Abstract
Over the last decade, advancements in stem cell biology have yielded a variety of sources for stem cell-based cardiovascular investigation. Stem cell behavior, whether to maintain its stable state of pluripotency or to prime toward the cardiovascular lineage is governed by a set of coordinated interactions between epigenetic, transcriptional, and translational mechanisms. The science of incorporating genes (genomics), RNA (transcriptomics), proteins (proteomics), and metabolites (metabolomics) data in a specific biological sample is known as systems biology. Integrating systems biology in progression with stem cell biologics can contribute to our knowledge of mechanisms that underlie pluripotency maintenance and guarantee fidelity of cardiac lineage specification. This review provides a brief summarization of OMICS-based strategies including transcriptomics, proteomics, and metabolomics used to understand stem cell fate and to outline molecular processes involved in heart development. Additionally, current efforts in cardioregeneration based on the “one-size-fits-all” principle limit the potential of individualized therapy in regenerative medicine. Here, we summarize recent studies that introduced systems biology into cardiovascular clinical outcomes analysis, allowing for predictive assessment for disease recurrence and patient-specific therapeutic response.
Keywords: Systems Biology, OMICS, Stem Cells, Cardiac Regeneration
Stem Cells in the Post-Genomic Era
Stem cell biology has entered the post-genomic era, allowing for a holistic understanding of developmental molecular events through epigenetic, transcriptional, and post-transcriptional signaling. The science of integrating genes (genomics), RNA (transcriptomics), proteins (proteomics), and metabolites (metabolomics) data in a specific biological sample is known as systems biology [1]. Systems-based approaches utilize the combination of gene and protein expression array analyses, gene-gene and protein-protein interactions, and intracellular metabolite levels to understand simultaneous occurrence of molecular processes [2]. Deciphering critical molecular interactions through systems biology-based strategies could guide our goal to achieve functional and safe stem cell-based therapies.
Stem cells are characterized by an inherent ability to self-renew and differentiate into cell types from the three primary germ layers, providing a source for tissue regeneration. Stem cell behavior, whether to maintain its stable state of pluripotency or to prime toward a potential cell fate is governed by a set of coordinated interactions between epigenetic, transcriptional, and translational mechanisms. To fulfill the therapeutic potential retained in pluripotency, an understanding of molecular properties of self-renewal and commitment is required. Several investigators have addressed this challenge with systems biology approaches [3–6]. Specifically, Boyer et al used chromatin immunoprecipitation (ChIP) with DNA microarrays (Chips) also known as ChIP-Chip analysis [7] to identify the network of transcription factors that regulate stem cell fate [3]. Other studies have used in vivo biotinylation mediated ChIP (bioChIP) for global target mapping (bioChIP-Chip) and reported an expanded set of factors associated with pluripotency maintenance [4]. Compared to ChIP-Chip analysis, the bioChIP-Chip relies on streptavidin affinity capture of tagged proteins and circumvents issues related to antibody availability [8]. By combining this technique with whole-lane liquid chromatography–tandem mass spectrometry (LC–MS/MS), a commonly used method to measure nuclear protein levels, Wang et al studied the protein interaction network and identified factors with critical roles in stem cell pluripotency [5]. Systems-level analyses such as ChIP-chip and LC-MS/MS have been used to measure global change in histone acetylation and nuclear protein levels to understand stem cell fate change [6]. Similar studies assessed stem cell development on the basis of chromatin structure and its epigenetic modifications [9,10]. Indeed, integrating systems biology in progression with stem cell biologics can contribute to our knowledge of mechanisms that underlie pluripotency maintenance and guarantee fidelity of lineage specification.
Advancements in Stem Cell Biology
Natural and bioengineered stem cell populations have been identified including hematopoietic stem cells (HSCs), mesenchymal stem cells (MSCs), embryonic stem cells (ESCs), tissue-specific adult stem cells, and induced pluripotent stem cells (iPSCs) (Figure 1).
Figure 1. Initial developments in stem cell biology in parallel with the evolution of systems biology.
Unraveling DNA structure, technological development of DNA sequencing, emergence of genomics, proteomics, and the completion of the genome project has coincided with critical discoveries in stem cell isolation and reprogramming strategies, resulting in cutting-edge efforts to utilize these findings for translational applications.
Human HSCs are found in the bone marrow, peripheral blood, and placenta, and give rise to all lineages of the blood [11]. Adult bone marrow-derived cells (Lin−CD34+/−CD45+/−c-kit+) have been shown to modestly augment cardiac function recovery by contributing to de novo myocardium in the post-infarcted heart [12]. Alternatively, studies have used cell fate assays to report that HSCs do not transdifferentiate into cardiac myocytes in myocardial infarcts [13]. To better understand the molecular characterization of HSC microenvironments and the core genetic network responsible for HSC differentiation, systems-based approaches using messenger RNA (mRNA) and microRNA (miRNA) transcriptomes have determined a comprehensive list of hematopoietic regulators [14].
Human MSCs are found in the bone marrow, adipose tissue, and the umbilical cord [15]. They have a propensity for multipotent differentiation into osteoblasts, chondrocytes, and adipocytes [16]. Bone marrow-derived MSCs were shown to be beneficial in the treatment of chronic ischemic cardiomyopathy [17,18]. Behfar et al primed bone marrow-derived MSCs with recombinant trophic factors including transforming growth factor-β (TGF-β) or bone morphogenetic protein (BMP), allowing for entry into the cardiac program [17]. Similarly, adipose tissue-derived MSCs, from minimally invasive liposuction [19,20], can transdifferentiate into cells with characteristics of cardiomyocytes and neovascular tissue [21]. However, recent studies observed a lack of spontaneous cell contraction in adipose MSC-derived cardiomyocytes [22]. Comparative analyses of MSCs from bone marrow, cartilage, and adipose tissue have been assessed for osteogenic, chrondrogenic, and adipogenic differentiation potential [23], yet it remains to be elucidated for cardiomyocyte differentiation. Indeed, advances in systems biology provide the tools to evaluate global molecular differences in MSCs due to variability in patient age, sex, and location of cell isolation. Recent studies utilized microarray technology for genomic profiling of bone marrow-derived MSCs and determined key molecules regulating stem cell survival, growth, and development [24]. Prior to harnessing their clinical benefit, the ability to track MSC regulatory pathway on a molecular level by transcriptomic, proteomic, and metabolomic analysis is required.
Embryonic stem cells are derived from the inner cell mass of a blastocyst and are pluripotent, giving rise to endoderm, mesoderm, and ectoderm lineages [25]. Multiple mouse and human ESC lines have been established [26–28]. Beyond the unrestricted growth potential, ESCs create an immunological challenge for regenerative medicine [29], limiting therapeutic applications to preclinical studies. Systems biology approaches have been actively applied to study ESC properties including self-renewal maintenance and lineage commitment [30].
The adult myocardium has a modest intrinsic regenerative capacity based on the presence of cardiac stem-progenitor cells [31–34]. This endogenous cardiac regeneration following injury is a highly debated event. Although some findings support the concept of the adult heart as a suitable target for regenerative intervention [35–37], the contribution of endogenous stem cells to restoring cardiac function is limited [38]. Despite the existence of c-kit+ population and the potential ability of bone marrow cells to facilitate cardiac repair, intrinsic mechanisms alone are inadequate to restore cardiac function to a failing heart [39]. Thus, strategies for guiding cells toward the cardiac lineage and stimulating proliferation of post-mitotic cardiomyocytes are needed. For this purpose, systems-based approaches affords the ability to target genomic, proteomic, and metabolomic influences that direct in vivo and in vitro cardiomyocyte differentiation.
Bioengineered stem cells, or induced pluripotent stem cells (iPSC), are generated by reprogramming somatic tissue, namely skin fibroblasts, using ectopic expression of defined factors [40,41]. Particularly, transcription factor sets Oct4, Sox2, c-Myc, and Klf4 or alternatively Oct4, Sox2, Nanog, and Lin28 are described to reprogram human somatic cells to the pluripotent state [42,43]. Studies show that iPSC-based transplantation into the adult infarcted heart modestly improves post-ischemic cardiac performance [44,45]. Despite the immunocompatible nature of iPSCs [46,47], their potential for clinical translation is currently hindered by the risk of dysregulated cell growth known as tumorigenicity [48]. Recent findings show that pharmacological purging with DNA-damaging agent, etoposide can reduce this threat of tumorigenicity [49]. While iPSC reprogramming event produces an embryonic stem cell (ESC)-like pluripotent state, human ESCs and their pluripotent transcriptome remain the standard for understanding lineage-specific differentiation [28,50]. Given the advantage of studying patient-specific and mutation-defined diseases using the iPSC platform, high throughput technologies and bioinformatics analyses could be progressively utilized to expand our knowledge on iPSC maturation potential and lineage specification.
Thus, a variety of sources for stem cell-based cardiovascular investigation are available. Prior to clinical-grade application, comprehensively mapping molecular signaling events that orchestrate different stages of stem cell differentiation into cardiac lineage is necessary. These molecular processes and regulatory mechanisms involved in heart development can be exploited to repair the injured heart and achieve tissue regeneration [51]. Additionally, the molecular and genetic composition of fully differentiated and functional cardiomyocytes needs to be defined. To meet these challenges, high-throughput technologies such as whole genome shotgun sequencing, deep sequencing, and CHiP-chip/CHiP-Seq are useful tools. Instead of focusing on the role of individual genes, proteins, and pathways in biological processes, the systems biology method allows for characterizing how molecular parts interact with each other to determine the collective intracellular dynamics as a whole [52]. Thus, the systems biology approach to understand commonalities of evolutionarily conserved regulatory networks provides new insight to controlling aspects of regeneration with transcriptomics, proteomics, and metabolomics.
Transcriptomics
Embryonic and adult stem cells possess a diverse developmental transcriptome, which is defined as the total set of RNA transcripts [53]. Recent findings suggest that stem cells vary in their differentiation potential towards highly specialized cells including cardiomyocytes [54]. Cardiogenesis is the developmental process of the embryonic heart and is tightly regulated by signaling pathways and networks of transcription factors [55]. Martinez-Fernandez et al demonstrate that transcriptome analysis can discern the maturation potential of bioengineered stem cells [54]. For instance, stage-specific cardiogenesis was assessed by genome-wide transcriptome analysis using distinctive mouse embryonic time points. Based on this referential gene expression guideline, it was determined that pluripotent cell lines possess variant cardiogenic potentials depending on three-factor versus four-factor reprogramming method.
Systems-based approaches have been explored in similar studies to understand the cardiac developmental program. Faustino et al investigated the developing mesoderm transcriptome in mouse ESCs and identified over 8,000 genes underlying cardiac specification [56]. Incorporating this data with bioinformatics analysis streamlined upregulated and downregulated signaling components implicated in inducing the cardiac muscle fate [56]. Similarly, system-expression profiling, in conjunction with bioinformatics network analysis, allowed for identifying specific biomarkers (CXCR4/FLK-1) primed for cardiogenic specification [57]. Nelson et al used genome-wide microarrays on ESC-derived cardiac progenitors and identified a distinct transcriptome profile of 11,272 transcripts. Based on this information, bioinformatics dissection of exposed surface biomarkers prioritized CXCR4 chemokine receptor cluster as the most over-represented gene receptor family during pre-cardiac induction. Subsequently, CXCR+/FLK-1+ subpopulation was isolated from the ESC pool and differentiated to yield an enriched Mesp-1, GATA-4, and Tbx5 population, indicative of pre-cardiac mesoderm [57]. Indeed, identification of novel biomarkers and secreted cardio-inductive signals at each phase of myocyte differentiation can be achieved by genomic profiling of cardiogenesis [58,59].
Mapping transcriptome networks accelerates our understanding of stem cell-based cardiogenesis and its biological underpinnings. Embryonic heart formation occurs through strict combinations of extracellular signaling and structured patterns of timing that guide pluripotent cells through mesoderm induction into specialized cardiac cells, including atrial and ventricular cardiomyocytes, conducting cells, fibroblasts, and vascular endothelial and smooth muscle cells [60,51]. In the developing embryo, WNT and NODAL signaling networks are evolutionary conserved molecular cascades that induce mesoderm and endoderm, and are widely used to initiate cardiogenesis in ESC cultures [61,62]. Instructive guidance from NOTCH and FGF play an important role in the proliferation and fate selection of cardiac progenitors and is directed by a temporal window that is either inductive or inhibitive [63]. Global changes in the transcriptome of newborn cardiomyocytes revealed that myocardial NOTCH signaling drives cardiomyocytes to a conduction-prone phenotype [64].
Recent transcriptome studies identified factors that differentially regulate cardiogenesis from murine ESCs. Cai et al showed that both NODAL and TGFβ signaling induced early cardiac progenitor formation, yet NODAL expression declined due to feedback inhibition and TGFβ expression continued [65]. At later stages of cardiogenesis, TGFβ suppressed cardiomyocyte formation and stimulated vascular smooth muscle and endothelial differentiation [65]. Indeed, specification of cardiogenic cells is highly regulated by methodical exposure to these signaling factors. To comprehensively analyze the dynamic gene expression profile from the stem cell stage to adult cardiac structures, Li et al used a time-course transcriptome analysis of innate murine cardiogenesis [66]. Stage-specific analysis of the cardiogenic interactome identified developmental disturbances in epithelial-to-mesenchmyal transition (EMT), BMP signaling, NF-AT signaling, TGFβ-dependent EMT, and NOTCH signaling, elucidating regulatory networks at the boundaries of health and disease (87). Such initial gene expression trends of cardiovascular development concur with the dynamism of lineage specification as each stage of cardiac differentiation assumes a discrete molecular fingerprint.
Identifying pro-cardiogenic genes by means of transcriptome analysis could allow for targeted execution of the cardiac program. High throughput approaches such as DNA microarrays have been used to elucidate cardiogenic instructive signaling in mouse ESCs [67]. Several studies agree that transcription factors including Nkx2.5, MEF2C, and GATA4 are upregulated with cardiac-specific commitment [56,68,67]. In addition, cellular binding small molecules including cytoskeletal, polysaccharide, and metal-ion-binding factors are upregulated during cardiovascular development [69,56]. Conversely, there is a decline of pluripotency markers including Oct4, Sox2, and Nanog, implying that differentiation downregulates components of DNA replication, cell cycling, and cancer mechanisms [56]. Gene Ontology Consortium studies of the differentiating transcriptome also found that downregulated transcripts exhibited significant RNA binding activity, ribosome structure, and translation regulators, defining a loss of oncogenicity associated with pluripotency and an acquisition of cardiac tissue-specificity [70]. This phenomenon of losing pluripotent expression prior to gaining mesoderm-specific markers suggests mutual exclusivity among each phase of cardiogenesis. Integration of gene expression profiles with proteomic and/or metabolic profiles could increase the precision of conserved cardiogenic transcripts.
Proteomics
Proteomics is defined as the large-scale study of protein abundance and its variations at specific time point [71]. Intracellular signaling events and post-translational modifications that direct self-renewal and differentiation events can be largely understood by mass-spectrometry-based proteomic technologies [72]. For example, 2D electrophoresis comparing the proteomic profile of spontaneous mouse ESC-derived cardiomyocytes and neonatal-derived cardiomyocytes yielded a 95% similarity of the proteins [73]. Similarly, human ESC-derived cardiomyocyte transcriptome revealed a gene expression profile at levels corresponding to 20-week fetal heart cells [74]. In contrast, Yin et al showed that ESC-derived smooth muscle cells (SMCs) expressed identical smooth muscle markers yet their proteome was vastly different from aortic SMCs [75]. This suggests that marker proteins used for characterizing mature cell populations may not be sufficient to classify stem cell-derived cell populations [75,76]. Discerning the divergent expression pattern between the naturally differentiated cell types and stem cell derived-mature cell types at both transcriptomic and proteomic levels could be considered an important step in advancing cell therapy-based regenerative medicine.
Proteomic methodologies used for lineage specification can reveal growth factor signaling relationships [77,78]. Although extracellular cues such as cytokines and matrix factors generate cell behavioral responses such as proliferation and differentiation, it is challenging to assign cue-response relationships in a direct manner [77]. Prudhomme et al used the computational partial-least-squares (PLS) analysis in combination with Western blot to delineate this cue-response relationship. Using this method, strongly correlated phosphorylated proteins involved in the signaling network that governs self-renewal versus differentiation were identified [77]. Similarly, comparative proteomics by tandem mass spectrometry (MS/MS) revealed candidate effectors of mouse ESC cardiac differentiation [79], contributing to our understanding of guided developmental cardiogenesis.
Network hub analyses, a commonly used technique to map nodes and interactions of proteins, have cross-referenced subsets of stem cell-derived cardiac progenitors to reveal a robust pro-cardiogenic network en route to cardiomyocyte differentiation [80]. Targeted treatment with SDF-1, VEGF, and BMP2 was shown to activate signaling networks that guide cardiac determination [80]. Further network analysis of cardiogenesis-associated signaling cascades identified integrin, WNT/β-catenin, IL6, IGF-1 and cardiovascular hypoxia pathways as prominent in cardiac progenitor cells [56,81]. Such signaling networks converged with TGF-β, JAK/STAT, granulocyte-macrophage colony stimulating factor/colony stimulating factor 2 (GM-CSF/CSF2), and calcium signaling in stem cell-derived cardiomyocytes [82]. In addition to factors that accelerate cardiomyocyte differentiation, studies also described network decelerants that postpone cardiogenesis [56,83]. Identifying promoting and rate-limiting signaling hubs in the cardiopoietic framework could be advantageous in directing cardiac lineage specification.
Metabolomics
Integrating OMICS data in cardiovascular research could guide our understanding of global regulation of signal transduction and cellular metabolism [76]. In particular, the study of metabolomics captures the chemical fingerprint behind cellular processes. Bioengineered stem cells undergo a metabotype conversion from oxidative metabolism to glycolysis during nuclear reprogramming [84]. Conversely, during cardiogenesis, the metabolomic module acquires a mandatory switch from glycolysis to oxidative phosphorylation that drives ESC cardiac differentiation [85,86]. Chung et al assessed the glycolytic phosophotransfer network during mouse ESC cardiogenesis by high-throughput arrays [86]. Lactate-generating capacity of ESCs, quantified by uncoupling agents and respiratory chain inhibition, was measured during stem cell cardiac differentiation and correlated with array findings [86]. Linking alteration of cellular metabolism and function to both proteomics and transcriptomics could facilitate our goal in engineering specialized cardiovascular tissues.
Metabolomics allows for identification of unique biomarkers that are informative of cellular status during stem cell differentiation and dedifferentiation [87]. Specifically, intracellular (fingerprint) and extracellular (footprint) metabolomes have defined baseline stem cell metabolic landscape and its changes in the diseased state [88,89]. Studies have utilized metabolomic techniques to investigate the role of energy metabolism in controlling stem cell fate [90,91]. Mohyeldin et al showed that hypoxia-mediated activation of glycolytic metabolism increased the efficiency of nuclear reprogramming and maintained the pluripotent ground state [92]. Conversely, upregulation of the electron transport chain subunits and tri-carboxylic acid enzymes with decreased expression of glycolytic enzymes propagated cardiomyocyte differentiation [93,94]. Metabolic profiling techniques allow for the resolution of energy-dependent pathways that control lineage specification and could be utilized to enrich the examination of metabolic dynamics in different physiological and pathological states.
The main question arising from current studies in systems biology remains how these molecular regulatory mechanisms interact during cardiac differentiation. Investigating the transcriptional, translational, and epigenetic interplay would provide a holistic perspective in understanding cardiovascular development. One systems biology approach combined cardiac mRNA profiles with cardiac transcription factors (Gata4, Mef2a, Nkx2.5, and Srf), activating histone modifications and miRNA profiles [70]. In this study, Schlesinger et al showed that combinatorial regulation of mouse HL-1 cardiomyocytes exhibited interdependency among the three levels of molecular regulation, suggesting a capacity for mutual and reciprocal regulation [70]. Future studies directed at leveraging system interdependence may extrapolate novel therapeutic avenues in the context of regenerative cardiology.
OMICS Approach to Cardioregenerative Medicine
Introducing OMICS-based strategies into clinical outcomes analysis may allow for predictive assessment for disease recurrence and patient-specific therapeutic response. Recent studies using whole-genome expression microarray on blood samples from first-time acute myocardial infarction (AMI) patients elucidated differentially regulated genes and modulate pathways associated with recurrent cardiovascular outcomes [95]. Furthermore, corresponding bioinformatics analysis revealed increasing disease severity was associated with decreased expression of genes involved in the developmental epithelial-to-mesenchymal transition pathway, providing a cell-based approach for risk stratification in patients following AMI [95]. Disease anticipation prior to symptomatic presentation provides opportunities for proactive and preventative clinical management.
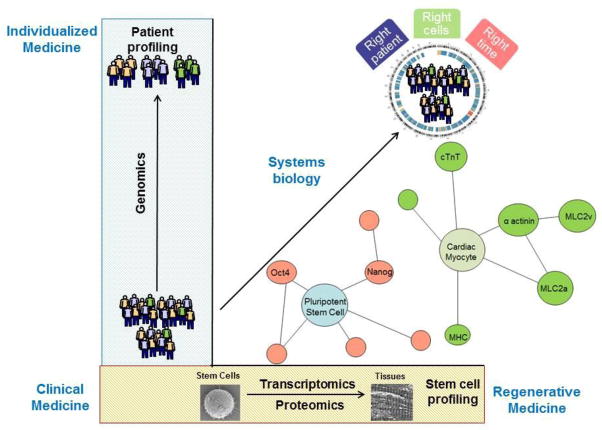
In the current era of cell therapy approaches for ischemic heart disease, clinical trials continue to evaluate the use of multipotent stem cells for cardiac regeneration. Investigators of the Phase I SCIPIO trial studied the delivery of mast/stem cell growth factor receptor Kit (SCFR; also known as c-Kit) positive cardiac stem cells and reported therapeutic benefit in LVEF of 12.3% ejection fraction units in the initial eight patients treated compared to baseline values [96]. Similarly, the Phase II C-CURE trial determined the safety of cardiopoietically-induced hMSCs in the context of heart failure and established a benefit in LVEF of 7% over baseline values [97]. Among the patients receiving C-CURE cell therapy that had 6-month follow-up (n=21) the reported change in LVEF varied from 2% to 10% over baseline values, suggesting a variability in patient response [97]. Genomic and proteomic analysis of patient-specific response could allow for stratification of therapeutic responders versus non-responders in cardiac regeneration (Figure 2). Additionally, current therapeutic benefit varies between clinical trials due to differences in transplanted cell types, differing in their paracrine potency, anti-apoptotic properties, tissue engraftment, and regenerative efficacy [98,99]. Such discrepancies could be analyzed using systems-based technologies to propose the most effective individualized cell therapy on a patient-specific basis.
Figure 2. Integration of systems biology algorithms towards clinical application.
Genomics, proteomics, metabolomics, and others allow for the elucidation of novel pathways involved in the step-wise differentiation of pluripotent stem cells to cardiomyocytes. Such bioinformatics analyses in the context of clinical translation can lead to patient-specific risk stratification and therapeutic response characterization.
Clinical Implications of Integrated Network Modeling
Novel insights into the biological networks governing cardiac regenerative mechanisms, including signaling factor and cytokine interaction, present innovative and effective strategies for applying stem cell therapeutics in acute regeneration of the infarcted ventricle [98]. Preclinical studies have validated the ability of cell therapy to increase myocardial functionality in various animal models, yet significant challenges preclude immediate clinical standardization of stem cell therapy [97]. The main challenges prior to achieving clinically meaningful regeneration include: (1) obtaining optimal amounts of primed cardiac progenitor cells and (2) assuring functional integration into the myocardium without inducing arrhythmic complications [35,100]. Systematic assessment of the signaling and genetic networks that target cardiac differentiation would advance our understanding of the genomic and proteomic regulators involved in enhancing cardiac repair. Utilizing OMICS-based network studies to standardize stem cell expansion and their cardiogenic guidance may facilitate implementation of safe and effective clinical translation (Figure 2).
The promise of cellular cardiogenesis and neovascularization using the regenerative platform necessitates thorough knowledge of the myocardial transcriptome, proteome, and metabolome. Genome-wide studies could elucidate key developmental circuits that might be involved in the transition of cardiac progenitor-stem cells into the cardiac muscle fate. Indeed, interdependent genomic and proteomic evaluation of adult stem cells demonstrating efficacy is necessary to devise an approach that maximizes cell capacity for repair prior to transplantation [101].
With the multiplicity of available stem and progenitor cell populations, the scope of clinical cardiac trials is continuously expanding [102,98]. Yet, a key concern in implementing stem cell transplantation therapy for heart disease is the selection of a particular developmental cell stage for post-engraftment safety and efficacy. Many studies have evaluated the generation and characterization of stem cell-derived cardiac progenitor cells, however little is known about the interdependent molecular signaling that subsequently converts committed cells into cardiomyocytes. Moreover, functional cardiomyocyte maturation including the acquisition of electrophysiological properties for conduction deserves further investigation. Understanding the cardiac transcriptome produces beneficial preclinical research regarding how each cell type affects cardiac performance and pathology. High-throughput systems biology approaches provide an unbiased holistic means to identify crucial signaling pathways involved in functional cardiomyocyte maturation and cardioprotective mechanisms. Integrated OMICS studies could yield global perspectives that view human development as a composition of highly interlinked cellular networks and bridge current knowledge gaps in myocardial lineage specification. Many investigators concur that a combination of tissue engineering, pharmacological approaches, and cellular transplantation, with appropriate quality control and validation of inter-trial consistency, would produce the safest and most promising therapies [103–105]. Indeed, system biology approaches characterizing the physiological and pathological cardiac blueprint can progress our ability to clinically regenerate the heart in situ and in vivo.
Conclusion
Current efforts in cardioregeneration based on the “one-size-fits-all” principle limits the potential of individualized therapy in regenerative medicine. Integrated systems biology presents the opportunity to match molecular defects in patients to the regenerative product required for safe and effective therapeutic benefit. Additionally, next generation systems biology approaches define with increased resolution the means for risk stratification in patients. In the era of translating stem cell advances into the clinic, bioinformatics serves as a critical tool to better inform our clinical practice and elevate patient care.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Westerhoff HV, Palsson BO. The evolution of molecular biology into systems biology. Nature Biotechnology. 2004;22(10):1249–1252. doi: 10.1038/nbt1020. [DOI] [PubMed] [Google Scholar]
- 2.Joyce AR, Palsson BO. The model organism as a system: integrating ‘omics’ data sets. Nature Reviews Molecular cell biology. 2006;7(3):198–210. doi: 10.1038/nrm1857. [DOI] [PubMed] [Google Scholar]
- 3.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444(7117):364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 6.Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma’ayan A, Boyer LA, Troyanskaya OG, Whetton AD, Lemischka IR. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462(7271):358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horak CE, Snyder M. ChIP-chip: a genomic approach for identifying transcription factor binding sites. Methods Enzymology. 2002;350:469–483. doi: 10.1016/s0076-6879(02)50979-4. [DOI] [PubMed] [Google Scholar]
- 8.de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(13):7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung H, Sidhu KS. Epigenetic modifications of embryonic stem cells: current trends and relevance in developing regenerative medicine. Stem Cells Cloning. 2008;1:11–21. doi: 10.2147/sccaa.s3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher CL, Fisher AG. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Current Opinions in Genetics & Development. 2011;21(2):140–146. doi: 10.1016/j.gde.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Smith C, Storms B. Hematopoietic stem cells. Clinical Orthopaedics and Related Research. 2000;379(Suppl):S91–97. doi: 10.1097/00003086-200010001-00012. [DOI] [PubMed] [Google Scholar]
- 12.Orlic D, Kajstura J, Chimenti S, Jakoniuk I, Anderson SM, Li B, Pickel J, McKay R, Nadal-Ginard B, Bodine DM, Leri A, Anversa P. Bone marrow cells regenerate infarcted myocardium. Nature. 2001;410(6829):701–705. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- 13.Murry CE, Soonpaa MH, Reinecke H, Nakajima H, Nakajima HO, Rubart M, Pasumarthi KB, Virag JI, Bartelmez SH, Poppa V, Bradford G, Dowell JD, Williams DA, Field LJ. Haematopoietic stem cells do not transdifferentiate into cardiac myocytes in myocardial infarcts. Nature. 2004;428(6983):664–668. doi: 10.1038/nature02446. [DOI] [PubMed] [Google Scholar]
- 14.Charbord P, Pouget C, Binder H, Dumont F, Stik G, Levy P, Allain F, Marchal C, Richter J, Uzan B, Pflumio F, Letourneur F, Wirth H, Dzierzak E, Traver D, Jaffredo T, Durand C. A Systems Biology Approach for Defining the Molecular Framework of the Hematopoietic Stem Cell Niche. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Wagner W, Wein F, Seckinger A, Frankhauser M, Wirkner U, Krause U, Blake J, Schwager C, Eckstein V, Ansorge W, Ho AD. Comparative characteristics of mesenchymal stem cells from human bone marrow, adipose tissue, and umbilical cord blood. Experimental Hematology. 2005;33(11):1402–1416. doi: 10.1016/j.exphem.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Augello A, De Bari C. The regulation of differentiation in mesenchymal stem cells. Human Gene Therapy. 2010;21(10):1226–1238. doi: 10.1089/hum.2010.173. [DOI] [PubMed] [Google Scholar]
- 17.Behfar A, Terzic A. Derivation of a cardiopoietic population from human mesenchymal stem cells yields cardiac progeny. Nature Clinical Practice Cardiovascular Medicine. 2006;3(Suppl 1):S78–82. doi: 10.1038/ncpcardio0429. [DOI] [PubMed] [Google Scholar]
- 18.Liechty KW, MacKenzie TC, Shaaban AF, Radu A, Moseley AM, Deans R, Marshak DR, Flake AW. Human mesenchymal stem cells engraft and demonstrate site-specific differentiation after in utero transplantation in sheep. Nature Medicine. 2000;6(11):1282–1286. doi: 10.1038/81395. [DOI] [PubMed] [Google Scholar]
- 19.Madonna R, De Caterina R. Adipose tissue: a new source for cardiovascular repair. Journal of Cardiovascular Medicine (Hagerstown) 2010;11(2):71–80. doi: 10.2459/JCM.0b013e328330e9be. [DOI] [PubMed] [Google Scholar]
- 20.Meliga E, Strem BM, Duckers HJ, Serruys PW. Adipose-derived cells. Cell Transplant. 2007;16(9):963–970. doi: 10.3727/096368907783338190. [DOI] [PubMed] [Google Scholar]
- 21.Planat-Benard V, Silvestre JS, Cousin B, Andre M, Nibbelink M, Tamarat R, Clergue M, Manneville C, Saillan-Barreau C, Duriez M, Tedgui A, Levy B, Penicaud L, Casteilla L. Plasticity of human adipose lineage cells toward endothelial cells: physiological and therapeutic perspectives. Circulation. 2004;109 (5):656–663. doi: 10.1161/01.CIR.0000114522.38265.61. [DOI] [PubMed] [Google Scholar]
- 22.Carvalho PH, Daibert AP, Monteiro BS, Okano BS, Carvalho JL, Cunha DN, Favarato LS, Pereira VG, Augusto LE, Del Carlo RJ. Differentiation of adipose tissue-derived mesenchymal stem cells into cardiomyocytes. Arquivos Brasileiros de Cardiologia. 2013;100(1):82–89. doi: 10.1590/s0066-782x2012005000114. [DOI] [PubMed] [Google Scholar]
- 23.Peng L, Jia Z, Yin X, Zhang X, Liu Y, Chen P, Ma K, Zhou C. Comparative analysis of mesenchymal stem cells from bone marrow, cartilage, and adipose tissue. Stem Cells and Development. 2008;17(4):761–773. doi: 10.1089/scd.2007.0217. [DOI] [PubMed] [Google Scholar]
- 24.Menicanin D, Bartold PM, Zannettino AC, Gronthos S. Genomic profiling of mesenchymal stem cells. Stem Cell Reviews and Reports. 2009;5(1):36–50. doi: 10.1007/s12015-009-9056-2. [DOI] [PubMed] [Google Scholar]
- 25.Solter D. From teratocarcinomas to embryonic stem cells and beyond: a history of embryonic stem cell research. Nature Reviews Genetics. 2006;7(4):319–327. doi: 10.1038/nrg1827. [DOI] [PubMed] [Google Scholar]
- 26.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292 (5819):154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 27.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(12):7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 29.Wu DC, Boyd AS, Wood KJ. Embryonic stem cells and their differentiated derivatives have a fragile immune privilege but still represent novel targets of immune attack. Stem Cells. 2008;26(8):1939–1950. doi: 10.1634/stemcells.2008-0078. [DOI] [PubMed] [Google Scholar]
- 30.Macarthur BD, Ma’ayan A, Lemischka IR. Systems biology of stem cell fate and cellular reprogramming. Nature Reviews Molecular Cell Biology. 2009;10(10):672–681. doi: 10.1038/nrm2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, Leri A, Kajstura J, Nadal-Ginard B, Anversa P. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114(6):763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 32.Smith RR, Barile L, Cho HC, Leppo MK, Hare JM, Messina E, Giacomello A, Abraham MR, Marban E. Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation. 2007;115(7):896–908. doi: 10.1161/CIRCULATIONAHA.106.655209. [DOI] [PubMed] [Google Scholar]
- 33.Chong JJ, Chandrakanthan V, Xaymardan M, Asli NS, Li J, Ahmed I, Heffernan C, Menon MK, Scarlett CJ, Rashidianfar A, Biben C, Zoellner H, Colvin EK, Pimanda JE, Biankin AV, Zhou B, Pu WT, Prall OW, Harvey RP. Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell Stem Cell. 2011;9(6):527–540. doi: 10.1016/j.stem.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellison GM, Torella D, Dellegrottaglie S, Perez-Martinez C, Perez de Prado A, Vicinanza C, Purushothaman S, Galuppo V, Iaconetti C, Waring CD, Smith A, Torella M, Cuellas Ramon C, Gonzalo-Orden JM, Agosti V, Indolfi C, Galinanes M, Fernandez-Vazquez F, Nadal-Ginard B. Endogenous cardiac stem cell activation by insulin-like growth factor-1/hepatocyte growth factor intracoronary injection fosters survival and regeneration of the infarcted pig heart. Journal of the American College of Cardiology. 2011;58(9):977–986. doi: 10.1016/j.jacc.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 35.Mercola M, Ruiz-Lozano P, Schneider MD. Cardiac muscle regeneration: lessons from development. Genes & Development. 2011;25(4):299–309. doi: 10.1101/gad.2018411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellison GM, Torella D, Karakikes I, Nadal-Ginard B. Myocyte death and renewal: modern concepts of cardiac cellular homeostasis. Nature Clinical Practice Cardiovascular Medicine. 2007;4(Suppl 1):S52–59. doi: 10.1038/ncpcardio0773. [DOI] [PubMed] [Google Scholar]
- 37.Torella D, Ellison GM, Nadal-Ginard B. Adult c-kit(pos) cardiac stem cells fulfill Koch’s postulates as causal agents for cardiac regeneration. Circulation Research. 2014;114 (4):e24–26. doi: 10.1161/CIRCRESAHA.113.303313. [DOI] [PubMed] [Google Scholar]
- 38.van Berlo JH, Kanisicak O, Maillet M, Vagnozzi RJ, Karch J, Lin SC, Middleton RC, Marban E, Molkentin JD. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509(7500):337–341. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olson EN. A decade of discoveries in cardiac biology. Nature Medicine. 2004;10(5):467–474. doi: 10.1038/nm0504-467. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 42.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451(7175):141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 43.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Fernandez A, Nelson TJ, Ikeda Y, Terzic A. c-MYC independent nuclear reprogramming favors cardiogenic potential of induced pluripotent stem cells. Journal of Cardiovascular Translational Research. 2010;3(1):13–23. doi: 10.1007/s12265-009-9150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson TJ, Martinez-Fernandez A, Yamada S, Perez-Terzic C, Ikeda Y, Terzic A. Repair of acute myocardial infarction by human stemness factors induced pluripotent stem cells. Circulation. 2009;120(5):408–416. doi: 10.1161/CIRCULATIONAHA.109.865154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson TJ, Behfar A, Yamada S, Martinez-Fernandez A, Terzic A. Stem cell platforms for regenerative medicine. Clinical and Translational Science. 2009;2(3):222–227. doi: 10.1111/j.1752-8062.2009.00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1(1):39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 48.Miura K, Okada Y, Aoi T, Okada A, Takahashi K, Okita K, Nakagawa M, Koyanagi M, Tanabe K, Ohnuki M, Ogawa D, Ikeda E, Okano H, Yamanaka S. Variation in the safety of induced pluripotent stem cell lines. Nature Biotechnology. 2009;27(8):743–745. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]
- 49.Wyles SP, Yamada S, Oommen S, Maleszewski JJ, Beraldi R, Martinez-Fernandez A, Terzic A, Nelson TJ. Inhibition of DNA topoisomerase II selectively reduces the threat of tumorigenicity following induced pluripotent stem cell-based myocardial therapy. Stem Cells and Development. 2014 doi: 10.1089/scd.2014.0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunt SA, Baker DW, Chin MH, Cinquegrani MP, Feldman AM, Francis GS, Ganiats TG, Goldstein S, Gregoratos G, Jessup ML, Noble RJ, Packer M, Silver MA, Stevenson LW, Gibbons RJ, Antman EM, Alpert JS, Faxon DP, Fuster V, Jacobs AK, Hiratzka LF, Russell RO, Smith SC., Jr ACC/AHA Guidelines for the Evaluation and Management of Chronic Heart Failure in the Adult: Executive Summary A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Committee to Revise the 1995 Guidelines for the Evaluation and Management of Heart Failure): Developed in Collaboration With the International Society for Heart and Lung Transplantation; Endorsed by the Heart Failure Society of America. Circulation. 2001;104(24):2996–3007. doi: 10.1161/hc4901.102568. [DOI] [PubMed] [Google Scholar]
- 51.Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nature Reviews Molecular Cellular Biology. 2013;14(8):529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aderem A. Systems biology: its practice and challenges. Cell. 2005;121(4):511–513. doi: 10.1016/j.cell.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 53.Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P, Hermus MC, van Asperen R, Boon K, Voute PA, Heisterkamp S, van Kampen A, Versteeg R. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291(5507):1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- 54.Martinez-Fernandez A, Li X, Hartjes KA, Terzic A, Nelson TJ. Natural cardiogenesis-based template predicts cardiogenic potential of induced pluripotent stem cell lines. Circulation Cardiovascular Genetics. 2013;6(5):462–471. doi: 10.1161/CIRCGENETICS.113.000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruneau BG. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harbor Perspectives in Biology. 2013;5(3):a008292. doi: 10.1101/cshperspect.a008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Faustino RS, Behfar A, Perez-Terzic C, Terzic A. Genomic chart guiding embryonic stem cell cardiopoiesis. Genome Biology. 2008;9(1):R6. doi: 10.1186/gb-2008-9-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nelson TJ, Faustino RS, Chiriac A, Crespo-Diaz R, Behfar A, Terzic A. CXCR4+/FLK-1+ biomarkers select a cardiopoietic lineage from embryonic stem cells. Stem Cells. 2008;26(6):1464–1473. doi: 10.1634/stemcells.2007-0808. [DOI] [PubMed] [Google Scholar]
- 58.Behfar A, Perez-Terzic C, Faustino RS, Arrell DK, Hodgson DM, Yamada S, Puceat M, Niederlander N, Alekseev AE, Zingman LV, Terzic A. Cardiopoietic programming of embryonic stem cells for tumor-free heart repair. Journal of Experimental Medicine. 2007;204(2):405–420. doi: 10.1084/jem.20061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gersh BJ, Simari RD, Behfar A, Terzic CM, Terzic A. Cardiac cell repair therapy: a clinical perspective. Mayo Clinic Proceedings. 2009;84(10):876–892. doi: 10.4065/84.10.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olson EN, Schneider MD. Sizing up the heart: development redux in disease. Genes & Development. 2003;17 (16):1937–1956. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- 61.Xu C, Police S, Hassanipour M, Gold JD. Cardiac bodies: a novel culture method for enrichment of cardiomyocytes derived from human embryonic stem cells. Stem Cells & Development. 2006;15(5):631–639. doi: 10.1089/scd.2006.15.631. [DOI] [PubMed] [Google Scholar]
- 62.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453(7194):524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 63.High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nature Reviews Genetics. 2008;9(1):49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- 64.Rentschler S, Yen AH, Lu J, Petrenko NB, Lu MM, Manderfield LJ, Patel VV, Fishman GI, Epstein JA. Myocardial Notch signaling reprograms cardiomyocytes to a conduction-like phenotype. Circulation. 2012;126(9):1058–1066. doi: 10.1161/CIRCULATIONAHA.112.103390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cai W, Guzzo RM, Wei K, Willems E, Davidovics H, Mercola M. A Nodal-to-TGFbeta cascade exerts biphasic control over cardiopoiesis. Circ Res. 2012;111 (7):876–881. doi: 10.1161/CIRCRESAHA.112.270272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li X, Martinez-Fernandez A, Hartjes KA, Kocher JP, Olson TM, Terzic A, Nelson TJ. Transcriptional Atlas of Cardiogenesis Maps Congenital Heart Disease Interactome. Physiology Genomics. 2014 doi: 10.1152/physiolgenomics.00015.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Terami H, Hidaka K, Shirai M, Narumiya H, Kuroyanagi T, Arai Y, Aburatani H, Morisaki T. Efficient capture of cardiogenesis-associated genes expressed in ES cells. Biochemical and Biophysical Research Communications. 2007;355(1):47–53. doi: 10.1016/j.bbrc.2007.01.109. [DOI] [PubMed] [Google Scholar]
- 68.Behfar A, Terzic A. Cardioprotective repair through stem cell-based cardiopoiesis. Journal of Applied Physiology. 2007;103(4):1438–1440. doi: 10.1152/japplphysiol.00713.2007. [DOI] [PubMed] [Google Scholar]
- 69.Ross RS, Borg TK. Integrins and the myocardium. Circulation Research. 2001;88(11):1112–1119. doi: 10.1161/hh1101.091862. [DOI] [PubMed] [Google Scholar]
- 70.Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, Lange M, Tonjes M, Dunkel I, Sperling SR. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genetics. 2011;7(2):e1001313. doi: 10.1371/journal.pgen.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang H, Zhang Q, Fang X. Transcriptomics and proteomics in stem cell research. Frontiers in Medicine. 2014 doi: 10.1007/s11684-014-0336-0. [DOI] [PubMed] [Google Scholar]
- 72.Van Hoof D, Heck AJ, Krijgsveld J, Mummery CL. Proteomics and human embryonic stem cells. Stem Cell Research. 2008;1(3):169–182. doi: 10.1016/j.scr.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 73.Baharvand H, Hajheidari M, Zonouzi R, Ashtiani SK, Hosseinkhani S, Salekdeh GH. Comparative proteomic analysis of mouse embryonic stem cells and neonatal-derived cardiomyocytes. Biochemical and Biophysical Research Communications. 2006;349(3):1041–1049. doi: 10.1016/j.bbrc.2006.08.151. [DOI] [PubMed] [Google Scholar]
- 74.Cao F, Wagner RA, Wilson KD, Xie X, Fu JD, Drukker M, Lee A, Li RA, Gambhir SS, Weissman IL, Robbins RC, Wu JC. Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS One. 2008;3(10):e3474. doi: 10.1371/journal.pone.0003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yin X, Mayr M, Xiao Q, Wang W, Xu Q. Proteomic analysis reveals higher demand for antioxidant protection in embryonic stem cell-derived smooth muscle cells. Proteomics. 2006;6(24):6437–6446. doi: 10.1002/pmic.200600351. [DOI] [PubMed] [Google Scholar]
- 76.Mayr M, Madhu B, Xu Q. Proteomics and metabolomics combined in cardiovascular research. Trends in Cardiovascular Medicine. 2007;17(2):43–48. doi: 10.1016/j.tcm.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 77.Prudhomme W, Daley GQ, Zandstra P, Lauffenburger DA. Multivariate proteomic analysis of murine embryonic stem cell self-renewal versus differentiation signaling. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(9):2900–2905. doi: 10.1073/pnas.0308768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arrell DK, Niederlander NJ, Perez-Terzic C, Chung S, Behfar A, Terzic A. Pharmacoproteomics: advancing the efficacy and safety of regenerative therapeutics. Clinical Pharmacology & Therapeutics. 2007;82(3):316–319. doi: 10.1038/sj.clpt.6100310. [DOI] [PubMed] [Google Scholar]
- 79.Arrell DK, Niederlander NJ, Faustino RS, Behfar A, Terzic A. Cardioinductive network guiding stem cell differentiation revealed by proteomic cartography of tumor necrosis factor alpha-primed endodermal secretome. Stem Cells. 2008;26(2):387–400. doi: 10.1634/stemcells.2007-0599. [DOI] [PubMed] [Google Scholar]
- 80.Chiriac A, Nelson TJ, Faustino RS, Behfar A, Terzic A. Cardiogenic induction of pluripotent stem cells streamlined through a conserved SDF-1/VEGF/BMP2 integrated network. PLoS One. 2010;5(4):e9943. doi: 10.1371/journal.pone.0009943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fraidenraich D, Stillwell E, Romero E, Wilkes D, Manova K, Basson CT, Benezra R. Rescue of cardiac defects in id knockout embryos by injection of embryonic stem cells. Science. 2004;306(5694):247–252. doi: 10.1126/science.1102612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Doss MX, Chen S, Winkler J, Hippler-Altenburg R, Odenthal M, Wickenhauser C, Balaraman S, Schulz H, Hummel O, Hubner N, Ghosh-Choudhury N, Sotiriadou I, Hescheler J, Sachinidis A. Transcriptomic and phenotypic analysis of murine embryonic stem cell derived BMP2+ lineage cells: an insight into mesodermal patterning. Genome Biology. 2007;8(9):R184. doi: 10.1186/gb-2007-8-9-r184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bader A, Al-Dubai H, Weitzer G. Leukemia inhibitory factor modulates cardiogenesis in embryoid bodies in opposite fashions. Circulation Research. 2000;86(7):787–794. doi: 10.1161/01.res.86.7.787. [DOI] [PubMed] [Google Scholar]
- 84.Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28(4):721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- 85.Folmes CD, Martinez-Fernandez A, Perales-Clemente E, Li X, McDonald A, Oglesbee D, Hrstka SC, Perez-Terzic C, Terzic A, Nelson TJ. Disease-causing mitochondrial heteroplasmy segregated within induced pluripotent stem cell clones derived from a patient with MELAS. Stem Cells. 2013;31(7):1298–1308. doi: 10.1002/stem.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chung S, Arrell DK, Faustino RS, Terzic A, Dzeja PP. Glycolytic network restructuring integral to the energetics of embryonic stem cell cardiac differentiation. Journal of Molecular and Cellular Cardiology. 2010;48(4):725–734. doi: 10.1016/j.yjmcc.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nature Reviews Molecular Cellular Biology. 2012;13(4):263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metabolism. 2011;14(2):264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, Herrerias A, Batchelder EM, Plongthongkum N, Lutz M, Berggren WT, Zhang K, Evans RM, Siuzdak G, Izpisua Belmonte JC. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Research. 2012;22(1):168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(13):4783–4788. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Westfall SD, Sachdev S, Das P, Hearne LB, Hannink M, Roberts RM, Ezashi T. Identification of oxygen-sensitive transcriptional programs in human embryonic stem cells. Stem Cells and Development. 2008;17(5):869–881. doi: 10.1089/scd.2007.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7(2):150–161. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 93.Chung S, Dzeja PP, Faustino RS, Perez-Terzic C, Behfar A, Terzic A. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nature Clinical Practice Cardiovascular Medicine. 2007;4(Suppl 1):S60–67. doi: 10.1038/ncpcardio0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park do J, Park KS, Lee HK. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochemical and Biophysical Research Communications. 2006;348(4):1472–1478. doi: 10.1016/j.bbrc.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 95.Suresh R, Li X, Chiriac A, Goel K, Terzic A, Perez-Terzic C, Nelson TJ. Transcriptome from circulating cells suggests dysregulated pathways associated with long-term recurrent events following first-time myocardial infarction. Journal of Molecular and Cellular Cardiology. 2014;74C:13–21. doi: 10.1016/j.yjmcc.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bolli R, Chugh AR, D’Amario D, Loughran JH, Stoddard MF, Ikram S, Beache GM, Wagner SG, Leri A, Hosoda T, Sanada F, Elmore JB, Goichberg P, Cappetta D, Solankhi NK, Fahsah I, Rokosh DG, Slaughter MS, Kajstura J, Anversa P. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378(9806):1847–1857. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 97.Bartunek J, Vanderheyden M, Hill J, Terzic A. Cells as biologics for cardiac repair in ischaemic heart failure. Heart. 2010;96(10):792–800. doi: 10.1136/hrt.2007.139394. [DOI] [PubMed] [Google Scholar]
- 98.Behfar A, Crespo-Diaz R, Terzic A, Gersh BJ. Cell therapy for cardiac repair--lessons from clinical trials. Nature Reviews Cardiology. 2014;11(4):232–246. doi: 10.1038/nrcardio.2014.9. [DOI] [PubMed] [Google Scholar]
- 99.Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LE, Berman D, Czer LS, Marban L, Mendizabal A, Johnston PV, Russell SD, Schuleri KH, Lardo AC, Gerstenblith G, Marban E. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 2012;379(9819):895–904. doi: 10.1016/S0140-6736(12)60195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Terzic A, Behfar A. Regenerative heart failure therapy headed for optimization. European Heart Journal. 2014;35(19):1231–1234. doi: 10.1093/eurheartj/ehu117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dimmeler S, Zeiher AM. Cell therapy of acute myocardial infarction: open questions. Cardiology. 2009;113(3):155–160. doi: 10.1159/000187652. [DOI] [PubMed] [Google Scholar]
- 102.Wollert KC, Drexler H. Cell therapy for the treatment of coronary heart disease: a critical appraisal. Nature Reviews Cardiology. 2010;7 (4):204–215. doi: 10.1038/nrcardio.2010.1. [DOI] [PubMed] [Google Scholar]
- 103.Behfar A, Crespo-Diaz R, Nelson TJ, Terzic A, Gersh BJ. Stem cells: clinical trials results the end of the beginning or the beginning of the end? Cardiovascular & Hematological Disorders-Drug Targets. 2010;10(3):186–201. doi: 10.2174/1871529x11006030186. [DOI] [PubMed] [Google Scholar]
- 104.Ellison GM, Nadal-Ginard B, Torella D. Optimizing Cardiac Repair and Regeneration Through Activation of the Endogenous Cardiac Stem Cell Compartment. Journal of Cardiovascular Translational Research. 2012 doi: 10.1007/s12265-012-9384-5. [DOI] [PubMed] [Google Scholar]
- 105.Nadal-Ginard B, Ellison GM, Torella D. The cardiac stem cell compartment is indispensable for myocardial cell homeostasis, repair and regeneration in the adult. Stem Cell Research. 2014 doi: 10.1016/j.scr.2014.04.008. [DOI] [PubMed] [Google Scholar]