Abstract
Protein-protein interactions (PPIs) are emerging as attractive targets for drug design because of their central role in directing normal and aberrant cellular functions. These interactions were once considered “undruggable” because their large and dynamic interfaces make small molecule inhibitor design challenging. However, landmark advances in computational analysis, fragment screening and molecular design have enabled development of a host of promising strategies to address the fundamental molecular recognition challenge. An attractive approach for targeting PPIs involves mimicry of protein domains that are critical for complex formation. This approach recognizes that protein subdomains or protein secondary structures are often present at interfaces and serve as organized scaffolds for the presentation of side chain groups that engage the partner protein(s). Design of protein domain mimetics is in principle rather straightforward but is enabled by a host of computational strategies that provide predictions of important residues that should be mimicked. Herein we describe a workflow proceeding from interaction network analysis, to modeling a complex structure, to identifying a high-affinity sub-structure, to developing interaction inhibitors. We apply the design procedure to peptidomimetic inhibitors of Ras-mediated signaling.
Keywords: protein-protein interaction, computational tools, inhibitor design, peptidomimetic, protein structure
1. Introduction
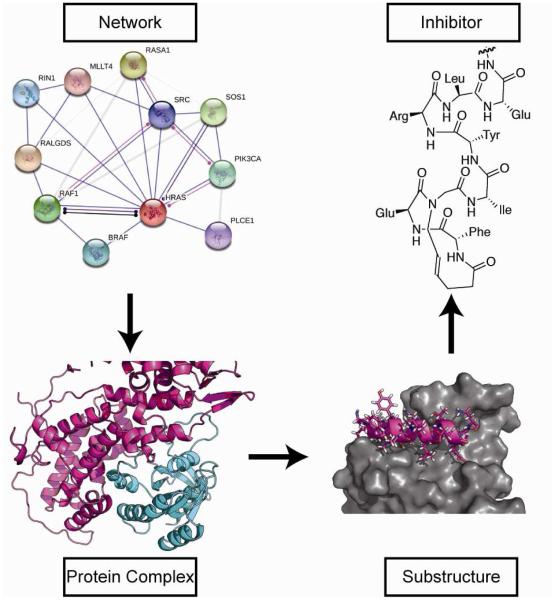
The centrality of protein-protein interaction (PPI) networks in regulating cellular function offers attractive opportunities for drug discovery [1, 2]. PPIs are considered fertile yet challenging targets for inhibitor design [3]; however, advances in molecular and structural biology as well as computational chemistry and molecular design have afforded potent inhibitors for previously intractable targets [4]. Mimicry of protein domains that are critical for the formation of native protein-protein complexes offers an attractive approach for the design of PPI inhibitors [5-8]. This protein domain mimicry approach is complementary to small molecule high throughput and fragment based screening approaches, with each method offering distinct advantages [9, 10]. While protein domain mimetics employ quaternary structure information to imitate native bound states, small molecule high-throughput screens can reveal binding pockets and molecules that allosterically modulate the binding surface [11-14]. Fragment-based methods identify small molecule binders and employ an iterative approach to recombine them to produce a potent ligand for the target interface [15]. These different yet complementary approaches have yielded orthosteric and allosteric inhibitors while revealing general principles that can be extended to broad classes of PPIs [16]. In this review, we outline steps to the design of protein domain or protein secondary structure mimetics as inhibitors of chosen PPIs. We focus on the design of PPI inhibitors that modulate Ras/Sos and Ras/Raf complexes as model systems, with a focus on description of the in silico resources available to guide a project from target selection to compound design. An overview of the process is depicted in Figure 1.
Figure 1.
Analysis of a diseased signaling network gives rise to specific protein complexes of interest, after which that complex is analyzed to identify minimal units of structure relevant for mimicry. Finally, a specific inhibitor molecule is designed based on that sub-structure.
2. Computational Methods to Target Protein-Protein Interactions
Commonly, efforts to design novel inhibitors begin with a disease state in mind, rather than a specific protein or a specific protein complex. Disease states can be distinguished from healthy states by comparing the signaling networks present in each; for example, cancers typically exhibit upregulated proliferation signaling circuits. Thus, before arriving at a specific protein complex, one must examine the perturbations native to the disease state and determine what interactions within that signaling network might return it to health.
2.1. From Disease State to Protein-Protein Interaction
The majority of inter-species variation owes to differences in the interactions between gene products rather than differences in gene sequences [17]. The connectivity of nodes in PPI networks is often employed to distinguish types of targets for prospective modulation [18]. High connectivity nodes likely have more off-target effects, which can potentially produce toxicity; on the other hand, low-connectivity nodes may be unlikely to have a meaningful effect on the disease phenotype. Synthetic inhibitors may be designed to be “frequent hitters” that are intrinsically nonselective or to specifically engage more than one target [19, 20]. As an example of the latter case, tumors with wild-type p53 frequently overexpress two negative regulators, Mdm2 and Mdmx; drug molecules that promiscuously bind both negative regulators are highly desirable [21].
PPI networks are typically evaluated using gene knockdown strategies, such as RNAi, which result in total and irreversible abrogation of a protein’s effects. Under such conditions, high-connectivity nodes are likely to produce a strong toxic effect. A distinguishing feature of molecular interaction inhibitors is that they are uniquely capable of specifically disrupting one edge of a network where the impact of modulating high-connectivity nodes can be titrated in a concentration dependent manner [22-25]. Thus, synthetic inhibitors afford dose-dependent controlled inhibition of specific sets of interactions for a particular protein [25-27].
Given a network believed to describe the interactions relevant to a certain disease state, the identification and analysis of the most important and inhibition-amenable interaction nodes is critical to develop useful PPI inhibitors. Several network analysis tools have been described. Network metrics beyond node connectivity can aid in target selection; for example, the pairwise disconnectivity index measures how essential a given protein is for sustaining the connection between two others [28]. Networks can even be used as inferential tools to support the existence of protein-protein interactions for which there exists no direct experimental evidence [29]. Johnson’s interface interaction network, or IIN, describes which protein interfaces are commonly bound by multiple proteins and thus permits the early identification of potential off-target effects [30].
2.2. From Protein-Protein Interaction to a Structural Model
Though there are hundreds of thousands of protein-protein interactions predicted in humans, there are fewer than twenty thousand non-redundant multiprotein complexes in the Protein Data Bank [31]. High resolution structures are invaluable for structure-based design of inhibitors. However, in the absence of experimental structures, homology models and mutagenesis data often provide sufficient information for preliminary design. Homology models are produced by performing multiple sequence alignments and threading the novel sequence along the backbone of template structures. The result is then refined, especially where the novel sequence is likely to differ structurally from the template. SWISS-MODEL provides a webserver to perform homology modeling and an annotated database of previously constructed homology models [32, 33]. Broadly speaking, models arising from at least 90-95% sequence identity tend to have equivalent resolution to the experimental structures from which they are derived; they exhibit errors mostly due to side-chain packing and are suitable for ligand docking and structure-based drug design [34]. For more precise and residue-level quality assessment, one may employ algorithms like QMEAN [35]. Many utilities for subsequent inhibitor docking and design studies can use an ensemble of top-scoring homology models or all the models of an NMR structure [36-38].
2.3. From Structural Model to Inhibitory Substructure
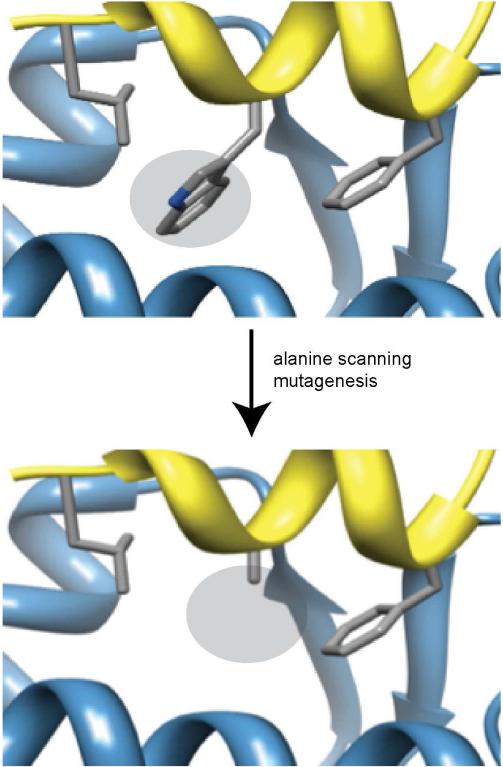
The typical protein comprises several hundred amino acids, and a surprisingly large proportion of those residues contribute to the interface of protein-protein interactions. One study suggests that almost a quarter of all residues in protein dimers appear at the interface [39]. Specifically, a sample of over 35,000 two-protein interfaces from the Protein Data Bank [31] suggests that a protein at a protein-protein interface typically contributes dozens of residues, often in disconnected segments [40]. Typical orthosteric inhibitors must mimic a minimal subset of interface residues. Two complementary metrics for judging the relative importance of different residues are ΔΔG and ΔSASA. ΔΔG refers to the change in binding energy upon mutation of a residue to alanine (Figure 2) and may be found by modeling or expressing point mutants of the protein of interest; generally, mutating important interface residues to alanine abrogates binding and results in high positive ΔΔG [41, 42]. ΔSASA, in contrast, is a description of the change in solvent-accessible surface area upon binding and may easily be decomposed on a per-residue basis. While ΔSASA is more straightforward to compute and requires fewer subjective choices of parameters and algorithms, its relationship to a corresponding Kd or IC50 is more distant; in contrast, ΔΔG is more difficult to compute but bears an immediate relationship to Kd Identifying hotspot residues (ΔΔG greater than some threshold, often 1–2 kcal/mol) and anchor residues (ΔSASA greater than 100 Å2) is a first step towards limiting the search space for desired interface substructures. A selection of databases cataloguing protein-protein interactions, along with useful biophysical data, is presented in Table 1.
Figure 2.
Alanine scanning mutagenesis of interfacial residues reveals the importance of each residue to complex formation. The example depicts mutation of a key tryptophan from the p53 (yellow ribbon) activation helix in complex with Mdm2 (blue).
Table 1.
Examples of protein-protein interaction databases
| Database Name | Description |
|---|---|
| ASEdb[43] | Experimental alanine scanning values; no longer maintained, though the raw data is still available |
| Relibase[44] | Protein-ligand interfaces, tools for binding site alignment and visual comparison |
| 2P2Idb[16] | PPIs with known orthosteric modulators where both the protein- protein and protein-ligand complex are known |
| TIMBAL[45] | PPIs, their small molecule inhibitors or stabilizers with experimental affinities and assay descriptions |
| PrePPI[46] | High-confidence predicted PPIs |
| ComSin[47] | Analysis of differences between proteins in bound and unbound states |
| PIFACE[48] | Cluster of PPIs by their interface structures |
| Dr. PIAS[49] | Predicts PPI druggability by similarity to known druggable PPIs |
| PINT[50, 51] | Experimental thermodynamic parameters (e.g. Kd, the corresponding ΔG, and the temperature and pH of the experiment) |
| SKEMPI[50, 51] | Binding kinetics; unification of PINT and ASEdb data |
| PepCyber:P~PEP[52] | Phosphoprotein binding domains (like SH2, WW) |
| 3D-interologs[53, 54] | Scores novel interactions via homology to Uniprot |
| PocketQuery[55] | Clusters of residues with favorable ΔSASA |
| HippDB. SippDB[56, 57] | Identification of PPIs by secondary structure elements, alanine scanning |
ΔΔG may be computed by using Rosetta or via MM/PBSA or MM/GBSA analysis of molecular dynamics trajectories [58, 59]. In situations where alanine scanning would either be laborious or prohibited by a structural feature that a specific computational package cannot model well, ΔΔG values may be computed indirectly by machine learning [60]. Evolutionary conservation, selecting the most often buried residues from a global docking study, or even feature analysis purely on the primary sequence can indicate important residues [61-65]. Influential interactions can also be identified and quantified by formalizations of traditional visual analysis. For example, buried hydrophobic surface area is worth about 2-2.5 kcal/mol per 100 Å2, lysine cation-pi interactions contribute around 0.4-1.1 kcal/mol, and buried neutral hydrogen bonds are valued anywhere from 0.5-1.8 kcal/mol [66-69]. Important substructures may also be identified by examining the target protein; viable binding pockets may be determined from apo structures [70]. In fact, while the receptor-centric notion of a hotspot for ligand binding lines up well with the native interaction-based notion of a hotspot residue, not all hotspot residues are bound in receptor hotspots, and thus not all hotspot residues can be used as handles for design [71].
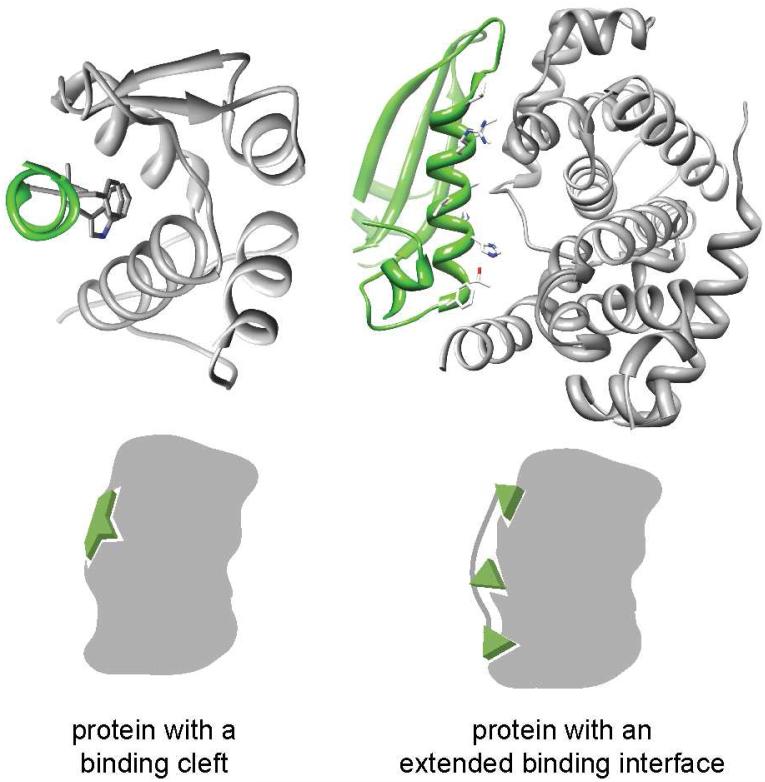
Assessing the shape of inhibitory substructures can guide design efforts by defining the types of inhibitors that are typically appropriate. We have classified helical interfaces as “binding clefts” where the important residues are concentrated within a binding pocket and as “extended interfaces” where the critical residues are diffused over the surface (Figure 3). Interfaces with a binding cleft are likely more susceptible to small molecule inhibition than especially large interfaces [72]. However, extended interfaces cannot be addressed by simply employing the native interface peptide. First, the ordered conformation the peptide adopts in the protein context is unlikely to be substantially populated in aqueous buffer. Second, that peptide is likely to be proteolytically cleaved and incapable of entering the desired cell type. The reason that one might consider only an inhibitory substructure becomes particularly plain for Ras/Sos. While protein-protein interactions generally present a prohibitively large interface, typically around 1600±400□Å2, the Ras/Sos interaction is an extreme case [73, 74]. Sos is a 150 kDa protein; its interface with chain R contains over a hundred disconnected residues, wildly more than would be feasible for direct mimicry.
Figure 3.
Helical interfaces can be divided between those that feature clefts for binding and those with extended interfaces. The cleft interfaces feature a high density of important contacts in a small region. The p53/MDM2 (left: p53 in green, MDM2 in grey; PDB code: 1YCR) and cyclin-dependent kinase6/D-type viral cyclin (right: cdk6 in green, D-type viral cyclin in grey; PDB code: 1G3N) complexes are representative examples of binding cleft and extended interfaces, respectively.
Substructures can indicate approaches to modulate specificity as much as affinity. In a seminal study, Milletti and Vulpetti screened pockets occupied by ATP and drug compounds against nearly two hundred thousand pockets between 300 and 4000 Å2 from other proteins in the Protein Databank [75]. Strikingly, E. coli phosphoenolpyruvate carboxykinase (PCKA) possesses an ATP-binding pocket similar to regions of a variety of proteins with entirely different folds, including the GTP-binding pocket of RAN and the catalytic domain of Ras-GDP; in contrast, applying the same procedure to pyridoxal kinase only returned alternative crystal structures of the same protein. Using this method, the pockets of a desired partner protein can be evaluated for their intrinsic promiscuity and likelihood to produce off-target effects; further design efforts can focus on a protein’s most unique interface regions.
2.4. From Inhibitory Substructure to Inhibitor Design
Many specific scaffolds are available to mimic interface secondary structure elements. The rational design of such native-inspired scaffolds provides a complementary approach to screening methods. Because the structures of native complexes are unlikely to describe the entire space of possible binders, screening can provide hits that structural design would be unable to predict. For example, interfaces that lack high-affinity secondary structure elements may nonetheless contain a number of medium-affinity pockets and thus be good candidates for fragment-based discovery methods. Small molecule drugs simply require a geometrically small interface with spatially dense hotspots. There are around 11,500 helical interfaces listed in HippDB that contain exactly two hotspots within one helical turn (i.e. i, i+1 through i, i+4); such interfaces are amenable to small molecule inhibition. The rational design of secondary structure mimetics, however, provides a unique opportunity to combine the modularity of peptides with the desired biological properties of small molecules.
Broadly speaking, secondary structure mimetics may be described as either peptidic or nonpeptidic. The former class possesses a native backbone but contains essential alterations that affect its conformational equilibrium, while the latter mimics the residue presentation of the native ensemble despite a very different distribution of conformations, owing to nonpeptidic backbone. The decision to employ peptidic or nonpeptidic domain mimetics is determined by a balance of predicted efficacy and other desirable properties. A nonpeptidic mimetic may only be able to present a few side chains or may only be able to mimic one helix or strand face; on the other hand, it may possess superior metabolic stability [76, 77]. Furthermore, peptidomimetics are naturally subject to conformational considerations, even if the sequence-based trends differ from native peptides. Peptidic scaffolds are far more likely to exhibit similar conformational distributions to native peptides than are nonpeptidic scaffolds [78].
Only a subset of known and validated peptidomimetic scaffolds are compatible with common structural modeling packages. Generally speaking, molecular mechanics-based packages like Schrödinger’s MacroModel[79] are only affected if a scaffold employs an unfamiliar atom type, which is particularly uncommon. Thus, most scaffolds can be simulated using such packages, as though they were small molecule ligands, without further effort. Often, however, it is desirable to use a package that is endowed with particular information about inter-residue interactions and packing, or a force field that is known to reproduce specific conformations of the desired ligand; in particular, while many force fields may do an acceptable job on a relatively small peptidomimetic, one may seek a modeling methodology that also models the protein well. Rosetta is one such modeling suite; it is considerably powerful at both ab initio structural prediction and at protein design [80, 81] and it recently was extended with support for a variety of peptidomimetics, such as the hydrogen bond surrogate, oligooxopiperazines, and peptoids [82]. Where such integration is not already available, it can be developed; a non-native residue, such as the hydrocarbon staple, can be incorporated into a molecular dynamics force field like AMBER simply by using high level geometry optimization (for example, using Gaussian) and providing the results to a restrained electrostatic potential charge-fitting service like the R.E.D. server [83, 84]. In parallel to efforts to incorporate non-native scaffolds, parameter sets have been developed to incorporate non-canonical amino acid residues into both Rosetta and molecular dynamics forcefields like Amber [85, 86].
3. Targeting the Ras/Sos and Ras/Raf interactions
To illustrate the prior summary of computational methods, we discuss computational approaches to targeting the Ras pathway, to illustrate decision points encountered in the rational design of synthetic ligands for protein-protein interactions. We identify two key protein-protein interactions that may modulate the downstream effects of the Ras pathway, find complexes that serve as viable structural models of those interactions, isolate minimal regions of those complexes that may be amenable to mimicry, and propose inhibitor candidates based on those substructures.
3.1. Network analysis
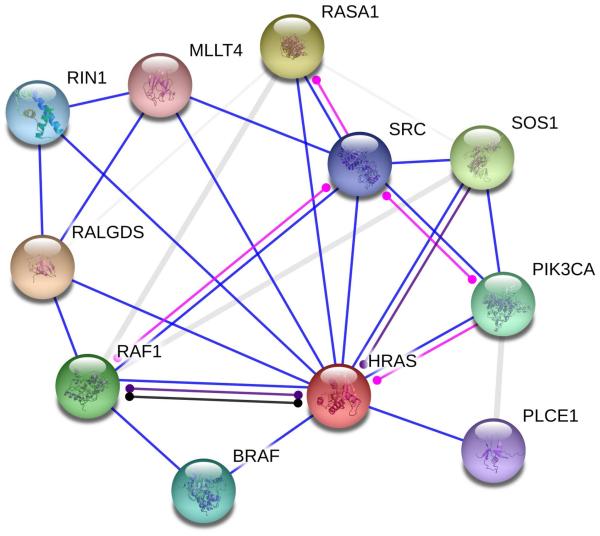
Ras is a small GTPase strongly implicated in cell proliferation in cancer [87-89]. Ras is a high-connectivity target, and its downstream effectors include a kinase cascade that affects nearly a hundred different targets.[90] Figure 4 depicts an annotated map of the interaction network surrounding HRas, produced using string-db.org.[91]
Figure 4.
Ras is a highly connected protein important to cancer proliferation circuits. Depicted are string-db.org functional associations of binding (blue), post-translational modification (magenta), reaction (black), and catalysis (purple). Links of unclear nature are shown in grey.
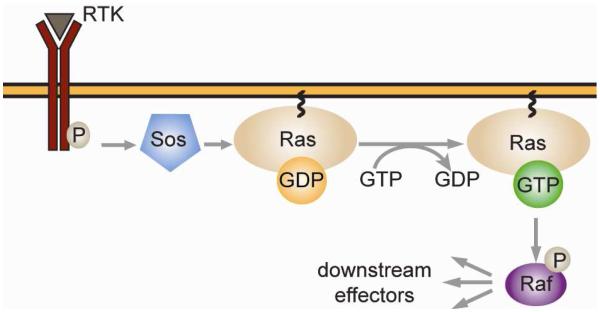
For the purpose of this investigation, we will consider modulating this signaling pathway via the inhibition of either of two interactions: Ras-Sos and Ras-Raf. Ras signaling is activated upon the conversion of GDP-bound Ras to GTP-bound Ras, a process catalyzed by the Ras-specific guanine nucleotide exchange factor Sos. Subsequently, GTP-Ras binds Raf, the entry point into the MAP/ERK pathway (Figure 5). Broadly, Ras signaling has been targeted by preventing receptor tyrosine kinase activation, Ras membrane localization, Ras/Sos complex formation, and activation of downstream kinases.[92] Small molecule inhibitors that bind allosteric sites on Ras and disrupt Ras-Sos complexation have been developed using SAR by NMR and fragment based drug design using thiol tethering [11, 93, 94]. In contrast, inhibition of the Ras-Raf complex has not been investigated as intensely. Successful inhibitors of this interaction include sulindac sulfide and MCP compounds discovered in yeast two-hybrid screens [95, 96].
Figure 5.
Ras/Sos and Ras/Raf are implicated in the MAP/ERK signaling pathway. Ras interacts with Sos only when a receptor tyrosine kinase binds its substrate and recruits Sos to the membrane. Following the Ras/Raf association depicted, Raf phosphorylates Mek, which phosphorylates MAPK, which modulates the activity of many downstream effectors.
3.2. Structure identification
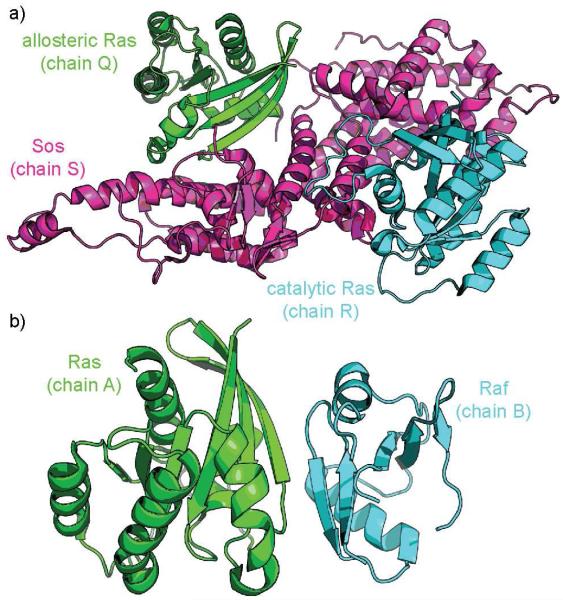
In the Ras-Sos case (Figure 6a), there exist four high-quality crystal structures of Ras in complex with Sos (PDB: 1NVU, 1NVV, 1NVW, 1NVX) [97]. The primary biological assembly and the asymmetric unit in all four cases contains two Ras molecules (chains Q and R) and one Sos molecule (chain S), but they differ in crucial ways. While chain R is wild-type in each case, only in 1NVW is chain Q wild-type; 1NVU and 1NVX contain A59G Ras, a mutation that is not at the SOS interface, and 1NVV contains Y62A Ras, a mutation close to the interface with SOS. Moreover, the chain Q Ras unit in 1NVW is bound to GNP, as desired, while chain Q in 1NVX is bound to GTP. 1NVW and 1NVV are clearly the most attractive structures for our purposes; for this narrative we will continue with 1NVV, keeping in mind the Y62A point mutation, simply because its resolution is somewhat better than that of 1NVW. For a full study, it would be instructive to perform the same procedure on all structures available and compare the results. We employed Rosetta’s “relax” protocol for structural refinement to optimize hydrogen placement and side-chain rotamer packing; we generated 200 decoys and used the lowest-energy resulting structure.
Figure 6.
a. Catalytic or orthosteric Ras (chain R, cyan) and allosteric RAS (chain Q, green) in complex with Sos (chain S, magenta), from PDB code 1NVV. b. Ras (chain A, green) and the Raf analogue Byr2 (chain B, cyan), from PDB code 1K8R.
For Ras-Raf, there exist four crystal structures that may be employed (Figure 6b). One is wild-type (PDB: 4G0N); one includes a mutant Ras (PDB: 4G3X); one examines a mutant Raf (PDB: 3KUD); finally, one employs in place of Raf a homologous kinase found in S. pombe, Byr2 (PDB: 1K8R). For this review, we utilized the Byr2 complex, which has been available since 2001 and has provided a foundation for past Ras/Raf inhibitor designs.
3.3. Finding an inhibitory substructure
We extended the investigations into Ras/Raf and Ras/Sos by conducting Rosetta’s alanine scanning protocol using the online server Robetta [98]. We used this data to evaluate the interactions of both the orthosteric (chain R) and allosteric (chain Q) Ras units with the Sos molecule, and similarly for the Ras (chain A)/Raf (chain B) complex. Both Ras/Sos interfaces contain a number of residues with sizable ΔΔG values (reported as Rosetta energy units, or REU, which are approximately 1 kcal/mol in magnitude). In each case, substantial ΔΔG is present across a wide range of primary sequence and Cartesian space. In the Ras/Sos complex, residues in the low 800s contribute 5 REU and residues in the low 1000s contribute almost 2 REU, while in Ras/Raf, residues in the upper 600s contribute over 11 REU while residues in the upper 900s contribute about 3 REU. Unquestionably, only a fraction of all the residues with inhibitory potential might be mimicked.
3.3.1. The Ras–Sos complex
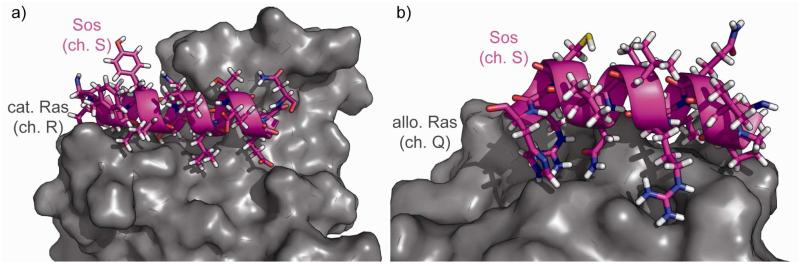
Ras–Sos interfaces present attractive starting points for Sos mimicry and thus Ras inhibition. Range 683-695, at the S-Q interface, and range 929-944, at the R-Q interface, are both alpha helices, which is an encouraging result due to the wide variety of α-helix mimetics available (Figure 7). We chose to corroborate our results using the PocketQuery server, imagining that similar indications of promising regions for mimicry would be strong evidence for our two high-affinity residue ranges. Using PocketQuery on 1NVV provides a list of many different combinations of residues that, taken together, make for strong inhibitor design candidates. Most are two or three residues in all, and top candidates include His911+Thr935+Lys939 and Arg694+Trp729. These results might be particularly instructive to explore from a fragment-based or small-molecule-screening approach.
Figure 7.
a. The substructure from Sos’s interface with the orthosteric Ras molecule (chain R), residues 683S-695S. b. The substructure from Sos’s interface with the allosteric Ras molecule (chain Q), residues 929S-944S.
3.3.2. Ras–Raf complex
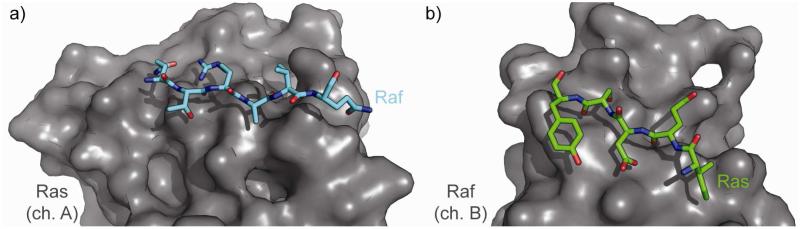
In contrast to the Ras–Sos complex, residues to mimic either from Ras or Raf are fewer and further between. Ras’s highest-affinity single hotspot residue is isolated, and 5 REU of ΔΔG is available in the 36-40 range. The same goes for Raf, with residues 81-86 similarly representing a limited amount of ΔΔG, though any strategy capable of mimicking 72 and 74 as well might have more use. The Ras/Raf interface presents few attractive handles for inhibitor design in the native structure; no connected segment presents a large amount of total ΔΔG. In each case, the secondary structure present is a beta strand. Furthermore, the residue sets present on each protein are part of pairs of antiparallel strands, which could be mimicked well by beta hairpins. Particularly due to the low-affinity results of this analysis, we explored the 1K8R structure using PocketQuery. The best-scoring pockets on chain B include some number of residues from the 81B-86B range as well as Lys101. The best-scoring pockets on Chain A include a subset of residues from the 36A-40A range. These results are relatively encouraging, as they suggest that secondary structure mimetics may be near-optimal approaches to inhibiting this interface (Figure 8). The analysis also suggests an improvement to the native secondary structure by finding a way to incorporate a pendant cation to mimic the interaction of Raf’s Lys101 with Ras.
Figure 8.
a. The substructure from Raf’s interface with Ras, residues 36A-40A. b. The substructure from Ras’s interface with Raf, residues 81B-86B.
3.4. From substructure to inhibitor design
3.4.1. A stabilized helix mimic as an inhibitor of the Ras/Sos interaction
Analysis of the SOS αH helix supports the experimental observation that four residues (F929, T235, E942, and N944) are essential for binding, with residues F929 and N944 making critical contacts with Ras. However, these two critical residues are located on the N- and C-termini of the helix, spanning 16 residues. The length of the helix and the positioning of these residues suggest that a stabilized α-helix rather than small molecule mimics may provide a better starting point for inhibitor design.[99] Based on this consideration, we described a stabilized helix mimic of the Sos αH sequence 929FFGIYLTNILKTEEGN944, with hot spot residues shown in bold) to inhibit association of Ras with Sos in cell free and cellular contexts (Figure 9) [100]. The Sos mimic was constructed using the hydrogen bond surrogate (HBS) constraint. The HBS constraint reduces conformational entropy by covalently locking the characteristic i to i+4 hydrogen bond and nucleates a stable α-helical conformation in the attached peptide [101]. The Sos HBS helix derived from the former peptide (Figure 9a) proved to be the first orthosteric inhibitor of the Ras/Sos interaction [100]. The Ras-binding site for the mimic was evaluated using 1H-15N HSQC NMR titration experiments with the Sos HBS and uniformly 15N-labeled recombinant Ras. The Sos HBS reduced Ras activation and Ras signaling, as demonstrated by downregulation of ERK phosphorylation, in cells.
Figure 9.
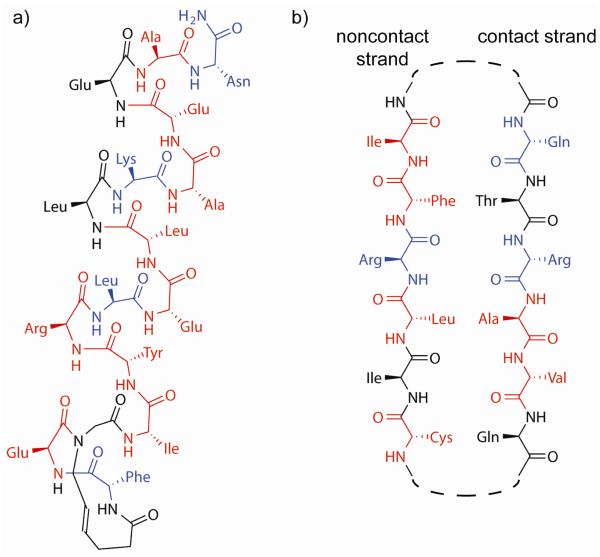
a. The Sos HBS helix developed as the first orthosteric Ras/Sos inhibitor. b. The Raf beta strands are natively organized in an antiparallel fashion, suggesting a hairpin mimetic (whose linker would appear at the dashed line). Residues highlighted in red may be especially well suited to further sequence design and optimization because they do not make optimal contacts, while residues highlighted in blue are designed to mimic hotspot residues (ΔΔG > 1 REU).
3.4.2. Potential β-hairpins as inhibitors of the Ras/Raf interaction
Similar design considerations would predominate in proposing a Ras/Raf inhibitor. The hairpins on either side of the interface are prominent options for mimicry; in particular, several approaches to develop β-hairpin mimics have been described [102-107]. It is important to note the orientation of the two interface hairpins in this complex: they are making edge-on-edge interactions, mediated by hydrogen bond formation, thus forming an inter-chain β-sheet [57]. This geometry indicates that nonpeptidic peptidomimetics that do not preserve hydrogen bond donors and acceptors will be greatly disadvantaged. A possible Raf hairpin mimetic to bind to Ras is depicted in Figure 6b.
Four of the six residues on the non-contact strand strand do not directly interact with the partner protein, permitting mutations to optimize solubility. The contact strand contains two hotspots and two moderate-affinity residues. Likely mutations to evaluate would include Val85 to a charged residue, particularly lysine or arginine because of the nearby chain B residues Asp33 and Asp38, or Ala84 to a larger aliphatic residue, such as methionine or norleucine.
5. Conclusion
Protein-protein interactions have emerged as attractive drug targets but offer a substantial challenge in biomolecular recognition. Analysis of forces that guide protein-protein complex formation can lead to rational design of interaction inhibitors. Such inhibitor discovery approaches are greatly aided by novel computational strategies. Herein we outline a workflow for peptidomimetic inhibitor design to demonstrate the breadth of computational options available for analyzing PPIs. New methods for interaction network analysis permit the precise identification of the most valuable, and most easily targeted, components of a signaling cascade. Increasingly powerful machine learning algorithms and more accurate scoring functions permit the rapid evaluation of structural models and their analysis. Finally, methods for docking, design, and refinement are expanding their scope to include peptidomimetic scaffolds and streamline inhibitor design.
Highlights.
Protein-protein interactions are fertile and challenging targets for inhibition
Protein-protein interaction analysis and classification aids in inhibitor selection
Mimicry of critical protein domains offers an attractive strategy for inhibitor design
Computational tools support every phase of inhibitor design
Table 2.
Sos segments that may serve as plausible starting points for inhibitor design.1
| Residue Range | Interface With | Secondary Structure? | Total ΔΔG |
|---|---|---|---|
| 825-829 | R | Yes | 4.85 |
| 880-884 | R | No | 2.58 |
| 908-912 | R | No | 4.48 |
| 929-944 | R | Yes | 14.1 |
| 1007-1010 | R | Yes | 1.88 |
| 683-695 | Q | Yes | 10.47 |
| 729-739 | Q | Yes | 5.79 |
| 750-755 | Q | No | 3.05 |
| 973-974 | Q | Yes | 3.02 |
Table 3.
Ras possesses one (β-strand sequence and one lone residue that may serve as plausible starting points for inhibitor design, while Raf possesses two low-affinity sequences.
| Residue Range | Interface With | Secondary Structure? | Total ΔΔG |
|---|---|---|---|
| 36A-40A | B | Yes | 5.68 |
| 54A | B | No | 3.68 |
| 72B-74B | A | Yes | 2.25 |
| 81B-86B | A | Yes | 5.02 |
Acknowledgements
We thank the National Institutes of Health (R01GM073943) and the National Science Foundation (CHE-1151554) for financial support. A.M.W. thanks New York University for a Kramer Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Nero TL, Morton CJ, Holien JK, Wielens J, Parker MW. Oncogenic protein interfaces: small molecules, big challenges. Nat. Rev. Cancer. 2014;14:248–262. doi: 10.1038/nrc3690. [DOI] [PubMed] [Google Scholar]
- [2].Zinzalla G, Thurston DE. Targeting protein-protein interactions for therapeutic intervention: a challenge for the future. Future Med. Chem. 2009;1:65–93. doi: 10.4155/fmc.09.12. [DOI] [PubMed] [Google Scholar]
- [3].Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- [4].Morelli X, Bourgeas R, Roche P. Chemical and structural lessons from recent successes in protein-protein interaction inhibition (2P2I) Curr. Opin. Chem. Biol. 2011;15:475–481. doi: 10.1016/j.cbpa.2011.05.024. [DOI] [PubMed] [Google Scholar]
- [5].Milroy L-G, Grossmann TN, Hennig S, Brunsveld L, Ottmann C. Modulators of Protein–Protein Interactions. Chem. Rev. 2014;114:4695–4748. doi: 10.1021/cr400698c. [DOI] [PubMed] [Google Scholar]
- [6].Jayatunga MKP, Thompson S, Hamilton AD. α-Helix mimetics: Outwards and upwards. Biorg. Med. Chem. Lett. 2014;24:717–724. doi: 10.1016/j.bmcl.2013.12.003. [DOI] [PubMed] [Google Scholar]
- [7].Azzarito V, Long K, Murphy NS, Wilson AJ. Inhibition of alpha-helix-mediated protein-protein interactions using designed molecules. Nat. Chem. 2013;5:161–173. doi: 10.1038/nchem.1568. [DOI] [PubMed] [Google Scholar]
- [8].Kushal S, Lao BB, Henchey LK, Dubey R, Mesallati H, Traaseth NJ, Olenyuk BZ, Arora PS. Protein domain mimetics as in vivo modulators of hypoxia-inducible factor signaling. Proc. Natl. Acad. Sci. USA. 2013;110:15602–15607. doi: 10.1073/pnas.1312473110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Murray CW, Rees DC. The rise of fragment-based drug discovery. Nat. Chem. 2009;1:187–192. doi: 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- [10].Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat. Rev. Drug Discov. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- [11].Maurer T, Garrenton LS, Oha A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, Ludlam MJC, Bowman KK, Wu JS, Giannetti AM, Starovasnik MA, Mellman I, Jackson PK, Rudolph J, Wang WR, Fang G. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. USA. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Feldman T, Kabaleeswaran V, Jang SB, Antczak C, Djaballah H, Wu H, Jiang X. A class of allosteric caspase inhibitors identified by high-throughput screening. Molecular cell. 2012;47:585–595. doi: 10.1016/j.molcel.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mueller R, Dawson ES, Niswender CM, Butkiewicz M, Hopkins CR, Weaver CD, Lindsley CW, Conn PJ, Meiler J. Iterative experimental and virtual high-throughput screening identifies metabotropic glutamate receptor subtype 4 positive allosteric modulators. Journal of molecular modeling. 2012;18:4437–4446. doi: 10.1007/s00894-012-1441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Burns MC, Sun Q, Daniels RN, Camper D, Kennedy JP, Phan J, Olejniczak ET, Lee T, Waterson AG, Rossanese OW, Fesik SW. Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange. Proc. Natl. Acad. Sci. USA. 2014;111:3401–3406. doi: 10.1073/pnas.1315798111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Murray CW, Rees DC. The rise of fragment-based drug discovery. Nat. Chem. 2009;1:187–192. doi: 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- [16].Basse MJ, Betzi S, Bourgeas R, Bouzidi S, Chetrit B, Hamon V, Morelli X, Roche P. 2P2Idb: a structural database dedicated to orthosteric modulation of protein-protein interactions. Nucleic Acids Res. 2013;41:D824–827. doi: 10.1093/nar/gks1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Valencia A, Pazos F. Computational methods for the prediction of protein interactions. Curr Opin Struct Biol. 2002;12:368–373. doi: 10.1016/s0959-440x(02)00333-0. [DOI] [PubMed] [Google Scholar]
- [18].Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, Maniatis T, Califano A, Honig B. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Roche O, Schneider P, Zuegge J, Guba W, Kansy M, Alanine A, Bleicher K, Danel F, Gutknecht EM, Rogers-Evans M, Neidhart W, Stalder H, Dillon M, Sjogren E, Fotouhi N, Gillespie P, Goodnow R, Harris W, Jones P, Taniguchi M, Tsujii S, von der Saal W, Zimmermann G, Schneider G. Development of a virtual screening method for identification of "frequent hitters" in compound libraries. J. Med. Chem. 2002;45:137–142. doi: 10.1021/jm010934d. [DOI] [PubMed] [Google Scholar]
- [20].Schorpp K, Rothenaigner I, Salmina E, Reinshagen J, Low T, Brenke JK, Gopalakrishnan J, Tetko IV, Gul S, Hadian K. Identification of Small-Molecule Frequent Hitters from AlphaScreen High-Throughput Screens. J. Biomol. Screen. 2013;19:715–726. doi: 10.1177/1087057113516861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Potuzak JS, Moilanen SB, Tan DS. Discovery and applications of small molecule probes for studying biological processes. Biotechnology & genetic engineering reviews. 2004;21:11–78. doi: 10.1080/02648725.2004.10648049. [DOI] [PubMed] [Google Scholar]
- [23].Koh B, Crews CM. Chemical genetics: a small molecule approach to neurobiology. Neuron. 2002;36:563–566. doi: 10.1016/s0896-6273(02)01059-0. [DOI] [PubMed] [Google Scholar]
- [24].Schreiber SL. Chemical genetics resulting from a passion for synthetic organic chemistry. Bioorg. Med. Chem. 1998;6:1127–1152. doi: 10.1016/s0968-0896(98)00126-6. [DOI] [PubMed] [Google Scholar]
- [25].Stockwell BR. Chemical genetics: ligand-based discovery of gene function. Nat. Rev. Genet. 2000;1:116–125. doi: 10.1038/35038557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Stockwell BR. Exploring biology with small organic molecules. Nature. 2004;432:846–854. doi: 10.1038/nature03196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].O' Connor CJ, Laraia L, Spring DR. Chemical genetics. Chem. Soc. Rev. 2011;40:4332–4345. doi: 10.1039/c1cs15053g. [DOI] [PubMed] [Google Scholar]
- [28].Potapov AP, Goemann B, Wingender E. The pairwise disconnectivity index as a new metric for the topological analysis of regulatory networks. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Huang TW, Tien AC, Huang WS, Lee YC, Peng CL, Tseng HH, Kao CY, Huang CY. POINT: a database for the prediction of protein-protein interactions based on the orthologous interactome. Bioinformatics. 2004;20:3273–3276. doi: 10.1093/bioinformatics/bth366. [DOI] [PubMed] [Google Scholar]
- [30].Johnson ME, Hummer G. Interface-Resolved Network of Protein-Protein Interactions. PLoS Comp. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucl. Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bordoli L, Schwede T. Automated protein structure modeling with SWISS-MODEL Workspace and the Protein Model Portal. Methods Mol. Biol. 2012;857:107–136. doi: 10.1007/978-1-61779-588-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Peitsch MC. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- [34].Schwede T, Sali A, Honig B, Levitt M, Berman HM, Jones D, Brenner SE, Burley SK, Das R, Dokholyan NV, Dunbrack RL, Jr., Fidelis K, Fiser A, Godzik A, Huang YJ, Humblet C, Jacobson MP, Joachimiak A, Krystek SR, Jr., Kortemme T, Kryshtafovych A, Montelione GT, Moult J, Murray D, Sanchez R, Sosnick TR, Standley DM, Stouch T, Vajda S, Vasquez M, Westbrook JD, Wilson IA. Outcome of a workshop on applications of protein models in biomedical research. Structure. 2009;17:151–159. doi: 10.1016/j.str.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Benkert P, Kunzli M, Schwede T. QMEAN server for protein model quality estimation. Nucl. Acids Res. 2009;37:W510–514. doi: 10.1093/nar/gkp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Novoa EM, de Pouplana LR, Barril X, Orozco M. Ensemble Docking from Homology Models. Journal of Chemical Theory and Computation. 2010;6:2547–2557. doi: 10.1021/ct100246y. [DOI] [PubMed] [Google Scholar]
- [37].Oh DY, Gray JB. GA-Ensemble: a genetic algorithm for robust ensembles. Comp. Stat. 2013;28:2333–2347. [Google Scholar]
- [38].Chaudhury S, Gray JJ. Conformer selection and induced fit in flexible backbone protein-protein docking using computational and NMR ensembles. J. Mol. Biol. 2008;381:1068–1087. doi: 10.1016/j.jmb.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wu FH, Towfic F, Dobbs D, Honavar V. Analysis of protein protein dimeric interfaces. 2007 Ieee International Conference on Bioinformatics and Biomedicine, Proceedings.2007. pp. 35–41. [Google Scholar]
- [40].Janin J, Bahadur RP, Chakrabarti P. Protein-protein interaction and quaternary structure. Quarterly reviews of biophysics. 2008;41:133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- [41].Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. Journal of molecular biology. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- [42].Clackson T, Wells JA. A Hot-Spot of Binding-Energy in a Hormone-Receptor Interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- [43].Thorn KS, Bogan AA. ASEdb: a database of alanine mutations and their effects on the free energy of binding in protein interactions. Bioinformatics. 2001;17:284–285. doi: 10.1093/bioinformatics/17.3.284. [DOI] [PubMed] [Google Scholar]
- [44].Hendlich M, Bergner A, Gunther J, Klebe G. Relibase: design and development of a database for comprehensive analysis of protein-ligand interactions. J Mol Biol. 2003;326:607–620. doi: 10.1016/s0022-2836(02)01408-0. [DOI] [PubMed] [Google Scholar]
- [45].Higueruelo AP, Jubb H, Blundell TL. TIMBAL v2: update of a database holding small molecules modulating protein-protein interactions. Database (Oxford) 2013;2013 doi: 10.1093/database/bat039. bat039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhang QC, Petrey D, Garzon JI, Deng L, Honig B. PrePPI: a structure-informed database of protein-protein interactions. Nucleic acids research. 2013;41:D828–D833. doi: 10.1093/nar/gks1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Fong JH, Shoemaker BA, Garbuzynskiy SO, Lobanov MY, Galzitskaya OV, Panchenko AR. Intrinsic disorder in protein interactions: insights from a comprehensive structural analysis. PLoS computational biology. 2009;5:e1000316. doi: 10.1371/journal.pcbi.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cukuroglu E, Gursoy A, Nussinov R, Keskin O. Non-Redundant Unique Interface Structures as Templates for Modeling Protein Interactions. PloS one. 2014;9 doi: 10.1371/journal.pone.0086738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sugaya N, Kanai S, Furuya T. Dr. PIAS 2.0: an update of a database of predicted druggable protein-protein interactions. Database (Oxford) 2012;2012 doi: 10.1093/database/bas034. bas034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kumar MD, Gromiha MM. PINT: Protein-protein Interactions Thermodynamic Database. Nucleic Acids Res. 2006;34:D195–198. doi: 10.1093/nar/gkj017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Moal IH, Fernandez-Recio J. SKEMPI: a Structural Kinetic and Energetic database of Mutant Protein Interactions and its use in empirical models. Bioinformatics. 2012;28:2600–2607. doi: 10.1093/bioinformatics/bts489. [DOI] [PubMed] [Google Scholar]
- [52].Gong W, Zhou D, Ren Y, Wang Y, Zuo Z, Shen Y, Xiao F, Zhu Q, Hong A, Zhou X, Gao X, Li T. PepCyber:P~PEP: a database of human protein protein interactions mediated by phosphoprotein-binding domains. Nucleic Acids Res. 2008;36:D679–683. doi: 10.1093/nar/gkm854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lo YS, Chen YC, Yang JM. 3D-interologs: an evolution database of physical protein-protein interactions across multiple genomes. BMC genomics. 2010;11(Suppl 3):S7. doi: 10.1186/1471-2164-11-S3-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh LS. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Koes DR, Camacho CJ. PocketQuery: protein-protein interaction inhibitor starting points from protein-protein interaction structure. Nucleic Acids Res. 2012;40:W387–392. doi: 10.1093/nar/gks336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bergey CM, Watkins AM, Arora PS. HippDB: a database of readily targeted helical protein-protein interactions. Bioinformatics. 2013;29:2806–2807. doi: 10.1093/bioinformatics/btt483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Watkins AM, Arora PS. Anatomy of beta-Strands at Protein-Protein Interfaces. ACS Chem. Biol. 2014 doi: 10.1021/cb500241y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Science's STKE : signal transduction knowledge environment. 2004;2004:pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- [59].Fogolari F, Brigo A, Molinari H. Protocol for MM/PBSA molecular dynamics simulations of proteins. Biophys J. 2003;85:159–166. doi: 10.1016/S0006-3495(03)74462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lise S, Buchan D, Pontil M, Jones DT. Predictions of Hot Spot Residues at Protein-Protein Interfaces Using Support Vector Machines. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Walker R, Bond JP, Tarone RE, Harris CC, Makalowski W, Boguski MS, Greenblatt MS. Evolutionary conservation and somatic mutation hotspot maps of p53: correlation with p53 protein structural and functional features. Oncogene. 1999;18:211–218. doi: 10.1038/sj.onc.1202298. [DOI] [PubMed] [Google Scholar]
- [62].Guharoy M, Chakrabarti P. Conservation and relative importance of residues across protein-protein interfaces. Proc. Natl. Acad. Sci. USA. 2005;102:15447–15452. doi: 10.1073/pnas.0505425102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Higa RH, Tozzi CL. Prediction of binding hot spot residues by using structural and evolutionary parameters. Genetics and Molecular Biology. 2009;32:626–633. doi: 10.1590/S1415-47572009000300029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Fernandez-Recio J, Totrov M, Abagyan R. Identification of protein-protein interaction sites from docking energy landscapes. J. Mol. Biol. 2004;335:843–865. doi: 10.1016/j.jmb.2003.10.069. [DOI] [PubMed] [Google Scholar]
- [65].Nguyen QT, Fablet R, Pastor D. Protein interaction hotspot identification using sequence-based frequency-derived features. IEEE Trans Biomed Eng. 2013;60:2993–3002. doi: 10.1109/TBME.2011.2161306. [DOI] [PubMed] [Google Scholar]
- [66].Chothia C. Hydrophobic Bonding and Accessible Surface-Area in Proteins. Nature. 1974;248:338–339. doi: 10.1038/248338a0. [DOI] [PubMed] [Google Scholar]
- [67].Finkelstein AV, Janin J. The Price of Lost Freedom - Entropy of Bimolecular Complex-Formation. Protein Eng. 1989;3:1–3. doi: 10.1093/protein/3.1.1. [DOI] [PubMed] [Google Scholar]
- [68].Prajapati RS, Sirajuddin M, Durani V, Sreeramulu S, Varadarajan R. Contribution of cation-pi interactions to protein stability. Biochemistry-Us. 2006;45:15000–15010. doi: 10.1021/bi061275f. [DOI] [PubMed] [Google Scholar]
- [69].Fersht AR, Shi JP, Knilljones J, Lowe DM, Wilkinson AJ, Blow DM, Brick P, Carter P, Waye MMY, Winter G. Hydrogen-Bonding and Biological Specificity Analyzed by Protein Engineering. Nature. 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- [70].Kozakov D, Hall DR, Chuang GY, Cencic R, Brenke R, Grove LE, Beglov D, Pelletier J, Whitty A, Vajda S. Structural conservation of druggable hot spots in protein-protein interfaces. Proc Natl Acad Sci U S A. 2011;108:13528–13533. doi: 10.1073/pnas.1101835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zerbe BS, Hall DR, Vajda S, Whitty A, Kozakov D. Relationship between Hot Spot Residues and Ligand Binding Hot Spots in Protein–Protein Interfaces. J. Chem. Inf. Model. 2012;52:2236–2244. doi: 10.1021/ci300175u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Jochim AL, Arora PS. Systematic analysis of helical protein interfaces reveals targets for synthetic inhibitors. ACS Chem. Biol. 2010;5:919–923. doi: 10.1021/cb1001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Janin J, Chothia C. The structure of protein-protein recognition sites. The Journal of biological chemistry. 1990;265:16027–16030. [PubMed] [Google Scholar]
- [74].Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- [75].Milletti F, Vulpetti A. Predicting polypharmacology by binding site similarity: from kinases to the protein universe. J Chem Inf Model. 2010;50:1418–1431. doi: 10.1021/ci1001263. [DOI] [PubMed] [Google Scholar]
- [76].Che Y, Brooks BR, Marshall GR. Protein recognition motifs: design of peptidomimetics of helix surfaces. Biopolymers. 2007;86:288–297. doi: 10.1002/bip.20744. [DOI] [PubMed] [Google Scholar]
- [77].Chongsiriwatana NP, Patch JA, Czyzewski AM, Dohm MT, Ivankin A, Gidalevitz D, Zuckermann RN, Barron AE. Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2794–2799. doi: 10.1073/pnas.0708254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hruby VJ, Balse PM. Conformational and topographical considerations in designing agonist peptidomimetics from peptide leads. Current medicinal chemistry. 2000;7:945–970. doi: 10.2174/0929867003374499. [DOI] [PubMed] [Google Scholar]
- [79].Schrödinger Release 2014-2: MacroModel, version 10.4, in, Schrödinger, LLC. 2014. New York, NY.
- [80].Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods in enzymology. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- [82].Drew K, Renfrew PD, Craven TW, Butterfoss GL, Chou FC, Lyskov S, Bullock BN, Watkins A, Labonte JW, Pacella M, Kilambi KP, Leaver-Fay A, Kuhlman B, Gray JJ, Bradley P, Kirshenbaum K, Arora PS, Das R, Bonneau R. Adding diverse noncanonical backbones to rosetta: enabling peptidomimetic design. PLoS One. 2013;8:e67051. doi: 10.1371/journal.pone.0067051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Vanquelef E, Simon S, Marquant G, Garcia E, Klimerak G, Delepine JC, Cieplak P, Dupradeau FY. R.E.D. Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011;39:W511–517. doi: 10.1093/nar/gkr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lama D, Quah ST, Verma CS, Lakshminarayanan R, Beuerman RW, Lane DP, Brown CJ. Rational Optimization of Conformational Effects Induced By Hydrocarbon Staples in Peptides and their Binding Interfaces. Scientific Reports. 2013;3 doi: 10.1038/srep03451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Renfrew PD, Choi EJ, Bonneau R, Kuhlman B. Incorporation of noncanonical amino acids into Rosetta and use in computational protein-peptide interface design. PLoS One. 2012;7:e32637. doi: 10.1371/journal.pone.0032637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Khoury GA, Smadbeck J, Tamamis P, Vandris AC, Kieslich CA, Floudas CA. Forcefield_NCAA: Ab Initio Charge Parameters to Aid in the Discovery and Design of Therapeutic Proteins and Peptides with Unnatural Amino Acids and Their Application to Complement Inhibitors of the Compstatin Family. ACS Synthetic Biology. 2014 doi: 10.1021/sb400168u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- [88].Gysin S, Salt M, Young A, McCormick F. Therapeutic strategies for targeting ras proteins. Genes Cancer. 2011;2:359–372. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Marshall CJ. Specificity of Receptor Tyrosine Kinase Signaling - Transient Versus Sustained Extracellular Signal-Regulated Kinase Activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- [91].Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Spiegel J, Cromm PM, Zimmermann G, Grossmann TN, Waldmann H. Small-molecule modulation of Ras signaling. Nat. Chem. Biol. 2014 doi: 10.1038/nchembio.1560. [DOI] [PubMed] [Google Scholar]
- [93].Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, Lee T, Rossanese OW, Fesik SW. Discovery of Small Molecules that Bind to K-Ras and Inhibit Sos-Mediated Activation. Angewandte Chemie-International Edition. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Herrmann C, Block C, Geisen C, Haas K, Weber C, Winde G, Moroy T, Muller O. Sulindac sulfide inhibits Ras signaling. Oncogene. 1998;17:1769–1776. doi: 10.1038/sj.onc.1202085. [DOI] [PubMed] [Google Scholar]
- [96].Kato-Stankiewicz J, Hakimi I, Zhi G, Zhang J, Serebriiskii I, Guo L, Edamatsu H, Koide H, Menon S, Eckl R, Sakamuri S, Lu YC, Chen QZ, Agarwal S, Baumbach WR, Golemis EA, Tamanoi F, Khazak V. Inhibitors of Ras/Raf-1 interaction identified by two-hybrid screening revert Ras-dependent transformation phenotypes in human cancer cells. Proc. Natl. Acad. Sci. USA. 2002;99:14398–14403. doi: 10.1073/pnas.222222699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Margarit SM, Sondermann H, Hall BE, Nagar B, Hoelz A, Pirruccello M, Bar-Sagi D, Kuriyan J. Structural evidence for feedback activation by Ras-GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112:685–695. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- [98].Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic acids research. 2004;32:W526–531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Bullock BN, Jochim AL, Arora PS. Assessing helical protein interfaces for inhibitor design. J. Am. Chem. Soc. 2011;133:14220–14223. doi: 10.1021/ja206074j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Patgiri A, Yadav KK, Arora PS, Bar-Sagi D. An orthosteric inhibitor of the Ras-Sos interaction. Nat. Chem. Biol. 2011;7:585–587. doi: 10.1038/nchembio.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Chapman RN, Dimartino G, Arora PS. A highly stable short alpha-helix constrained by a main-chain hydrogen-bond surrogate. J. Am. Chem. Soc. 2004;126:12252–12253. doi: 10.1021/ja0466659. [DOI] [PubMed] [Google Scholar]
- [102].Khakshoor O, Demeler B, Nowick JS. Macrocyclic beta-sheet peptides that mimic protein quaternary structure through intermolecular beta-sheet interactions. J. Am. Chem. Soc. 2007;129:5558–5569. doi: 10.1021/ja068511u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Shankaramma SC, Athanassiou Z, Zerbe O, Moehle K, Mouton C, Bernardini F, Vrijbloed JW, Obrecht D, Robinson JA. Macrocyclic hairpin mimetics of the cationic antimicrobial peptide protegrin I: a new family of broad-spectrum antibiotics. ChemBiochem. 2002;3:1126–1133. doi: 10.1002/1439-7633(20021104)3:11<1126::AID-CBIC1126>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- [104].Hoffmann RW, Schopfer U, Muller C, Brandl T. Synthesis of a conformationally flexible beta-hairpin mimetic. Helv Chim Acta. 2002;85:4424–4441. [Google Scholar]
- [105].Gardner RR, Liang GB, Gellman SH. An Achiral Dipeptide Mimetic That Promotes Beta-Hairpin Formation. Journal of the American Chemical Society. 1995;117:3280–3281. [Google Scholar]
- [106].Erdelyi M, Karlen A, Gogoll A. A new tool in peptide engineering: A photoswitchable stilbene-type beta-hairpin mimetic. Chem-Eur J. 2006;12:403–412. doi: 10.1002/chem.200500648. [DOI] [PubMed] [Google Scholar]
- [107].Lingard H, Han JT, Thompson AL, Leung IKH, Scott RTW, Thompson S, Hamilton AD. Diphenylacetylene-Linked Peptide Strands Induce Bidirectional β-Sheet Formation. Angew. Chem. Int. Ed. 2014;53:3650–3653. doi: 10.1002/anie.201309353. [DOI] [PubMed] [Google Scholar]