SUMMARY
BS69 (aka ZMYND11) contains tandemly arranged PHD, BROMO and PWWP domains, which are chromatin recognition modalities. Here we show that BS69 selectively recognizes histone variant H3.3 lysine 36 trimethylation (H3.3K36me3) via its chromatin-binding domains. We further identify BS69 association with RNA splicing regulators including the U5 snRNP components of the spliceosome such as EFTUD2. Remarkably, RNA-seq shows that BS69 mainly regulates intron retention (IR), which is the least well-understood RNA alternative splicing event in mammalian cells. Biochemical and genetic experiments demonstrate that BS69 promotes IR by antagonizing EFTUD2 through physical interactions. We further show that regulation of IR by BS69 also depends on its binding to H3K36me3-decorated chromatin. Taken together, our study identifies an H3.3K36me3-specific reader and a regulator of IR, and reveals a novel and unexpected role of BS69 in connecting histone H3.3K36me3 to regulated RNA splicing, providing significant new insights into chromatin regulation of pre-mRNA processing.
INTRODUCTION
Chromatin structure is organized by nucleosomes, each of which consists of an octamer composed of four core histones (H2A, H2B, H3 and H4) and ~147 bp DNA, which wraps around the octamer. The N-terminal tails of histones are extensively modified post-translationally, including phosphorylation, acetylation and methylation, which contribute to the modulation of gene expression. For instance, histone H3K4 trimethylation marks promoters that are either active or poised for transcription while H3K36 trimethylation is often associated with gene bodies and has been shown to suppress spurious gene transcription and promote transcriptional elongation (Carrozza et al., 2005; Li et al., 2002). More recently, H3K36 trimethylation has also been suggested to regulate RNA splicing (Luco et al., 2010; Pradeepa et al., 2012), but by and large regulation of RNA splicing by chromatin modifications remains poorly understood. RNA splicing is involved in the expression of most human genes, playing key roles in differentiation, cell cycle progression, and development. Mis-regulation of RNA splicing is frequently associated with various human diseases. Emerging evidence suggests that splicing is often co-transcriptional, thus splicing regulation is believed to be dependent not only on the core splicing factors binding to their pre-mRNA target sites but also on transcription-associated features such as local chromatin structure (Bentley, 2014). Out of the five known alternative splicing events (exon skipping, mutually exclusive exons, alternative donor site, alternative acceptor site and intron retention), intron retention (IR) is the least well understood. Gene transcripts whose introns are retained are subject to RNA nonsense-mediated decay (NMD) therefore increased IR is often correlated with reduced level of expression. Recent evidence suggests that IR may be deployed as a potential mechanism for coordinated regulation of gene expression during differentiation and tumorigenesis (Wong et al., 2013; Yap et al., 2012). However the molecular mechanism by which IR is regulated is essentially unknown.
It’s generally accepted that once histone modification patterns are laid down, they are recognized by various so called “reader’ proteins, which help to execute the transcriptional and possibly RNA splicing programs. Perhaps not surprisingly, cells have developed a complex network of proteins that carry distinct protein motifs dedicated to recognizing histone modifications, including acetylation and methylation on canonical histones such as histone H3.1 and H3.2 (Ruthenburg et al., 2007; Taverna et al., 2007). The variant histone H3.3 differs from H3.1 by only five amino acids, but unlike H3.1 and H3.2, H3.3 is incorporated into chromatin in a DNA replication independent manner (Tagami et al., 2004), and is localized to active genes as well as telomeric regions (Goldberg et al., 2010; Jin and Felsenfeld, 2006; Li et al., 2002; Mito et al., 2005; Wong et al., 2009). Recently, H3.3 has gained significant attention owing to the identification of recurrent H3.3 point mutations in pediatric brain tumors (Behjati et al., 2013; Lewis et al., 2013; Schwartzentruber et al., 2012; Wu et al., 2012). But it remains unclear whether there are specific readers for H3.3 and if so what roles these readers may play.
Here we report the identification of BS69/ZMYND11, originally identified as an interacting protein of adenoviral E1A proteins (Hateboer et al., 1995), an H3.3-specific reader and a regulator of IR. BS69 possesses multiple histone reader modalities, including a plant homeodomain (PHD), a bromodomian and a PWWP domain, suggesting a potential role in recognizing histone modification(s). We demonstrate that BS69 functions as an H3.3-specific reader; preferentially recognizing H3.3K36me3 over H3.1K36me3 in vitro through its “reader” modules, in particular the PWWP domain. We show that BS69 localizes mainly to gene bodies in a manner that is dependent on the H3K36 trimethyltransferase, SETD2 (Edmunds et al., 2008), consistent with the in vitro binding data. Importantly, our biochemical and proteomics analyses revealed an association of BS69 with RNA spliceosome machinery, including the U5 snRNP such as EFTUD2, which is critical for spliceosome activation (Bartels et al., 2002). As predicated by the biochemical results, genome-wide RNA profiling analyses revealed hundreds of altered splicing events as a result of BS69 knockdown. Among them, IR is the main event that appears to be impacted by the BS69 loss, indicating that BS69 is a regulator of IR. ChIP-seq data show a statistically significant correlation between BS69 occupancy and IR regulation, suggesting a direct regulation. Further genetic rescue experiments demonstrate that binding of BS69 to EFTUD2 as well as H3K36me3 are necessary for BS69 to antagonize the activity of EFTUD2 and to promote IR. Our investigation thus discovered BS69 as a novel H3.3 specific reader, which regulates IR by targeting a specific step during spliceosome activation, connecting BS69-mediated IR regulation to H3.3K36me3-decorated chromatin.
RESULTS
The Genome-wide Distribution of BS69 Resembles That of H3K36me3
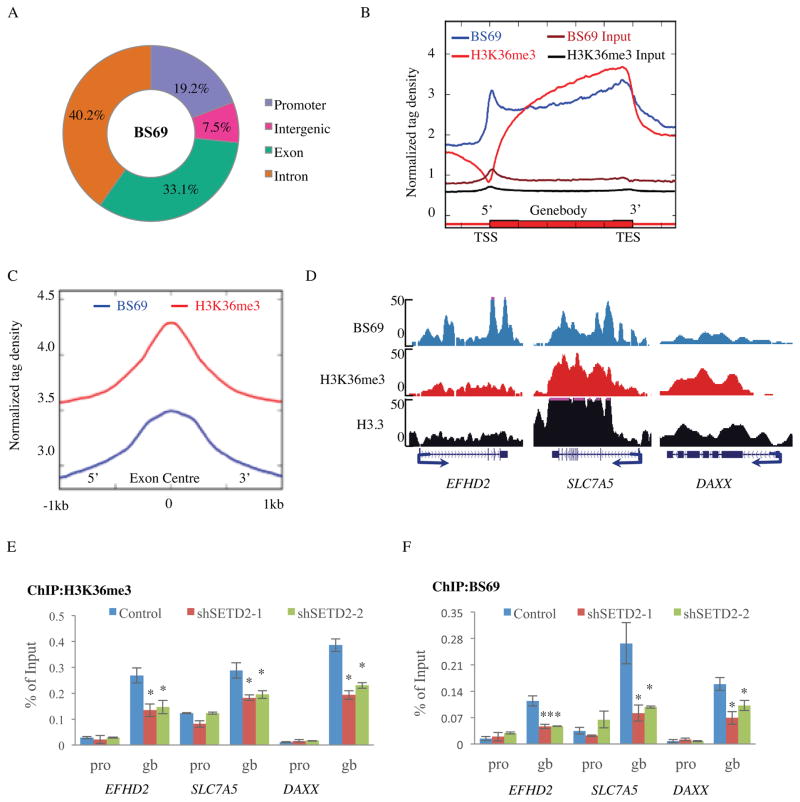
To better understand the function of BS69 during transcription, we sought to first identify BS69 genomic locations. ChIP-seq of endogenous BS69 yielded 18,406 potential BS69 binding events (covering 8,080 genes) compared with input, which appear to be enriched at both promoters (p-value < 2.2e-16, binomial test) and gene bodies (p-value < 2.2e-16, binomial test) relative to the distribution of random peaks in HeLa cells (Figure 1A and Figure S1A). Consistently, analysis of the genomic distributions of BS69 across an average Refseq gene also shows a high density of BS69 at promoters and gene bodies (Figure 1B), indicating that BS69 binds both promoters and gene bodies. Further bioinformatics analysis showed that BS69 binding to gene body represents the majority of BS69 genomic distribution events (73.3%, 13,492/18,406). Taken together, these findings suggest that BS69 mainly localizes to gene bodies but it may also play a role at promoters.
Figure 1. BS69 shows a similar genomic distribution pattern as H3K36me3.
(A) Genomic distribution of BS69 peaks (18,406) in HeLa cells. (B) Normalized tag density of BS69 and H3K36me3 along the transcription unit. Each gene body is represented from 0% (TSS) to 100% (Jorgensen et al.) on the X-axis. Normalized Tag density is plotted from 30% upstream of TSS to 30% downstream of TES. (C) Averaged exonic occupancies of BS69 and H3K36me3. The centers of exons are aligned at position 0 with 1kb of both upstream and downstream sequences included in the analysis. (D) Representatives of BS69, H3K36me3 and H3.3-FLAG ChIP-seq peaks in HeLa cells. Arrows denote TSS and transcription orientation. (E) and (F) Occupancies of H3K36me3 and BS69 at selected gene bodies (gb), but not promoters (pro), are reduced in SETD2 KD HeLa cells. Data are represented as mean ± SEM from 3 biological replicates (E, F), *P<0.05, **P<0.01. See also Figure S1.
Exons are known to be preferentially marked with H3K36me3 relative to introns (Kolasinska-Zwierz et al., 2009). The ChIP-seq profiles of BS69 and H3K36me3 display similarities at gene bodies and exons (Figure 1B and 1C) but also differences at promoters (Figure 1B). Indeed, 91% (7,332/8,080) of the BS69-bound genes are also marked by H3K36me3 (Figure S1B), while only 31% (17457/56223) of all genes are marked by H3K36me3 in our ChIP-seq analyses. Representatives of such an overlap between H3K36me3 and BS69 binding are shown over three BS69 target genes (taken from the ChIP-seq data) (Figure 1D). This was confirmed by ChIP-qPCR, which showed BS69 gene body enrichment similar to that of H3K36me3 (Figure S1C). These findings suggest that BS69 binding to gene bodies may be dependent on H3K36me3. To address this possibility, we carried out ChIP analysis upon knockdown of SETD2, the main methyltransferase that mediates H3K36 trimethylation in mammalian cells (Edmunds et al., 2008; Kolasinska-Zwierz et al., 2009; Strahl et al., 2002). Knockdown of SETD2 not only reduced H3K36me3 signals, as expected, but also reduced BS69 gene body (not promoter) occupancy on a number of BS69 target genes (Figure 1E, F). As an additional control, knockdown of SETD2 did not affect BS69 expression (Figure S1D). These findings support the notion that BS69 gene body association is dependent on H3K36 trimethylation mediated by SETD2. However, H3K36me3 is known to primarily mark gene bodies, thus the mechanism by which BS69 is recruited to promoter remains unclear but may involve sequence-specific DNA binding factors, such as E2F6 reported previously (Velasco et al., 2006).
BS69 Preferentially Binds H3.3K36me3 In Vitro
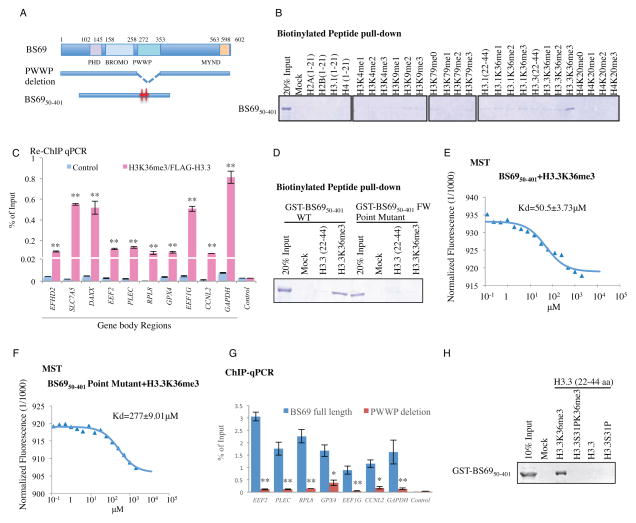
BS69 carries three, tandemly arranged, putative chromatin modification recognition modalities (readers) including PHD, BROMO and PWWP domains (Figure 2A). The PWWP domains of BRPF1, DNMT3A, MSH6, and NPAC all have recently been shown to recognize H3K36me3 (Dhayalan et al., 2010; Li et al., 2013; Vezzoli et al., 2010), raising the possibility that BS69 dependency on SETD2 for gene body association may be accomplished by direct recognition of H3K36me3 through its PWWP domain. To address this possibility, we carried out in vitro binding assays using recombinant BS69 carrying all three putative reader modalities (BS6950-401) and various histone H3.1 and H3.3 peptides. We found that BS6950-401 (as well as full length BS69, Figure S2A) does not bind unmodified N-terminal tails of H2A, H2B, H3 or H4 (Figure 2B). Interestingly, it seems to preferentially bind H3.3 when lysine (K) 36 is tri-methylated (H3.3K36me3) (not mono- or dimethylated) but bind H3.1K36me3 much more poorly, if at all (Figure 2B and Figure S2A). As additional specificity controls, we show that BS6950-401 failed to bind H3 peptides methylated on H3K4, H3K9 and H3K79 as well as H4K20 methylated substrates (Figure 2B). These findings suggest that BS69 mainly recognizes H3K36 trimethylation on H3.3 in vitro. To explore this further, we established a HeLa cell line stably expressing a FLAG-tagged H3.3 and carried out ChIP-seq of FLAG tagged H3.3 as well as H3K36me3. Consistently, a comparison of the genome-wide distributions of H3.3, H3K36me3 and BS69 showed strong correlation, especially at TES (transcription termination site) regions (Figure S2B). By sequential ChIP assay, we have further confirmed that BS69 and H3K36me3 likely co-occupy regions in the gene bodies of BS69 target genes (Figure 2C).
Figure 2. BS69 binds H3.3K36me3 in vitro via its PWWP domain.
(A) Schematic diagrams of human BS69 and mutants carrying PWWP deletion and point mutations denoted by red stars. (B) Recombinant GST-BS6950-401 specifically recognizes H3.3K36me3 peptide in the biotinylated histone peptide pull-down assays. (C) Sequential ChIP using H3K36me3 and anti-FLAG antibodies in HeLa cells overexpressing H3.3-FLAG shows co-occupancy of H3K36me3 and H3.3-FLAG at indicated locales of BS69 target genes. (D) GST-BS6950-401 FW point mutant (F293A, W294A) loses the ability to bind H3.3K36me3 in vitro. (E) and (F) MicroScale Thermophoresis analysis determined the Kd of GST-BS6950-401 WT (E) interaction with H3.3K36me3 to be 50 μM. The Kd value of the interaction between the BS69 PWWP point mutant and H3.3K36me3 (F) was determined to be 277 μM. (G) HA-ChIP and q-PCR demonstrated that the ectopically expressed WT BS69 but not the BS69 PWWP deletion mutant binds the BS69 targets (identified through BS69 ChIP). (H) The binding of GST-BS6950-401 to H3.3K36me3 peptide is abolished by phosphorylation at position S31. Data are represented as mean ± SEM from 3 biological replicates (C,G), *P<0.05, **P<0.01. See also Figure S2.
The PWWP Domain is Critical for BS69 Binding to H3.3K36me3
Previous studies have identified amino acids critical for the PWWP domain of MSH6 and BRPF1 to bind H3K36me3 (Li et al., 2013; Vezzoli et al., 2010), which are conserved in the PWWP domain of BS69 (Figure S2C). Importantly, mutations of these amino acids, F293 and W294, to alanines abolished binding of BS6950-401 to H3.3K36 trimethylated peptide as judged both by in vitro pull-down assays (Figure 2D), as well as by MST (Wienken et al., 2010) (Microscale Thermophoresis). Specifically, the Kd value for wildtype BS6950-401 binding to H3.3K36me3 is 50.5 μM (Figure 2E), while in contrast, the Kd value for the interaction between the BS6950-401 PWWP point mutant and the H3.3K36me3 histone peptide was significantly higher (Figure 2F, Kd=277 μM), supporting the pulldown result that the PWWP point mutant has a significantly reduced ability to bind H3.3K36me3. Consistently, the Kd values for wildtype BS6950-401 binding to H3.1K36me3, H3.1K36me0 and H3.3K36me0 were undetectable (Figure S2D). These results suggest that BS69 preferentially binds H3.3K36me3 in a manner that is dependent on the PWWP domain. In addition to histone peptides, full length BS69 also binds nucleosome purified from HeLaS cells (Figure S2E, top panel). Consistently, we found chromatin association of BS69 to the gene bodies of 7 target genes in vivo is essentially abrogated when its PWWP domain is deleted (Figure 2A, 2G) (BS69 ΔPWWP was expressed at a similar level and localized to the nucleus, Figure S2F and data not shown). Taken together, these findings indicate that BS69 directly binds H3.3K36me3 and associates with chromatin in a PWWP domain-dependent manner. Interestingly, deletion of the PHD or the Bromo domain also resulted in a loss of BS69 association with its target genes (assayed by ChIP-qPCR of their binding to the same 7 BS69 target genes) (Figure S2G), suggesting that in addition to PWWP, the PHD and Bromo domains of BS69 may also participate in the recognition of H3.3K36me3 and BS69 chromatin association in vivo. Of course, we cannot exclude the possibility that deletions could have resulted in improper folding of the mutant proteins.
Phosphorylation at Serine 31 Disrupts the Binding of BS69 to H3.3K36me3
The exquisite specificity of BS69 binding to H3.3K36me3 was unexpected given the fact that the entire H3.3 differs from the canonical H3.1 by only five amino acids (Hake and Allis, 2006). Furthermore, the only difference between the H3.3 and H3.1 histone peptides used in our binding assays is the amino acid at position 31 where H3.3 has a serine (S) and H3.1 contains an alanine (A) residue. We speculated that S31 must be playing an important role in this highly specific interaction between H3.3 and BS69 and therefore modifications such as phosphorylation may impact BS69 binding to H3.3K36me3. Indeed, the H3.3K36me3 peptide with S31 phosphorylation (H3.3S31PK36me3) was significantly compromised in its ability to bind BS69 (Figure 2H and Figure S2H, Kd ~ 1.5mM). Interestingly, phosphorylation of H3.3S31 has been found to be enriched during mitosis at telomere ends in mES cells and pericentric heterochromatin after differentiation (Hake et al., 2005; Wong et al., 2009), suggesting that BS69 binding H3.3 may be subject to regulation in vivo, and may play a role in these specific biological processes.
Chromatin-Bound BS69 Associates with Multiple Components of Spliceosome
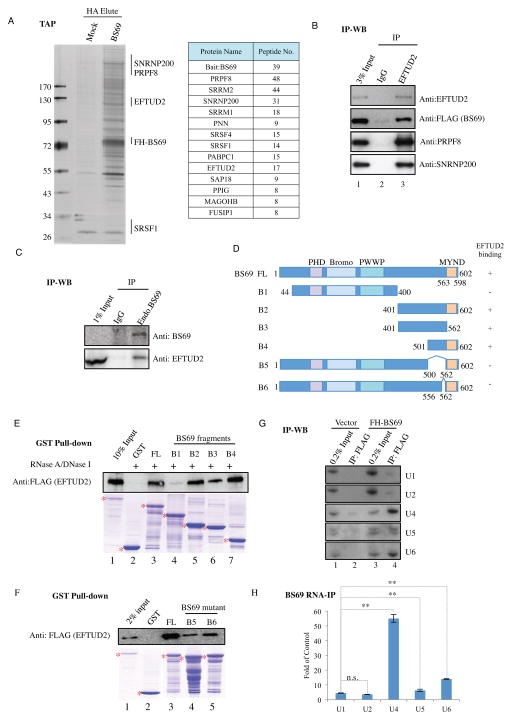
H3K36 trimethylation has recently been demonstrated to play a role in regulating transcriptional elongation and alternative splicing (Li et al., 2002; Luco et al., 2010; Pradeepa et al., 2012; Schaft et al., 2003) but the underlying mechanisms remain incompletely understood. To investigate a potential role of BS69 in these processes, we established a HeLa cell line expressing FLAG-HA-tagged BS69 and purified the tagged BS69 by sequential immunoprecipitation using FLAG and HA antibodies, respectively. The BS69 purified from soluble nuclear fraction appears to mainly associate with factors involved in transcriptional regulation, including DNA binding transcription factors and cofactors, as well as chromatin regulators, such as E2F6, MLL, NSD1, KDM3B and BRG1, suggesting that BS69 may directly regulate transcription by working with these proteins (Table S1). In addition to purifying BS69 from the soluble nuclear fraction, we also purified chromatin-bound BS69 by dissociating it from the nuclear pellet with micrococcal nuclease (MNase). Interestingly, the chromatin-bound BS69 mainly associates with a very different set of proteins that are involved in regulating RNA splicing (Figure 3A). The presence of many of these proteins in the purified BS69 from chromatin fraction can be further confirmed by Western blot analysis (Figure S3A). These BS69 associated proteins can be grouped into at least two classes, i.e., the U5 snRNP proteins (EFTUD2, PRPF8, and SNRNP200) and Serine and Arginine-rich (SR) proteins (SRSF1, PNN). Reciprocal immunoprecipitation using an EFTUD2 antibody, incubating with FLAG immunoprecipants purified from HeLa cells stably expressing FLAG-HA-tagged BS69, not only brought down EFTUD2 and tagged BS69, but also PRPF8 and SNRNP200, suggesting that these proteins are likely to be in the same complex (Figure 3B). We further demonstrated by co-immunoprecipitation that endogenous BS69 also interacts with EFTUD2 (Figure 3C).
Figure 3. BS69 interacts with splicing factors.
(A) Left panel: TAP purified FLAG-HA-BS69 protein complex was resolved by gradient SDS-PAGE and visualized by silver staining. The positions of the tagged BS69 (FH-BS69) as well as some of the RNA splicing factors are indicated on the right. Right panel: polypeptides identified by tandem mass spectrometry. (B) Reciprocal immunoprecipitation by EFTUD2 antibody incubated with FLAG immunoprecipitants purified from HeLa cells stably expressing FLAG-HA-BS69 brought down FLAG-HA-BS69 as well PRPF8 and SNRNP200. (C) Interaction between endogenous BS69 and EFTUD2 was confirmed by co-immunoprecipitation using HeLa nuclear extract. (D) Schematic representation of full length and various truncated forms of BS69. Amino acid positions are indicated. (E) and (F) Interactions between EFTUD2 and full length and various truncated forms of BS69 were determined by GST pull-down assays. Recombinant BS69 proteins and FLAG-EFTUD2 were purified from E. coli and insect cells, respectively. FL: full length. (G) and (H), Interactions between FLAG-BS69 and snRNAs were determined by Northern blotting (G) and RT-qPCR (H). Data are represented as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01; n.s., not significant. See also Figure S3, Table S1.
BS69 Directly Binds EFTUD2, a component of the U5 snRNP
We next wished to determine which splicing regulators directly bind BS69. Using recombinant splicing factors purified from insect Sf9 cells, we only detected interactions of SRSF1 and EFTUD2 with BS69 in vitro (Figure S3B, left panels). We further demonstrated that EFTUD2, but not SRSF1, directly binds BS69 in a RNA and DNA independent manner (Figure S3B, right panels). Using various deletion mutants, we identified that the C-terminal region of BS69 (aa 401-602 (B2)) is important to mediate this interaction (Figure 3D and 3E, compare B1 and B2 with BS69 FL). Further analyses narrowed the interacting region to amino acids 556-562 whose removal from the full length BS69 significantly reduced BS69 interaction with EFTUD2 (Figure 3D, F, compare B6 with BS69 FL), suggesting that the region encompassing aa 556-562 is necessary for BS69 interaction with EFTUD2.
RNA splicing is a multi-step process involving many splicing factors and U1, U2, U4, U5 and U6 snRNPs (Wahl et al., 2009). EFTUD2 is involved in unwinding U4/U6 RNA during spliceosome activation, an essential step for spliceosome activation (Bartels et al., 2002). The fact that BS69 interacts with EFTUD2 and other splicing factors predicts that BS69 may also interact with the spliceosomal snRNPs. We immunoprecipitated FLAG-tagged BS69 and then interrogated the presence of the spliceosomal U1, U2, U4, U5, and U6 snRNAs by Northern blotting. As shown in Figure 3G, tagged BS69 co-immunoprecipitated U4, U5, and U6 snRNA (compare lane 4 with 3). We next used real-time PCR to quantify each of the co-precipitated snRNA and found that BS69 mainly associates with U4 snRNA, but also to a lesser extent with U5 and U6 (Figure 3H). In contrast, EFTUD2 antibody mainly co-precipitated U5 snRNA (Figure S3C). Collectively these findings support a potential role for BS69 in regulating RNA splicing.
BS69 Primarily Promotes Intron Retention
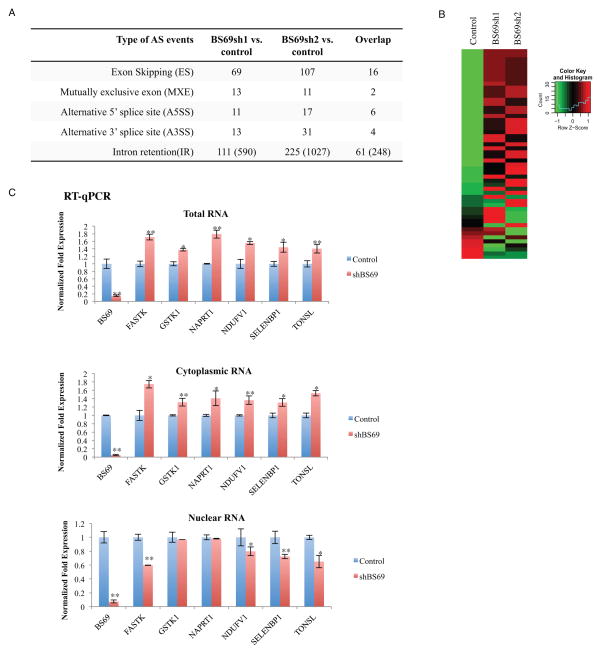
To determine whether BS69 regulates splicing, we performed RNA-seq using mRNA samples isolated from two independent BS69 shRNA treated HeLa cells (Table S2). MATS (Multivariate Analysis of Transcript Splicing) program detected a number of differential alternative splicing (AS) events upon BS69 knockdown. As shown in Figure 4A, we identified 217 significant AS events in BS69 shRNA-1 versus control, and 391 significant AS events in BS69 shRNA-2 versus control (FDR<10%). The higher number of events detected in the BS69 shRNA-2-treated sample is likely to be due to better knockdown efficiency of BS69 shRNA-2 (Figure S4A and S4B). Importantly, we find a high degree of data overlap between the two independent RNA-seq experiments (with two independent BS69 shRNAs), i.e., a total of 89 AS events passed the genome-wide significance cutoff (FDR<10%) in both RNA-seq experiments (Figure 4A). Among the 89 AS events regulated by BS69, we identified 61 intron retention (IR) and 16 exon skipping (ES) events. Given that the overall frequency of IR is the lowest among the different types of AS events in mammalian genes (Sakabe and de Souza, 2007), it is striking that more than 2/3 of the AS events regulated by BS69 are IR events.
Figure 4. BS69 mainly regulates intro retention.
(A) Summary of altered splicing (AS) events in HeLa cells upon BS69 KD detected by RNA-seq from three biological replicates. Two independent BS69 shRNAs were used. (B) Heatmap comparison of the mRNA levels of genes whose IR events are regulated by BS69 in the presence and absence of BS69. (C) RT-qPCR analyses of mRNA levels of six BS69-regulated IR genes in different cellular compartments. Data are represented as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01; n.s., not significant. See also Figure S4, Table S2.
The analysis described above on BS69-regulated IR events was done for ~6,000 documented IR events (Ensembl transcript database). To perform a truly genome-wide, unbiased search for more BS69-regulated IR events, we expanded our analysis to ~92,000 introns, which are human introns without partial overlap with exons of sense or antisense transcripts, in order to avoid confounding signals from overlapping transcripts. This new analysis identified 590 and 1,027 BS69-regulated IR events, respectively, from the same two RNA-seq experiments (Figure 4A, numbers are in parenthesis). Importantly, a total of 248 BS69-regulated IR events are shared by both RNA-seq experiments, which are distributed among 219 genes (after correction for genome-wide multiple testing, FDR<10%) (Figure 4A). It should be noted that RNA-seq detection of differential alternative splicing is known to have substantial false negatives especially for moderately or lowly expressed genes(Dittmar et al., 2012), as we need high sequencing coverage for the events of interest to call statistically significant changes in splicing levels. Here we adopted a conservative criterion to define BS69-regulated IR events, by requiring that the events were called significant in both shRNA experiments, and this likely would underestimate the impact of BS69 on IR regulation. To evaluate this possibility further, we examined the concordance of the two shRNA experiments (Figure S4C). As expected, the 248 IR events called significant in both shRNA experiments showed a strong correlation in their shRNA-induced changes in IR levels (Pearson correlation r=0.96). Interestingly, targets called significant in only one of the two shRNA experiments also showed a strong correlation between the two shRNA experiments (n=342, r=0.70; and n=779, r=0.79, respectively; see Figure S4C), which were further supported by additional RT-qPCR confirmation (Figure S4D). These findings suggest that the actual number of IR events regulated by BS69 is likely to be significant higher than 248. Strikingly, among the 248 BS69-regulated IR events, 238 (96.0%) had decreased levels of IR upon BS69 knockdown, suggesting that BS69 functions primarily to promote retention of specific introns.
BS69-Regulated IR Events are Significantly Enriched for BS69 Binding
Further analysis showed that the 248 BS69-regulated IR events are significantly enriched for BS69 binding (38.3%) as compared to BS69-independent IR events (28.9%) (Fisher’s Exact Test P=5.4e−3) (Figure S4E). Binding of BS69 to ten of these genes (whose IR events are regulated by BS69) is further demonstrated by ChIP-qPCR (Figure S4F), supporting a direct involvement of BS69 in their IR regulation. As the sensitivity of ChIP-seq may be limited, we employed ChIP-qPCR to determine whether some BS69 binding signals at its regulated IR events may have been missed as a result. Indeed, we were able to find BS69 binding at 5 out of 6 randomly selected BS69-regulated IR events that are not detected by ChIP-seq (Figure S4G, Left panel), while no BS69 binding was detected at another set of 6 randomly selected, BS69-independent IR sites (Figure S4G, Right panel). These results suggest that ChIP-seq may have only detected strong BS69 binding events, and as a result we are underestimating the actual BS69 binding events as well as the correlation of BS69 binding with BS69 regulated IR events.
In addition to identifying BS69 enrichment at its regulated IR, about 62.1% of BS69 regulated IR introns are also H3.3 enriched (assessed by ectopic expressing H3.3-FLAG) as opposed to the control (53%) (Fisher’s Exact Test p=1.2e−2) (Figure S4H), and 88.7% BS69 regulated IR introns are decorated by H3K36 trimethylation as to the control (79.9%) (Fisher’s Exact Test p=1.2e−3, Figure S4I). Consistent with the idea that BS69 is recruited to specific genomic locations to regulate alternative splicing events by binding H3.3K36me3 via its PWWP domain, the average ChIP-seq densities of BS69, H3K36me3 and H3.3 at the 248 BS69 regulated introns are significantly higher than those at the control BS69-independent set (Figure S4J) further supporting the notion that BS69 regulates IR events directly at genomic regions that are enriched for trimethylated histone H3.3K36.
We next asked whether there are specific features associated with BS69-regulated IR events that are directly bound by BS69, we compared the 95 BS69-bound IR events that respond to BS69 depletion with the 272 BS69-bound IR events that do not. Our bioinformatics analyses found that BS69-bound IR introns that respond to BS69 depletion are significantly longer (median length 1,025bp vs 548bp, P= 0.0003 by two-sided Wilcoxon test) and have significantly stronger 5′ splice sites (median 5′ splice site score 8.94 vs 8.05, P= 0.004 by two-sided Wilcoxon test, calculated by MAXENT)(Yeo and Burge, 2004). In contrast, we did not observe any other unique features at the 3′ splice sites and any significant reported motifs for RNA-binding proteins and splicing factors in the first 250bp and the last 250bp of the introns (Anderson et al., 2012; Ray et al., 2013). How do such features contribute to BS69 regulation of IR remains an interesting question for future investigation.
BS69 Knockdown Results in an Increase of the Steady-State mRNA Level of Its Target Genes Containing BS69-regulated Introns
Intron-retaining transcripts often contain a premature stop codon, which triggers nonsense mediated decay pathway (NMD) (Braunschweig et al., 2013; Lejeune and Maquat, 2005). As shown by the heatmaps (Figure 4B and Figure S4K), most of the genes that contain BS69-regulated introns showed an increase in their steady-state mRNA levels upon knockdown of BS69. This was further confirmed by ChIP-qPCR of six selected genes (Figure 4C, top panel). Consistently, the BS69-regulated RNA transcripts by and large showed an increased level in the cytoplasm and a reduced level in the nucleus, respectively (Figure 4C, middle and lower panels), suggesting that the unspliced RNAs are retained/degraded in the nucleus while the spliced RNAs are exported to the cytoplasm, resulting in an overall increase in gene expression. This finding supports the notion that BS69 regulates IR and further suggests that BS69 may coordinately regulate a select group of genes by regulating their IR. Importantly, among different types of RNA alternative splicing, including ES and alternative splice site usage, IR is the least well understood, and therefore identification of BS69 as a regulator that primarily controls IR represents an important step towards understanding the molecular mechanisms of IR regulation. Given the vast number of introns in the genome, the finding that BS69 impacts a limited number of introns indicates a highly specific role of BS69 in regulated RNA splicing.
BS69 Antagonizes EFTUD2 in IR regulation Dependent on Its Physical Interaction with EFTUD2
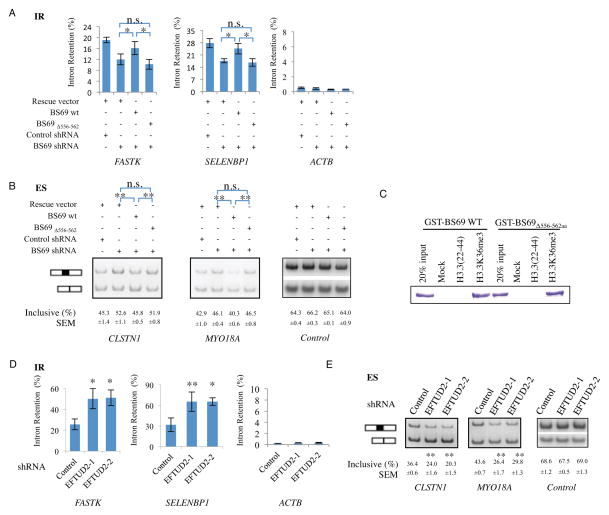
To confirm RNA-seq findings, we carried out RT-qPCR and RT-PCR assays for BS69-regulated alternative splicing events, including 14 selected from the original 61 IR events identified from the ~6,000 introns and an additional 4 from the expanded analysis (248 IR events), as well as 4 ES events, respectively. As shown in Figure S5A–C, these AS events are readily detectable upon BS69 knockdown. The regulation of IR by BS69 is not limited to HeLa cells as we have observed the same impact of BS69 on IR events in a number of other cell lines, including the human lung cancer cell line A549 and human lung fibroblast HFL-1 (Figure S5D). To rule out potential shRNA off-target effects, we carried out rescue experiments by adding back RNAi-resistant BS69. Indeed, wildtype BS69 readily rescued these AS events (Figure 5A, B), indicating that the observed change in splicing is due to the knockdown of BS69. Importantly, reintroduction of an EFTUD2-interaction defective mutant of BS69 (BS69Δ556-562aa), which was expressed at a comparable level to that of wildtype BS69, localized in the nucleus (Figure S5E and S5F) and retained the ability of H3.3K36me3 binding (Figure 5C), failed to do so (Figure 5A, B), suggesting that EFTUD2 interaction is critical for BS69 to regulate RNA splicing.
Figure 5. BS69 regulates intron retention and exon skipping events by antagonizing EFTUD2 through physical interaction.
(A) and (B) Wild type but not EFTUD2 interaction defective mutant (Δ556-562) rescued the alteration of IR (Intron Retention, panel A) and ES (Exon Skipping, panel B) caused by BS69 knockdown. (C) EFTUD2-interaction defective mutant of BS69 (BS69Δ556-562aa) retains H3.3K36me3 binding ability at a comparable level as that of wildtype in histone peptide pull-down assay in vitro. (D) and (E) Knockdown of EFTUD2 caused an increase in IR (D) and an altered ratio of ES (panel E) of target genes in HeLa cells. Data are represented as mean ± SEM from 3 biological replicates (A, B, D, E), *P<0.05, **P<0.01; n.s., not significant. See also Figure S5.
We next examined whether these splicing events were also regulated by EFTUD2. Strikingly, knockdown of EFTUD2 had an opposite effect to that of BS69 knockdown on either the IR or ES events. Specifically, we examined the same 14 IR events and found that loss of EFTUD2 resulted in an increase in IR as opposed to a decrease when BS69 was knocked down (Figure 5D, Figure S5G). Similarly, we also examined the same 4 ES events and found that knockdown of EFTUD2 likewise had the opposite effects (Figure 5E, Figure S5H). These findings suggest that BS69 and EFTUD2 may function antagonistically in regulating RNA splicing and raises the possibility that BS69 may promote IR by suppressing the core splicing machinery.
SETD2 Knockdown affects BS69-Dependent AS Events Similar to That of BS69 Knockdown
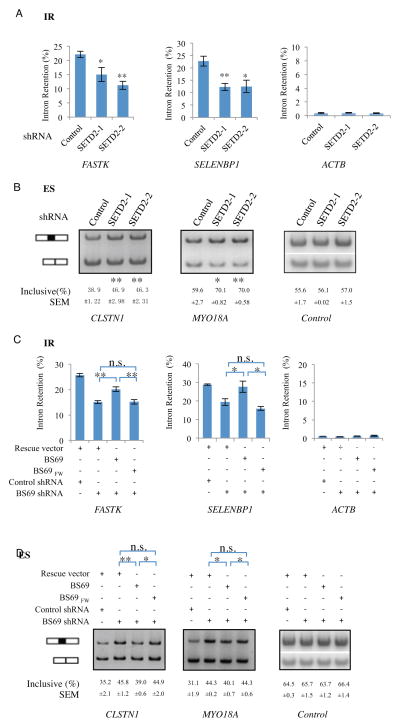
As mentioned earlier, H3K36 trimethylation has been suggested to play a role in RNA splicing but exactly how it regulates splicing remains incompletely understood. Since BS69 binds its target genes in a SETD2-dependent manner, which mediates H3K36 trimethylation, we asked whether the same AS events regulated by BS69 are similarly regulated by SETD2. In our analyses, the same set of 14 IR events (Figure 6A and Figure S6A) and 2 ES events (Figure 6B) regulated by BS69 showed a similar regulation (12 out of 14 IR events) by SETD2 knockdown. Furthermore, SETD2 knockdown also reduced H3K36me3 levels, as expected, as well as BS69 binding to these genes (Figure S6B, and S6C). These findings suggest that BS69 may be an important downstream player in mediating the function of H3K36me3 to regulate RNA splicing. To investigate this possibility further, we carried out genetic rescue using the wildtype BS69 and a PWWP point mutant (BS69FW) defective in binding to H3K36me3 but expressed at a comparable level to that of the wildtype BS69 and localized to the nucleus (Figure S6D and 6E). As shown in Figure 6C and 6D, while wildtype BS69 restored the normal splicing patterns of both the IR and ES events, the BS69FW mutant failed to do so, indicating that the ability of BS69 to regulate RNA splicing is also dependent on its binding to chromatin via H3K36me3.
Figure 6. Binding H3K36me3 is important for BS69 to regulate alternative splicing.
(A) and (B) Knockdown of SETD2 by two independent shRNAs in HeLa cells caused a decrease in IR (A) and an altered ratio of ES (B) of BS69 target genes. (C) and (D) Wild type but not the PWWP FW point mutant rescued the alteration of IR (C) and ES (D) caused by BS69 knockdown. Data are represented mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01; n.s., not significant. See also Figure S6.
DISCUSSION
We have shown that BS69 regulation of RNA splicing occurs in a SETD2-dependent manner, which mediates H3K36 trimethylation. The specificity that BS69 exhibits towards H3.3K36me3 is quite remarkable as the entire H3.3 and H3.1 differ only by five amino acids with only one amino acid difference (amino acid 31) in the vicinity of K36. Amino acid 31 is a serine in H3.3 and alanine in H3.1, suggesting that phosphorylation may play a role in regulating BS69 recognition of H3.3K36me3. Indeed our result shows that S31 phosphorylation significantly impedes BS69 binding to H3.3K36me3 in vitro. Given that BS69 mainly binds H3.3K36me3 in vitro and that the ability of BS69 to regulate splicing is dependent on its ability to bind H3K36me3, our findings strongly suggest a role for BS69 downstream of H3.3K36 trimethylation, thus linking K36 trimethylation of the histone variant H3.3 to RNA splicing regulation. Although binding of BS69 to H3.1K36me3 in vitro was barely detectable, we cannot formally exclude the possibility that in vivo, BS69 may bind H3.1H36me3 in some genomic regions and therefore the effects of BS69 on RNA splicing could also be in part mediated by H3.1K36me3. Importantly, our genome wide ChIP-seq location analysis of BS69 and H3.3 showed that not all H3.3 are bound by BS69, predicting the existence of additional H3.3K36me3-specific readers. Furthermore, not all BS69 directly bound targets displayed alterations in IR regulation in response to BS69 knockdown, suggesting that additional unknown factors may influence the ability of BS69 to regulate IR. Our study further identified an antagonistic relationship between BS69 and the core RNA splicing machinery, supporting the model whereby BS69 promotes IR by antagonizing the activity of core splicing machinery through physical interactions with snRNPs such as EFTUD2. Interestingly, while physically associated with the U5 snRNA protein EFTUD2, unexpectedly, BS69 mainly interacts with U4 snRNA, suggesting that BS69 may suppress splicing by sequestering U4 snRNA, whose release is a prerequisite for spliceosome activation (Wahl et al., 2009).
A very recent study by Wen et al (Wen et al., 2014) reported binding of BS69/ZMYND11 to H3.3K36me3. In that study, the authors generated and analyzed the co-crystal structure of BS69 reader domains bound to H3.3K36me3 and identified critical contacts that involve both the PWWP and Bromo domains, as well as S31, thus providing atomic level understanding of the interaction between BS69 and H3.3K36me3. Wen et al also reported that loss of BS69 impacts transcriptional elongation, consistent with the previously proposed model that elongation rates regulates alternative splicing (Wen et al., 2014). We compared our BS69-regulated IR event genes (both BS69-bound and unbound IR genes) with the BS69 elongation-regulated genes (268 up-regulated genes) but found no significant overlap. Since these two studies were conducted in two different cell lines, HeLa (this study) and U2OS (Wen), it remains to be determined whether there is a functional relationship between BS69-regulated elongation versus IR or other RNA alternative splicing events. However, our genetic rescue result demonstrating that the direct interaction of BS69 with EFTUD2 is critical for BS69 to regulate IR favors the model that BS69 directly participates in IR regulation.
Another recent study investigated the mechanism by which H3K36me3 regulates RNA splicing and identified MRG15, which binds H3.1K36me3 via its chromodomain and recruits the Polypyrimidine Tract Binding protein (PTB) to regulate alternative splicing of the human fibroblast growth factor receptor 2 gene, thus making the first connection between chromatin and regulated RNA splicing (Luco et al., 2010). Interestingly, a separate study found that PTB coordinates neuronal gene expression through regulation of IR events specific to a subset of neuronal genes (Yap et al., 2012). However, whether this regulation is connected to chromatin via MRG15 remains unclear. Another reported connection between H3K36me3 and alternative splicing is the PSIP1 short isoform (p52) (Pradeepa et al., 2012). Similar to our findings, PSIP1 (p52) was reported to be associated with a large number of splicing factors, including SR proteins, U5 snRNP proteins, hnRNP proteins and DEAD/H box helicases. Knockdown of PSIP1 (p52) resulted in 95 alternative exon usage events from a survey of 40,443 exons in 7,715 genes with one or more predicted alternative transcripts. However, whether PSIP1 (p52) regulates IR has not been investigated. Thus, our findings identify for the first time a novel regulator that controls IR, mediated primarily by K36 trimethylated histone variant H3.3. Collectively, these findings suggest that MRG15, PSIP1 (p52) and BS69 may represent separate pathways that link chromatin to RNA splicing regulation.
In yeast, regulated IR has been shown to be important for coordinated expression of meiotic and ribosomal protein genes (Averbeck et al., 2005; Cremona et al., 2011; Moldon et al., 2008; Munding et al., 2010; Parenteau et al., 2011). However, the biological function of regulated IR in mammals is unclear at the present time. Interestingly, recent studies showed that regulated IR helps to coordinate gene expression during neuronal differentiation and granulopoiesis suggesting a possible role for IR in differentiation (Wong et al., 2013; Yap et al., 2012). Alterations in IR and ES have also been identified as major RNA splicing events in breast cancer (Eswaran et al., 2013), suggesting that altered IR and ES events may also contribute to tumorigenesis. In this context, it’s interesting to note that recent studies identified histone H3.3 point mutations, which either directly (K36M) or indirectly (G34R/V) abolished H3.3K36 trimethylation, as driver mutations in a number of different tumors (Behjati et al., 2013; Lewis et al., 2013; Schwartzentruber et al., 2012; Wu et al., 2012). Furthermore, SETD2 and H3K36me3 have also been reported to be frequently mutated and lost in renal cell carcinoma and pediatric high-grade glioma (Duns et al., 2010; Fontebasso et al., 2013). Additionally, our data that H3.3S31 phosphorylation impedes BS69 binding suggests the importance of S31 in regulating BS69 function, and raises the question of whether S31 may play a role in tumorigenesis and is also subject to mutational events in cancers. The ongoing efforts of deep sequencing of cancer tissues and identifying the signaling pathway that regulates H3.3S31 phosphorylation will provide insight into these issues. We speculate that BS69 may participate in tumorigenesis through regulation of RNA splicing events such as IR and ES and that an impaired RNA splicing regulation as a result of inactivation of the SETD2-H3K36me3-BS69 axis and/or the H3.3S31 signaling pathway may contribute to tumorigenesis of various types of cancers.
METHODS
The Microscale Thermophoresis (MST) analysis
The determination of the binding capacity of BS69 to histone peptides was performed by microscale thermophoresis (MST) according to manufacturer’s instruction with 20% LED and 20% MST power (Wienken et al., 2010) (Nano Temper, Monolith NT.115). Fluorescently labeled BS6950-401 or BS6950-401 mutant were used as tracer. The final concentrations of peptides ranged from 122 nM to 2 mM.
BS69 protein complex purification and reciprocal immunoprecipitation
Tandem affinity purification was performed as described previously (Ogawa et al., 2002) except that HeLaS nuclear extract was treated with MNase at 37°C for 3 min. For reciprocal immunoprecipitation in Figure 2b, FLAG-purified BS69 complex was precipitated using either anti-EFTUD2 or IgG antibody. The immunoprecipitants were analyzed by Western blotting using corresponding antibodies.
snRNA Association analyses
HeLa cells transfected with FLAG-BS69 plasmid grown in 150 cm plates were covered with ice-cold PBS buffer and subjected to UV-irradiation (150 mJ/cm2) for crosslinking. Immunoprecipitation was performed using anti-FLAG beads. After washing, RNA was extracted with TRIzol, and analyzed by qRT-PCR and northern blot.
Chromatin immunoprecipitation (ChIP) and ChIP-seq
ChIP assays were carried out as previously described (Lan et al., 2007). ChIP-Seq was performed according to Illunima’s protocol. The FASTQ data were mapped to the human genome (hg19) using Bowtie, and significant enrichments were identified by MACS2.0 using Broad Peak mode with Q-value = 0.05 as a cutoff to call peaks from the aligned results (Zhang et al., 2008). Gene lists bound by BS69 or H3K36me3 were generated based on Ensembl version 65 gene annotation.
RNA-seq and bioinformatics analysis
HeLa cells were infected with lentiviruses carrying either control or BS69 shRNA. After puromycin selection for 7 days, RNA samples were prepared using TRIZOL and subjected to 100bp × 2 non-strand-specific paired-end RNA-seq by Genome Center, WuXi App Tec. We used MATS(Shen et al., 2012) (http://rnaseq-mats.sourceforge.net/; version 3.0.7) to identify BS69-regulated differential alternative splicing events corresponding to all five types of alternative splicing patterns.
Splicing Assay
Total RNA was subjected to gDNA eraser treatment for 5 min, followed by reverse transcription using random primers for each sample (Takara, PrimeScript™ RT reagent Kit with gDNA Eraser). For intron retention analyses, the cDNA samples were subjected to qPCR analyses with primers designed for the retained introns (In) and one if the adjacent exons (All), the IR ratios (Inclusive %) were calculated as 100% * 2 [Ct(All) − Ct(In)]. For exon skipping analyses, the same cDNA samples were subjected to PCR amplification with primers corresponding to the flanking exons of the alternatively spliced exons and the PCR products were analyzed by TBE gel electrophoresis and quantitated by Kodak Image Station 4000R, the exon skipping ratios (Inclusive %) were calculated as 100% * Intensity(Inclusive)/[Intensity(Inclusive) + Intensity(Exclusive)].
Figure 1. BS69 shows a similar genomic distribution pattern as H3K36me3. Related to Figure 1.
Supplementary Material
Figure S1A. Genomic distribution of random peaks in HeLa cells.
Figure S1B. BS69 bound genes are significantly enriched for H3K36me3. P-value is calculated by hypergeometric test.
Figure S1C. BS69 co-localizes with H3K36me3 in gene bodies assessed by ChIP-qPCR. Primers for each gene were designed for the promoter (pro) and gene body (gb) regions, respectively. Data are represented as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S1D. H3K36me3 and BS69 levels in two independent SETD2 shRNA knockdown HeLa cells.
Figure 2. BS69 binds H3.3K36me3 in vitro via its PWWP domain. Related to Figure 2.
Figure S2A. GST tagged BS69 full length preferentially binds H3.3K36me3 in a histone peptide pull-down assay in vitro.
Figure S2B. Heatmap analysis of H3.3-FLAG and H3K36me3 distributions at BS69-bound TES regions. The TES regions are sorted by H3.3-FLAG ChIP-seq signal intensity. TESs are also grouped into “Plus strand” and “Minus strand” based on the orientation of transcription.
Figure S2C. Amino acid sequence alignment of the PWWP domains from human BS69, MSH6, DNMT3A, HDGF and WHSC1 proteins. Red arrows indicate the mutated amino acids, F and W, in the BS69 PWWP FW point mutant proteins used in this study.
Figure S2D. MST analyses of BS6950-401 binding to various histone peptides. No detectable interaction between BS6950-401 and the H3.1K36me3 (a), H3.1K36me0 (b) and H3.3K36me0(c) histone peptides, was observed in MST analyses.
Figure S2E. HA tagged BS69 full length protein purified from insect cells was used to bind HeLa nucleosome followed by histone H3 Western blot. Mock was performed using HA beads to inoculate with HeLa nucleosome in parallel. Immobilized BS69 protein was stained by Coomassie brilliant blue in the lower panel.
Figure S2F. Full length BS69 and the PWWP deletion mutant were expressed at comparable levels in HeLaS cells.
Figure S2G. Ectopically expressed wildtype BS69 but not the PHD and BROMO deletion mutants binds the selected BS69 targets (identified through endogenous BS69 ChIP) in HeLaS cells. Chromatin binding was determined by FLAG-ChIP and q-PCR. Data are represented as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S2H. MST analyses of BS6950-401 binding to H3.3S31PK36me3. The Kd of BS6950- 401 binding to H3.3S31PK36me3 was determined to be 1540±43.6μM.
Figure 3. BS69 interacts with splicing factors. Related to Figure 3.
Figure S3A. The associations of splicing factors with FLAG-HA-BS69 were confirmed by FLAG IP followed by Western Blot analyses using indicated antibodies.
Figure S3B. The interactions between GST-BS69 and FLAG-EFTUD2 and FLAG-SRSF1 were verified by GST pull-down assay with or without RNase A/DNase I treatment. Recombinant GST-BS69 was purified from E. coli, and recombinant FLAG-EFTUD2 and FLAG-SRSF1 were purified from Sf9 cells.
Figure S3C. Association of EFTUD2 with snRNAs. EFTUD2 was immunoprecipitated from HeLa cell extract using anti-EFTUD2 antibody and the associated RNA species were analyzed by RT-qPCR. Data are represented as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01; n.s., not significant.
Figure 4. BS69 regulates RNA splicing and induced NMD. Related to Figure 4.
Figure S4A. Two independent shRNAs effectively knocked down BS69 protein in HeLa cells.
Figure S4B. BS69 mRNA levels in the samples for RNA-seq analyses. Data from 3 biological replicates were shown.
Figure S4C. The correlation of the IR changes between two BS69 shRNAs analyzed by the scatterplot of the deltaPSI. The red dots are for 248 common events, green dots are for 342 events significant only in BS69-1, and blue dots are 779 events significant only in BS69-2.
Figure S4D: q-PCR confirmation of IR events affected by shRNA1 (a) or shRNA2 (b), respectively, but are not called by bioinformatics analysis (MATS: Multivariate Analysis of Transcript Splicing).
Figure S4E. BS69-dependent IR events are significantly enriched for BS69 binding.
Figure S4F. ChIP-qPCR confirmation of endogenous BS69 occupancies at 10 selected targets with altered AS upon BS69 KD. Data are are represented mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S4G: Left panel, BS69 binding events are identified at 5 out the 6 randomly selected IR events that regulated by BS69 KD by ChIP-q-PCR, but not identified by ChIP-seq. Right panel, in contrast, no BS69 binding was detected at another set of 6 randomly selected, BS69-independent IR sites by ChIP-q-PCR.
Figure S4H. BS69-dependent IR events are significantly enriched for H3.3-FLAG binding.
Figure S4I. BS69-dependent IR events are significantly enriched for H3K36me3 binding.
Figure S4J. ChIP-seq tag densities of BS69, H3K36me3 and H3.3-FLAG over introns that are BS69-dependent and BS69-independent. Significant differences of tag densities of all three factors were identified between BS69-dependent and -independent IR events. P-values were calculated using two-way unpaired Student’s t-test.
Figure S4K. Heatmap analysis of the mRNA levels of 219 BS69 dependent IR genes identified from genome-wide intron survey.
Figure 5. BS69 regulates intron retention and exon skipping events by antagonizing EFTUD2 through physical interaction. Related to Figure 5.
Figure S5A. Knockdown of BS69 caused decreased intron retention (IR) of target genes in HeLa cells. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S5B. Knockdown of BS69 altered the ratio of exon skipping (ES) of target genes in HeLa cells. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S5C. Validation of 4 randomly chosen IR events identified from genome-wide intron survey in HeLa. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S5D. Knockdown of BS69 caused a decrease of IR in A549 and HFL1 cell lines. The target introns were selected based on our HeLa cell data. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S5E. Western blot analysis of BS69 protein levels in cells transfected with indicated constructs. BS69 RNAi-resistant rescue constructs directed BS69 expression in BS69 KD cells to a comparable level of the endogenous BS69.
Figure S5F. Both wildtype BS69 and EFTUD2 binding defective mutant (BS69 Δ556-562) showed nuclear localization when overexpressed in HeLa cells. M2 FLAG antibody was used in immunofluorescence assay to detect FLAG tagged BS69 proteins. Data shown are representatives of n=30 cells.
Figure S5G. Knockdown of EFTUD2 caused an increase in intron retention of BS69 target genes in HeLa cells. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S5H. Knockdown of EFTUD2 altered the ratio of exon skipping (ES) of target genes in HeLa cells. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure 6. Binding H3K36me3 is important for BS69 to regulate AS. Related to Figure 6.
Figure S6A. Knockdown of SETD2 cause the alteration of intron retention (IR) of BS69 target genes in HeLa cells. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S6B and 6C. The occupancies of H3K36me3 and BS69 at selected genes with IR change were reduced in SETD2 shRNA treated HeLa cells. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S6D. The BS69 PWWP FW point mutant is expressed at a comparable level to that of the wildtype BS69 in HeLa cells.
Figure S6E. The BS69 PWWP FW point mutant showed nuclear localization when overexpressed in HeLa cells. M2 Flag antibody was used in immunofluorescence assay to detect FLAG tagged BS69 protein. Data shown are representatives of n=30 cells.
Table S1. Polypeptides identified by tandem mass spectrometry from TAP purified BS69 complex from soluble HeLaS nuclear fraction without MNase treatment. Related to Figure 3.
Table S2. Mapping Statistics of BS69 RNA-seq data. A total of 597 million pairs (199M BS69-1.KD, 204M BS69-2.KD,193M CTRL) were mapped. Sequencing mode: 100bp paired-end read, 25bp seed, 2 bp mismatch per seed allowed, anchor Length 8bp. Related to Figure 4.
Table S3. List of DNA oligonucleotides used in this study. Related to Figure 1, 2, 3, 4, 5 and 6.
Acknowledgments
We thank Dr. Zhenguo Wu (Hong Kong University of Science &Technology) for reagents. We thank Tom Misteli, Peter Stoilov, Doug Black, Gang Wang, Bing Ren, Matthew Meyerson, Jeff Settleman, Zhemin Zhang, Florian Gnad and Hank Qi for helpful discussion and suggestions. We thank Ying Zhang and Di Hu for drawing the graphical abstract. This work was supported by the “985” Program from the Chinese Ministry of Education and “973” State Key Development Program of Basic Research of China (2009CB825602, 2009CB825603) to the Epigenetics Laboratory at the Institutes of Biomedical Sciences, Fudan University Medical School, Shanghai, China. This work was also supported in part by a grant (CA118487) from the NIH and Boston Children’s Hospital funds to YS and a grant to XDF (GM049369). F.L. was supported in part by China “Thousand Youth Telants” (KHH1340001) and Shanghai “Oriental Scholar” (SHH129002) grants. R.G. was supported in part by grants from “National Natural Science Foundation of China Young Program (31000567)” H.C. was supported in part by grants from “Mingdao Plan” of Fudan University (MDJH2012029). YX was supported by an Alfred Sloan Research Fellowship. XBS was supported by CPRIT RP110471 and Welch G1719. Y.S, is American Cancer Society Research Professor.
Footnotes
Author contributions:
RG, LJZ, HC, FFJ, FL, SRM, HW, ZTW carried out essentially all the experiments described in this manuscript. JWP and YX were responsible for the bioinformatics analysis of the RNA-seq data. FFJ carried out part of IR and ES analyses. PYY was responsible for proteomics analyses while RTL, WQX and FZW carried out bioinfomatics analyses of the ChIP-seq data. HC carried out the MST experiments; HW performed BS69 ChIP-seq; ZTW expressed recombinant proteins from insect cells; SRM, JSQ, JYH and XDF contributed to RNA splicing regulation experimentations and discussion. XBS provided discussion and unpublished information on BS69 chromatin binding. FL, GYJS and YS directed all the experiments; YS conceived the project and co-wrote the manuscript with RG, FL and YX with input from GYJS, XDF and XBS.
Conflict of interest statement:
Yang Shi is a co-fonder of Constellation Pharmaceuticals, Inc. and a member of its scientific advisory board.
Accession Numbers
ChIP-seq data of BS69, H3K36me3 and H3.3-FLAG are deposited at NCBI under accession numbers GSE51672. RNA-seq data has been deposited to the NCBI Gene Expression Omnibus under the accession number GSE51261.
Supplemental information includes four figures and three tables and can be found with this article online at http://dx.doi.org/xxxxxx
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson ES, Lin CH, Xiao X, Stoilov P, Burge CB, Black DL. The cardiotonic steroid digitoxin regulates alternative splicing through depletion of the splicing factors SRSF3 and TRA2B. Rna. 2012;18:1041–1049. doi: 10.1261/rna.032912.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averbeck N, Sunder S, Sample N, Wise JA, Leatherwood J. Negative control contributes to an extensive program of meiotic splicing in fission yeast. Molecular cell. 2005;18:491–498. doi: 10.1016/j.molcel.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Bartels C, Klatt C, Luhrmann R, Fabrizio P. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO reports. 2002;3:875–880. doi: 10.1093/embo-reports/kvf172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, Wedge DC, Cooke SL, Gundem G, Davies H, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45:1479–1482. doi: 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. Coupling mRNA processing with transcription in time and space. Nature reviews Genetics. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152:1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Cremona N, Potter K, Wise JA. A meiotic gene regulatory cascade driven by alternative fates for newly synthesized transcripts. Molecular biology of the cell. 2011;22:66–77. doi: 10.1091/mbc.E10-05-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Jiang P, Park JW, Amirikian K, Wan J, Shen S, Xing Y, Carstens RP. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Molecular and cellular biology. 2012;32:1468–1482. doi: 10.1128/MCB.06536-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duns G, van den Berg E, van Duivenbode I, Osinga J, Hollema H, Hofstra RM, Kok K. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer research. 2010;70:4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. The EMBO journal. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran J, Horvath A, Godbole S, Reddy SD, Mudvari P, Ohshiro K, Cyanam D, Nair S, Fuqua SA, Polyak K, et al. RNA sequencing of cancer reveals novel splicing alterations. Scientific reports. 2013;3:1689. doi: 10.1038/srep01689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontebasso AM, Schwartzentruber J, Khuong-Quang DA, Liu XY, Sturm D, Korshunov A, Jones DT, Witt H, Kool M, Albrecht S, et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta neuropathologica. 2013;125:659–669. doi: 10.1007/s00401-013-1095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6428–6435. doi: 10.1073/pnas.0600803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Garcia BA, Kauer M, Baker SP, Shabanowitz J, Hunt DF, Allis CD. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6344–6349. doi: 10.1073/pnas.0502413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hateboer G, Gennissen A, Ramos YF, Kerkhoven RM, Sonntag-Buck V, Stunnenberg HG, Bernards R. BS69, a novel adenovirus E1A-associated protein that inhibits E1A transactivation. The EMBO journal. 1995;14:3159–3169. doi: 10.1002/j.1460-2075.1995.tb07318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. Distribution of histone H3.3 in hematopoietic cell lineages. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:574–579. doi: 10.1073/pnas.0509974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R, Yates SP, Teal DJ, Nilsson J, Prentice GA, Merrill AR, Andersen GR. Crystal structure of ADP-ribosylated ribosomal translocase from Saccharomyces cerevisiae. The Journal of biological chemistry. 2004;279:45919–45925. doi: 10.1074/jbc.M406218200. [DOI] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature genetics. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Mao G, Tong D, Huang J, Gu L, Yang W, Li GM. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. The Journal of biological chemistry. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nature genetics. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Moldon A, Malapeira J, Gabrielli N, Gogol M, Gomez-Escoda B, Ivanova T, Seidel C, Ayte J. Promoter-driven splicing regulation in fission yeast. Nature. 2008;455:997–1000. doi: 10.1038/nature07325. [DOI] [PubMed] [Google Scholar]
- Munding EM, Igel AH, Shiue L, Dorighi KM, Trevino LR, Ares M., Jr Integration of a splicing regulatory network within the meiotic gene expression program of Saccharomyces cerevisiae. Genes Dev. 2010;24:2693–2704. doi: 10.1101/gad.1977410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Parenteau J, Durand M, Morin G, Gagnon J, Lucier JF, Wellinger RJ, Chabot B, Elela SA. Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell. 2011;147:320–331. doi: 10.1016/j.cell.2011.08.044. [DOI] [PubMed] [Google Scholar]
- Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nature reviews Molecular cell biology. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe NJ, de Souza SJ. Sequence features responsible for intron retention in human. BMC genomics. 2007;8:59. doi: 10.1186/1471-2164-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaft D, Roguev A, Kotovic KM, Shevchenko A, Sarov M, Shevchenko A, Neugebauer KM, Stewart AF. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic acids research. 2003;31:2475–2482. doi: 10.1093/nar/gkg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- Shen S, Park JW, Huang J, Dittmar KA, Lu ZX, Zhou Q, Carstens RP, Xing Y. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic acids research. 2012;40:e61. doi: 10.1093/nar/gkr1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Molecular and cellular biology. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature structural & molecular biology. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco G, Grkovic S, Ansieau S. New insights into BS69 functions. The Journal of biological chemistry. 2006;281:16546–16550. doi: 10.1074/jbc.M600573200. [DOI] [PubMed] [Google Scholar]
- Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Gottgens B, Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nature structural & molecular biology. 2010;17:617–619. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wen H, Li Y, Xi Y, Jiang S, Stratton S, Peng D, Tanaka K, Ren Y, Xia Z, Wu J, et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508:263–268. doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nature communications. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- Wong JJ, Ritchie W, Ebner OA, Selbach M, Wong JW, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- Wong LH, Ren H, Williams E, McGhie J, Ahn S, Sim M, Tam A, Earle E, Anderson MA, Mann J, et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009;19:404–414. doi: 10.1101/gr.084947.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes & development. 2012;26:1209–1223. doi: 10.1101/gad.188037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. Journal of computational biology : a journal of computational molecular cell biology. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1A. Genomic distribution of random peaks in HeLa cells.
Figure S1B. BS69 bound genes are significantly enriched for H3K36me3. P-value is calculated by hypergeometric test.
Figure S1C. BS69 co-localizes with H3K36me3 in gene bodies assessed by ChIP-qPCR. Primers for each gene were designed for the promoter (pro) and gene body (gb) regions, respectively. Data are represented as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S1D. H3K36me3 and BS69 levels in two independent SETD2 shRNA knockdown HeLa cells.
Figure 2. BS69 binds H3.3K36me3 in vitro via its PWWP domain. Related to Figure 2.
Figure S2A. GST tagged BS69 full length preferentially binds H3.3K36me3 in a histone peptide pull-down assay in vitro.
Figure S2B. Heatmap analysis of H3.3-FLAG and H3K36me3 distributions at BS69-bound TES regions. The TES regions are sorted by H3.3-FLAG ChIP-seq signal intensity. TESs are also grouped into “Plus strand” and “Minus strand” based on the orientation of transcription.
Figure S2C. Amino acid sequence alignment of the PWWP domains from human BS69, MSH6, DNMT3A, HDGF and WHSC1 proteins. Red arrows indicate the mutated amino acids, F and W, in the BS69 PWWP FW point mutant proteins used in this study.
Figure S2D. MST analyses of BS6950-401 binding to various histone peptides. No detectable interaction between BS6950-401 and the H3.1K36me3 (a), H3.1K36me0 (b) and H3.3K36me0(c) histone peptides, was observed in MST analyses.
Figure S2E. HA tagged BS69 full length protein purified from insect cells was used to bind HeLa nucleosome followed by histone H3 Western blot. Mock was performed using HA beads to inoculate with HeLa nucleosome in parallel. Immobilized BS69 protein was stained by Coomassie brilliant blue in the lower panel.
Figure S2F. Full length BS69 and the PWWP deletion mutant were expressed at comparable levels in HeLaS cells.
Figure S2G. Ectopically expressed wildtype BS69 but not the PHD and BROMO deletion mutants binds the selected BS69 targets (identified through endogenous BS69 ChIP) in HeLaS cells. Chromatin binding was determined by FLAG-ChIP and q-PCR. Data are represented as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S2H. MST analyses of BS6950-401 binding to H3.3S31PK36me3. The Kd of BS6950- 401 binding to H3.3S31PK36me3 was determined to be 1540±43.6μM.
Figure 3. BS69 interacts with splicing factors. Related to Figure 3.
Figure S3A. The associations of splicing factors with FLAG-HA-BS69 were confirmed by FLAG IP followed by Western Blot analyses using indicated antibodies.
Figure S3B. The interactions between GST-BS69 and FLAG-EFTUD2 and FLAG-SRSF1 were verified by GST pull-down assay with or without RNase A/DNase I treatment. Recombinant GST-BS69 was purified from E. coli, and recombinant FLAG-EFTUD2 and FLAG-SRSF1 were purified from Sf9 cells.
Figure S3C. Association of EFTUD2 with snRNAs. EFTUD2 was immunoprecipitated from HeLa cell extract using anti-EFTUD2 antibody and the associated RNA species were analyzed by RT-qPCR. Data are represented as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01; n.s., not significant.
Figure 4. BS69 regulates RNA splicing and induced NMD. Related to Figure 4.
Figure S4A. Two independent shRNAs effectively knocked down BS69 protein in HeLa cells.
Figure S4B. BS69 mRNA levels in the samples for RNA-seq analyses. Data from 3 biological replicates were shown.
Figure S4C. The correlation of the IR changes between two BS69 shRNAs analyzed by the scatterplot of the deltaPSI. The red dots are for 248 common events, green dots are for 342 events significant only in BS69-1, and blue dots are 779 events significant only in BS69-2.
Figure S4D: q-PCR confirmation of IR events affected by shRNA1 (a) or shRNA2 (b), respectively, but are not called by bioinformatics analysis (MATS: Multivariate Analysis of Transcript Splicing).
Figure S4E. BS69-dependent IR events are significantly enriched for BS69 binding.
Figure S4F. ChIP-qPCR confirmation of endogenous BS69 occupancies at 10 selected targets with altered AS upon BS69 KD. Data are are represented mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S4G: Left panel, BS69 binding events are identified at 5 out the 6 randomly selected IR events that regulated by BS69 KD by ChIP-q-PCR, but not identified by ChIP-seq. Right panel, in contrast, no BS69 binding was detected at another set of 6 randomly selected, BS69-independent IR sites by ChIP-q-PCR.
Figure S4H. BS69-dependent IR events are significantly enriched for H3.3-FLAG binding.
Figure S4I. BS69-dependent IR events are significantly enriched for H3K36me3 binding.
Figure S4J. ChIP-seq tag densities of BS69, H3K36me3 and H3.3-FLAG over introns that are BS69-dependent and BS69-independent. Significant differences of tag densities of all three factors were identified between BS69-dependent and -independent IR events. P-values were calculated using two-way unpaired Student’s t-test.
Figure S4K. Heatmap analysis of the mRNA levels of 219 BS69 dependent IR genes identified from genome-wide intron survey.
Figure 5. BS69 regulates intron retention and exon skipping events by antagonizing EFTUD2 through physical interaction. Related to Figure 5.
Figure S5A. Knockdown of BS69 caused decreased intron retention (IR) of target genes in HeLa cells. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S5B. Knockdown of BS69 altered the ratio of exon skipping (ES) of target genes in HeLa cells. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S5C. Validation of 4 randomly chosen IR events identified from genome-wide intron survey in HeLa. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S5D. Knockdown of BS69 caused a decrease of IR in A549 and HFL1 cell lines. The target introns were selected based on our HeLa cell data. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S5E. Western blot analysis of BS69 protein levels in cells transfected with indicated constructs. BS69 RNAi-resistant rescue constructs directed BS69 expression in BS69 KD cells to a comparable level of the endogenous BS69.
Figure S5F. Both wildtype BS69 and EFTUD2 binding defective mutant (BS69 Δ556-562) showed nuclear localization when overexpressed in HeLa cells. M2 FLAG antibody was used in immunofluorescence assay to detect FLAG tagged BS69 proteins. Data shown are representatives of n=30 cells.
Figure S5G. Knockdown of EFTUD2 caused an increase in intron retention of BS69 target genes in HeLa cells. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S5H. Knockdown of EFTUD2 altered the ratio of exon skipping (ES) of target genes in HeLa cells. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure 6. Binding H3K36me3 is important for BS69 to regulate AS. Related to Figure 6.
Figure S6A. Knockdown of SETD2 cause the alteration of intron retention (IR) of BS69 target genes in HeLa cells. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S6B and 6C. The occupancies of H3K36me3 and BS69 at selected genes with IR change were reduced in SETD2 shRNA treated HeLa cells. Data are shown as mean ± SEM from 3 biological replicates, *P<0.05, **P<0.01.
Figure S6D. The BS69 PWWP FW point mutant is expressed at a comparable level to that of the wildtype BS69 in HeLa cells.
Figure S6E. The BS69 PWWP FW point mutant showed nuclear localization when overexpressed in HeLa cells. M2 Flag antibody was used in immunofluorescence assay to detect FLAG tagged BS69 protein. Data shown are representatives of n=30 cells.
Table S1. Polypeptides identified by tandem mass spectrometry from TAP purified BS69 complex from soluble HeLaS nuclear fraction without MNase treatment. Related to Figure 3.
Table S2. Mapping Statistics of BS69 RNA-seq data. A total of 597 million pairs (199M BS69-1.KD, 204M BS69-2.KD,193M CTRL) were mapped. Sequencing mode: 100bp paired-end read, 25bp seed, 2 bp mismatch per seed allowed, anchor Length 8bp. Related to Figure 4.
Table S3. List of DNA oligonucleotides used in this study. Related to Figure 1, 2, 3, 4, 5 and 6.