to the +1 state and is similar to our observations using sodium dithionite to reduce the cluster (Figure 4, orange line). To our knowledge, the use of Ti citrate to reduce [4Fe-4S] clusters of radical SAM enzymes has not been reported elsewhere. Thus, this compound represents a promising, low-potential reducing agent for other Cbl-dependent radical SAM methyltransferases. In vivo, the cluster reductant for SD_1168 is most likely a ferredoxin or flavodoxin; multiple forms of these electron carriers can be found in the genome of S. denitrificans OS217. Although others have reported that flavodoxin is a suitable reductant [20], we have tested recombinant flavodoxin and have not successfully observed activity for any Cbl-dependent radical SAM methyltransferase [58].
We also tested SD_1168 for methylation activity upon DMPT, another proposed intermediate in the PT biosynthetic pathway [1]. This activity is not catalyzed by PhpK, the “original” P-methyltransferase, which appears to require the N-acetyl modification for substrate recognition and binding. This modification, added by the Bar or Pat acetyltransferase depending on the microbe, confers resistance and is removed by a deacetylase immediately before export of the antibiotic [59]. There is currently no genomic evidence for the existence of either the acetyltransferase or the deacetylase in S. denitrificans OS217.
By gHSQC, the phosphorus-coupled C-4 methylene protons of DMPT give rise to a cross-peak centered at 1.45 ppm (1H) and 26 ppm (31P) (Figure 6B). Due to long-range coupling, the C-3 methylene protons are also observed as a cross-peak at 1.90 ppm (1H) and 26 ppm (31P). When SD_1168 was incubated with Ti citrate, SAM, DMPT, and CH3Cbl, approximately 3.5% turnover was observed as evidenced by the new cross-peak corresponding to the methyl group protons of PT at 1.12 ppm (1H) and 40.5 ppm (31P) (Table 1, Figure 6B). Unlike PhpK, SD_1168 does not require the N-acetyl modification for substrate recognition. This highlights an important difference in the active sites of these two enzymes and may provide a clue as to the structure of the physiological substrate for SD_1168. Although approximately twice as much product was formed from methylation of NAcDMPT compared to DMPT (Table 1), it currently remains unclear which phosphinate substrate is preferred by SD_1168.
3.4 Cofactor and substrate requirements and potential catalytic mechanism
Control experiments were performed in which one of each hypothesized required component was omitted. In the absence of SD_1168, reductant, or CH3Cbl, NAcPT formation was not observed (Table 1, Figures S4A-C). However, when SAM was omitted from the reactions, a cross-peak corresponding to ~8.4% NAcPT formation was observed (Table 1, Figure S4D). We hypothesized that this result was due to SD_1168 binding of SAM from cellular extracts during refolding and purification. Therefore, we partially unfolded purified SD_1168a, which had originally been refolded without SAM and without HOCbl, with urea to release any small molecules bound by the protein. The resulting preparation was refolded and reconstituted and was denoted SD_1168 (−SAM). UV-Vis spectrophotometry and EPR spectroscopy showed that SD_1168 (−SAM) remained able to bind a reducible [4Fe-4S] cluster (Figures S5 and 5E). After incubation of SD_1168 (−SAM) with all hypothesized reaction components except for SAM, no methylation activity was observed (Table 1, Figure S6). Thus, as expected for a radical SAM enzyme, SAM is required for SD_1168-catalyzed P-methylation.
Previous in vivo labeling experiments, knockout studies, and in vitro enzymatic studies have highlighted the role of CH3Cbl as the methyl group donor for other Cbl-dependent radical SAM methyltransferases (13,15,60-62). To investigate the methyl donor for the SD_1168-catalyzed P-methylation of NAcDMPT, we performed the reaction in the presence of [methyl-13C]Cbl. Under these conditions, the previously observed cross-peak (indicated by the asterisk *) corresponding to the methyl protons of unlabeled NAcPT was observed at 1.20 ppm (1H) and 56.4 ppm (31P, Figure 6C). Two new cross-peaks (indicated by the double daggers ‡) centered at 1.30 ppm (1H) and 56.5 ppm (31P) and at 1.09 ppm (1H) and 56.3 ppm (31P) were also observed. The splitting of these cross-peaks result from passive couplings of 1H and 31P with the newly added 13CH3 group from [methyl-13C]Cbl to produce [methyl-13C]NAcPT [15]. This result suggests that an alternative methyl group donor beyond CH3Cbl is used by the enzyme. We suspect that, as with other characterized radical SAM methyltransferases, this donor is a second molecule of SAM [19, 20]. The approximate ratio of 13C to 12C product was 1:1, indicating that SD_1168 was able to remethylate once prior to NMR analysis. Since Ti citrate is not capable of reducing 13CH3Cbl to allow for methylation of Cbl(I) with unlabeled SAM (Figure S7), the 12C product must arise from a second turnover and not from non-enzymatic side reactions. This observed result differs from our previous work with PhpK which suggested that the K. phosalacinea P-methyltransferase was only capable of a single turnover [15, K. Hu, W.J. Werner, K.D. Allen, and S.C. Wang, submitted for publication]. However, our previous PhpK studies used dithionite as the reducing agent [15]. As described earlier, dithionite is likely not the ideal reductant to use with SD_1168 and, perhaps, other related methyltransferases in vitro.
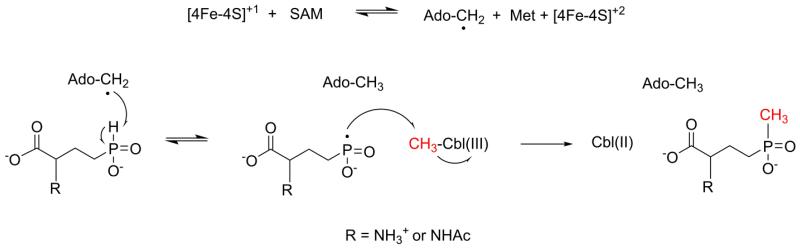
A hypothetical radical SAM P-methylation mechanism supported by our data is depicted in Figure 7 [1,18]. First, the [4Fe-4S]+1 cluster donates an electron to the required molecule of SAM to produce Ado-CH2•. Next, Ado-CH2• abstracts the phosphinyl hydrogen of NAcDMPT to generate a phosphonyl radical that reacts with CH3Cbl to produce the methylated product and Cbl(II), a dead end that cannot support further turnover. Therefore, the enzyme must either bind a new molecule of CH3Cbl or it must regenerate CH3Cbl to proceed with further catalysis. To date, the use of CH3Cbl strictly as a substrate is unprecedented [63,64]. Thus, it is far more likely that CH3Cbl is a regenerated cofactor. Remethylation of Cbl in Cbl-dependent radical SAM methyltransferases may occur in a manner similar to the reactivation cycle of Cbl-dependent methionine synthase [18]. In this cycle, flavodoxin reduces Cbl(II) to Cbl(I), which is then remethylated after nucleophilic attack on SAM to form CH3Cbl [64]. Our hypothetical remethylation mechanism for SD_1168 is supported by data from experiments with the Cbl-dependent radical SAM methyltransferase GenK in which both [methyl-13C]product and [methyl-13C]Cbl were observed after incubation with [methyl-13C]SAM, indicating that the methyl group from SAM was transferred to both Cbl and the final GenK product [20].
Figure 7.
Proposed radical SAM P-methylation mechanism.
To further test the hypothesis that CH3Cbl is not truly a substrate for SD_1168 and instead is generated as an intermediate, we examined SD_1168 for activity in the presence of Ti citrate, SAM, NAcDMPT, and HOCbl. Under these conditions, the new cross-peak was equivalent to approximately 8.5% NAcPT formation (Table 1, Figure 6D), indicating that CH3Cbl is not directly required. Together with our [methyl-13C]Cbl results, these data provide the first evidence for methylation of Cbl within a P-methyltransferase enzyme. SD_1168 is apparently able to form CH3Cbl using a methyl group from another source, then transfer this methyl group to the final NAcPT product. Ti citrate can reduce any Cbl(II) formed to Cbl(I), which can then attack a methyl group donor such as SAM to generate CH3Cbl. This function of Ti citrate has been well established in Cbl- and corrinoid-dependent enzymes [65-67]. Ti citrate can also reduce HOCbl to Cbl(I) (Figure S8), so we cannot yet rule out the possibility that the observed remethylation of Cbl may be due to a non-enzymatic process. Additionally, further studies using [methyl-13C]SAM are required to confirm whether SD_1168 remethylates Cbl using SAM as part of its catalytic cycle.
3.4 Potential biological role for SD_1168, conclusions, and future directions
Our work with SD_1168 has provided further insight into the unusual reactions catalyzed by and the diversity of Cbl-dependent radical SAM methyltransferases. SD_1168 requires a strong reductant, SAM, and Cbl for P-methytransferase activity. This is consistent with the previously hypothesized radical SAM mechanism for P-methylation (Figure 7), our laboratory’s previous work on PhpK, and the recent study by Liu and coworkers on the distantly-related GenK methyltransferase [15,18,20]. Our results also support the hypothesis that Cbl functions as a coenzyme, acting as an intermediate methyl group carrier that must be regenerated for subsequent turnover.
The observed P-methylation activity of SD_1168 raises the obvious question: what is the biological role of this protein? Though the P-methyltransferase activity of SD_1168 upon both DMPT and NAcDMPT is reasonable and, for the latter compound, comparable to that of PhpK, it is unlikely that either phosphinate is the true physiological substrate for this enzyme. We currently do not know whether SD_1168 or PhpK is a “better” P-methyltransferase as evidenced by kcat and/or Km since sensitivity and separation difficulties have prevented us from performing kinetic analyses of SD_1168 or PhpK. To date, the only putative Cbl-dependent radical SAM methyltransferase for which any kinetic information has been reported is TsrM, which apparently does not perform traditional radical SAM-dependent chemistry since Ado-CH2• is reportedly not required for activity [19]. This lack of kinetic characterization is presumably due to the inherent complexity of these systems as well as limitations in separation and detection technologies.
Since the genome of S. denitrificans encodes the first two genes (sden_1161/pepM and sden_1162/ppd) required for most known C-P compound biosynthetic pathways [23], it is likely that this bacterium produces a C-P natural product. A comprehensive study recently investigated the variety and prevalence of C-P biosynthetic genes among microbes [23], illustrating a variety of pathways spanning phosphonolipid and phosphonoglycan biosynthesis as well as production of biologically-active C-P compounds [1,23,68-71]. The genes in the immediate neighborhood of SD_1168 show striking similarity to a group of genes identified in 2013 in an actinomycete, Kitasatospora sp. NRRL F-6133 (compare Figure 3, top and bottom) [23]. In fact, SD_1168 shares higher sequence identity and similarity (61% and 77%, respectively) with the putative Cbl-dependent radical SAM methyltransferase encoded by orf11 from F-6133 than with either Fom3 or PhpK. To date, Orf11 has not been investigated. However, the high similarity suggests that the biological roles of SD_1168 and Orf11 may be very similar, whether for the production of bioactive C-P compounds or for another biosynthetic purpose. In addition, a preliminary analysis of S. denitrificans cells using 2-D high-resolution magic angle spinning (HR-MAS) NMR spectroscopy suggests that this microbe may produce a phosphinate as a cellular component [G. Helms and S.C. Wang, unpublished data]. The relatively inert nature of the C-P bond to hydrolysis may be an important way for S. denitrificans to retain phosphorus from its marine environment. Thus, we currently speculate that the true substrate for SD_1168 is an alkyl phosphinate. Knockout of the sden_1168 gene in S. denitrificans OS217, synthesis of potential substrates and, ultimately, the development of a kinetic assay for SD_1168 and other Cbl-dependent radical SAM methyltransferases will shed light on these questions and ultimately reveal the purpose of this intriguing enzyme.
Supplementary Material
Highlights.
Cloning and purification of a radical SAM methyltransferase
Direct P-methylation forming phosphinothricin
Cobalamin coenzyme required for catalysis
Acknowledgments
We thank Dr. William H. Johnson Jr., for assistance with organic synthesis; Dr. Gregory Helms, Dr. William Hiscox, and Dr. Kaifeng Hu for assistance with NMR; Drs. Eva Schinko and Wolfgang Wohlleben for assistance with expression in S. lividans; and Williard Werner and Drs. Catherine Drennan, Perry Frey, and Christian Whitman for insightful discussions. The Washington State University NMR Center equipment was supported by the NIH (RR0631401 and RR12948), the NSF (CHE-9115282 and DBI-9604689), and the Murdock Charitable Trust. K.D.A. was supported by NIH Training Grant T32GM008336 and a Bank of America Poncin Trust Fellowship. This research was funded by Washington State University and a Faculty Early Career Development Award (CAREER) from the NSF to S.C.W. (CHE-0953721).
Abbreviations
- 2-HEP
2-hydroxyethylphosphonate
- 2-HPP
2-hydroxypropylphosphonate
- [4Fe-4S]
four-iron, four-sulfur
- Ado-CH2•
5′-deoxyadenosyl radical
- Ado-CH3
5′-deoxyadenosine
- Cbl
cobalamin
- CH3Cbl
methylcobalamin
- DMPT
L-2-amino-4-hydroxyphosphinylbutanoate or demethylphosphinothricin
- DMPTT
DMPT tripeptide
- EPPS
4-(2-hydroxyethyl)piperazine-1-propanesulfonic acid
- EPR
electron paramagnetic resonance
- gHSQC
gradient heteronuclear single quantum correlation
- HOCbl
hydroxocobalamin
- HPLC
high performance liquid chromatography
- NAcDMPT
N-acetyldemethylphosphinothricin
- NAcDMPTT
NAcDMPT tripeptide
- NAcPT
N-acetylphosphinothricin
- NAcPTT
NAcPT tripeptide
- NMR
nuclear magnetic resonance
- PT
L-2-amino-4-hydroxymethylphosphinylbutanoate or L-phosphinothricin
- PTT
phosphinothricin tripeptide
- SAH
S-adenosylhomocysteine
- SAM
S-adenosyl-L-methionine
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- UV-Vis
ultraviolet-visible
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Metcalf WW, van der Donk WA. Biosynthesis of phosphonic and phosphinic acid natural products. Annu. Rev. Biochem. 2009;78:65–94. doi: 10.1146/annurev.biochem.78.091707.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinrucken HC, Amrhein N. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvyl-shikimic acid-3-phosphate synthase. Biochem. Biophys. Res. Commun. 1980;94:1207–1212. doi: 10.1016/0006-291x(80)90547-1. [DOI] [PubMed] [Google Scholar]
- 3.De Clercq E, Holy A. Acyclic nucleoside phosphonates: a key class of antiviral drugs. Nat. Rev. Drug. Discov. 2005;4:928–940. doi: 10.1038/nrd1877. [DOI] [PubMed] [Google Scholar]
- 4.Leadbetter MR, Adams SM, Bazzini B, Fatheree PR, Karr DE, Krause KM, Lam BM, Linsell MS, Nodwell MB, Pace JL, Quast K, Shaw JP, Soriano E, Trapp SG, Villena JD, Wu TX, Christensen BG, Judice JK. Hydrophobic vancomycin derivatives with improved ADME properties: discovery of telavancin (TD-6424) J. Antibiot. (Tokyo) 2004;57:326–336. doi: 10.7164/antibiotics.57.326. [DOI] [PubMed] [Google Scholar]
- 5.Thompson CJ, Seto H. Bialaphos. Biotechnology. 1995;28:197–222. doi: 10.1016/b978-0-7506-9095-9.50014-9. [DOI] [PubMed] [Google Scholar]
- 6.Gill HS, Eisenberg D. The crystal structure of phosphinothricin in the active site of glutamine synthetase illuminates the mechanism of enzymatic inhibition. Biochemistry. 2001;40:1903–1912. doi: 10.1021/bi002438h. [DOI] [PubMed] [Google Scholar]
- 7.Abell LM, Villafranca JJ. Investigation of the mechanism of phosphinothricin inactivation of Escherichia coli glutamine synthetase using rapid quench kinetic technique. Biochemistry. 1991;30:6135–6141. doi: 10.1021/bi00239a008. [DOI] [PubMed] [Google Scholar]
- 8.Ogawa H, Tsuruoka T, Inouye S, Niida T. Studies on a new antibiotic SF-1293. Sci. Rep. Meiji Seika Kaisha. 1973;13:42–48. [Google Scholar]
- 9.Omura S, Murata M, Hanaki H, Hinotozawa K, Oiwa R, Tanaka H. Phosalacine, a new herbicidal antibiotic containing phosphinothricin. Fermentation, isolation, biological activity and mechanism of action. J. Antibiot. (Tokyo) 1984;37:829–835. doi: 10.7164/antibiotics.37.829. [DOI] [PubMed] [Google Scholar]
- 10.Bayer E, Gugel KH, Hagele K, Hagenmaier H, Jessipow S, Konig WA, Zahner H. Metabolic products of microorganisms. 98. Phosphinothricin and phosphinothricyl-alanyl-alanine. Helv. Chim. Acta. 1972;55:224–239. doi: 10.1002/hlca.19720550126. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz D, Berger S, Heinzelmann E, Muschko K, Welzel K, Wohlleben W. Biosynthetic gene cluster of the herbicide phosphinothricin tripeptide from Streptomyces viridochromogenes Tü494. Appl. Environ. Microbiol. 2004;70:7093–7102. doi: 10.1128/AEM.70.12.7093-7102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blodgett JA, Zhang JK, Metcalf WW. Molecular cloning, sequence analysis, and heterologous expression of the phosphinothricin tripeptide biosynthetic gene cluster from Streptomyces viridochromogenes DSM 40736. Antimicrob. Agents Chemother. 2005;49:230–240. doi: 10.1128/AAC.49.1.230-240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamigiri K, Hidaka T, Imai S, Murakami T, Seto H. Studies on the biosynthesis of bialaphos (SF-1293) 12. C-P bond formation mechanism of bialaphos: discovery of a P-methylation enzyme. J. Antibiot. (Tokyo) 1992;45:781–787. doi: 10.7164/antibiotics.45.781. [DOI] [PubMed] [Google Scholar]
- 14.Imai S, Seto H, Sasaki T, Tsuruoka T, Ogawa H, Satoh A, Inouye S, Niida T, Otake N. Studies on the biosynthesis of bialaphos (SF-1293). 6. Production of N-acetyl-demethylphosphinothricin and N-acetylbialaphos by blocked mutants of Streptomyces hygroscopicus SF-1293 and their roles in the biosynthesis of bialaphos. J. Antibiot. (Tokyo) 1985;38:687–690. doi: 10.7164/antibiotics.38.687. [DOI] [PubMed] [Google Scholar]
- 15.Werner WJ, Allen KD, Hu K, Helms GL, Chen BS, Wang SC. In vitro phosphinate methylation by PhpK from Kitasatospora phosalacinea. Biochemistry. 2011;50:8986–8988. doi: 10.1021/bi201220r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frey PA, Hegeman AD, Ruzicka FJ. The Radical SAM Superfamily. Crit. Rev. Biochem. Mol. Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q, van der Donk WA, Liu W. Radical-mediated enzymatic methylation: a tale of two SAMs. Acc. Chem. Res. 2011;45:555–564. doi: 10.1021/ar200202c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierre S, Guillot A, Benjdia A, Sandstrom C, Langella P, Berteau O. Thiostrepton tryptophan methyltransferase expands the chemistry of radical SAM enzymes. Nat. Chem. Biol. 2012;8:957–959. doi: 10.1038/nchembio.1091. [DOI] [PubMed] [Google Scholar]
- 20.Kim HJ, McCarty RM, Ogasawara Y, Liu YN, Mansoorabadi SO, Levieux J, Liu HW. The GenK-catalyzed C-6′ methylation in the biosynthesis of gentamicin: isolation and characterization of a cobalamin-dependent radical SAM enzyme. J. Am. Chem. Soc. 2013;135:8093–8096. doi: 10.1021/ja312641f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen KD, Wang SC. Initial characterization of Fom3 from Streptomyces wedmorensis, the methyltransferase in fosfomycin biosynthesis. Arch. Biochem. Biophys. 2014;543:67–73. doi: 10.1016/j.abb.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brettar I, Christen R, Hofle MG. Shewanella denitrificans sp. nov., a vigorously denitrifying bacterium isolated from the oxic-anoxic interface of the Gotland Deep in the central Baltic Sea. Int. J. Sys. Evol. Microbiol. 2002;52:2211–2217. doi: 10.1099/00207713-52-6-2211. [DOI] [PubMed] [Google Scholar]
- 23.Yu X, Doroghazi JR, Janga SC, Zhang JK, Circello B, Griffin BM, Labeda DP, Metcalf WW. Diversity and abundance of phosphonate biosynthetic genes in nature. Proc. Natl. Acad. Sci. U.S.A. 2013;110:20759–20764. doi: 10.1073/pnas.1315107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hidaka T, Goda M, Kuzuyama T, Takei N, Hidaka M, Seto H. Cloning and nucleotide sequence of fosfomycin biosynthetic genes of Streptomyces wedmorensis. Mol. Gen. Genet. 1995;249:274–280. doi: 10.1007/BF00290527. [DOI] [PubMed] [Google Scholar]
- 25.Woodyer RD, Li G, Zhao H, van der Donk WA. New insight into the mechanism of methyl transfer during the biosynthesis of fosfomycin. Chem. Commun. (Camb) 2007:359–361. doi: 10.1039/b614678c. [DOI] [PubMed] [Google Scholar]
- 26.Zehnder AJ, Wuhrmann K. Titanium (III) citrate as a nontoxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science. 1976;194:1165–1166. doi: 10.1126/science.793008. [DOI] [PubMed] [Google Scholar]
- 27.Tollinger M, Derer T, Konrat R, Krautler B. An efficient method for the preparation of methylcobalamin, nature’s organometallic methyl transfer catalyst. J. Mol. Catal. 1997;116:147–155. [Google Scholar]
- 28.Lu WP, Schiau I, Cunningham JR, Ragsdale SW. Sequence and expression of the gene encoding the corrinoid/iron-sulfur protein from Clostridium thermoaceticum and reconstitution of the recombinant protein to full activity. J. Biol. Chem. 1993;268:5605–5614. [PubMed] [Google Scholar]
- 29.Waddell WJ. A simple ultraviolet spectrophotometric method for the determination of protein. J. Lab Clin. Med. 1956;48:311–314. [PubMed] [Google Scholar]
- 30.Beinert H. Micro methods for the quantitative determination of iron and copper in biological material. Methods Enzymol. 1978;54:435–445. doi: 10.1016/s0076-6879(78)54027-5. [DOI] [PubMed] [Google Scholar]
- 31.Beinert H. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal. Biochem. 1983;131:373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- 32.Kennedy MC, Kent TA, Emptage M, Merkle H, Beinert H, Munck E. Evidence for the formation of a linear [3Fe-4S] cluster in partially unfolded aconitase. J. Biol. Chem. 1984;259:14463–14471. [PubMed] [Google Scholar]
- 33.Kulzer R, Pils T, Kappl R, Huttermann J, Knappe J. Reconstitution and characterization of the polynuclear iron-sulfur cluster in pyruvate formate-lyase-activating enzyme. Molecular properties of the holoenzyme form. J. Biol. Chem. 1998;273:4897–4903. doi: 10.1074/jbc.273.9.4897. [DOI] [PubMed] [Google Scholar]
- 34.Ugulava NB, Frederick KK, Jarrett JT. Control of adenosylmethionine-dependent radical generation in biotin synthase: a kinetic and thermodynamic analysis of substrate binding to active and inactive forms of BioB. Biochemistry. 2003;42:2708–2719. doi: 10.1021/bi0261084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haddy A. EPR spectroscopy of the manganese cluster of photosystem II. Photosynth. Res. 2007;92:357–368. doi: 10.1007/s11120-007-9194-9. [DOI] [PubMed] [Google Scholar]
- 36.Palmer G. The electron paramagnetic resonance of metalloproteins. Biochem. Soc. Trans. 1985;13:548–560. doi: 10.1042/bst0130548. [DOI] [PubMed] [Google Scholar]
- 37.Duschene KS, Broderick JB. The antiviral protein viperin is a radical SAM enzyme. FEBS Lett. 2010;581:1263–1267. doi: 10.1016/j.febslet.2010.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wecksler SR, Stoll S, Tran H, Magnusson OT, Wu SP, King D, Britt RD, Klinman JP. Pyrroloquinoline quinone biogenesis: demonstration that PqqE from Klebsiella pneumoniae is a radical S-adenosyl-L-methionine enzyme. Biochemistry. 2009;48:10151–10161. doi: 10.1021/bi900918b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broderick JB, Henshaw TF, Cheek J, Wojtuszewski K, Smith SR, Trojan MR, McGhan RM, Kopf A, Kibbey M, Broderick WE. Pyruvate formate-lyase-activating enzyme: strictly anaerobic isolation yields active enzyme containing a [3Fe-4S](+) cluster. Biochem. Biophys. Res. Commun. 2000;269:451–456. doi: 10.1006/bbrc.2000.2313. [DOI] [PubMed] [Google Scholar]
- 40.Petrovich RM, Ruzicka FJ, Reed GH, Frey PA. Characterization of iron-sulfur clusters in lysine 2,3-aminomutase by electron paramagnetic resonance spectroscopy. Biochemistry. 1992;31:10774–10781. doi: 10.1021/bi00159a019. [DOI] [PubMed] [Google Scholar]
- 41.Lim A, Grasland A. Electron paramagnetic resonance evidence for a novel interconversion of [3Fe-4S]+ and [4Fe-4S]+ clusters with endogenous iron and sulfide in anaerobic ribonucleotide reductase activity in vitro. J. Biol. Chem. 2000;275:12367–12373. doi: 10.1074/jbc.275.17.12367. [DOI] [PubMed] [Google Scholar]
- 42.Wang SC, Frey PA. Binding energy in the one-electron reductive cleavage of S-adenosylmethionine in lysine 2,3-aminomutase, a radical SAM enzyme. Biochemistry. 2007;46:12889–12895. doi: 10.1021/bi701745h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsby CJ, Ortillo D, Yang J, Nnyepi MR, Broderick WE, Hoffman BM, Broderick JB. Spectroscopic approaches to elucidating novel iron-sulfur chemistry in the “radical-SAM” protein superfamily. Inorg. Chem. 2005;44:727–741. doi: 10.1021/ic0484811. [DOI] [PubMed] [Google Scholar]
- 44.Krebs C, Broderick WE, Henshaw TF, Broderick JB, Huynh BH. Coordination of adenosylmethionine to a unique iron site of the [4Fe-4S] of pyruvate formate-lyase activating enzyme: a Mossbauer spectroscopic study. J. Am. Chem. Soc. 2002;124:912–913. doi: 10.1021/ja017562i. [DOI] [PubMed] [Google Scholar]
- 45.Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ. A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science. 2011;332:604–607. doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
- 46.Cicchillo RM, Iwig DF, Jones AD, Nesbitt NM, Baleanu-Gogonea C, Souder MG, Tu L, Booker SJ. Lipoyl synthase requires two equivalents of S-adenosyl-L-methionine to synthesize one equivalent of lipoic acid. Biochemistry. 2004;43:6378–6386. doi: 10.1021/bi049528x. [DOI] [PubMed] [Google Scholar]
- 47.Grove TL, Ahlum JH, Sharma P, Krebs C, Booker SJ. A consensus mechanism for Radical SAM-dependent dehydrogenation? BtrN contains two [4Fe-4S] clusters. Biochemistry. 2010;49:3783–3785. doi: 10.1021/bi9022126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ugulava NB, Gibney BR, Jarrett JT. Biotin synthase contains two distinct iron-sulfur cluster binding sites: chemical and spectroelectrochemical analysis of iron-sulfur cluster interconversions. Biochemistry. 2001;40:8343–51. doi: 10.1021/bi0104625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jellinek K. Concerning the electrolytic potential of hydrosulphite reactions. Z. Elektrochem. Angew. Phys. Chem. 1911;17:157–176. [Google Scholar]
- 50.McMillan WG, Jr, Roberts JD, Coryell CD. The thermodynamic constants of the dithionite (hydrosulfite) ion. J. Am. Chem. Soc. 1942;64:398–399. [Google Scholar]
- 51.Watt GD, Burns A. The thermochemical characterization of sodium dithionite, flavin mononucleotide, flavin-adenine dinucleotide and methyl and benzyl viologens as low-potential reductants for biological systems. Biochem. J. 1975;152:33–37. doi: 10.1042/bj1520033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayhew SG. The redox potential of dithionite and SO2− from equilibrium reactions with flavodoxins, methyl viologen and hydrogen plus hydrogenase. Eur. J. Biochem. 1978;85:535–547. doi: 10.1111/j.1432-1033.1978.tb12269.x. [DOI] [PubMed] [Google Scholar]
- 53.Salnikov DS, Silaghi-Dumitrescu R, Makarov SV, van Eldik R, Boss GR. Cobalamin reduction by dithionite. Evidence for the formation of a six-coordinate cobalamin(II) complex. Dalton Trans. 2012;40:9831–9834. doi: 10.1039/c1dt10219b. [DOI] [PubMed] [Google Scholar]
- 54.McKenna CE, Gutheil WG, Song W. A method for preparing analytically pure sodium dithionite. Dithionite quality and observed nitrogenase-specific activities. Biochimica et Biophysica Acta – General Subjects. 1991;1075:109–117. doi: 10.1016/0304-4165(91)90082-r. [DOI] [PubMed] [Google Scholar]
- 55.Lindahl PA, Munck E, Ragsdale SW. CO dehydrogenase from Clostridium thermoaceticum. EPR and electrochemical studies in CO2 and argon atmospheres. J. Biol. Chem. 1990;265:3873–3879. [PubMed] [Google Scholar]
- 56.Bender G, Pierce E, Hill JA, Darty JE, Ragsdale SW. Metal centers in the anaerobic microbial metabolism of CO and CO2. Metallomics. 2011;3:797–815. doi: 10.1039/c1mt00042j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Menon S, Ragsdale SW. Role of the [4Fe-4S] cluster in reductive activation of the cobalt center of the corrinoid iron-sulfur protein from Clostridium thermoaceticum during acetate biosynthesis. Biochemistry. 1998;37:5689–5698. doi: 10.1021/bi9727996. [DOI] [PubMed] [Google Scholar]
- 58.Allen KD. Dissertation: Characterization of Cobalamin-dependent Radical S-Adenosyl-L-Methionine Methyltransferases Involved in Antibiotic Biosynthesis. 2013. [Google Scholar]
- 59.Thompson CJ, Movva NR, Tizard R, Crameri R, Davies JE, Lawereys M, Botterman J. Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J. 1987;6:2519–2523. doi: 10.1002/j.1460-2075.1987.tb02538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seto H, Kuzuyama T. Bioactive natural products with carbon-phosphorus bonds and their biosynthesis. Nat. Prod. Rep. 1999;16:589–596. doi: 10.1039/a809398i. [DOI] [PubMed] [Google Scholar]
- 61.Kuzuyama T, Hidaka T, Kamigiri K, Imai S, Seto H. Studies on the biosynthesis of fosfomycin. 4. The biosynthetic origin of the methyl group of fosfomycin. J. Antibiot. (Tokyo) 1992;45:1812–1814. doi: 10.7164/antibiotics.45.1812. [DOI] [PubMed] [Google Scholar]
- 62.Seto H, Hidaka T, Kuzuyama T, Shibahara S, Usui T, Sakanaka O, Imai S. Studies on the biosynthesis of fosfomycin. 2. Conversion of 2-hydroxypropyl-phosphonic acid to fosfomycin by blocked mutants of Streptomyces wedmorensis. J. Antibiot. (Tokyo) 1991;44:1286–1288. doi: 10.7164/antibiotics.44.1286. [DOI] [PubMed] [Google Scholar]
- 63.Frey PA. Cobalamin coenzymes in enzymology. Comprehensive Natural Products II: Chemistry and Biology. 2010;7:501–546. [Google Scholar]
- 64.Matthews RG. Cobalamin-dependent methyltransferases. Acc. Chem. Res. 2001;34:681–689. doi: 10.1021/ar0000051. [DOI] [PubMed] [Google Scholar]
- 65.Kengen SWM, Mosterd JJ, Nelissen RLH, Keltjens JT, van der Drift C, Vogels GD. Reductive activation of the methyl-tetrahydromethanopterin:coenzyme M methyl-transferase from Methanobacterium thermoautotrophicum strain H. Arch. Microbiol. 1988;150:405–421. [Google Scholar]
- 66.Jarrett JT, Goulding CW, Fluhr K, Huang S, Matthews RG. Purification and assay of cobalamin-dependent methionine synthase from Escherichia coli. Methods Enzymol. 1997;281:196–213. doi: 10.1016/s0076-6879(97)81026-9. [DOI] [PubMed] [Google Scholar]
- 67.Gartner P, Weiss DS, Harms U, Thauer RK. N5-methyltetrahydromethanopterin:coenzyme M methyltransferase from Methanobacterium thermoautotrophicum. Catalytic mechanism and sodium ion dependence. Eur. J. Biochem. 1994;226:465–472. doi: 10.1111/j.1432-1033.1994.tb20071.x. [DOI] [PubMed] [Google Scholar]
- 68.Baumann H, Tzianabos AO, Brisson JR, Kasper DL, Jennings HJ. Structural elucidation of two capsular polysaccharides from one strain of Bacteroides fragilis using high-resolution NMR spectroscopy. Biochemistry. 1992;31:4081–4089. doi: 10.1021/bi00131a026. [DOI] [PubMed] [Google Scholar]
- 69.Watanabe Y, Nakajima M, Hoshino T, Jayasimhulu K, Brooks EE, Kaneshiro ES. A novel sphingophosphonolipid head group 1-hydroxy-2-aminoethyl phosphonate in Bdellovibrio stolpii. Lipids. 2001;36:513–519. doi: 10.1007/s11745-001-0751-3. [DOI] [PubMed] [Google Scholar]
- 70.Shao Z, Blodgett JA, Circello BT, Eliot AC, Woodyer R, Li G, van der Donk WA, Metcalf WW, Zhao H. Biosynthesis of 2-hydroxyethylphosphonate, an unexpected intermediate common to multiple phosphonate biosynthetic pathways. J.Biol.Chem. 2008;283:23161–23168. doi: 10.1074/jbc.M801788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu X, Price NPJ, Evans BS, Metcalf WW. Purification and characterization of phosphonoglycans from Glycomyces sp. Strain NRRL B-16210 and Stackebrandtia nassauensis NRRL B-16338. J.Bact. 2014;196:1768–1779. doi: 10.1128/JB.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.