Summary
Cells deficient in mitochondrial fusion have been shown to have defects linked to the exchange of innermembrane and matrix components. Because outer-mitochondrial membrane (OMM) constituents insert directly from the cytoplasm, a role for fusion in their inter-mitochondrial transfer was unanticipated. Here we show that fibroblasts lacking the GTPases responsible for OMM fusion, Mitofusins1/2 (MFN1/2), display more heterogeneous distribution of OMM proteins. Proteins with different modes of OMM association display varying degrees of heterogeneity in Mfn1/2−/− cells and different kinetics of transfer during fusion in fusion-competent cells. Pro-apoptotic Bak exhibits marked heterogeneity, which is normalized upon expression of MFN2. Bak is critical for Bid-induced OMM permeabilization and cytochrome c release and Mfn1/2−/− cells show dysregulation of Bid-dependent apoptotic signaling. Bid sensitivity of Bak-deficient mitochondria is regained upon fusion with Bak-containing mitochondria. Thus, OMM protein distribution depends on mitochondrial fusion and is a locus of apoptotic dysfunction in conditions of fusion deficiency.
Keywords: MFN1, MFN2, mitofusin, VDAC2, Bak, Bid, hemifusion
Introduction
The various functions of mitochondria in cells are intimately connected to their shape, size, positioning and composition. Both morphology and composition are dependent on mitochondrial fusion. The primary actors in fusion are dynamin-family GTPases. In mammals, MFN1 and 2 mediate outer mitochondrial membrane (OMM) fusion (Santel and Fuller, 2001). OPA1 both maintains cristae structure and performs fusion of the inner membrane (IMM) in an MFN1- but not MFN2-dependent manner (Cipolat et al., 2004; Frezza et al., 2006; Meeusen et al., 2006; Tondera et al., 2009).
The physiological importance of properly functioning mitochondrial fusion machinery is well established. Genetic links to inherited neuropathies have been found: mutations in Mfn2 lead to Charcot-Marie Tooth types 2A and 6 (Zuchner et al., 2006; Zuchner et al., 2004) and Opa1 mutations to dominant optic atrophy (Alexander et al., 2000; Delettre et al., 2000). Deletion of either Mfn isoform or Opa1 is embryonic lethal in mice (Chen et al., 2003), and fibroblasts isolated from the Mfn1/2−/− knockout embryos display metabolic defects similar to those found in wild-type cells after down-regulation of OPA1 by siRNA (Chen et al., 2005). Mfn double-knockout in skeletal muscle results in atrophy and accumulation of mutations in mitochondrial DNA (Chen et al., 2010) and organ-specific knockouts for Mfn2 have revealed its importance in several tissues (Chen et al., 2007; Sebastian et al., 2012). Since perturbation of OPA1 is similarly detrimental as interference with MFN1 and MFN2, and because in both cases dysfunction of matrix and IMM components have been shown, mitochondrial fusion has been considered to be primarily important for matrix and IMM homeostasis.
It has been established in vivo and in vitro that OMM fusion precedes IMM fusion and that OMM fusion is separable in mammalian cells (Liu et al., 2009; Malka et al., 2005; Meeusen et al., 2004; Song et al., 2009), however the specific implications of OMM fusion remain unknown. OMM proteins are synthesized in the cytosol and associate with the mitochondria either through protein-protein interactions or by insertion into the lipid membrane through one of a number of pathways (Chacinska et al., 2009; Dukanovic and Rapaport, 2011; Walther and Rapaport, 2009). Figure 1A shows schematically the various modes of OMM association with specific examples of proteins examined in this work: A-kinase anchoring proteins (AKAPs) interact with the membrane via a hydrophobic segment at the N-terminal which may intercalate into the outer leaflet of the OMM (Ma and Taylor, 2002). Several Bcl-2 family proteins, including Bak and Bcl-xL are constitutive tail-anchored proteins of the OMM that have hydrophobic transmembrane helices near their Ctermini which insert into the membrane (Shore et al., 1995). Bak is unique in requiring the β-barrel protein, VDAC2 to unmask the hydrophobic helix for insertion, where OMP25, another tail-anchored protein is able to insert spontaneously (Setoguchi et al., 2006).
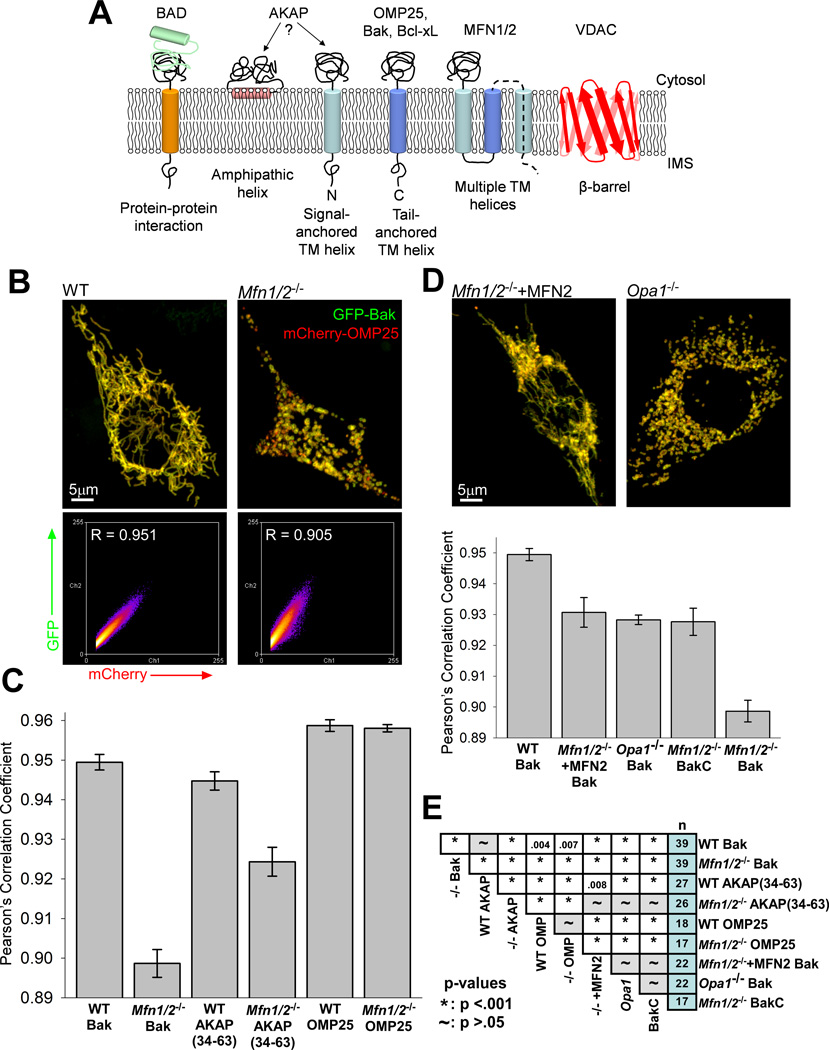
Figure 1. Fusion-deficient cells show heterogeneous distributions of OMM proteins.
(A) Various modes of anchorage of proteins at the OMM, with specific proteins discussed in this work noted (adapted from (Walther and Rapaport, 2009)). (B) Representative reconstructions of 3D confocal micrographs of WT and Mfn1/2−/− cells expressing GFP-Bak and mCherry-OMP25 and the fluorescence frequency scatter plots generated from the corresponding image stacks. Automatic thresholding of the mCherry-OMP25 signal (x-axis) was used to select the relevant pixels (see Experimental Procedures). (C) Summary of the calculated Pearson’s correlation coefficients in WT and Mfn1/2−/− cells of the GFP-tagged version of the noted constructs compared with mCherry-OMP25. (D) Representative reconstructions of an MFN2-overexpressing Mfn1/2−/− cell and an Opa1−/− cell, and a summation of mean correlation coefficients for noted conditions, as in (C). (E) Table showing the significance (p-value) of each pair of the conditions shown above, and the number of cells for each condition. See also Figure S1.
OMM proteins and associated complexes are implicated in numerous signaling pathways including metabolic regulation, physical interaction with the ER which allows inter-organellar coordination and anchoring motor complexes that dynamically distribute the organelles within the cell (de Brito and Scorrano, 2010; Eisner et al., 2013; Merrill and Strack, 2014; Rowland and Voeltz, 2012; Schwarz, 2013). Many OMM proteins are central to the regulation of cell survival and death by controlling the integrity of the OMM and the release of soluble intermembrane space contents, leading to apoptosis (Sarosiek et al., 2013; Tait and Green, 2013). Since OMM constituents can directly insert to mitochondria from the cytoplasm, the role of mitochondrial fusion has never been studied in the distribution and function of the OMM proteins.
Results
Distribution of OMM proteins in Mfn1/2−/− and wild type (WT) MEFs
To explore the effects of fusion on the distribution of OMM proteins, we employed transient expression of several fluorescent protein-tagged OMM proteins—AKAP, an N-terminal anchored protein, and OMP25 and Bak which are tail-anchored—in Mfn1/2−/− and WT MEFs. (Fig1A). Because the distribution of any particular protein of the OMM is unknown, we co-expressed GFP-tagged versions of the various proteins with mCherry-OMP25 and assessed the relation between their distributions. OMP25 has been reported to insert into the OMM in the absence of ATP, mitochondrial membrane potential (ΔΨm) and cytosolic factors (Setoguchi et al., 2006), thus it was expected to be homogeneously distributed and therefore was chosen as reference.
Mfn1/2−/− MEFs transiently expressing mCherry-OMP25 and GFP-Bak together revealed a mottled appearance at 24 hours after transfection with different organelles showing varying fluorescence intensities of GFP-Bak compared to the uniform appearance in WT cells (Fig1B). A similar difference in AKAP-GFP localization was observed between Mfn1/2−/− and WT MEFs (not shown). From high-resolution, 3D confocal micrographs of these cells we could perform a quantitative analysis of the correlation of the fluorescence intensities of the two proteins and determine Pearson’s correlation coefficient using the Colocalization Threshold plugin in ImageJ (Collins, 1997–2012). The spatial distribution of AKAP-GFP and especially GFP-Bak fluorescence was significantly less well correlated with mCherry-OMP25 in Mfn1/2−/− MEFs than in WT cells (Fig1C and E). Co-expression of mCherry-OMP25 and GFP-OMP25 revealed an identically high level of correlation between the two fluorophores in both cell types (Fig1C).
Bak is normally anchored to the OMM, but requires VDAC2 to unmask its C-terminal hydrophobic sequence for membrane insertion (Setoguchi et al., 2006). To further explore the impact of membrane association type on the distribution of OMM proteins, we used a GFP construct of only the C-terminal, transmembrane segment of Bak (BakC), which does not require VDAC2 to insert into the OMM (Setoguchi et al., 2006). This construct was more evenly distributed in the Mfn1/2−/− cells than full-length Bak, though still less than Bak in WT cells, (Fig1D and E), suggesting that the distribution of VDAC2 is a compounding factor in the highly heterogeneous appearance of GFP-Bak. Collectively, the above results support a membrane-association dependent heterogeneity of the distribution of OMM proteins in cells lacking fusion.
Heterogeneity of the OMM protein distribution might result from either heterogeneity in protein import or lack of interorganellar exchange in Mfn1/2−/− MEFs or both. Previous work has indicated that GFP-tagged Bak emulates the insertion of endogenous Bak (Roy et al., 2009a; Setoguchi et al., 2006). We tested whether differences in the expression level of GFP-Bak contributed to its distinct distribution in Mfn1/2−/− and WT MEFs. The correlation coefficient shows a weak dependence on the expression levels of GFP-Bak and mCherry-OMP25 in both Mfn1/2−/− and WT cells, but expression was, on average, comparable (FigS1C). The difference in GFP-Bak distribution between Mfn1/2−/− and WT MEFs is not likely to arise from differences in protein import. GFP-Bak expression was, on average, ~10% of endogenous Bak (FigS1A&B), though cells chosen for colocalization evaluation were brighter than average and may have expressed as much as 30–50+% relative to endogenous Bak. Finally, though some mitochondria in Mfn1/2−/− cells lack ΔΨm, those organelles were also able to import GFP-Bak (FigS1D) (Setoguchi et al., 2006).
Bak distribution heterogeneity is reversed by OMM fusion activity
To confirm the importance of OMM fusion to the heterogeneity of OMM protein distribution, we performed genetic rescue in Mfn1/2−/− MEFs. MFN2 was able to largely reproduce the elongated mitochondrial morphology of the WT cells, and also to significantly rescue the distribution of GFP-Bak (Fig1D and E). Furthermore, cells lacking only one MFN isoform maintain some fusion activity (Chen et al., 2005) and also show increased homogeneity in GFP-Bak distribution compared with Mfn1/2−/− (FigS1E). We also studied Opa1−/− cells, which retain OMM but not IMM fusion activity (Song et al., 2009). These cells also showed a more homogeneous distribution of Bak than Mfn1/2−/− cells, further demonstrating that a significant fraction of the heterogeneity observed can be attributed specifically to the lack of OMM fusion (Fig1D and E).
Bak-mediated OMM permeabilization in Mfn1/2−/− and WT cells
Bak is a pro-apoptotic Bcl-2 family protein that acts downstream of Bid truncation to mediate OMM permeabilization during apoptosis, releasing cytochrome c (cyto c) and other intermembrane space factors (Wei et al., 2000). It is constitutively present in the OMM in cells containing VDAC2 and mediates the acute phase of Bid-induced OMM permeabilization, while Bax, which is commonly cytosolic in healthy cells, inserts and oligomerizes at the OMM more slowly (Roy et al., 2009a; Sarosiek et al., 2013).
To test if the observed heterogeneity of Bak distribution in Mfn1/2−/− cells has an effect on OMM permeabilization induced by truncated Bid (tBid), we transfected Mfn1/2−/− and WT cells with cyto c-GFP and performed imaging in permeabilized cells to observe the kinetics of cyto c release after tBid addition. Both Mfn1/2−/− and WT cells showed complete release of cyto c-GFP from the intermembrane space within 10 min after addition of 25 or 75nM tBid, but the kinetics differed substantially (Fig2A). Synchronizing the traces of cyto c-GFP release in response to 75nM tBid to the points of 50% release reveals the faster average kinetic of tBid response in WT cells (Fig2B). Moreover, the maximum rate of cyto c-GFP release was greater in WT cells in response to both 25 and 75nM tBid (Fig2C). Overall Bak and VDAC2 levels are somewhat reduced in Mfn1/2−/− cells (~70% of WT) (FigS2A), but the rate at which Bak reached its activated oligomeric state in response to tBid was comparable (FigS2B). The fraction of total Bak that was oligomerized at maximum response (25nM tBid, 500s) was also quite similar (WT: 73%, Mfn1/2−/−: 71%), as was the quantity of released cyto c (FigS2C), similar to previous results with Mfn1−/− cells (Frezza et al., 2006). Bax was not a factor in cyto c release: it is similarly expressed in both cell types and showed no detectable insertion to the membrane before or after maximum tBid (FigS2D).
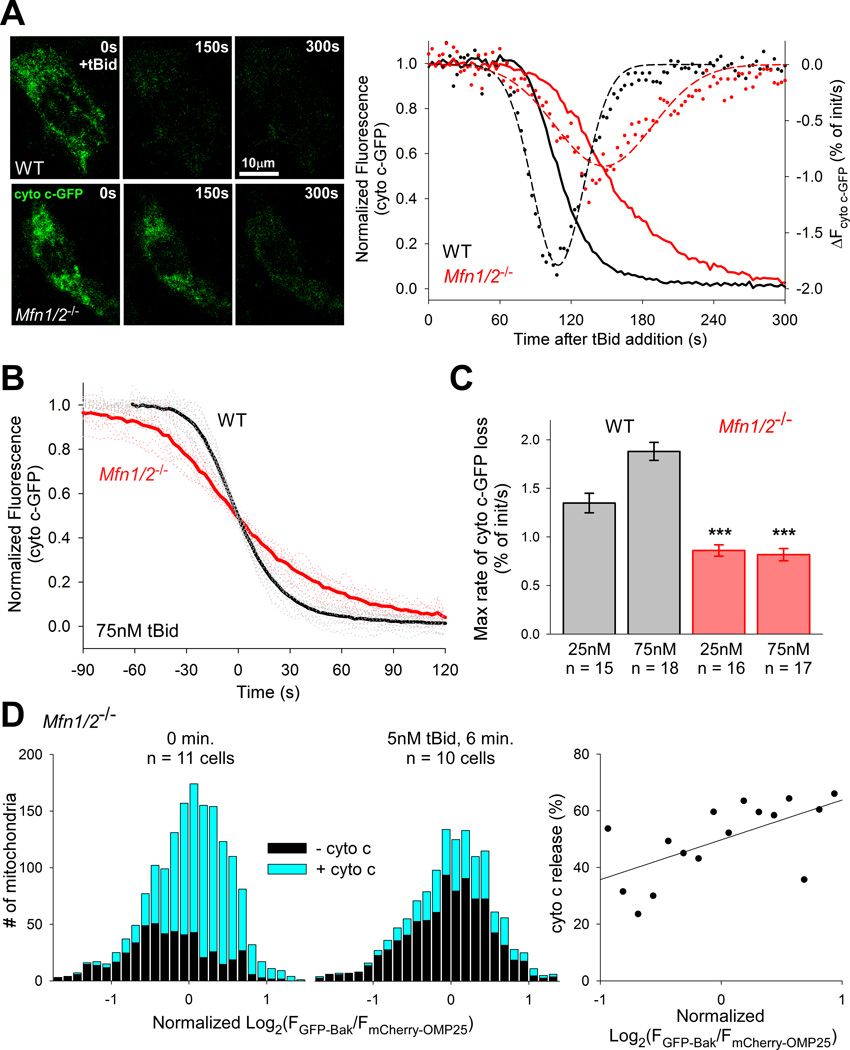
Figure 2. tBid-induced OMM permeabilization depends on the even distribution of Bak maintained by fusion.
(A) Representative still images from a time-course of cyto c-GFP release induced by 75nM tBid in WT and Mfn1/2−/− cells and graphs for these cells. Solid lines (left axis) show normalized fluorescence traces. Dots show the calculated rates of cyto c release, and dashed lines Gaussian fits to this data (right axis, R2= .955 and .858 for WT and Mfn1/2−/− respectively). (B) Cell-wise, mean traces of cyto c-GFP release from permeabilized WT and Mfn1/2−/− MEFs induced by 75nM tBid, synchronized to the 50% release point. (C) Cell-wise means of the maximum rates of cyto c-GFP release calculated as shown in (A) for the noted celltypes and tBid concetrations. ***p < .001 compared with the same concentration of tBid in WT cells. (D) Histograms of individual mitochondria from Mfn1/2−/− MEFs binned by the logarithm (base 2) of the GFPBak to mCherry-OMP25 fluorescence ratio, normalized by the mean for that cell and grouped by the presence/absence of cyto c, determined by immunofluorescence: left, 0 min. (1,622 mitochondria from 11 cells) and center, 6 min. treatment with 5nM tBid (1,279 mitochondria from 10 cells). At right, a scatter plot of the fractional change in cyto c-negative mitochondria by bin of the histograms, covering two standard deviations from the mean. A linear regression indicates an increasing probability of cyto c release with increasing GFP:mCherry ratio (slope: 14 ± 5%, p = 0.01; R2 = .382). See also Figure S2.
To confirm whether the heterogeneity in cyto c-GFP release kinetic reflects Bak distribution, we also quantified the release of cyto c by immunocytochemistry. Permeabilized Mfn1/2−/− cells expressing GFP-Bak and mCherry-OMP25 were fixed immediately or 6 min. after addition of 5nM tBid and immunostained for cyto c, then imaged as in the colocalization experiments described above. Each mitochondrion was scored for the presence/absence of cyto c immunoreaction and binned into histograms according to the ratio of GFP:mCherry. At 0 minutes, 67% of mitochondria were cyto c positive which was reduced by half after 6 minutes exposure to tBid (p < .001) (Fig2D, left, center). Comparing the fraction of cyto c positive organelles at 0 and 6 minutes in each bin of the histograms, a linear regression indicates that the likelihood of cyto c release after exposure to tBid increases with the expression of GFP-Bak (p = 0.01) (Fig2D, right). While the effect is moderate—an approximately 14% increase in likelihood of cyto c release per two-fold increase in GFP-Bak—the result is complicated by the presence of relatively large amounts of endogenous Bak. We attempted to circumvent this difficulty by acutely silencing mitofusins in Bax/Bak−/− MEFs. However, silencing of one mitofusin, MFN1 was insufficient to result in heterogeneous GFP-Bak distribution (FigS2F) and silencing of both MFN1 and MFN2 was not effective (not shown). Nonetheless, the above results support that heterogeneity in Bak distribution is coupled with an attenuated response to tBid-induced OMM permeabilization.
Rescue of tBid sensitivity of Vdac2−/− mitochondria by fusion
To demonstrate directly the effect of Bak transfer during fusion in tBid-induced OMM permeabilization, we employed polyethylene glycol (PEG) cell fusion of WT and Vdac2−/− MEFs expressing different mitochondrial-targeted fluorescent proteins. Vdac2−/− cells are unable to insert Bak to the OMM, which leaves them insensitive to tBid (Roy et al., 2009a). Hybrids of WT and Vdac2−/− cells that showed partly hybridized mitochondria were recorded after permeabilization and addition of 25nM tBid to visualize membrane depolarization by TMRE fluorescence, which closely follows cyto c release in the presence of oligomycin (Madesh et al., 2002). There were clear differences in the tBid sensitivity of the three populations of mitochondria in such cells (Fig3A). Looking at mitochondria that could be identified clearly as individual units (a small fraction of the total) from six such cell hybrids, 31 of 33 hybridized mitochondria were depolarized at 10 minutes post-tBid addition, compared with just 7 of 37 pure Vdac2−/− mitochondria and 30 of 30 pure WT (Fig3B). In addition to Bak, other OMM factors relevant for the Bid pathway such as VDAC2 or MTCH2 are likely to be transferred during fusion. However, Bak−/− MEFs are also completely insensitive to tBid (FigS3A) while expressing comparable levels of VDAC2 as WT cells (FigS3B), indicating that VDAC2 cannot support tBid-induced OMM permeabilization independent of Bak. Furthermore, MTCH2, another important factor in the tBid pathway (Shamas-Din et al., 2013; Zaltsman et al., 2010), is found at comparable levels in Vdac2−/− and WT cells (FigS3C). Thus, Bak transfer by fusion likely is the primary cause of the rescue of tBid sensitivity in the hybrid mitochondria.
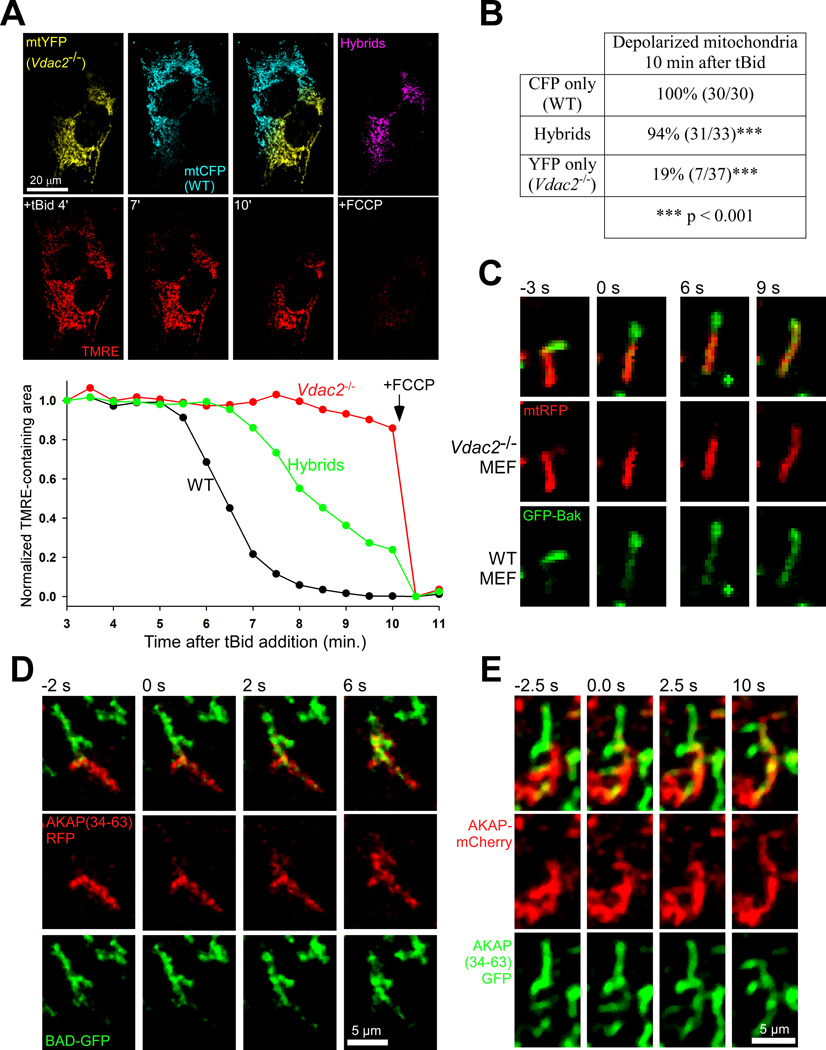
Figure 3. Rescue of tBid sensitivity to Vdac2−/− mitochondria by Bak transfer during fusion.
(A) Representative example of a PEG-induced cell hybrid of Vdac2−/− and WT MEFs expressing mtYFP and mtCFP, respectively. The accompanying graph shows the time course of ΔΨm loss from each mitochondrial population as the normalized area of mitochondria containing TMRE signal. (B) Summary of individual mitochondria identified in six WT- Vdac2−/− cell hybrids. (C) Image sequence showing the transfer of GFPBak during a fusion event in a WT- Vdac2−/− cell hybrid, expressing GFP-Bak and mtRFP, respectively. OMM fusion and Bak transfer, initiated by 0s, precedes matrix fusion by at least 6 seconds. (D) Time lapse images of a fusion event showing the transfer of fluorescent proteins between mitochondria expressing BAD-GFP and AKAP(34–63)-RFP in a cell fusion experiment with H9c2 myoblasts; fusion can first be observed at 0s. (E) Transfer of a full-length AKAP construct during a fusion event in a hybrid of H9c2 cells expressing (full-length) AKAP-mCherry and AKAP(34–63)-GFP. See also Figure S3.
Transfer of Bak and other signaling proteins by OMM fusion
To directly validate Bak transfer, we fused WT and Vdac2−/− cells expressing GFP-Bak and matrix-targeted mRFP (mtRFP), respectively and recorded the process of fusion in intact cells. By this method, we were able to directly observe the transfer of GFP-Bak during a fusion of WT and Vdac2−/− mitochondria (Fig3C). In summary, we have shown that Bak is efficiently transferred during OMM fusion, maintaining an even distribution of Bak across the mitochondria and that the evenness of Bak (and possibly of other OMM proteins) has functional consequences for the OMM-permeabilization stage of apoptosis.
Besides VDAC2 and Bak, numerous other signaling proteins and complexes reside at the OMM. One such complex contains BAD, another proapoptotic, Bcl-2 family protein that also interacts with glucose metabolism enzymes and AKAPs that likely provide a scaffold for the phospho-regulation of BAD (Danial et al., 2003; Roy et al., 2009b). We were able to observe the rapid transfer of these signaling molecules during individual fusion events by confocal microscopy: H9c2 myoblasts expressing BAD-GFP were fused by PEG with cells expressing RFP attached to the 30 amino-acid targeting sequence of AKAP, amino acids 34–63. (Fig3D) and H9c2 expressing full-length AKAP1-mCherry were fused with AKAP(34–63)-GFP expressing cells (Fig3E). Thus, OMM fusion supports the transfer of several signaling molecules and regulators of multiple OMM permeabilization pathways.
Dynamics of protein transfer during fusion in human skeletal muscle cells
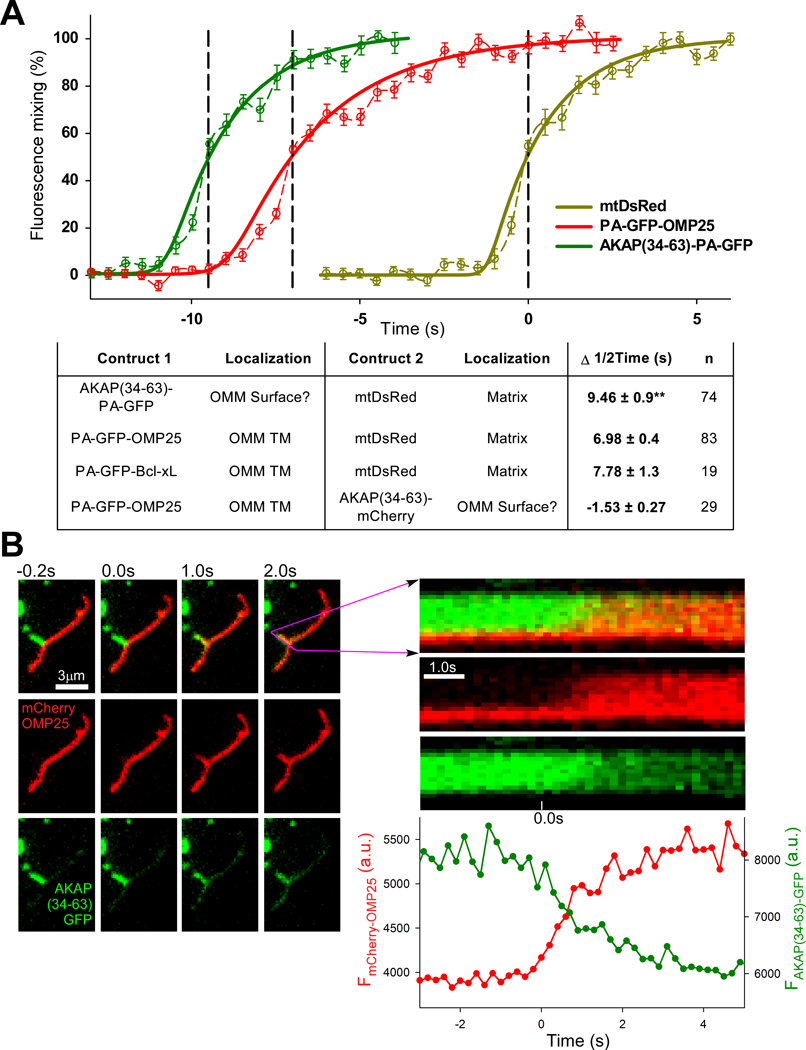
The proteins transferred among individual mitochondria during OMM fusion have different membrane anchorage (Fig1A). This might result in differential lateral mobility and transfer, especially during short lasting fusion events (kiss-and-run) (Liu et al., 2009). Therefore, we sought to visualize the mixing dynamics of proteins with different OMM attachments during fusion. Employing photoactivateable-GFP (PA-GFP) fusion proteins (OMP25 and AKAP(34–63)) at the OMM in combination with DsRed in the matrix allows for simultaneous photobleaching of DsRed and photoactivation of PA-GFP. With this technique we are able to clearly separate the events of OMM and matrix fusion FigS4). We found that, of the cell types tested, human skeletal muscle derived cells (HUSM) showed the longest average gap between the transfer of OMM and matrix contents during fusion, making it a useful system for examination of the mixing dyamics. By measuring the fluorescence in the PA-GFP-accepting mitochondrion for each construct during fusion, we generated average traces that revealed distinct transfer kinetics of each membrane attachment (Fig4A). mtDsRed, which is freely diffusing in the matrix, evinced noticeably faster transfer than the membrane-bound OMM constructs, though the transfer of the OMM proteins was also completed within 10s. Interestingly, the various OMM proteins showed distinct kinetics of transfer and also different characteristic durations before matrix mixing of between 6 and 10 seconds. Combining PA-GFP-OMP25 with AKAP(34–63)-mCherry in a similar experiment also revealed a small gap of 1.5s between the 50% transfer times.
Figure 4. Comparison of the dynamics of transfer of different OMM proteins.
(A) Graph showing the mean kinetics of transfer (dashed lines) of AKAP(34–63)-PA-GFP (green) and PA-GFP-OMP25 (red) measured in separate photoactivation/bleaching experiments with mtDsRed (yellow) in HUSM cells. For each event the interval between 50% transfer of the OMM construct and mtDsRed was determined and the averages were used to merge the graphs together. Sigmoidal fits for each probe are also shown (solid lines). Average intervals for tested combinations of OMM and matrix-targeted proteins are summarized in the table. ** p =.007 comparing AKAP(34–63)-PA-GFP and PA-GFP-OMP25. (B) Time-lapse images of a single fusion event from a cell fusion experiment with mitochondria expressing AKAP(34–63)-GFP and mCherry-OMP25 at high temporal resolution, with accompanying line-scan of the fluorescence profile along the length of the AKAP-donor mitochondrion (length, vertical axis; time, horizontal axis), as well as a graph of the mean fluorescence in the AKAP-donor. The initiation points of OMP and AKAP(34–63) are indistinguishable. See also Figure S4.
Observing these differences between the midpoint transfer times of OMP25 and AKAP in the HUSM cells raised the question of whether this could be the result of AKAP binding to the OMM by intercalating into the outer leaflet only, as has been proposed (Ma and Taylor, 2002). In this scenario, the difference could be the result of fusion proceeding through a hemifusion intermediate, whereby the outer leaflets of the two fusing organelles become continuous before the inner leaflets (Chernomordik and Kozlov, 2005; Yang and Huang, 2002). To distinguish such a hemifusion step, we combined AKAP(34–63) PA-GFP with mCherry-OMP25 in a single cell and fused AKAP-GFP and mCherry-OMP25 expressing cells.
Two ways to resolve a possible potential OMM hemifusion step were explored. First, hemifusion may sometimes be reversed without proceeding to fusion of the inner leaflet. In that case, during a hemifusion event between a mitochondrion expressing AKAP and one with OMP25 only AKAP would be transferred, but we were unable to find such an occurrence either by photoactivation or the cell fusion assay. Second, by performing imaging at high temporal resolution, we attempted to see a separation of the initiations of AKAP and OMP25 transfer. Our highest practical temporal resolution of 5s−1 proved unable to resolve such a separation in a cell fusion model (Fig4B). Based on the relatively small variation in transfer kinetics of OMM proteins we observed, most proteins are exchanged within 10s of mitochondria merging. Therefore most OMM proteins are effectively equilibrated by fusion events that last several seconds.
Discussion
In this study we have directly demonstrated the transfer of signaling molecules at the OMM during fusion and showed for the first time that mitochondrial fusion plays an important role in maintaining the distribution of such proteins. That the uneven distribution of Bak in cells lacking fusion activity leads to dysregulated Bid-induced OMM permeabilization is likely one example among numerous deleterious effects associated with the heterogeneity resulting from a lack of fusion. AKAP, for instance, is also less-evenly distributed in Mfn1/2−/− MEFs than WT. Further, we have shown that the extent of the heterogeneity of OMM protein distribution in Mfn1/2−/− cells varies among different proteins and that various OMM proteins also show different kinetics of transfer during fusion events in fusion-competent cells.
We employed an established model of Bak-dependent OMM permeabilization (Roy et al., 2009a) to reveal a functional defect caused by the misdistribution of OMM proteins in fusion-deficient cells. Our data implicate heterogeneity not only of Bak in apoptotic dysregulation, but also of VDAC2, a β-barrel protein of the OMM and essential factor for Bak import. β-barrel proteins, though few in number, serve essential roles, including in the import of mitochondrial proteins through the TOM complex (Dukanovic and Rapaport, 2011). It is not difficult to imagine that a lack of fusion activity could lead to a cascading loss of factors necessary for protein import, especially considering that IMM proteins also depend on the TOM complex. It has been shown that Mfn1/2−/− and Opa1−/− cells have mitochondria lacking ΔΨm and evince other bioenergetic defects (Chen et al., 2005; Chen et al., 2003) which can reduce or eliminate the organelle’s ability to import various factors, the lack of which factors may cause further degradation of the viability of the organelle, and so on. Interestingly, the data presented here indicates that for the case of Bak distribution, the energetic defects common to Mfn1/2−/− and Opa1−/− MEFs are partly overcome by the OMM fusion activity in Opa1−/− cells.
Our data also add to the understanding of the links between mitochondrial dynamics and apoptosis. Fragmentation of the mitochondria via activation of Drp1 and inhibition of MFN-mediated fusion is a well described step preceding OMM permeabilization in several models of apoptosis (Frank et al., 2001; Karbowski et al., 2004). Direct interactions of MFNs and DRP1 with Bak and Bax have been described where the functions of the dynamics-involved proteins are modulated by the apoptotic proteins and vice versa (Brooks et al., 2007; Hoppins et al., 2011; Karbowski et al., 2006). Here we show the somewhat unexpected result that the completely fragmented mitochondria of Mfn1/2−/− cells respond on average more slowly to tBid than those of WT cells, which are predominantly elongated. This appears to stem from the relative incapacity of some organelles to import Bak in Mfn1/2−/− cells, which we correlated to a delayed release of cyto c. We also showed directly that tBid sensitivity is conferred by the transfer Bak to formerly insensitive mitochondria during OMM fusion, representing a previously unrecognized signaling pathway.
Finally, we attempted to address the potential presence of a hemifusion phase that may exist either as a first step in the OMM fusion process or on its own as a separate type of fusion that involves only the outer leaflet and associated, peripheral proteins of the OMM. A hemifused state generated during DRP1-mediated fission has been proposed to promote Bax oligomerization and OMM permeabilization (Montessuit et al., 2010). While we were able to show that different OMM proteins’ membrane attachments show different kinetics of transfer during fusion, we were unable to directly observe a hemifusion step with the available tools. There remain several reasons that we are unable to resolve the hemifusion intermediate: our model membrane attachment, AKAP(34–63), may not interact exclusively with the outer leaflet, the lifetime of the hemifusion phase may be too short for our current imaging capabilities, or, even if the hemifusion phase exists that allows for diffusion of lipids, there may be a barrier to protein diffusion during this time. To address the possibility of hemifusion would require better-characterized peripheral OMM markers. However, it seems likely that the difference observed in HUSM cells between AKAP and OMP25 is the result of differences in the diffusivity of each protein in the OMM and across the fusion interface and not a discernible hemifusion stage.
Experimental Procedures
Cells
H9c2 cells (ATCC) and transformed MEFs: Mfn-KO, respective WT (provided by David Chan, and Opa1−/− (Addgene) were grown in DMEM (ATCC), 10%FBS (Gibco), Pen/Strep, L-Glutamine, Sodium Pyruvate. Vdac2−/− and respective WT MEFs (rom William Craigen) and Bak−/− and Bax/Bak−/− (from Stanley Korsmeyer) were grown in DMEM, 10% FBS, Pen/Strep, L-Glutamine, and NEAA. HUSM cultures were established from the medical waste of skeletal muscle biopsies (kindly provided by Dr. Henry Rosenberg) taken for in vitro contracture test with the approval of the Institutional Review Board (protocol #01.0601). The cells were cultured in DMEM (ATCC), 20%FBS, Pen/Strep, L-Glutamine, Sodium Pyruvate, once reaching 80% confluency, the media was replaced by identical medium supplemented with 2% FBS to allow differentiation into myotubes.
Image analysis
Image analysis and processing was performed in Spectralyzer and/or ImageJ
Calculation of Pearson’s correlation coefficient, R, for colocalization analysis was performed on 3D image stacks acquired as described above in ImageJ. Image stacks were median filtered and the mCherry-OMP25 image was automatically thresholded using the Otsu method applied to the total stack fluorescence (‘use stack histogram’ option) to create a mask image stack of the mitochondrial voxels to be considered in the colocalization analysis. The mask was applied to each median-filtered image stack and the ‘colocalization threshold’ method was run to calculate R excluding zero-zero (unmasked) voxels.
See also ‘Image Analysis’ in Supplemental Experimental Procedures.
See also Supplemental Experimental Procedures.
Supplementary Material
Highlights.
OMM proteins are more heterogeneously distributed in cells lacking OMM fusion.
OMM fusion supports the transfer of signaling molecules, including Bak, BAD and AKAP.
Bak transfer by fusion is needed for tBid-induced OMM permeabilization.
OMM protein distribution is a locus of apoptotic dysfunction in fusion deficiency.
Acknowledgements
We thank Drs. David Chan, William Craigen, Stanley Korsmeyer, Jean-Claude Martinou, Heidi McBride, Katsuyoshi Mihara, Shamim Naghdi, Manuel Rojo, Donna George and Henry Rosenberg for reagents and advice. V.E. is an awardee of the PEW Latin American Fellows Program in the Biomedical Sciences. (2008-000411-003). P.V. was supported by the Hungarian Scientific Research Fund (OTKA K105006). This work was supported by a Louis and Fannie Tolz Award to A.G. & G.H. and by an NIH grant AA017773 to G.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- Brooks C, Wei Q, Feng L, Dong G, Tao Y, Mei L, Xie ZJ, Dong Z. Bak regulates mitochondrial morphology and pathology during apoptosis by interacting with mitofusins. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11649–11654. doi: 10.1073/pnas.0703976104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. The Journal of biological chemistry. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM. Membrane hemifusion: crossing a chasm in two leaps. Cell. 2005;123:375–382. doi: 10.1016/j.cell.2005.10.015. [DOI] [PubMed] [Google Scholar]
- Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins T. Colocalization Threshold. In: Rasband WS, editor. ImageJ. Bethesda, Maryland USA: U.S. National Institutes of Health; 1997–2012. [Google Scholar]
- Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, Datta SR, Greenberg ME, Licklider LJ, Lowell BB, et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003;424:952–956. doi: 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. An intimate liaison: spatial organization of the endoplasmic reticulum-mitochondria relationship. The EMBO journal. 2010;29:2715–2723. doi: 10.1038/emboj.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- Dukanovic J, Rapaport D. Multiple pathways in the integration of proteins into the mitochondrial outer membrane. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2011;1808:971–980. doi: 10.1016/j.bbamem.2010.06.021. [DOI] [PubMed] [Google Scholar]
- Eisner V, Csordas G, Hajnoczky G. Interactions between sarco-endoplasmic reticulum and mitochondria in cardiac and skeletal muscle - pivotal roles in Ca(2)(+) and reactive oxygen species signaling. Journal of cell science. 2013;126:2965–2978. doi: 10.1242/jcs.093609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Developmental cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Hoppins S, Edlich F, Cleland MM, Banerjee S, McCaffery JM, Youle RJ, Nunnari J. The soluble form of Bax regulates mitochondrial fusion via MFN2 homotypic complexes. Molecular cell. 2011;41:150–160. doi: 10.1016/j.molcel.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Arnoult D, Chen H, Chan DC, Smith CL, Youle RJ. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J Cell Biol. 2004;164:493–499. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- Liu X, Weaver D, Shirihai O, Hajnoczky G. Mitochondrial 'kiss-and-run': interplay between mitochondrial motility and fusion-fission dynamics. The EMBO journal. 2009;28:3074–3089. doi: 10.1038/emboj.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Taylor S. A 15-residue bifunctional element in D-AKAP1 is required for both endoplasmic reticulum and mitochondrial targeting. The Journal of biological chemistry. 2002;277:27328–27336. doi: 10.1074/jbc.M201421200. [DOI] [PubMed] [Google Scholar]
- Madesh M, Antonsson B, Srinivasula SM, Alnemri ES, Hajnoczky G. Rapid kinetics of tBid-induced cytochrome c and Smac/DIABLO release and mitochondrial depolarization. The Journal of biological chemistry. 2002;277:5651–5659. doi: 10.1074/jbc.M108171200. [DOI] [PubMed] [Google Scholar]
- Malka F, Guillery O, Cifuentes-Diaz C, Guillou E, Belenguer P, Lombes A, Rojo M. Separate fusion of outer and inner mitochondrial membranes. EMBO Rep. 2005;6:853–859. doi: 10.1038/sj.embor.7400488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S, DeVay R, Block J, Cassidy-Stone A, Wayson S, McCaffery JM, Nunnari J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Meeusen S, McCaffery JM, Nunnari J. Mitochondrial fusion intermediates revealed in vitro. Science. 2004;305:1747–1752. doi: 10.1126/science.1100612. [DOI] [PubMed] [Google Scholar]
- Merrill RA, Strack S. Mitochondria: A kinase anchoring protein 1, a signaling platform for mitochondrial form and function. The international journal of biochemistry & cell biology. 2014;48C:92–96. doi: 10.1016/j.biocel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montessuit S, Somasekharan SP, Terrones O, Lucken-Ardjomande S, Herzig S, Schwarzenbacher R, Manstein DJ, Bossy-Wetzel E, Basanez G, Meda P, et al. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell. 2010;142:889–901. doi: 10.1016/j.cell.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nature reviews. Molecular cell biology. 2012;13:607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SS, Ehrlich AM, Craigen WJ, Hajnoczky G. VDAC2 is required for truncated BID-induced mitochondrial apoptosis by recruiting BAK to the mitochondria. EMBO Rep. 2009a;10:1341–1347. doi: 10.1038/embor.2009.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SS, Madesh M, Davies E, Antonsson B, Danial N, Hajnoczky G. Bad targets the permeability transition pore independent of Bax or Bak to switch between Ca2+-dependent cell survival and death. Molecular cell. 2009b;33:377–388. doi: 10.1016/j.molcel.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. Journal of cell science. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- Sarosiek KA, Chi X, Bachman JA, Sims JJ, Montero J, Patel L, Flanagan A, Andrews DW, Sorger P, Letai A. BID preferentially activates BAK while BIM preferentially activates BAX, affecting chemotherapy response. Molecular cell. 2013;51:751–765. doi: 10.1016/j.molcel.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz TL. Mitochondrial trafficking in neurons. Cold Spring Harbor perspectives in biology. 2013;5 doi: 10.1101/cshperspect.a011304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian D, Hernandez-Alvarez MI, Segales J, Sorianello E, Munoz JP, Sala D, Waget A, Liesa M, Paz JC, Gopalacharyulu P, et al. Mitofusin 2 (Mfn2) links mitochondrial and endoplasmic reticulum function with insulin signaling and is essential for normal glucose homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5523–5528. doi: 10.1073/pnas.1108220109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi K, Otera H, Mihara K. Cytosolic factor- and TOM-independent import of C-tail-anchored mitochondrial outer membrane proteins. The EMBO journal. 2006;25:5635–5647. doi: 10.1038/sj.emboj.7601438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamas-Din A, Bindner S, Zhu W, Zaltsman Y, Campbell C, Gross A, Leber B, Andrews DW, Fradin C. tBid undergoes multiple conformational changes at the membrane required for Bax activation. The Journal of biological chemistry. 2013;288:22111–22127. doi: 10.1074/jbc.M113.482109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore GC, McBride HM, Millar DG, Steenaart NA, Nguyen M. Import and insertion of proteins into the mitochondrial outer membrane. European journal of biochemistry / FEBS. 1995;227:9–18. doi: 10.1111/j.1432-1033.1995.tb20354.x. [DOI] [PubMed] [Google Scholar]
- Song Z, Ghochani M, McCaffery JM, Frey TG, Chan DC. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell. 2009;20:3525–3532. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait SW, Green DR. Mitochondrial regulation of cell death. Cold Spring Harbor perspectives in biology. 2013;5 doi: 10.1101/cshperspect.a008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, Da Cruz S, Clerc P, Raschke I, Merkwirth C, et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. The EMBO journal. 2009;28:1589–1600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DM, Rapaport D. Biogenesis of mitochondrial outer membrane proteins. Biochimica et biophysica acta. 2009;1793:42–51. doi: 10.1016/j.bbamcr.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M, Thompson CB, Korsmeyer SJ. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 2000;14:2060–2071. [PMC free article] [PubMed] [Google Scholar]
- Yang L, Huang HW. Observation of a membrane fusion intermediate structure. Science. 2002;297:1877–1879. doi: 10.1126/science.1074354. [DOI] [PubMed] [Google Scholar]
- Zaltsman Y, Shachnai L, Yivgi-Ohana N, Schwarz M, Maryanovich M, Houtkooper RH, Vaz FM, De Leonardis F, Fiermonte G, Palmieri F, et al. MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nature cell biology. 2010;12:553–562. doi: 10.1038/ncb2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchner S, De Jonghe P, Jordanova A, Claeys KG, Guergueltcheva V, Cherninkova S, Hamilton SR, Van Stavern G, Krajewski KM, Stajich J, et al. Axonal neuropathy with optic atrophy is caused by mutations in mitofusin 2. Annals of neurology. 2006;59:276–281. doi: 10.1002/ana.20797. [DOI] [PubMed] [Google Scholar]
- Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.