Abstract
The study of chromosome segregation is currently one of the most exciting research frontiers in cell biology. In this review, we discuss our current knowledge of the chromosome segregation process in Vibrio cholerae, based primarily on findings from fluorescence microscopy experiments. This bacterium is of special interest because of its eukaryotic feature of having a divided genome, a feature shared with 10% of known bacteria. We also discuss how the segregation mechanisms of V. cholerae compare with those in other bacteria, and highlight some of the remaining questions regarding the process of bacterial chromosome segregation.
Introduction
The three main events of the cell cycle are chromosome replication, chromosome segregation, and cell division. Coordination of these processes is essential for efficient maintenance of the genome during cell proliferation. Because these processes are functionally integrated, an effective understanding of any one of them is only possible in the context of the other events of the cell cycle. Therefore, in our discussion of chromosome segregation we try to make connections to chromosome replication and cell division.
In bacteria, chromosome segregation is not as well understood as chromosome replication and cell division. In eukaryotes, chromosomes can be visualized using a simple microscope, and mitosis was described more than a century ago. In contrast, visualization of chromosome dynamics in bacteria was realized only a decade or two ago with the introduction of fluorescence microscopy, which allowed visualization of smaller objects such as specific proteins and DNA loci [Webb et al., 1997]. It is now clear that even in apparently compartment-less organisms such as bacteria, there is considerable spatio-temporal order in the organization and segregation of chromosomes [Niki H et al., 2000; Possoz et al., 2012; Wang et al., 2013; Wiggins et al., 2010]. These cytological studies are also contributing to the understanding of how the segregation process is coordinated with chromosome replication and cell division. Just as great strides in molecular biology and biochemistry owe a great deal to X-ray crystallography for the discovery of DNA structure, the illumination of the inner workings of the bacterial cell in space and time, and “bacterial mitosis” in particular, owe a great deal to fluorescence microscopy.
Segregation of V. cholerae chromosomes
V. cholerae is closely related to Escherichia coli except that the V. cholerae genome is divided into two chromosomes, chr1 and chr2. The close relationship has contributed both conceptually and experimentally to studies in V. cholerae and has made it the leading system for studying chromosome dynamics in bacteria with divided genomes.
The two chromosomes of V. cholerae are circular, ~3 (chr1) and 1.1 (chr2) Mbp long. The replication systems of chr1 and chr2 are distinct from each other and regulated by replicon-specific initiators, similarly to the situation for the E. coli chromosome and one of its plasmid, P1 [Egan and Waldor, 2003]. Both chromosomes encode the three-component parABS segregation system. This system was first discovered and studied in plasmids, and is found in the chromosomes of about 70% of sequenced bacteria [Gerdes et al., 2000; Livny et al., 2007]. The parABS system allows active mobilization of centromeric sites, which comprise the parS component. ParB is a parS-specific DNA- binding protein and ParA is an ATPase that activates segregation. Although the par systems in general are highly homologous, including the ones in V. cholerae (parABS1 and parABS2) [Gerdes et al., 2000], they function in a chromosome-specific fashion [Dubarry et al., 2006; Kadoya et al., 2011; Saint-Dic et al., 2006]. Underlying this specificity are differences in parS sequences that ensure binding to cognate ParB proteins [Yamaichi et al., 2007a]. The ParB-parS complexes in turn also recognize their cognate ParA partner through specific protein-protein interactions [Radnedge et al., 1998].
The choreography of chromosome dynamics in V. cholerae has been studied in live cells by tagging chromosomal loci with either operator arrays or the plasmid ParB-parS systems [David et al., 2014; Fiebig et al., 2006; Fogel and Waldor, 2006; Srivastava and Chattoraj, 2007]. The localization patterns of parS sites (and the nearby origin region) are different for the two chromosomes. In newborn cells that are obtained under slow growth conditions, the chr1 origin ori1 is found near the old pole. After duplication, ori1 copies segregate asymmetrically; one ori1 stays at the pole of duplication and the other moves to the opposite pole in a directed fashion, a process requiring hydrolysis of ParA1-bound ATP (Figure 1) [Fogel and Waldor, 2006]. In contrast, the chr2 origin ori2 is found at midcell. After duplication, ori2 copies segregate symmetrically to cell-quarter positions in a parAB2-dependent fashion [Yamaichi et al., 2007b]. Under rapid growth conditions, cells are born with two copies of ori1, one at each pole, but with one ori2 at midcell [Srivastava and Chattoraj, 2007]. Initiation from ori1 occurs from both poles and segregation of the duplicated copies appears similar in the two halves of the cell. The old and the new pole therefore appear equally efficient in initiating replication and segregating the sisters. This is noteworthy because in Caulobacter crescentus, whose chromosomal origin also migrates from one pole to the other pole, only the origin at the old pole fires in the next rounds [Sliusarenko et al., 2011]. This is rationalized from the fact that two cell-halves of C. crescentus have distinct behavioral characteristics, which is not the case in V. cholerae. The higher number of ori1 over ori2 (ori1:ori2::2:1) in V. cholerae at fast growth rates is expected because the time to replicate the larger chr1 but not the smaller chr2 exceeds the cell generation time [Skarstad et al., 1985; Srivastava and Chattoraj, 2007; Stokke et al., 2011].
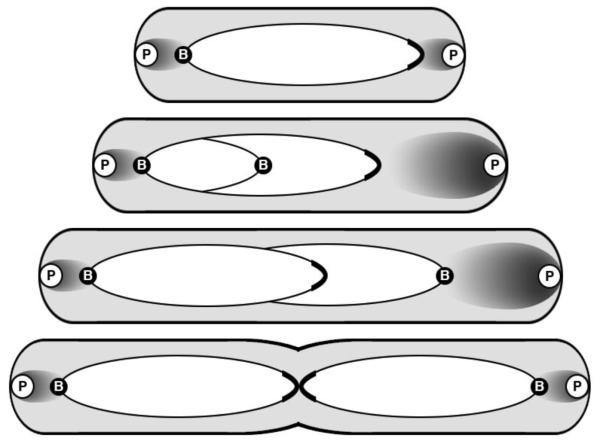
Figure 1.
Model of parABS-mediated segregation of V. cholerae chr1. The chromosome is represented by an ellipse, whose major axis has the ParB1 bound centromere (B) at one end and the replication terminus region (capped) at the other. In newborn cells, the centromere is found anchored to the old pole by the HubP (P) trans-membrane polar protein via ParB1, ParA1 and HubP interactions. ParA1 is shown as a cloud whose density is maximal near the pole. After replication initiation and duplication of the nearby centromeric region, one of the daughter centromeres migrates towards the new pole where ParA1 is present at higher concentration, shown by a larger cloud. The cloud meets the ParB1/parS1 centromeric complex when replication elongation has progressed further. The complex is then rapidly pulled to the pole by retraction of the ParA1 cloud. In predivisional (mother) cells, the terminus region is found together at midcell. Adapted from [Fogel and Waldor, 2006; Srivastava et al., 2006; Yamaichi et al., 2012].
Chromosome segregation studies have revealed that the origin is the first region that segregates, even in bacteria that do not have parABS system, and this is believed to set the course of separation for the remainder of the chromosome [Wang et al., 2013]. The region where replication terminates (ter) has also drawn special attention because its segregation makes room for cell division. Complete segregation of sister chromosomes clears the midcell area and allows septum closure without steric hindrance from the nucleoid [Bernard et al., 2010; Cambridge et al., 2013]. In newborn V. cholerae cells, the chr1 ter (ter1) is found at the new pole opposite to the pole where ori1 is found (Figure 1) [Srivastava et al., 2006]. The ter1 region then gradually migrates to the midcell in older cells, where following replication the copies remain together until the time of septum closure. In contrast, the duplicated copies of chr2 ter (ter2), which are formed at about the same time as ter1 [Rasmussen et al., 2007], can segregate away from the midcell significantly before septum formation. However, in a significant fraction of cells, ter2 copies also remain together at the midcell until the time of septum closure, similar to the ter1 copies.
The reason for the heterogeneous behavior of ter2 and the basis for the different timing of separation of ter1 and ter2 appear to be due to the differential role that the MatP/matS system plays at the two termini [Demarre et al., 2014]. The MatP/matS system was discovered in E. coli and is specifically involved in organizing the ter region [Mercier et al., 2008]. The E. coli chromosome appears to be divided into a few large domains (macrodomains), (connect to the Waldminshaus review) one of which encompasses the ter region [Mercier et al., 2008; Valens et al., 2004]. Special sites (matS) in the ter macrodomain bind MatP, which causes compaction of the region, and in E. coli, tethers the domain to the closing division septum [Bailey et al., 2014]. V. cholerae has MatP, and both ter1 and ter2 regions have matS sites [Demarre et al., 2014]. Without MatP both ter regions show segregation earlier than the onset of septal closing. The significance of the differential behavior of ter1 and ter2 remains to be understood. It is an intriguing issue for future investigation, given the timing and locations of replication termination are so similar for chr1 and chr2.
The segregation of the bulk of the chromosome has also been followed in some bacterial species, including V. cholerae [David et al., 2014]. The location of markers between ori and ter determines the overall topology of the chromosomes both during and between replication phases. In both chr1 and chr2, markers on the two replichores (the halves of the chromosome between ori and ter loci) are arranged along the longitudinal axis of the cell (Figure 2) [David et al., 2014]. The order of location of the markers follows the order in which they are present on the chromosome, and the markers segregate sequentially as they are replicated. Orientation of chr1 is dictated by parS1. When parS1 is displaced to an ectopic locus, it still migrates to the pole, causing rotation of the bulk of the chromosome. An exception is the ter1 region that retains its normal midcell location when parS1 is misplaced. As mentioned earlier, the ter region is specifically controlled by the MatP/matS system, which probably confines it to the midcell irrespective of the pull from parS1. Displacing parS also rotates the C. crescentus chromosome, including the ter region [Umbarger et al., 2011]. The rotation of ter was possibly because there is no MatP/matS system in C. crescentus.
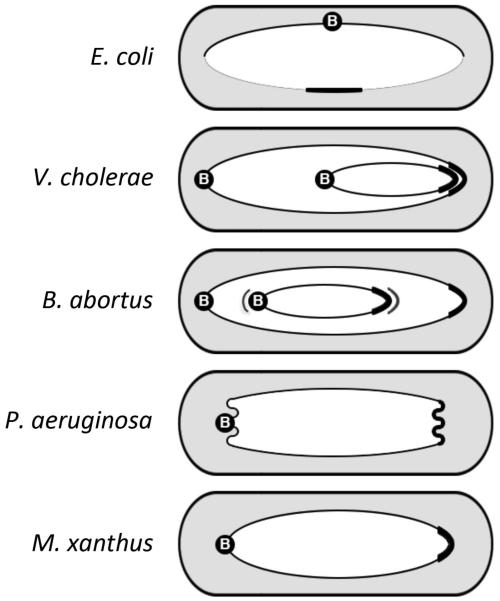
Figure 2.
Spatial arrangement of chromosomes in newborn bacterial cells. Note that the locations of origin/parS region (B) and the terminus region (capped) can vary in different bacteria. In E. coli, the terminus region is compacted by the MatP/matS system but the region also must extend to join the two arms (replichores) of the chromosome, which do not overlap under slow growth conditions. The junctions are indicated by thinner lines. In other examples, the replichores overlap over their entire length. Some other noteworthy features are as follows. The second (smaller) chromosomes in V. cholerae and in B. abortus are located differently [Deghelt et al., 2014]. The extra lines in B. abortus next to the centromere and the terminus indicate that their locations are not that fixed and move around their mean positions a bit. In P. aeruginosa, the origin and terminus regions are more compacted than in other bacteria [Vallet-Gely and Boccard F, 2013]. Both regions are also more removed from the poles. In M. xanthus, the regions are also removed from the pole. It is worth noting that centromeres are poleward in all examples (an exception is E. coli which may not have a centromere).
The location of chromosome-wide markers also indicated the relative positions of the two V. cholerae chromosomes. In newborn cells, chr1 extends over the entire cell length, whereas chr2 occupies only the cell-half containing the new pole (Figure 2). Chr2 thus overlaps the terminal half of chr1. Naked DNA occupies <1% of cell space, although the nucleoid could occupy ~75% of the cell space [Fisher et al., 2013]. Whether the overlap between the chromosomes causes any steric hindrance for their independent segregation remains to be addressed. Demixing of the two chromosomes is expected from entropic reasons [Jun and Wright, 2010]. Other forces (e.g., anchoring of parS and ter) might be at work as well.
Generality in chromosome segregation systems
Chromosome segregation has been studied in many bacterial species, to varying depth: Bacillus subtilis [Wang et al., 2014a], C. crescentus [Viollier et al., 2004], E. coli [Nielsen et al., 2006; Wang et al., 2006], Helicobacter pylori [Sharma et al., 2014], Pseudomonas aeruginosa [Vallet-Gely and Boccard, 2013], Streptomyces coelicolor (only ori region)[Jakimowicz et al., 2007] and Myxococcus xanthus (only ori region)[Harms et al., 2013]. Although these bacteria belong to different phyla, their segregation systems appear to have a lot in common. First, wherever tested the chromosomal loci are found to have fixed addresses and the overall loci organization changes during replication/segregation in a reproducible manner. Second, the chromosomal loci segregate sequentially as they are replicated [David et al., 2014; Vallet-Gely and Boccard, 2013; Viollier et al., 2004; Wang et al., 2006]. (An exception is found in E. coli where some loci stay together longer than others [Bates and Kleckner, 2005; Joshi et al., 2013].) Third, the two chromosomal arms flanking ori (the replichores) overlap over their entire length. An exception is seen in E. coli, where the replicores stay away from each other in separate cell halves under slow growth conditions (Figure 2). However, in fast growth conditions, the replichores overlap even in E. coli [Youngren et al., 2014]. Recently, the two patterns of replichore orientations, overlapping vs. non-overlapping, were found to occur at different stages of the same replication cycle in B. subtilis [Wang et al., 2014a]. When overlapping, the daughter chromosomes show mirror symmetry in the mother cell, whereas when non-overlapping, they are directly repeated [Wang et al., 2006]. Fourth, the parAB genes show a remarkable degree of conservation [Gerdes et al., 2000]. The parS sites are also conserved or have undergone minor changes [Lin and Grossman, 1998; Livny et al., 2007; Passot et al., 2012]. When present, the parABS system makes at least some contribution to the segregation process. Fifth, the parAB genes as well as multiple parS sites (up to about 10) are found close to the replication origin. The origin region is the only one that is actively segregated in a directed manner, both in parABS dependent and independent systems. In dependent systems, the parS sites migrate ahead of the origin [Fogel and Waldor, 2006; Yamaichi et al., 2012]. Finally, the segregation process is interfaced with other cell cycle events, particularly cell division, discussed below under “Chromosome segregation and the cell cycle”.
The “spindle” pole
It is to be expected that the segregation process should not only move the chromosomal loci to their designated addresses, but also retain them after the segregation completes. Although no specific host factors have been found in plasmid segregation and positioning, in the case of chromosomes there are several examples of proteins that appear to tether segregated parS regions to cell poles, including the parS region of chr1 [Yamaichi et al., 2012]. A transmembrane polar protein, HubP, directly interacts with ParA1. Since ParA1 interacts with the ParB1-parS1 complexe, the HubP-ParA1 interaction helps to retain the parS region at the pole. A similar mechanism operates in C. crescentus, where a polar protein, PopZ, directly interacts with the ParB-parS complex and anchors parS to the pole [Bowman et al., 2008; Ebersbach et al., 2008]. An anchoring mechanism was described even earlier in sporulating B. subtilis, in which there are several centromeric sites in and around the origin where the ParB analog RacA binds. RacA interacts with the polar protein DivIVA, which localizes itself by recognizing the negative curvature of the pole [Ben-Yehuda et al., 2005; Lenarcic et al., 2009; Ramamurthi and Losick, 2009]. DivIVA also participates in chromosome segregation in another Gram-positive bacterium, Corynebacterium glutamicum [Donovan et al., 2012]. In S. coelicolor, ParA interacts with a polar protein, Scy, to coordinate segregation with growth [Ditkowski et al., 2013].
Models for segregation mechanism
Several models for active segregation of chromosomes have been proposed [Wang et al., 2013]. Here, we discuss the ones that are germane to centromere movement. The centromeres of V. cholerae chr1 appear to segregate by a mitosis-like pulling mechanism [Fogel and Waldor, 2006]. A similar mechanism seems to operate in C. crescentus, where chromosome segregation has been studied in the most detail [Lim et al., 2014; Ptacin et al., 2014; Shebelut et al., 2010]. In both organisms, the origins travel from one pole to the other with the help of parABS systems and are anchored by polar proteins, HubP in V. cholerae [Yamaichi et al., 2012], and PopZ and TipN in C. crescentus [Bowman et al., 2008; Ebersbach et al., 2008; Schofield et al., 2010] (Figure 1). The poleward movement requires hydrolysis of ParA-bound ATP [Fogel and Waldor, 2006; Ptacin et al., 2010; Schofield et al., 2010]. A model invoking pulling by ParA has also been described for M. xanthus, although in this system the origins stay a bit removed from the pole (Figure 2) [Harms et al., 2013].
The pulling mechanism appears to involve retraction of the ParA “cloud” towards the new pole. ParA-ATP concentrates (forms a “cloud”) at the pole, particularly the new pole, due to its attraction for proteins, such as PopZ and HubP, which are concentrated more at the new pole (Figure 1). Interaction of the ParB bound centromeres with ParA-ATP hydrolyzes ATP, and this is believed to dissolve the cloud frontier and the retracting cloud causes a directional movement of the centromeres towards the pole. The directional retraction mechanism includes features of the diffusion-ratchet model proposed for plasmid movement, where the ParA-ATP is attracted to the nucleoid (rather than to polar proteins) due to non-specific DNA binding activity of ParA-ATP [Hwang et al., 2013; Ptacin et al., 2014; Szardenings et al., 2011; Vecchiarelli et al., 2013]. The ParA-ATP cloud serves as a matrix for ParB-bound plasmid movement because interaction of plasmid-bound ParB with ParA-ATP stimulates the release of ParA-ATP from DNA, which is likely to be coupled to ParB-stimulated ATP-hydrolysis by ParA. Rebinding of ParA to DNA requires not only ATP binding but also a conformational change of ParA-ATP, which is a slow step. The time delay prevents ParA from rebinding the nucleoid in the vicinity of the ParB-bound plasmid. The resulting ParA gradient that surrounds the ParB-bound plasmid allows it to drift with bias towards locations on the nucleoid where ParA-ATP concentrations are higher. (Note that the time delay is directly related to forming the ParA gradient, not force generation. It is the ParA gradient that is directly related to the force generation mechanism.) The model adequately explains maximal separation of plasmids away from each other.
In principle, ParB-bound chromosomal centromeres can also surf on a nucleoid associated ParA gradient, depleting ParA from the old to the new pole direction as segregation proceeds. For this mechanism to be effective, the released ParA should not readily rebind (back track) to the depleted regions of the nucleoid towards the old pole. It appears that in C. crescentus, the polar proteins PopZ and TipN sequester released ParA molecules and thereby favor directional movement of the centromeres [Ptacin et al., 2014; Schofield et al., 2010]. In V. cholerae, it is not known whether HubP has a similar sequestering function. Backtracking would also be discouraged if the reactivation rate of released ParA to a DNA-binding competent state is slow, the key feature of the diffusion- ratchet model. Prevention of backtracking must be efficient because the ParB1/parS1 complex is found at the leading edge of the nucleoid, significantly ahead of the nearby chromosomal region, and the poleward retraction of ParA1 coincides with ParB1/parS1 movement [Fogel and Waldor, 2006]. These facts were initially accommodated in a model wherein ParA1 polymerizes to form filaments (rather than a cloud), which are bound to HubP at one end and depolymerize from the ParB1/parS1 bound end [Fogel and Waldor, 2006]. At present, the evidence for ParA polymerization is generally clear only in vitro, not in vivo [Vecchiarelli et al., 2012]. Also, filament formation may not be obligatory since a concentration gradient of ParA dimers or small oligomers with maximal density at the new pole could also explain the results. The latter scenario is more likely because ParA appears as a cloud in most cases, and a recent high-resolution study in C. crescentus has argued convincingly against the existence of filaments [Lim et al., 2014]. It is noteworthy that the movement of chromosomal parS sites in the polar region is unlikely to be due to cloud formation over the nucleoid because the polar regions are usually devoid of DNA due to exclusion from polysomes [Bakshi et al., 2012] and because parS1 moves significantly ahead of the nearby chromosomal regions in V. cholerae [Fogel and Waldor, 2006]. The ParA1 cloud formation in the DNA-free polar region most likely owes to polar proteins (Figure 1).
The recent C. crescentus study also found the cloud-based diffusion-rachet model to be inadequate to account for the speed of ParB-parS movement, and proposed that the movement is aided by the chromosomal DNA itself because of its elastic nature. The idea here is that ParA bound to the chromosome pushes the ParB-parS complex from one point to another (“DNA-relay”) when the stretched (elastic) chromosomal regions relax. Thus a combination of diffusion-ratchet and intrinsic chromosome dynamics seems to account for the speed, while the burden of providing the direction of movement still rests with the ParA concentration gradient.
Which aspects of these mechanisms are general and which are particular to certain systems need to be established. The DNA-relay model was proposed on the basis that ParA is static on DNA and the ParA concentration is particularly low (90 dimers per cell) in C. crescentus [Lim et al., 2014]. If these are not generally found, other mechanisms might exist. V. cholerae chr1 is less dependent on par than the C. crescentus chromosome. The Caulobacter origin travels no more than 40% of the cell length without par [Shebelut et al., 2010; Toro et al., 2008]. In contrast, V. cholerae chr1 origin travels about 80% of cell length without par1 [Fogel and Waldor, 2006; Kadoya et al., 2011]. In V. cholerae, the role of par1 appears to be only in anchoring the centromere to the pole at the final step of segregation. V. cholerae and C. crescentus also differ in another respect: Whereas C. crescentus depends on ParB to sequester MipZ, a potent cell division inhibitor [Thanbichler and Shapiro, 2006], par1 is not essential for V. cholerae. This bacterium does not have MipZ and therefore may not need ParB1 to allow cell division.
Unlike par1, par2 (parABS2) is essential for segregation of V. cholerae chr2 [Yamaichi et al., 2007b]. Although the overall organization and segregation program of chr2 are known, the mechanism of segregation is not [Demarre et al., 2014; Fiebig et al., 2006; Fogel and Waldor, 2006; Srivastava and Chattoraj, 2007]. In particular, the mechanisms underlying the directionality of centromere movement and retention of the centromeres once they reach their destination in cell-quarter positions remain to be determined (Figure 2). Since the location of markers in chr2 follows their order on the chromosome, it should be actively organized as in chr1. It is to be noted that although chr2 is believed to have originated from a plasmid, the maintenance mode of chr2, particularly its replication, is typical of chromosomes rather than plasmids. Whereas both the timing of replication initiation and centromere movement are random for plasmids [Leonard and Helmstetter, 1988; Sengupta et al., 2010], chr2 replication starts at a fixed time in the cell cycle that might well depend on the timing of chr1 replication, and the sister origins progressively separate away from each other [Baek and Chattoraj, 2014; Fiebig A et al., 2006; Fogel and Waldor, 2005; Rasmussen et al., 2007; Srivastava and Chattoraj, 2007]. Because the plasmid diffusion-ratchet mechanism accounts for directional movement over a short range only, it may not be adequate to explain the ordered movement of chr2 over a quarter of cell-length.
Competition (incompatibility) between chr1 and chr2 for segregation
Mis-segregation of chromosomes in eukaryotes is avoided through sister-chromatid-cohesion, and sensing of tension from microtubules pulling the sister centromeres towards the opposite spindle poles. There is no competition among the chromosomes because the cell-cycle specific factors that participate in segregation, such as cohesins and microtubules are apparently not limiting. From studies in a few bacteria with a multipartite genome (V. cholerae, Burkhoderia cenocepacia [Dubarry et al., 2006], B. cereus [Deghelt et al., 2014]) it is becoming clear that the factors that control chromosome replication and segregation are chromosome specific, whereas in eukaryotes they are not. It has been well established that by encoding specific control factors, plasmids avoid competition (incompatibility) with the host as well as with other plasmids if they happen to be present in the same bacterium. This principle of coexistence of different plasmids with each other and the host chromosome has been retained in bacteria with a multipartite genome. This is to be expected since in these bacteria the largest chromosome is equivalent to the single chromosome of bacteria with undivided genomes, and the other (smaller) chromosomes appear to have originated from plasmids [Egan et al., 2005]. In the plasmid to chromosome transition, the plasmid-like strategy for avoiding incompatibility has apparently been retained. It is worth noting that overlapping binding specificities have been discerned among the ParBs and centromeres of the Burkholderiales multipartite genomes [Passot et al., 2012]. The binding affinities, however, differ widely, and this might suffice to avoid incompatibility in this system.
Par-independent segregation of chr1
About 30% of sequenced bacteria do not have par genes, including E. coli. How chromosomes move in these bacteria is not clear. Even the bacteria that do have par genes (e.g., B. subtilis) may not require them for chromosome segregation. The same can be said for V. cholerae chr1. In the absence of par1, chr1 segregation appears normal except that the mean position of centromeres becomes ~10% further removed from the pole [Fogel and Waldor, 2006; Kadoya et al., 2011]. These results indicate that the separation of the duplicated ori1 and their pole-ward migration can occur independently of the par1 system. This initial par-independent and subsequent par-dependent migration is also true for the C. crescentus chromosome except that the par-dependent step is more extensive, as discussed above [Kadoya et al., 2011; Shebelut et al., 2010]. A recent study indicates that the origin region could provide the centromeric function in the absence of par1 in V. cholerae [David et al., 2014]. This was evident when an extra ori1 was placed in chr1 distal to the native ori1. Either one of the origins could go to the pole indicating that the origin activity suffices for polar segregation. Notably, parS1 is dominant over ori1: when both are present, parS1 occupies the polar position.
Chromosome segregation and the cell cycle
Accumulating evidence supports a connection between chromosome replication, segregation and cell division in many bacteria, including in V. cholerae.
Connection of replication to segregation is suggested by the proximity of parS sites to ori [Livny et al., 2007] and co-replicational segregation of replicated regions [David et al., 2014]. The free energy of DNA polymerization could be a source of motive force for bidirectional segregation of newly replicated sister DNA [Lemon and Grossman, 2001]. Similarly, the “snap”-mediated build-up of tension due to ongoing replication could be another source of force to push sisters apart when the tension is released [Joshi et al., 2013]. As discussed above, the best evidence for replication contributing to segregation has come from recent evidence that V. cholerae ori1 can fulfill centromeric function in the absence of par1 [David et al., 2014]. We note that the role of the origin as centromere was initially tested in E. coli and was considered unlikely [Gordon et al., 2002]. Another link between replication and segregation is the fact that the V. cholerae chr2 replication initiator, RctB, regulates the chr2 par operon [Yamaichi et al., 2011].
The evidence for segregation proteins influencing replication is more direct. In B. subtilis and Streptococcus pneumonia, ParB organizes the replication origin by loading a condensin protein in the vicinity the origin [Gruber and Errington, 2009; Minnen et al., 2011; Sullivan et al., 2009; Wang et al., 2014b]. ParA controls the activity of DnaA and thereby controls replication initiation in B. subtilis, and in V. cholerae for chr1 [Kadoya et al., 2011; Scholefield et al., 2012]. V. cholerae ParB2 also controls replication of chr2 using two separate mechanisms [Venkova-Canova et al., 2013; Yamaichi et al., 2011]. It is noteworthy that Par proteins control replication by targeting the most important regulators of replication, the DnaA initiator for chr1 and the B. subtilis chromosome, and the RctB initiator for chr2. The proximity of centromeres advances replication timing in several eukaryotes [Tanaka et al., 2013]. Recently, one of the replication initiator proteins (Orc1) was shown to control centriole duplication [Hossain and Stillman, 2012]. It is only a matter of time before more connections like these will be evident.
There is also considerable evidence for coordination between segregation and cell division. Nucleoid occlusion directly prevents cytokinesis in B. subtilis and E. coli [Bernard et al., 2010; Cambridge et al., 2013]. The ter macrodomain at the cell center may also serve as a landmark for the formation of the septal ring [Bailey et al., 2014]. The best evidence so far has come from studies in C. crescentus, where sequestration of the cell division inhibitor, MipZ, by ParB is required for cell viability [Thanbichler and Shapiro, 2006]. This has been one of the most insightful findings in understanding how the sequence of the three basic cell cycle processes can be coordinated. Only upon replication initiation and segregation of the origins to the poles can MipZ be sequestered away from the midcell, allowing FtsZ polymerization at the right place (midcell) and at the right time (only after ensuring that replication and segregation have initiated). Clear evidence has also come from the finding that the ter macrodomain organizer MatP interacts with ZapB, a component of the cell division apparatus of E. coli [Espeli et al., 2012]. Since V. cholerae has MatP and both ter1 and ter2 regions contain matS sites, both chromosomes might interface with the cell division apparatus [Demarre et al., 2014].
Future
Since the discovery in plasmids of ParA and ParB proteins and the finding that they serve motor function and kinetochore formation, respectively, the search has been on for the missing components of the mitotic apparatus, the spindle. It was expected that the host might provide an equivalent of the spindle to be used as tracks for plasmid segregation. However, no convincing evidence of host factor participation was obtained. The plasmid segregation models can now explain how segregation can be achieved without requiring additional proteins, although some plasmid segregation systems do require the help from the nucleoid [Hester and Lutkenhaus, 2007; Szardenings et al., 2011]. However, there is more order in chromosome segregation than is the case for plasmid segregation, and there is evidence that additional factors (MipZ, TipN, PopZ, HubP) play crucial roles in ordered segregation of the chromosome. Initial studies using bacterial and yeast two-hybrid systems in V. cholerae have indicated that there are proteins other than HubP that interact with both ParA and ParB [Baek et al., 2014]. The interactions need to be authenticated and their physiological significance determined. These studies are likely to help understand how the different cell cycle processes are coordinated.
Segregation falls in the general category of pattern formation and theories on biological pattern formation have been considered to explain plasmid segregation. The most recent among them is the diffusion-ratchet mechanism [Meinhardt and Gierer, 2000; Vecchiarelli et al., 2010]. As mentioned earlier, the diffusion-ratchet mechanism has also been invoked to explain chromosome segregation [Lim et al., 2014; Ptacin et al., 2014]. In this context, it would be of interest to know the reactivation rate of inactive ParA in vitro, which is the essence of the diffusion-ratchet model, and how the chromosomal Par proteins compare with their plasmid counterparts in making ParA-free zones on the nucleoid. In other words, can the nucleoid be used as scaffold for its own segregation?
Chromosome segregation is a multistep process that includes not only motion but condensation (keeping the chromosome bulk together), anchoring of centromeres, clearing the nucleoids from midcell and coordination with the cell cycle. Understanding how chromosomal markers move in a directional fashion is the most pressing problem confronting the chromosome segregation field. Tracking the mover ParA in space and time and at super-resolution is crucial for a deeper understanding of the dynamics. Use of superresolution microscopy to measure the diffusion rate of ParA molecules (free and nucleoid bound, as has been done for RNA polymerase [Bakshi et al., 2013]) is in order. Efforts have already begun in C. crescentus [Lim et al., 2014; Ptacin et al., 2014] and it remains to be seen how closely the V. cholerae segregation system resembles that of C. crescentus.
Understanding the basis of par-independent segregation is more challenging. From the examples in Figure 2, it appears that movement towards the pole is an inherent feature of the origin region. Determining the role of replication in segregation appears to be the first order of business. The exciting possibility that a completely new mechanism may await discovery is also there.
The levels of Par proteins are crucial in thinking about segregation models [Lim et al., 2014], especially as overexpression of par genes is often detrimental [Kadoya et al., 2011; Mohl and Gober, 1997; Schofield et al., 2010]. The regulation of chromosomal par genes are generally not known except in some isolated cases [Casart et al., 2008]. In plasmids, the parAB genes form an autoregulated operon, but this was not the case when chromosomal par genes were tested in P. aeruginosa and in V. cholerae for chr1 [Baek et al., 2014; Bartosik et al., 2014]. Whether and how par genes of chr1 are regulated remains to be determined. Studies in both P. aeruginosa and V. cholerae also showed that Par proteins directly or indirectly control several other genes apparently unrelated to segregation. The mechanism of this regulation remains totally unknown and its understanding may reveal how Par proteins play their wider role in bacteria.
Cohesins play major and variable roles in mitosis and meiosis [Tanaka et al., 2013]. Although cohesins per se have not been found in bacteria, a similar role of lesser duration could be played by SeqA [Joshi et al., 2013; Stokke et al., 2011]. SeqA has been found to affect segregation and cell division over and above its role in replication in V. cholerae [Demarre and Chattoraj DK, 2010; Saint-Dic et al., 2008]. The condensins on the other hand (e.g., MukB in E. coli and SMC in B. subtilis) could play significant role in cell cycle events (connect to Graumann and Rybenkov reviews)[Danilova et al., 2007; Gruber and Errington, 2009; Sullivan et al., 2009]. Unexpectedly, the role of MukB does not appear critical for V. cholerae, although this remains to be further studied [Davis et al., 2014].
Cell division requires clearing the midcell region of chromosomal DNA [Bernard et al., 2010; Cambridge et al., 2013]. This is a rather intricate process, requiring not only completion of replication but also decatenation of the sisters, resolution of dimers that occasionally form when sisters recombine, and organization of the ter macrodomain. These processes are fundamental prerequisites for cytokinesis (connect to Cornet review) and, although not yet demonstrated, must be operating on both chromosomes of V. cholerae.
V. cholerae provides the opportunity to examine the behavior of two distinct chromosomal maintenance systems operating in the same environment. It is not known whether the presence of one chromosome influences the segregation of the other. We know only that preventing chr2 replication does not influence chr1 replication or segregation [Kadoya and Chattoraj, 2012]. V. cholerae strains are available with largescale rearrangement of the genome [Val et al., 2012]. Segregation studies in these cells could reveal whether the chromosomal bulk causes any steric hindrance to segregation. These cells are also of interest for investigating whether a divided genome has helped in chromosome segregation, as has been argued [Srivastava and Chattoraj, 2007].
Acknowledgements
The authors are indebted to Christophe Possoz [David et al., 2014], Francois-Xavier Barre [Demarre et al., 2014] and Xavier De Bolle [Deghelt et al., 2014] for communicating results prior to publication. The authors are also indebted to Abhishek Goel for help in drawing the figures, and to David Lane, Christophe Possoz, Anthony Vecchiarelli, Zemer Gitai, Michael Lichten and Michael Yarmolinsky for thoughtful comments, and Jemima Barrowman for editing the manuscript. This work was supported by the Intramural Research Program, Center for Cancer Research, The National Cancer Institute.
References
- Baek JH, Chattoraj DK. Chromosome I controls chromosome II replication in Vibrio cholerae. PLoS Genet. 2014;10:e1004184. doi: 10.1371/journal.pgen.1004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek JH, Rajagopala SV, Chattoraj DK. Chromosome segregation proteins of Vibrio cholerae as transcription regulators. MBio. 2014;5:e01061–01014. doi: 10.1128/mBio.01061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MW, Bisicchia P, Warren BT, et al. Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli. PLoS Genet. 2014;10:e1004504. doi: 10.1371/journal.pgen.1004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi S, Dalrymple RM, Li W, et al. Partitioning of RNA polymerase activity in live Escherichia coli from analysis of single-molecule diffusive trajectories. Biophys J. 2013;105:2676–2686. doi: 10.1016/j.bpj.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi S, Siryaporn A, Goulian M, et al. Superresolution imaging of ribosomes and rna polymerase in live Escherichia coli cells. Mol Microbiol. 2012;85:21–38. doi: 10.1111/j.1365-2958.2012.08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosik AA, Glabski K, Jecz P, et al. Transcriptional profiling of parA and parB mutants in actively dividing cells of an opportunistic human pathogen Pseudomonas aeruginosa. PLoS One. 2014;9:e87276. doi: 10.1371/journal.pone.0087276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Kleckner N. Chromosome and replisome dynamics in Escherichia coli: Loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Fujita M, Liu XS, et al. Defining a centromere-like element in Bacillus subtilis by identifying the binding sites for the chromosome-anchoring protein RacA. Mol Cell. 2005;17:773–782. doi: 10.1016/j.molcel.2005.02.023. [DOI] [PubMed] [Google Scholar]
- Bernard R, Marquis KA, Rudner DZ. Nucleoid occlusion prevents cell division during replication fork arrest in Bacillus subtilis. Mol Microbiol. 2010;78:866–882. doi: 10.1111/j.1365-2958.2010.07369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Zhu J, et al. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambridge J, Blinkova A, Magnan D, et al. A replication-inhibited un-segregated nucleoid at mid-cell blocks Z-ring formation and cell division independently of SOS and the SlmA nucleoid occlusion protein in Escherichia coli. J Bacteriol. 2013 doi: 10.1128/JB.01230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casart Y, Gamero E, Rivera-Gutierrez S, et al. Par genes in Mycobacterium bovis and Mycobacterium smegmatis are arranged in an operon transcribed from "siggc" promoters. BMC Microbiol. 2008;8:51. doi: 10.1186/1471-2180-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova O, Reyes-Lamothe R, Pinskaya M, et al. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol Microbiol. 2007;65:1485–1492. doi: 10.1111/j.1365-2958.2007.05881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David A, Demarre G, Muresan L, et al. The two cis-acting sites, parS1 and oriC1, contribute to the longitudinal organisation of Vibrio cholerae chromosome i. PLoS Genet. 2014;10:e1004448. doi: 10.1371/journal.pgen.1004448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deghelt M, Mullier C, Sternon J-F, et al. G1-arrested newborn cells are the predominant infcctious form of the pathogen brucella abortus. Nature Communications. 2014;5:4366. doi: 10.1038/ncomms5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarre G, Chattoraj DK. DNA adenine methylation is required to replicate both Vibrio cholerae chromosomes once per cell cycle. PLoS Genet. 2010;6:e1000939. doi: 10.1371/journal.pgen.1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarre G, Galli E, Muresan L, et al. Differential management of the replication terminus regions of the two Vibrio cholerae chromosomes during cell division. PLoS Genet. 2014;10:e1004557. doi: 10.1371/journal.pgen.1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditkowski B, Holmes N, Rydzak J, et al. Dynamic interplay of ParA with the polarity protein, Scy, coordinates the growth with chromosome segregation in Streptomyces coelicolor. Open Biol. 2013;3:130006. doi: 10.1098/rsob.130006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan C, Sieger B, Kramer R, et al. A synthetic Escherichia coli system identifies a conserved origin tethering factor in actinobacteria. Mol Microbiol. 2012;84:105–116. doi: 10.1111/j.1365-2958.2012.08011.x. [DOI] [PubMed] [Google Scholar]
- Dubarry N, Pasta F, Lane D. Parabs systems of the four replicons of Burkholderia cenocepacia: New chromosome centromeres confer partition specificity. J Bacteriol. 2006;188:1489–1496. doi: 10.1128/JB.188.4.1489-1496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Briegel A, Jensen GJ, et al. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ES, Fogel MA, Waldor MK. Divided genomes: Negotiating the cell cycle in prokaryotes with multiple chromosomes. Mol Microbiol. 2005;56:1129–1138. doi: 10.1111/j.1365-2958.2005.04622.x. [DOI] [PubMed] [Google Scholar]
- Egan ES, Waldor MK. Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell. 2003;114:521–530. doi: 10.1016/s0092-8674(03)00611-1. [DOI] [PubMed] [Google Scholar]
- Espeli O, Borne R, Dupaigne P, et al. A MatP-divisome interaction coordinates chromosome segregation with cell division in Escherichia coli. EMBO J. 2012;31:3198–3211. doi: 10.1038/emboj.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig A, Keren K, Theriot JA. Fine-scale time-lapse analysis of the biphasic, dynamic behaviour of the two Vibrio cholerae chromosomes. Mol Microbiol. 2006;60:1164–1178. doi: 10.1111/j.1365-2958.2006.05175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JK, Bourniquel A, Witz G, et al. Four-dimensional imaging of Escherichia coli nucleoid organization and dynamics in living cells. Cell. 2013;153:882–895. doi: 10.1016/j.cell.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel MA, Waldor MK. Distinct segregation dynamics of the two Vibrio cholerae chromosomes. Mol Microbiol. 2005;55:125–136. doi: 10.1111/j.1365-2958.2004.04379.x. [DOI] [PubMed] [Google Scholar]
- Fogel MA, Waldor MK. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 2006;20:3269–3282. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Moller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: Surprises from phylogeny. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- Gordon GS, Shivers RP, Wright A. Polar localization of the Escherichia coli oriC region is independent of the site of replication initiation. Mol Microbiol. 2002;44:501–507. doi: 10.1046/j.1365-2958.2002.02901.x. [DOI] [PubMed] [Google Scholar]
- Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- Harms A, Treuner-Lange A, Schumacher D, et al. Tracking of chromosome and replisome dynamics in Myxococcus xanthus reveals a novel chromosome arrangement. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester CM, Lutkenhaus J. Soj (ParA) DNA binding is mediated by conserved arginines and is essential for plasmid segregation. Proc Natl Acad Sci U S A. 2007;104:20326–20331. doi: 10.1073/pnas.0705196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M, Stillman B. Meier-gorlin syndrome mutations disrupt an Orc1 Cdk inhibitory domain and cause centrosome reduplication. Genes Dev. 2012;26:1797–1810. doi: 10.1101/gad.197178.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LC, Vecchiarelli AG, Han YW, et al. ParA-mediated plasmid partition driven by protein pattern self-organization. EMBO J. 2013;32:1238–1249. doi: 10.1038/emboj.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimowicz D, Zydek P, Kois A, et al. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol Microbiol. 2007;65:625–641. doi: 10.1111/j.1365-2958.2007.05815.x. [DOI] [PubMed] [Google Scholar]
- Joshi MC, Magnan D, Montminy TP, et al. Regulation of sister chromosome cohesion by the replication fork tracking protein SeqA. PLoS Genet. 2013;9:e1003673. doi: 10.1371/journal.pgen.1003673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Wright A. Entropy as the driver of chromosome segregation. Nat Rev Microbiol. 2010;8:600–607. doi: 10.1038/nrmicro2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoya R, Baek JH, Sarker A, et al. Participation of chromosome segregation protein ParA1 of Vibrio cholerae in chromosome replication. J Bacteriol. 2011;193:1504–1514. doi: 10.1128/JB.01067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoya R, Chattoraj DK. Insensitivity of chromosome I and the cell cycle to blockage of replication and segregation of Vibrio cholerae chromosome II. MBio. 2012;3:00067–00012. doi: 10.1128/mBio.00067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD. The extrusion-capture model for chromosome partitioning in bacteria. Genes Dev. 2001;15:2031–2041. doi: 10.1101/gad.913301. [DOI] [PubMed] [Google Scholar]
- Lenarcic R, Halbedel S, Visser L, et al. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AC, Helmstetter CE. Replication patterns of multiple plasmids coexisting in Escherichia coli. J Bacteriol. 1988;170:1380–1383. doi: 10.1128/jb.170.3.1380-1383.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HC, Surovtsev IV, Beltran BG, et al. Evidence for a DNA-relay mechanism in parABS-mediated chromosome segregation. Elife. 2014:e02758. doi: 10.7554/eLife.02758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Livny J, Yamaichi Y, Waldor MK. Distribution of centromere-like parS sites in bacteria: Insights from comparative genomics. J Bacteriol. 2007:189, 8693–8703. doi: 10.1128/JB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt H, Gierer A. Pattern formation by local self-activation and lateral inhibition. Bioessays. 2000;22:753–760. doi: 10.1002/1521-1878(200008)22:8<753::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Mercier R, Petit MA, Schbath S, et al. The MatP/matS site-specific system organizes the terminus region of the Escherichia coli chromosome into a macrodomain. Cell. 2008;135:475–485. doi: 10.1016/j.cell.2008.08.031. [DOI] [PubMed] [Google Scholar]
- Minnen A, Attaiech L, Thon M, et al. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol Microbiol. 2011;81:676–688. doi: 10.1111/j.1365-2958.2011.07722.x. [DOI] [PubMed] [Google Scholar]
- Mohl DA, Gober JW. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- Nielsen HJ, Ottesen JR, Youngren B, et al. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol. 2006;62:331–338. doi: 10.1111/j.1365-2958.2006.05346.x. [DOI] [PubMed] [Google Scholar]
- Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14:212–223. [PMC free article] [PubMed] [Google Scholar]
- Passot FM, Calderon V, Fichant G, et al. Centromere binding and evolution of chromosomal partition systems in the Burkholderiales. J Bacteriol. 2012;194:3426–3436. doi: 10.1128/JB.00041-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possoz C, Junier I, Espeli O. Bacterial chromosome segregation. Front Biosci (Landmark Ed) 2012;17:1020–1034. doi: 10.2741/3971. [DOI] [PubMed] [Google Scholar]
- Ptacin JL, Gahlmann A, Bowman GR, et al. Bacterial scaffold directs pole-specific centromere segregation. Proc Natl Acad Sci U S A. 2014;111:E2046–2055. doi: 10.1073/pnas.1405188111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Lee SF, Garner EC, et al. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12:791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radnedge L, Youngren B, Davis M, et al. Probing the structure of complex macromolecular interactions by homolog specificity scanning: The P1 and P7 plasmid partition systems. EMBO J. 1998;17:6076–6085. doi: 10.1093/emboj/17.20.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci U S A. 2009;106:13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T, Jensen RB, Skovgaard O. The two chromosomes of Vibrio cholerae are initiated at different time points in the cell cycle. EMBO J. 2007;26:3124–3131. doi: 10.1038/sj.emboj.7601747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Dic D, Frushour BP, Kehrl JH, et al. A ParA homolog selectively influences positioning of the large chromosome origin in Vibrio cholerae. J Bacteriol. 2006;188:5626–5631. doi: 10.1128/JB.00250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Dic D, Kehrl J, Frushour B, et al. Excess SeqA leads to replication arrest and a cell division defect in Vibrio cholerae. J Bacteriol. 2008;190:5870–5878. doi: 10.1128/JB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield WB, Lim HC, Jacobs-Wagner C. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J. 2010;29:3068–3081. doi: 10.1038/emboj.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholefield G, Errington J, Murray H. Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 2012;31:1542–1555. doi: 10.1038/emboj.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta M, Nielsen HJ, Youngren B, et al. P1 plasmid segregation: Accurate redistribution by dynamic plasmid pairing and separation. J Bacteriol. 2010;192:1175–1183. doi: 10.1128/JB.01245-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Kamran M, Verma V, et al. Intracellular locations of replication proteins and the origin of replication during chromosome duplication in the slowly growing human pathogen Helicobacter pylori. J Bacteriol. 2014;196:999–1011. doi: 10.1128/JB.01198-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shebelut CW, Guberman JM, van Teeffelen S, et al. Caulobacter chromosome segregation is an ordered multistep process. Proc Natl Acad Sci U S A. 2010;107:14194–14198. doi: 10.1073/pnas.1005274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K, Steen HB, Boye E. Escherichia coli DNA distributions measured by flow cytometry and compared with theoretical computer simulations. J Bacteriol. 1985;163:661–668. doi: 10.1128/jb.163.2.661-668.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T, et al. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava P, Chattoraj DK. Selective chromosome amplification in Vibrio cholerae. Mol Microbiol. 2007;66:1016–1028. doi: 10.1111/j.1365-2958.2007.05973.x. [DOI] [PubMed] [Google Scholar]
- Srivastava P, Fekete RA, Chattoraj DK. Segregation of the replication terminus of the two Vibrio cholerae chromosomes. J Bacteriol. 2006;188:1060–1070. doi: 10.1128/JB.188.3.1060-1070.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokke C, Waldminghaus T, Skarstad K. Replication patterns and organization of replication forks in Vibrio cholerae. Microbiology. 2011;157:695–708. doi: 10.1099/mic.0.045112-0. [DOI] [PubMed] [Google Scholar]
- Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParP-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szardenings F, Guymer D, Gerdes K. ParA ATPases can move and position DNA and subcellular structures. Curr Opin Microbiol. 2011;14:712–718. doi: 10.1016/j.mib.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Tanaka TU, Clayton L, Natsume T. Three wise centromere functions: See no error, hear no break, speak no delay. EMBO Rep. 2013;14:1073–1083. doi: 10.1038/embor.2013.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Toro E, Hong SH, McAdams HH, et al. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci U S A. 2008;105:15435–15440. doi: 10.1073/pnas.0807448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbarger MA, Toro E, Wright MA, et al. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell. 2011;44:252–264. doi: 10.1016/j.molcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Val ME, Skovgaard O, Ducos-Galand M, et al. Genome engineering in Vibrio cholerae: A feasible approach to address biological issues. PLoS Genet. 2012;8:e1002472. doi: 10.1371/journal.pgen.1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valens M, Penaud S, Rossignol M, et al. Macrodomain organization of the Escherichia coli chromosome. EMBO J. 2004;23:4330–4341. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet-Gely I, Boccard F. Chromosomal organization and segregation in Pseudomonas aeruginosa. PLoS Genet. 2013;9:e1003492. doi: 10.1371/journal.pgen.1003492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli AG, Han YW, Tan X, et al. ATP control of dynamic P1 ParA-DNA interactions: A key role for the nucleoid in plasmid partition. Mol Microbiol. 2010;78:78–91. doi: 10.1111/j.1365-2958.2010.07314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli AG, Hwang LC, Mizuuchi K. Cell-free study of F plasmid partition provides evidence for cargo transport by a diffusion-ratchet mechanism. Proc Natl Acad Sci U S A. 2013;110:E1390–1397. doi: 10.1073/pnas.1302745110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli AG, Mizuuchi K, Funnell BE. Surfing biological surfaces: Exploiting the nucleoid for partition and transport in bacteria. Mol Microbiol. 2012;86:513–523. doi: 10.1111/mmi.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkova-Canova T, Baek JH, Fitzgerald PC, et al. Evidence for two different regulatory mechanisms linking replication and segregation of Vibrio cholerae chromosome II. PLoS Genet. 2013;9:e1003579. doi: 10.1371/journal.pgen.1003579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci U S A. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu X, Possoz C, et al. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 2006;20:1727–1731. doi: 10.1101/gad.388406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Montero Llopis P, Rudner DZ. Organization and segregation of bacterial chromosomes. Nat Rev Genet. 2013;14:191–203. doi: 10.1038/nrg3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Montero Llopis P, Rudner DZ. Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc Natl Acad Sci U S A. 2014a doi: 10.1073/pnas.1407461111. PMID: 25071173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tang OW, Riley EP, et al. The SMC condensin complex is required for origin segregation in Bacillus subtilis. Curr Biol. 2014b;24:287–292. doi: 10.1016/j.cub.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CD, Teleman A, Gordon S, et al. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- Wiggins PA, Cheveralls KC, Martin JS, et al. Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc Natl Acad Sci U S A. 2010;107:4991–4995. doi: 10.1073/pnas.0912062107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y, Bruckner R, Ringgaard S, et al. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev. 2012;26:2348–2360. doi: 10.1101/gad.199869.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y, Fogel MA, McLeod SM, et al. Distinct centromere-like parS sites on the two chromosomes of vibrio spp. J Bacteriol. 2007a;189:5314–5324. doi: 10.1128/JB.00416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y, Fogel MA, Waldor MK. Par genes and the pathology of chromosome loss in Vibrio cholerae. Proc Natl Acad Sci U S A. 2007b;104:630–635. doi: 10.1073/pnas.0608341104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y, Gerding MA, Davis BM, et al. Regulatory cross-talk links Vibrio cholerae chromosome II replication and segregation. PLoS Genet. 2011;7:e1002189. doi: 10.1371/journal.pgen.1002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren B, Nielsen HJ, Jun S, et al. The multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymer. Genes Dev. 2014;28:71–84. doi: 10.1101/gad.231050.113. [DOI] [PMC free article] [PubMed] [Google Scholar]