Abstract
A total of 568 normal feces from calves on a beef farm in Fukui Prefecture, Japan, in 2011–2012 were examined by RT-semi-nested PCR for rotavirus A (RVA) VP4 genes. Through partial sequencing and BLAST analyses of 84 VP4-positive specimens, we identified an avian-like RVA strain, N2342, which shares highest nucleotide identity (80.0%) with known avian-like bovine strain 993/83, in one specimen. Phylogenetic analysis also revealed a close genetic relationship between N2342 and avian RVAs, suggesting bird-to-cattle transmission. We observed frequent contact of wild birds with calves in the farm, suggesting that these birds were the source of the virus.
Keywords: avian-like, cattle, rotavirus A, VP4
Rotavirus A (RVA) is known as the major agent of severe acute gastroenteritis in infants and young humans and in animals worldwide [6]. RVA is nonenveloped and possesses a triple-layered protein capsid composed of an outer layer, an intermediate layer and an icosahedral inner core layer. The genome consists of 11 segments of double-stranded RNA. The two outer capsid proteins, VP7 and VP4, independently elicit neutralizing antibodies, which are used to classify the rotavirus strains into G (for glycoprotein) and P (for protease-sensitive) genotypes, respectively. So far, 27 G genotypes and 37 P genotypes have been identified in humans and animals [11, 12, 14, 20].
Although RVAs are generally believed to have host species specificity [6], there have been a number of reports that RVAs can be transmitted from the original mammalian hosts to heterologous species by direct transmission of the virus or by the contribution of one or several genes to reassortants [5, 7, 9,10,11, 16]. Notably, several mammalian animals including cattle, sheep and pigs are regarded as potential reservoirs for genetic diversity of human RVAs [5, 6, 10, 13]. Therefore, it is important to survey the genetic diversity of RVAs in animals not only for animal hygiene but also for public health.
In addition to mammalian RVAs, avian RVAs have the potential to be a threat to both animal hygiene and public health. It was previously reported that RVA strain 993/83, which had been isolated from a calf with diarrhea [3], has close antigenic and genetic relationships with avian RVAs [4, 17,18,19]: sequence analyses revealed that VP4, VP6 and VP7 genes of the 993/83 strain share highest homologies with the respective genes of avian RVA strain PO-13 at both nucleotide and amino acid levels. These findings highlight the risk of infection of mammalian species including humans with avian RVAs. However, very little is known about the interspecies transmission of avian RVAs. Here, we describe the detection of an avian-like RVA strain in feces from a healthy calf, strongly suggesting interspecies transmission from birds to cattle.
A total of 568 normal fecal specimens were collected from healthy calves under 12 months of age on a beef cattle farm in Fukui Prefecture of Japan from April 2011 to March 2012 for monitoring the genetic diversity of RVAs, which were prevalent among cattle. The stool samples were taken directly from calves before falling to the ground. The fecal samples were diluted with Eagle’s minimum essential medium to 20% suspensions and clarified by centrifugation at 1,500 × g for 10 min. The viral RNA was extracted from the supernatant by using a QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany). Synthesis of the cDNA was performed using a PrimeScript II 1st strand cDNA Synthesis Kit with random primers (TaKaRa BIO, Otsu, Japan). The cDNAs were amplified by semi-nested PCR using TaKaRa Ex Taq HS (TaKaRa BIO). For the detection of RVA, the VP4 gene was amplified by an outer PCR with the primers VP4-HeadF and VP4-1094R2 and by an inner PCR with the primers VP4-HeadF and VP4-887R as previously reported [2]. The amplicons of the second PCR were purified with Nucleo Spin ExtractII (MACHEREY-NAGEL, Duren, Gremany) and sequenced with a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, U.S.A) on an ABI PRISM 3100 DNA analyzer (Applied Biosystems). The second PCR primers were also used as sequencing primers. Initially, the partial nucleotide sequences of VP4 genes determined were analyzed using BLAST analyses. In the case of detection of an atypical bovine RVA VP4 sequence, the partial nucleotide sequences of the VP4 gene were compared with those of reference RVA strains available in GenBank. Phylogenetic analyses for partial VP4 genes were conducted using MEGA version 4.0. Genetic distances were calculated using the Kimura 2 correction parameter at the nucleotide level, and phylogenetic trees were constructed using the neighbor-joining method with 1,000 bootstrap replicates.
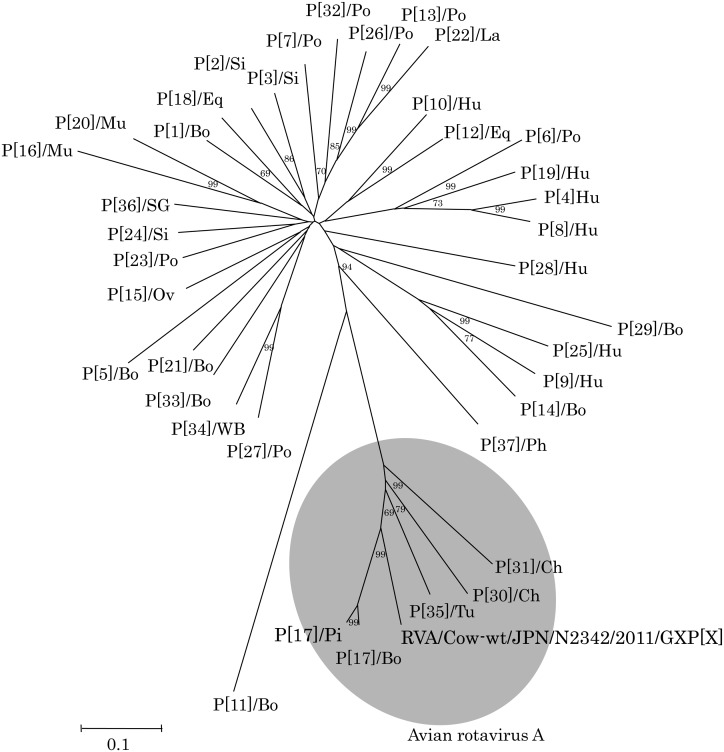
VP4 genes were detected in 84 of the 568 fecal samples from healthy calves. Based on BLAST and phylogenetic analyses of the partial VP4 gene, 83 of the 84 strains detected in calves were genetically closely related to those of known RVAs belonging to P[5], P[11], P[14] and P[29] genotypes (data not shown), which are generally prevalent in bovines [1, 2, 6, 10]. The remaining strain (N2342), which was detected in a 3.4-month-old healthy calf, shared the highest identity (80.0%) with avian-like RVA 993/83 belonging to P[17] in 769 bp corresponding to nucleotides 53–833 of the NCDV VP4 gene. The partial nucleotide sequence of the strain determined in this study was deposited in the GenBank database (accession No, AB846649). The partial sequence of the N2342 VP4 gene showed the second highest identity (79.2%) with the corresponding sequence of avian P[17] RVA PO-13 strain [8]. Comparative analysis of the partial VP4 nucleotide sequences revealed that the partial VP4 gene of N2342 strain shared higher homologies with those of avian RVAs (68.0–79.2%) than those of bovine RVAs (47.3–57.7%), except for avian-like bovine RVA 993/83 strain (Table 1). Phylogenetic analysis based on the partial VP4 gene also revealed that N2342 strain formed within branches of avian RVAs (Fig. 1). These findings clearly indicate that the partial VP4 gene of N2342 strain is genetically closely related to avian RVAs, strongly suggesting interspecies transmission of RVA from birds to cattle.
Table 1. Comparison of nucleotide sequence identities of partial VP4 (769 bp, corresponding to nucleotides 53-833 of the NCDV VP4 gene) of N2342 strain with bovine and avian RVAs.
| Strain | Origin | P genotype | Nucletotide identity with N2342 strain (%) | Accession No. |
|---|---|---|---|---|
| NCDV | Bovine | P[1] | 57.7 | AB119636 |
| UK | Bovine | P[5] | 51.3 | M22306 |
| B223 | Bovine | P[11] | 47.3 | D13394 |
| Sun9 | Bovine | P[14] | 54.6 | AB158430 |
| 993/83 | Bovine | P[17] | 80.0 | D16352 |
| Hg18 | Bovine | P[21] | 54.6 | AF237665 |
| AzuK-1 | Bovine | P[29] | 51.3 | AB454420 |
| Dai-10 | Bovine | P[33] | 56.2 | AB513836 |
| PO-13 | Pigeon | P[17] | 79.2 | AB009632 |
| Ch-2G3 | Chiken | P[30] | 71.3 | EU486956 |
| Ch-661G1 | Chiken | P[31] | 68.0 | EU486962 |
| 03V0002E10 | Turkey | P[35] | 73.5 | JX204825 |
Fig. 1.
Phylogenetic tree based on partial VP4 nucleotide sequences (769 bp, corresponding to nucleotides 53-833 of the NCDV VP4 gene) of 37 established reference P genotype strains and N2342 strains. Bootstrap values above 60% are shown at the branch nodes. Reference rotavirus strains and accession numbers for the VP4 genes used to construct the phylogenetic tree are as follows. P[1]: NCDV (AB119636), P[2]: SA11 (M23188), P[3]: RRV (M18736), P[4]: L26 (M58292), P[5]: UK (M22306), P[6]: Gottfried (M33516), P[7]: OSU (X13190), P[8]: Wa (L34161), P[9]: AU-1 (D10970), P[10]: 69M (M60600), P[11]: B223 (D13394), P[12]: H-2 (L04638), P[13]: MDR-13 (L07886), P[14]: Sun9 (AB158430), P[15]: Lp14 (L11599), P[16]: EDIM (AF039219), P[17]: PO-13 (AB009632), P[17]: 993/83 (D16352), P[18]: L338 (D13399), P[19]: Mc345 (D38054), P[20]: EHP (U08424), P[21]: Hg18 (AF237665), P[22]: 160/01 (AF526374), P[23]: 34461-4 (AY768809), P[24]: TUCH (AY596189), P[25]: Dhaka6 (AY773004), P[26]: 134/04-15 (DQ061053), P[27]: CMP034 (DQ534016), P[28]: Ecu534 (EU805773), P[29]: AzuK-1 (AB454420), P[30]: Ch-2G3 (EU486956), P[31]: Ch-661G1 (EU486962), P[32]: 61-07-ire (FJ492835), P[33]: Dai-10 (AB513836), P[34]: FGP51 (AB571047), P[35]: 03V0002E10 (JX204825), P[36]: SG385 (AB823215), P[37]: 10V0112H5 (JX204814). Bo, bovine; Ch, chicken; Eq, equine; Hu, human; La, lapine; Mu, murine; Ov, ovine; Ph, pheasant; Pi, pigeon; Po, porcine; SG, sugar glider; Si, simian; Tu, turkey; WB, wild boar.
For a better understanding of bird-to-cattle transmission of RVA, we attempted to analyze the complete open reading frame of VP4 and VP7 segments of N2342 strain. However, the VP4 and VP7 genes were not amplified using several primer sets, which we designed for N2342 strain. Since we also failed to isolate N2342 strain from the feces sample even after several trials with different methods, we were not able to obtain further genetic information on the strain.
The N2342 and 993/83 strains, which have been both detected/isolated from cattle under natural conditions, are genetically related or classified to P[17] RVAs (Table 1). In addition, we previously reported that avian P[17] RVA PO-13 strain can experimentally infect and induce disease in a mouse model [15]. These facts lead us to speculate that avian P[17](-like) RVAs might have a special property to infect mammalian species including cattle, suggesting the necessity for monitoring of P[17] RVAs in nature to evaluate their risk to animal and human health.
The fact that 993/83 strain was isolated from a calf with diarrhea [3] raises an important question about whether the genetically related N2342 strain has the potential to cause diarrhea in cattle. Experimental infection of cattle with the isolated N2342 strain would be required to answer this question. However, we could not implement such an experiment due to failure to isolate the virus. Although the N2342 strain was detected in a healthy calf, this fact is not sufficient to exclude the possibility that this strain is pathogenic in cattle, because the clinical outcome of RVA infection is generally believed to depend on a number of factors, such as infection dose, viral capacity to cause disease and host factors including the age and nutritional status [6].
Although our results strongly suggest that the N2342 strain originated from birds, the natural origin of this strain remains unknown. There was no poultry farm near the beef cattle farm from which fecal specimens were obtained for this study, suggesting that N2342 strain did not originate from poultry. We frequently observed wild birds including crows in the beef cattle farm. We also found crow’s feces not only on the bodies of calves but also in the bedding and feeder in the cowshed. These situations imply that the N2342 strain originated from wild birds, such as crows, suggesting the potential risk of infection of mammalian species with RVAs circulating among wild birds. However, information on avian RVAs circulating among wild birds is very limited. Active epidemiological surveillance in both wild birds and mammalian species will be crucial to evaluate the risk of interspecies transmission of RVAs from birds to mammals under natural conditions.
In this study, we identified an avian-like RVA, N2342 strain, in a calf through epidemiological surveillance of RVA in healthy calves. Genetic analyses showed close relationships between N2342 and avian RVA strains in the partial VP4 gene, suggesting interspecies transmission of RVA from birds to cattle. To our knowledge, there has been no report on detection/isolation of an avian-like RVA from cattle since the isolation of 993/83 strain in 1983, indicating that this is the second report.
Acknowledgments
This study was supported in part by Grant-in-Aids for Scientific Research (B) from Japan Society for the Promotion of Science (no. 26292148).
REFERENCES
- 1.Abe M., Ito N., Masatani T., Nakagawa K., Yamaoka S., Kanamaru Y., Suzuki H., Shibano K., Arashi Y., Sugiyama M.2011. Whole genome characterization of new bovine rotavirus G21P[29] and G24P[33] strains provides evidence for interspecies transmission. J. Gen. Virol. 92: 952–960. doi: 10.1099/vir.0.028175-0 [DOI] [PubMed] [Google Scholar]
- 2.Abe M., Ito N., Morikawa S., Takasu M., Murase T., Kawashima T., Kawai Y., Kohara J., Sugiyama M.2009. Molecular epidemiology of rotaviruses among healthy calves in Japan: isolation of a novel bovine rotavirus bearing new P and G genotypes. Virus Res. 144: 250–257. doi: 10.1016/j.virusres.2009.05.005 [DOI] [PubMed] [Google Scholar]
- 3.Brüssow H., Nakagomi O., Gerna G., Eichhorn W.1992. Isolation of an avianlike group A rotavirus from a calf with diarrhea. J. Clin. Microbiol. 30: 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brüssow H., Nakagomi O., Minamoto N., Eichhorn W.1992. Rotavirus 993/83, isolated from calf faeces, closely resembles an avian rotavirus. J. Gen. Virol. 73: 1873–1875. doi: 10.1099/0022-1317-73-7-1873 [DOI] [PubMed] [Google Scholar]
- 5.De Grazia S., Martella V., Rotolo V., Bonura F., Matthijnssens J., Bányai K., Ciarlet M., Giammanco G. M.2011. Molecular characterization of genotype G6 human rotavirus strains detected in Italy from 1986 to 2009. Infect. Genet. Evol. 11: 1449–1455. doi: 10.1016/j.meegid.2011.05.015 [DOI] [PubMed] [Google Scholar]
- 6.Estes M. K., Greenberg H. B.2013. Rotaviruses. pp. 1347–1401. In: Fields Virology, 6th ed. (Knipe, D. M. and Howley, P. M. eds.), Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- 7.Ghosh S., Varghese V., Samajdar S., Sinha M., Naik T. N., Kobayashi N.2007. Evidence for bovine origin of VP4 and VP7 genes of human group A rotavirus G6P[14] and G10P[14] strains. J. Clin. Microbiol. 45: 2751–2753. doi: 10.1128/JCM.00230-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito H., Sugiyama M., Masubuchi K., Mori Y., Minamoto N.2001. Complete nucleotide sequence of a group A avian rotavirus genome and a comparison with its counterparts of mammalian rotaviruses. Virus Res. 75: 123–138. doi: 10.1016/S0168-1702(01)00234-9 [DOI] [PubMed] [Google Scholar]
- 9.Martella V., Bányai K., Ciarlet M., Iturriza-Gómara M., Lorusso E., De Grazia S., Arista S., Decaro N., Elia G., Cavalli A., Corrente M., Lavazza A., Baselga R., Buonavoglia C.2006. Relationships among porcine and human P[6] rotaviruses: evidence that the different human P[6] lineages have originated from multiple interspecies transmission events. Virology 344: 509–519. doi: 10.1016/j.virol.2005.08.029 [DOI] [PubMed] [Google Scholar]
- 10.Martella V., Bányai K., Matthijnssens J., Buonavoglia C., Ciarlet M.2010. Zoonotic aspects of rotaviruses. Vet. Microbiol. 140: 246–255. doi: 10.1016/j.vetmic.2009.08.028 [DOI] [PubMed] [Google Scholar]
- 11.Matthijnssens J., Ciarlet M., Heiman E., Arijs I., Delbeke T., McDonald S. M., Palombo E. A., Iturriza-Gómara M., Maes P., Patton J. T., Rahman M., Van Ranst M.2008. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 82: 3204–3219. doi: 10.1128/JVI.02257-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthijnssens J., Ciarlet M., McDonald S. M., Attoui H., Bányai K., Brister J. R., Buesa J., Esona M. D., Estes M. K., Gentsch J. R., Iturriza-Gómara M., Johne R., Kirkwood C. D., Martella V., Mertens P. P., Nakagomi O., Parreño V., Rahman M., Ruggeri F. M., Saif L. J., Santos N., Steyer A., Taniguchi K., Patton J. T., Desselberger U., Van Ranst M.2011. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch. Virol. 156: 1397–1413. doi: 10.1007/s00705-011-1006-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthijnssens J., Potgieter C. A., Ciarlet M., Parreño V., Martella V., Bányai K., Garaicoechea L., Palombo E. A., Novo L., Zeller M., Arista S., Gerna G., Rahman M., Van Ranst M.2009. Are human P[14] rotavirus strains the result of interspecies transmissions from sheep or other ungulates that belong to the mammalian order Artiodactyla? J. Virol. 83: 2917–2929. doi: 10.1128/JVI.02246-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyazaki A., Kuga K., Suzuki T., Kohmoto M., Katsuda K., Tsunemitsu H.2011. Genetic diversity of group A rotaviruses associated with repeated outbreaks of diarrhea in a farrow-to-finish farm: identification of a porcine rotavirus strain bearing a novel VP7 genotype, G26. Vet. Res. 42: 112. doi: 10.1186/1297-9716-42-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori Y., Sugiyama M., Takayama M., Atoji Y., Masegi T., Minamoto N.2001. Avian-to-mammal transmission of an avian rotavirus: analysis of its pathogenicity in a heterologous mouse model. Virology 288: 63–70. doi: 10.1006/viro.2001.1051 [DOI] [PubMed] [Google Scholar]
- 16.Mukherjee A., Ghosh S., Bagchi P., Dutta D., Chattopadhyay S., Kobayashi N., Chawla-Sarkar M.2011. Full genomic analyses of human rotavirus G4P[4], G4P[6], G9P[19] and G10P[6] strains from North-eastern India: evidence for interspecies transmission and complex reassortment events. Clin. Microbiol. Infect. 17: 1343–1346. [DOI] [PubMed] [Google Scholar]
- 17.Rohwedder A., Hotop H., Minamoto N., Ito H., Nakagomi O., Brüssow H.1997. Bovine rotavirus 993/83 shows a third subtype of avian VP7 protein. Virus Genes 14: 147–151. doi: 10.1023/A:1007921418679 [DOI] [PubMed] [Google Scholar]
- 18.Rohwedder A., Irmak H., Werchau H., Brüssow H.1993. Nucleotide sequence of gene 6 of avian-like group A rotavirus 993/83. Virology 195: 820–825. doi: 10.1006/viro.1993.1437 [DOI] [PubMed] [Google Scholar]
- 19.Rohwedder A., Schütz K. I., Minamoto N., Brüssow H.1995. Sequence analysis of pigeon, turkey, and chicken rotavirus VP8* identifies rotavirus 993/83, isolated from calf feces, as a pigeon rotavirus. Virology 210: 231–235. doi: 10.1006/viro.1995.1338 [DOI] [PubMed] [Google Scholar]
- 20.Trojnar E., Sachsenröder J., Twardziok S., Reetz J., Otto P. H., Johne R.2013. Identification of an avian group A rotavirus containing a novel VP4 gene with a close relationship to those of mammalian rotaviruses. J. Gen. Virol. 94: 136–142. doi: 10.1099/vir.0.047381-0 [DOI] [PubMed] [Google Scholar]