Abstract
Activation of precursor 25-hydroxyvitamin D3 (25D) to hormonal 1,25-dihydroxyvitamin D3 (1,25D) is a pivotal step in vitamin D physiology, catalyzed by the enzyme 25-hydroxyvitamin D-1α-hydroxylase (1α-hydroxylase). To establish new models for assessing the physiological importance of the 1α-hydroxylase-25D-axis, we used Danio rerio (zebrafish) to characterize expression and biological activity of the gene for 1α-hydroxylase (cyp27b1). Treatment of day 5 zebrafish larvae with inactive 25D (5-150 nM) or active 1,25D (0.1-10 nM) induced dose responsive expression (15-95 fold) of the vitamin D-target gene cyp24a1 relative to larvae treated with vehicle, suggesting the presence of Cyp27b1 activity. A full-length zebrafish cyp27b1 cDNA was then generated using RACE and RT-PCR methods. Sequencing of the resulting clone revealed an open reading frame encoding a protein of 505 amino acids with 54% identity to human CYP27B1. Transfection of a cyp27b1 expression vector into HKC-8, a human kidney proximal tubular epithelial cell line, enhanced intracrine metabolism of 25D to 1,25D resulting in greater than 2-fold induction of CYP24A1 mRNA expression and a 25-fold increase in 1,25D production compared to empty vector. These data indicate that we have cloned a functional zebrafish CYP27B1, representing a phylogenetically distant branch from mammals of this key enzyme in vitamin D metabolism. Further analysis of cyp27b1 expression and activity in zebrafish may provide new perspectives on the biological importance of 25D metabolism.
Keywords: CYP27B1, vitamin D, cytochrome P450, CYP24A1, metabolism
Introduction
Optimal vitamin D status has long been recognized as a key factor in the prevention of bone diseases but in the last decade it has become increasingly clear that facets of extra-skeletal physiology may also be influenced by vitamin D [6,13]. Cholecalciferol (vitamin D3) is produced by UVB photoconversion of 7-dehydrocholesterol in skin or consumed in vitamin D3 containing foods or oral supplements. Vitamin D3 undergoes a hydroxylation step in the liver yielding prohormone 25-hydroxyvitamin D3 (25D) followed by a subsequent hydroxylation (in renal and extra-renal sites) yielding the active 1,25-dihydroxyvitamin D3 (1,25D) [30]. 1,25D produced by the kidney acts in an endocrine manner to regulate calcium metabolism and is vital to bone health while the actions of 1,25D in extra-skeletal physiology appear to be dependent on tissue-specific conversion of prohormone 25-hydroxyvitamin D3 (25D) to active 1,25-dihydroxyvitamin D3 (1,25D) [26,28], with target cells expressing the enzyme 25D-1α-hydroxylase (CYP27B1) that catalyzes this reaction [26,27,28]. Coincident expression of the nuclear receptor for 1,25D (vitamin D receptor, VDR) by these cells suggests that many effects of vitamin D are mediated via an intracrine, rather than an endocrine mechanism. In this setting, given that 25D is the major circulating form of vitamin D, it is likely that impaired vitamin D status will compromise local synthesis of 1,25D and associated physiological responses [5,25]. A role for vitamin D in non-classical, extra-skeletal physiology is supported by data from various studies using mouse models. These include mice with knockout of the murine genes for Cyp27b1 [35], Vdr [21,31,60], and mice with dietary deficiency of vitamin D [32].
The actions of vitamin D in zebrafish specifically, and fish in general, remains unclear. Vitamin D metabolism has been demonstrated in fish [44,46], and it has been assumed that vitamin D contributes to the skeletal homeostasis of these animals in much the same way as other vertebrates. Although no specific requirements have been set, standard diets for zebrafish routinely include vitamin D [1,2]. The zebrafish genome project has identified putative genes for various components of the vitamin D system such as: vdr (vdra, and vdrb), cyp2r1, cyp27a1 (cyp27a1.2, and cyp27a1.4), cyp27b1, cyp24a1 and gc. Despite this abundance of genome project data for zebrafish, only a handful of reports have documented functional responses to vitamin D in these animals. To date, these studies have focused exclusively on the effects of active 1,25D upon calcium handling, vdr expression and transcriptome analysis [16,17,18,34]. By contrast, actions of precursor 25D in zebrafish are much less well understood. In the current study, we have sought to expand this by cloning the zebrafish gene for CYP27B1, which may be important for both the classical (endocrine) and non-classical (intracrine) effects of vitamin D in this organism.
As a model system, zebrafish have the advantage of being relatively inexpensive, small, fast growing organisms with a larval state that can be easily visualized and genetically manipulated [7]. As such, zebrafish are a potentially attractive model system to study vitamin D’s role in bone development and immune function. In regards to immune response to infection in particular, zebrafish can be infected with, Mycobacterium marinum [47,49,56], a mycobacterial pathogen with similarities to the Mycobacterium tuberculosis pathogen that causes tuberculosis in humans. Thus, Danio rerio (zebrafish) may provide a useful alternative animal model for studies of vitamin D and its potential role in infection and immunity.
Material and methods
Animal care
Fish were raised on a 14/10 hour light/dark cycle at 28.5°C. Embryos were maintained in a 28.5°C incubator. Experiments were performed at larval stages when male and female zebrafish cannot be distinguished. The Chancellor’s Animal Research Care Committee at the University of California, Los Angeles, approved all experiments.
qRT-PCR analysis of mRNA expression in zebrafish
Zebrafish larvae (day 5) were incubated with 25D (0 - 150 nM) or 1,25D (0 - 10 nM) for 6 hours. In the inhibitor studies, itraconazole (0, 0.1, and 1.0 μM) was added one hour prior to the six hour incubation with vehicle, 5 nM 25D or 0.1 nM 1,25D. RNA from zebrafish larvae was extracted by Trizol (Life Technologies, Carlsbad, CA) and cDNA generated by Super Script III Reverse Transcriptase (Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol. After cDNA synthesis, samples were diluted three-fold with RNAse free water, and 2 μl aliquots were used in qPCR reactions. A master mix of SYBR-qPCR enzyme mix (Agilent, Santa Clara, CA) and 50 nM primer pairs (either reported or designed on Primer3 [53]) was prepared and added to templates to a final volume of 25 μl. qPCR analysis was performed on MX3005P instrument (Agilent, Santa Clara, CA) using the following amplification program: 10 min 95°C (1×), 30 sec 95°C, 1 min 55°C, 1 min 72°C (40×). Amplification program was followed by a dissociation program: 1 min 95°C, 30 sec 55°C, 0.2C°/sec ramp up until 95°C. Ct values were determined by instrument software. Ct values for the gene of interest, zebrafish 24-vitamin D hydroxylase (cyp24a1; Genbank NM_001089458; forward 5′ AAAAGTCAACGGC AAAATGG 3′; reverse 5′ GTGTGGTCCTTCCACGTCTT 3′) were subtracted by the Ct values or the calibrator gene, zebrafish elongation factor 1-alpha [39] (elfa; Genbank AY422992; forward 5′ CTTCTCAGGCTGACTGTGC 3′; reverse 5′ CCGCTAGCATTACCCTCC 3′) to yield ΔCt values. Data were expressed as either ΔΔCt or fold change (fold = 2ΔΔCt) relative to vehicle treated sample. Error bars were displayed as standard deviation (SD) unless indicated otherwise. Equal variance two-tailed Student’s t-test (Microsoft Excel) was used for statistical analysis and results with p < 0.05 were deemed significant.
Cloning of zebrafish cyp27b1
First Choice RLM-RACE Kit (Ambion, Austin, TX) was utilized to clone 5′ and 3′ ends. Following the manufacturer protocol, 5′ and 3′ RACE cDNA was generated from five-day old zebrafish RNA. The predicted cyp27b1 cDNA sequence (ENSDARP00000066177) was entered nto Primer3 software [53] to design primers that were synthesized (Life Technologies, Carlsbad, CA) for usage in RACE cDNA synthesis. Primers used are listed in Table 1. The cDNAs were then PCR-amplified with Ambion supplied RACE outer primers and cyp27b1 specific outer primers, followed by nested PCR with Ambion supplied RACE inner primers and cyp27b1 specific inner primers. Taq polymerase (Life Technologies, Carlsbad, CA) was used for nested PCR reactions and middle fragment PCR. To clone the overlapping middle fragment, cDNA rom 0.5 μg of RNA was synthesized with Super Script III Reverse Transcriptase (Life Technologies, Carlsbad, CA) followed by PCR with cyp27b1 specific primers. Reaction conditions for the PCR were: 94°C for 3 min followed by 35 cycles of 94°C for 45 sec, 60°C for 30 sec and 72°C for 2 min with a final 10 min extension at 72°C. Fragments were separated by agarose gel electrophoresis, and isolated with the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA). The fragments were then ligated into the pCR-TOPO vector (Life Technologies, Carlsbad, CA) and sequenced by the UCLA Sequencing and Genotyping Core. The 5′ RACE and 3′ RACE fragments were combined with the middle fragment through splicing by overlapping extension (SOE) PCR. Pfx polymerase (Life Technologies, Carlsbad, CA) was used for subsequent template generation. Reaction conditions for Pfx were: 94°C for 2 min followed by 35 cycles of 94°C for 15 sec, 61°C for 30 sec and 68°C for 2 min. The 5′ SOE and 3′ SOE fragments were combined by ligation at a common Kpn I site, yielding a full-length cDNA, which was cloned into the TOPO TA pcDNA3.1 vector (Life Technologies, Carlsbad, CA). The cDNA sequence has been deposited to Genbank (KM262796).
Table 1. Oligonucleotide primer sequences used for cloning.
| Cloning Step | Oligo name and sequence |
|---|---|
| Middle fragment PCR | (1F) 5′-CATGTGGAAGGCCAGTTTCG-3′ (4R) 5′-TACACCTCCAGCTCAGCGAT-3′ |
| Nested 5′ RACE PCR Reaction 1 | Forward 5′ RACE outer from Ambion 5′-GCTGATGGCGATGAATGAACACTG-3′ (2R) 5′-GGTCAGCAGAGTCATCACGA-3′ |
| Nested 5′ RACE PCR Reaction 2 | Forward 5′ RACE inner from Ambion 5′-CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG-3′ (1R) 5′-CGAAACTGGCCTTCCACATG-3′ |
| Nested 3′ RACE PCR Reaction 1 | (3F) 5′-CACGATCTCCAGCACACTGT-3′ Reverse 3′ RACE outer from Ambion 5′-GCGAGCACAGAATTAATACGACT-3′ |
| Nested 3′ RACE PCR Reaction 2 | (4F) 5′-ATCGCTGAGCTGGAGGTGTA-3′ Reverse 3′ RACE inner from Ambion 5′-CGCGGATCCGAATTAATACGACTCACTATAGG-3′ |
| Splice overlap Extension PCR 5′ RACE + Middle fragment as template |
Forward 5′ RACE inner from Ambion 5′-CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG-3′ (4R) 5′-TACACCTCCAGCTCAGCGAT-3′ |
| Splice overlap Extension PCR Middle fragment + 3′ RACE as template |
(1F) 5′-CATGTGGAAGGCCAGTTTCG-3′ Reverse 3′ RACE inner from Ambion 5′-CGCGGATCCGAATTAATACGACTCACTATAGG-3′ |
Zebrafish tissue analysis for cyp27b1 by qPCR
Two independent preparations of RNA by Trizol extraction were obtained from organs (kidney, heart and spleen) of two or three 18-month old male zebrafish. cDNA was synthesized as described above for qPCR analysis of ELFA for normalization purposes. To facilitate detection of the low expression of cyp27b1, a separate batch of cDNA was synthesized using cyp27b1 gene specific primer (5′ TGGTTTCCTCCGCTGTGTTT 3′) located near the 3′ end. qPCR analysis was conducted with cyp27b1 probe/primer set near the 5′ end: forward 5′ GTTCGCTAAAGGACACATTGAC 3′; reverse 5′ CCTGAGACAGAAAGTACGTGAG 3′; and probe 5′ CAGCAGGAGAAGCAGAAGCTGGAG 3′ modified with 5′ FAM, ZEN internal Quencher, and 3′ Iowa Black FQ (Integrated DNA Technologies, Coralville, Iowa). qPCR analysis was performed on MX3005P instrument (Agilent, Santa Clara, CA). ΔCt = Ct(cyp27b1) − Ct(elfa) was calculated for each sample and fold change relative to the lowest expressing organ was determined.
Transfection of HKC-8 for qPCR and HPLC analysis
Human proximal kidney tubule cells (HKC-8 cells) were detached by trypsin, diluted with Opti-MEM (Life Technologies, Carlsbad, CA) and 2.5 × 105 cells were seeded into each well of 12-well plates. 8 hours later, 0, 0.5 or 1.0 μg pcDNA3.1-zcyp27b1 were normalized to a total of 1.0 μg with pcDNA3.1 and diluted with 100 μl Opti-MEM for each well to be transfected. 4 μl of Lipofectamine 2000 (LF2000, Life Technologies, Carlsbad, CA) was diluted with 100 μl of Opti-MEM for each well to be transfected. The diluted plasmid DNA was combined with the diluted LF2000, mixed vigorously, incubated for 20 minutes to allow complex formation, and then seeded onto the plated cells. 18 hours later, LF2000/DNA containing media was removed by aspiration and fresh Opti-MEM media was added. 0 or 100 nM 25D dissolved in ethanol was added to each well at a 0.1% volume. Six hours later, media were removed, and 1 ml of Trizol (Life Technologies, Carlsbad, CA) was added to the cells. RNA extraction and cDNA synthesis was according to manufacturer’s protocol. CYP24A1 (Hs00167999_m1) and 18S rRNA (4310893E) probe/primers were used in TaqMan qPCR analysis (Life Technologies, Grand Island, NY). For HPLC analysis, LF2000 transfection of pcDNA3.1 and pcDNA3.1-zcyp27b1 was scaled up according to product protocol for 24 μg plasmid per 10cm dish. 48-hours after transfection, cells were detached with trypsin and one million cells were incubated with 300,000 cpm 3H-25-hydroxyvitamin D3 (3H-25D, 155 Ci/mmol; Perkin Elmer, Waltham, MA) per glass tube for two hours. Metabolites were extracted, purified and analyzed by HPLC as previously described [8]. Protein concentration of cellular material used in metabolism experiment was determined by Bradford assay (Bio-Rad, Hercules, CA) to normalize samples with results expressed as fmol/hr/mg.
Phylogenetic Analysis of DNA sequences
The evolutionary history of CYP27B1 protein sequences were inferred by using the Maximum Likelihood method as implemented in MEGA 5 [48]. CYP27B1 sequences analyzed were NCBI reference sequences from human (NP_000776.1), mouse (NP_034139.2), rat (NP_446215.1), cow (NP_001179213.1), pig (NP_999160.1), rhesus (NP_001181642.1), cat (XP_003989015.1), dog (XP_538254.3), chimpanzee (XP_509175.2), xenopus (NP_001006907.1), and horse (NP_001157429.1), and Ensembl sequences from zebrafish (ENSDARP00000066177), and anole (ENSACAP00000012941). NCBI reference sequences of CYP27A1 from chicken (XP_422056.3) and zebrafish (XP_001923080.3), and CYP27C1 from chicken (XP_422077.2) and zebrafish (NP_001106808.2) were also included in the analysis.
Results and Discussion
Effects of 25D and 1,25D on whole zebrafish gene expression
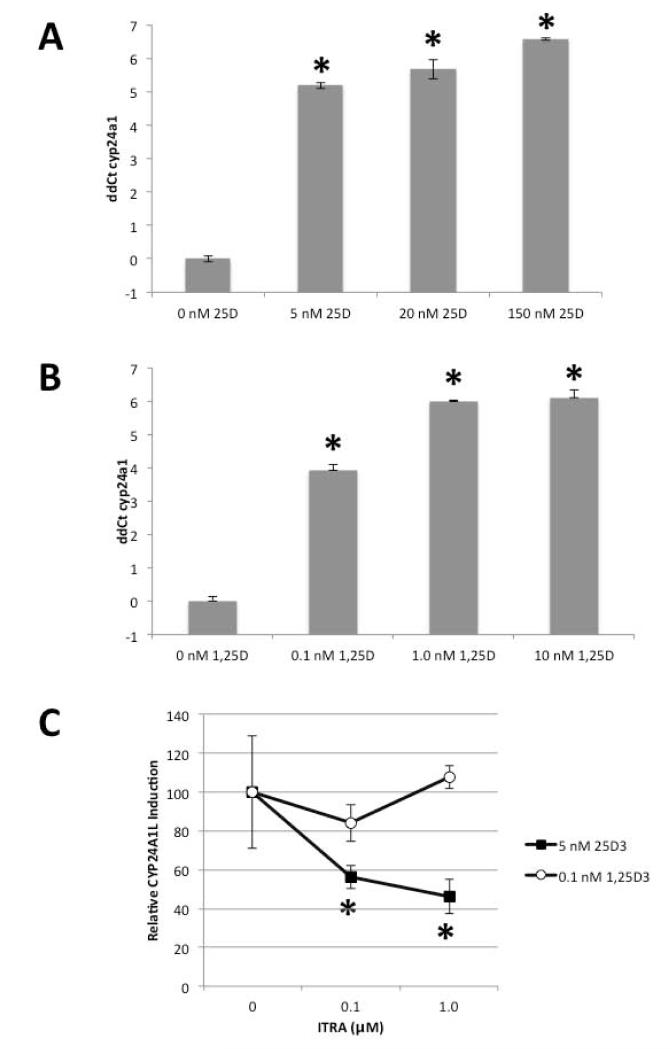
Day 5 zebrafish larvae were treated with varying doses of inactive precursor 25D (5-150 nM), active metabolite 1,25D (0.1-10 nM), or vehicle (0.1% ethanol) for six hours. Total RNA from the zebrafish was then used to quantify expression of mRNA for zebrafish equivalent of the 1,25D-inducible catabolic enzyme vitamin D-24-hydroxylase (cyp24a1). Data in Figure 1A and 1B indicate that both 25D (36 to 95-fold) and 1,25D (15 to 69-fold) induced cyp24a1 expression relative to ethanol-treated control larvae. The ability of pro-hormone 25D to induce this change in gene expression suggested that zebrafish express a CYP27B1 gene that is associated with synthesis of 1,25D in mammals. To confirm the presence of a functional 1α-hydroxylase activity in zebrafish, day 5 larvae were treated with vehicle (0.1% DMSO) or with the 1α-hydroxylase inhibitor itraconzole (ITRA; 0.1 or 1.0 μM) for one hour prior to initiation of six hour incubations with either 25D (5 nM) or 1,25D (0.1 nM). Subsequent analysis of mRNA expression in these larvae showed that ITRA pre-treatment decreased 25D-induction of cyp24a1 by 50%, but did not have any effect on 1,25D-induced cyp24a1 (Figure 1C). These data suggest that the effects of 25D on gene expression in zebrafish larvae involve a functional 1α-hydroxylase enzyme. Further studies were therefore carried out to clone the gene associated with this enzyme activity in zebrafish.
Figure 1. Induction of cyp24a1 mRNA expression by 25D and 1,25D identifies a functional 25-hydroxyvitamin D-1-hydroxylase in zebrafish.
Zebrafish larvae (day 5) were treated with increasing doses of: A) 25D (5-150 nM), B) 1,25D (0.1-10 nM) or vehicle (0.1% ethanol) for six hours, and expression of cyp24a1 mRNA determined by qRT-PCR. Data are expressed as ΔΔCt = ΔCt (cyp24a1) - ΔCt(elfa). C) Zebrafish larvae (day 5) were treated with increasing doses of itraconazole (ITRA, 0, 0.1, and 1.0 μM) one hour prior to six hour incubation with vehicle, 5 nM 25D or 0.1 nM 1,25D. Expression of cyp24a1 was then determined by qRT-PCR and expressed relative to vehicle ITRA set to 100%. Fold = 2ΔΔCt. Data are shown as n=3 replicates ± SD. * P < 0.05.
Cloning of zebrafish cyp27b1
Rapid amplification of cDNA ends (RACE) and sequence data from the zebrafish genome database were used to clone a cDNA for zebrafish cyp27b1. The full-length cDNA sequence (Genbank accession number KM262796) cloned contains an open reading frame (ORF) yielding a predicted 505-amino acid protein. The cloned sequence (Genbank accession number KM262796) shares 99% identity at the nucleotide level and 98% identity at the protein level with the predicted sequence in the zebrafish genome database (Genbank accession number XP_003199448.2). Amino acid sequence comparison analysis between zebrafish and human 1α-hydroxylases showed 54% identity between zebrafish 1α-hydroxylase and its human homolog (Figure 2). Functionally important regions of the human 1α-hydroxylase protein such as the heme-binding domain (bold) [45], ferredoxin binding domain (underlined) [40], and residues believed to be vital for enzymatic function (Q66 and T408; bold and asterisk) [59] were found in the zebrafish sequence with high identity.
Figure 2. Amino acid sequence comparison between zebrafish Cyp27b1 and human CYP27B1.
The amino acid sequence for Cyp27b1 was compared with human CYP27B1 using NCBI BLAST. Heme binding domain was marked in bold. Ferredoxin binding domain was marked with underline. Q66 and T408 (bold and asterik) have been identified as residues found mutated in human type I rickets suggesting functionally significant amino acids. The cDNA sequence has been deposited to Genbank (KM262796).
Zebrafish cyp27b1 is expressed in kidney, spleen and heart
Expression of CYP27B1 was found first and at high levels in kidney [33,61]; however, it has also been found expressed in extra-renal sites [3,24]. To determine relative tissue expression levels in zebrafish, kidney, spleen and heart tissues were obtained from adult male fish and qPCR analysis conducted. In Figure 3, the heart exhibited the lowest expression relative to the other organs analyzed while the kidney, as expected, had the highest (17 and 65 fold greater than heart) and the spleen had intermediate levels (4 to 7 fold higher compared to heart). These findings are consistent with reports from other animals that revealed low but detectable expression of CYP27B1 in spleen [11,42] and heart [11,12].
Figure 3. Expression of zebrafish cyp27b1 in kidney, spleen and heart.
Quantitative real-time PCR was conducted on cDNA prepared from heart (H1, H2) spleen (S1, S2) and kidney (K1, K2). The graphs display the data from triplicate qPCR assessments of two independent preparations of the respective organs. The data is presented as relative fold expression with H2 (lowest expression sample) set to one-fold and error bars ± SD.
Zebrafish cyp27b1 is biologically active
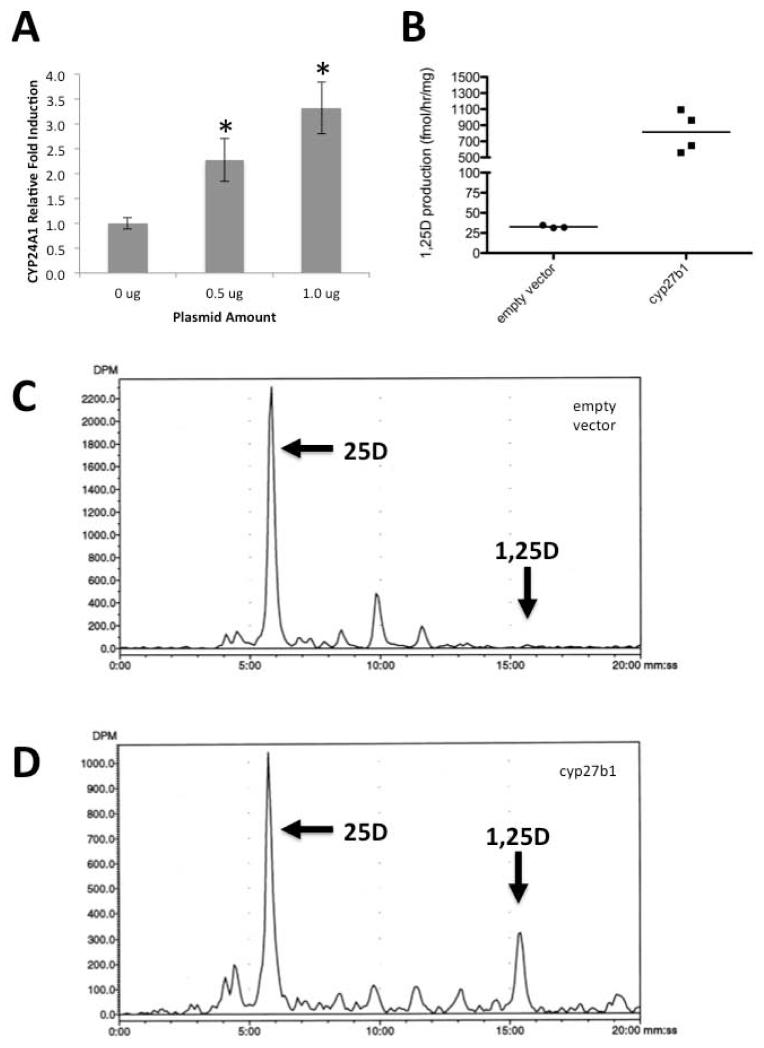
To determine whether the cyp27b1 identified in Figure 2 is biological active, a cDNA expression construct for cyp27b1 was transfected into HKC-8, human proximal tubule kidney cells, which are known to support a modest level of conversion of 25D to 1,25D [9,14]. Following transfection with either empty vector, 0.5 or 1 μg of pcDNA3.1-zcyp27b1, HKC-8 cells were incubated with vehicle (0.1% ethanol) or 25D (100 nM) for six hours. Subsequent RT-PCR analysis of mRNA from these cells showed that 25D-induced expression of mRNA for human CYP24A1 increased in a dose-dependent fashion in HKC-8 cells that received cyp27b1 cDNA relative to cells that received empty vector (Figure 4A). To further confirm the functionality of the cloned cyp27b1, HPLC analysis of 1,25D production was conducted. HKC-8 cells were transfected with pcDNA3.1 or pcDNA3.1-zcyp27b1 and 48-hours post-transfection, cells were detached and incubated for two hours with 3H-25D. Vitamin D metabolites were then extracted, purified and analyzed by HPLC (Figure 4B-4D) and results expressed as fmol/hr/mg protein. As shown in Figure 4B, HKC-8 transfected with pcDNA3.1-zcyp27b1 produced 25-fold more 1,25D relative to the empty vector transfected cells (815.3 vs. 32.6 fmol/hr/mg). These data confirm that the cloned zebrafish cyp27b1 encoded a functional 25-hydroxyvitamin D-1α-hydroxylase.
Figure 4. Zebrafish cyp27b1 cDNA encodes a functional 25-hydroxyvitamin D-1α-hydroxylase activity.
A) HKC-8, a human proximal tubule kidney cell line, was transfected with indicated amounts of pcDNA3.1-zcyp27b1 expression plasmid or empty vector. The resulting cells were incubated with vehicle (0.1% ethanol) or 100 nM 25D for six hours. qRT-PCR analysis was used to quantify expression of mRNA for the 1,25D-VDR-induced catabolic enzyme 24-hydroxylase (CYP24A1) in human HKC-8 cells. Data are expressed as fold-induction of CYP24A1 expression relative to 100 nM 25D incubated empty vector control cells set to a value of one. Data are shown as mean ± SE for n=3 replicates; * P < 0.05. B) HKC-8 transfected with plasmid expressing zebrafish cyp27b1 or empty vector was analyzed by HPLC for 1,25D3 synthesis after two-hour incubation with 300,000 cpm 3H-25D3. 3H-1,25D3 production was calculated and normalized to mg protein per sample and expressed as fmol/mg/hr. The graph displays the average of N=3 (empty vector) and N=4 (cyp27b1) HPLC runs. Representative traces are shown of analysis of HKC-8 transfected with C) pcDNA3.1 empty vector or D) pcDNA3.1-zcyp27b1.
Phylogenetic analysis of zebrafish cyp27b1
Based on sequence analysis, cytochrome P450 genes have been grouped into 11 clans [41] of which CYP27B1 belongs to the mitochondrial clan. The cladogram in Figure 5 shows a phylogenetic analysis of CYP27B1 protein sequences retrieved from thirteen vertebrates, along with two CYP27A1 and two CYP27C1 sequences as outgroups. The phylogenetic analysis indicates that the zebrafish CYP27B1 is derived from an ancient bifurcation from the lineage eading to mammals. As such, the most similar sequences to zebrafish 1α-hydroxylase are amphibian (xenopus) and reptile (anole) 1α-hydroxylases. Predictably, within the mammalian CYP27B1, sequences grouped as expected with most similarity within rodent and primate species with less confident clustering of the other mammalian species. Though zebrafish 1α-hydroxylase/cyp27b1 was distinct from the mammalian cluster, it remains firmly within the CYP27B1 family, clearly separated from the CYP27A1 and CYP27C1 lineages.
Figure. 5. Phylogenetic analysis of CYP27B1 found in various species.
CYP27B1 evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model. The tree with the highest log likelihood (-8036.2717) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 17 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 487 positions in the final dataset. Evolutionary analyses were conducted in MEGA5. Zebrafish Cyp27a1 and Cyp27c1 were included as outgroups. CYP27A1 and CYP27C1 from chicken were included since no CYP27B1 was found for that species.
In view of the fundamental importance of vitamin D for skeletal homeostasis, it seems likely that all genes such as VDR and CYP27B1 would be common to all species in the subphylum vertebrata. However, an unexpected result of our analysis was finding an absence of any avian members of the CYP27B1 sequence family within the genome database. Some of the early work on vitamin D metabolism utilized chickens [22,33] and they are known to synthesize 1,25D [4,23,57], so the absence of CYP27B1 in avian genome databases is surprising. Investigators in this field recognized that there is 5-10% of the chicken genome where sequence data remains absent or of poor quality [20,58] thus, perhaps, explaining the current absence of CYP27B1 in the chicken genome database. On the other hand, numerous avian species have CYP27A1 and CYP27C1 represented in the databases, thus it is unlikely that CYP27B1 has simply been missed by avian sequencing projects. This suggests the avian lineage uses an alternate, as yet unidentified, enzyme to fulfill the function of a vitamin D 1α-hydroxylase, possibly another member of the very large P450 superfamily. For example, although CYP2R1 is understood to be the major contributor to 25D production, it is not the only CYP with vitamin D-25-hydroxylase activity [62]. By contrast, Cyp27b1 knockout mice [19,43], and natural CYP27B1 mutations in humans [37] strongly suggest that this gene is solely responsible for production of 1,25D in these species. The conspicuous absence of CYP27B1 in avian genome databases is an area of active investigation (Wes Warren, personal communication) and firm conclusions about the presence or absence of CYP27B1 awaits further research.
Zebrafish as a potential model system for vitamin D and TB
In mammals, 25D is the major circulating form of vitamin D and the principal marker of vitamin D status [29]. In recent years the importance of 25D in vitamin D physiology has been elevated with the recognition that many effects of vitamin D may be mediated via local intracrine, conversion of 25D to 1,25D, with resulting tissue-specific actions of 1,25D bound to the VDR [6,13,15]. As a consequence there has been renewed interest in the expression and activity of the CYP27B1 gene product, 1α-hydroxylase, which catalyzes conversion of 25D to 1,25D [24,52]. The site-specific impact of 1α-hydroxylase/CYP27B1 on vitamin D physiology has been studied extensively in murine models [35,36,42,55], but has yet to be assessed in zebrafish.
Cloning of a functional cyp27b1 provides a useful complement to the previous characterization of zebrafish vdr [16] adding additional evidence for the existence of the vitamin D system in this organism. Future studies aimed at manipulating expression of these two components of the vitamin D system through morpholino knockdown or zinc-finger nuclease mutation will help clarify the relative activities of these gene products in diverse facets of vitamin D physiology. The functional 1α-hydroxylase enzyme activity demonstrated in this report further underlines the versatility of zebrafish as a model system. Of particular interest is the role of vitamin D in immune responses to infection [27]. Vitamin D-deficiency has been closely linked to the mycobacterial infectious disease tuberculosis (TB) [38,54], a major global health problem. However, current animal models for TB are poor and greatly limit the scope of studies aimed at defining the potential effect of molecules such as vitamin D as treatment for or prevention of this disease. By contrast zebrafish are a highly informative model for infection with Mycobacterium marinum, a mycobacterium related to the TB pathogen, Mycobacterium tuberculosis [10,49,50,51]. Thus, in future studies, zebrafish may be a useful model for assessing the role of vitamin D metabolism and signaling in the pathophysiology and treatment of diseases such as TB.
Acknowlegements
We thank Ann Cavanaugh (UCLA) for isolating organ tissue from adult zebrafish. We thank Jessica Sea (UCLA) for critical reading of the manuscript. Research reported in this publication was supported by Institutional Development Awards (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under grant numbers P20GM104318 (for COBRE) and P20GM103423 (INBRE), and by a Department of Defense award – USAMRAA (W81XWH-BAA).
Footnotes
Conflict of Interest
The authors have declared that there is no conflict of interest.
References
- 1. http://www.brineshrimpdirect.com/c1/c3/Spirulina-and-Kelp-Flake-c44.html.
- 2. http://www.aquaneer.com/pdf_files/scientific_hatcheries_diet.pdf.
- 3.Adams JS, Gacad MA. Characterization of 1 alpha-hydroxylation of vitamin D3 sterols by cultured alveolar macrophages from patients with sarcoidosis. J Exp Med. 1985;161:755–765. doi: 10.1084/jem.161.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams JS, Beeker TG, Hongo T, Clemens TL. Constitutive expression of a vitamin D 1-hydroxylase in a myelomonocytic cell line: a model for studying 1,25-dihydroxyvitamin D production in vitro. J Bone Miner Res. 1990;5:1265–1269. doi: 10.1002/jbmr.5650051212. [DOI] [PubMed] [Google Scholar]
- 5.Adams JS, Hewison M. Unexpected actions of vitamin D: new perspectives on the regulation of innate and adaptive immunity. Nat Clin Pract Endocrinol Metab. 2008;4:80–90. doi: 10.1038/ncpendmet0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams JS, Hewison M. Update in vitamin D. J Clin Endocrinol Metab. 2010;95:471–478. doi: 10.1210/jc.2009-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen JP, Neely MN. Trolling for the ideal model host: zebrafish take the bait. Future Microbiol. 2010;5:563–569. doi: 10.2217/fmb.10.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bacchetta J, Sea JL, Chun RF, Lisse TS, Wesseling-Perry K, et al. Fibroblast growth factor 23 inhibits extrarenal synthesis of 1,25-dihydroxyvitamin D in human monocytes. J Bone Miner Res. 2013;28:46–55. doi: 10.1002/jbmr.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bland R, Walker EA, Hughes SV, Stewart PM, Hewison M. Constitutive expression of 25-hydroxyvitamin D3-1alpha-hydroxylase in a transformed human proximal tubule cell line: evidence for direct regulation of vitamin D metabolism by calcium. Endocrinology. 1999;140:2027–2034. doi: 10.1210/endo.140.5.6683. [DOI] [PubMed] [Google Scholar]
- 10.Cambier CJ, Takaki KK, Larson RP, Hernandez RE, Tobin DM, et al. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature. 2014;505:218–222. doi: 10.1038/nature12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chanakul A, Zhang MY, Louw A, Armbrecht HJ, Miller WL, et al. FGF-23 regulates CYP27B1 transcription in the kidney and in extra-renal tissues. PLoS One. 2013;8:e72816. doi: 10.1371/journal.pone.0072816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Glenn DJ, Ni W, Grigsby CL, Olsen K, et al. Expression of the vitamin d receptor is increased in the hypertrophic heart. Hypertension. 2008;52:1106–1112. doi: 10.1161/HYPERTENSIONAHA.108.119602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christakos S, Hewison M, Gardner DG, Wagner CL, Sergeev IN, et al. Vitamin D: beyond bone. Ann N Y Acad Sci. 2013;1287:45–58. doi: 10.1111/nyas.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chun RF, Gacad M, Nguyen L, Hewison M, Adams JS. Co-chaperone potentiation of vitamin D receptor-mediated transactivation: a role for Bcl2-associated athanogene-1 as an intracellular-binding protein for 1,25-dihydroxyvitamin D3. J Mol Endocrinol. 2007;39:81–89. doi: 10.1677/JME-07-0042. [DOI] [PubMed] [Google Scholar]
- 15.Chun RF, Adams JS, Hewison M. Back to the future: a new look at ‘old’ vitamin D. J Endocrinol. 2008;198:261–269. doi: 10.1677/JOE-08-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig TA, Sommer S, Sussman CR, Grande JP, Kumar R. Expression and regulation of the vitamin D receptor in the zebrafish, Danio rerio. J Bone Miner Res. 2008;23:1486–1496. doi: 10.1359/JBMR.080403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craig TA, Zhang Y, McNulty MS, Middha S, Ketha H, et al. Research resource: whole transcriptome RNA sequencing detects multiple 1alpha,25-dihydroxyvitamin D(3)-sensitive metabolic pathways in developing zebrafish. Mol Endocrinol. 2012;26:1630–1642. doi: 10.1210/me.2012-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craig TA, Zhang Y, Magis AT, Funk CC, Price ND, et al. Detection of 1,25-Dihydroxyvitamin D-Regulated miRNAs in Zebrafish by Whole Transcriptome Sequencing. Zebrafish. 2014 doi: 10.1089/zeb.2013.0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dardenne O, Prud’homme J, Arabian A, Glorieux FH, St-Arnaud R. Targeted inactivation of the 25-hydroxyvitamin D(3)-1(alpha)-hydroxylase gene (CYP27B1) creates an animal model of pseudovitamin D-deficiency rickets. Endocrinology. 2001;142:3135–3141. doi: 10.1210/endo.142.7.8281. [DOI] [PubMed] [Google Scholar]
- 20.Dodgson JB, Delany ME, Cheng HH. Poultry genome sequences: progress and outstanding challenges. Cytogenet Genome Res. 2011;134:19–26. doi: 10.1159/000324413. [DOI] [PubMed] [Google Scholar]
- 21.Froicu M, Weaver V, Wynn TA, McDowell MA, Welsh JE, et al. A crucial role for the vitamin D receptor in experimental inflammatory bowel diseases. Mol Endocrinol. 2003;17:2386–2392. doi: 10.1210/me.2003-0281. [DOI] [PubMed] [Google Scholar]
- 22.Haussler MR, Myrtle JF, Norman AW. The association of a metabolite of vitamin D3 with intestinal mucosa chromatin in vivo. J Biol Chem. 1968;243:4055–4064. [PubMed] [Google Scholar]
- 23.Haussler MR, Rasmussen H. The metabolism of vitamin D 3 in the chick. J Biol Chem. 1972;247:2328–2335. [PubMed] [Google Scholar]
- 24.Hewison M, Burke F, Evans KN, Lammas DA, Sansom DM, et al. Extra-renal 25-hydroxyvitamin D3-1alpha-hydroxylase in human health and disease. J Steroid Biochem Mol Biol. 2007;103:316–321. doi: 10.1016/j.jsbmb.2006.12.078. [DOI] [PubMed] [Google Scholar]
- 25.Hewison M. Vitamin D and innate immunity. Curr Opin Investig Drugs. 2008;9:485–490. [PubMed] [Google Scholar]
- 26.Hewison M. Vitamin D and the intracrinology of innate immunity. Mol Cell Endocrinol. 2010;321:103–111. doi: 10.1016/j.mce.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hewison M. Antibacterial effects of vitamin D. Nat Rev Endocrinol. 2011;7:337–345. doi: 10.1038/nrendo.2010.226. [DOI] [PubMed] [Google Scholar]
- 28.Hewison M. Vitamin D and immune function: autocrine, paracrine or endocrine? Scand J Clin Lab Invest Suppl. 2012;243:92–102. doi: 10.3109/00365513.2012.682862. [DOI] [PubMed] [Google Scholar]
- 29.Holick MF. Vitamin D status: measurement, interpretation, and clinical application. Ann Epidemiol. 2009;19:73–78. doi: 10.1016/j.annepidem.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones G, Prosser DE. In: Vitamin D (Third Edition) Chapter 3 - The Activating Enzymes of Vitamin D Metabolism (25- and 1a-Hydroxylases) Feldman D, Pike JW, Adams JS, editors. Elsevier; London: 2011. pp. 23–42. [Google Scholar]
- 31.Kong J, Zhang Z, Musch MW, Ning G, Sun J, et al. Novel role of the vitamin D receptor in maintaining the integrity of the intestinal mucosal barrier. Am J Physiol Gastrointest Liver Physiol. 2008;294:G208–216. doi: 10.1152/ajpgi.00398.2007. [DOI] [PubMed] [Google Scholar]
- 32.Lagishetty V, Misharin AV, Liu NQ, Lisse TS, Chun RF, et al. Vitamin D deficiency in mice impairs colonic antibacterial activity and predisposes to colitis. Endocrinology. 2010;151:2423–2432. doi: 10.1210/en.2010-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawson DE, Fraser DR, Kodicek E, Morris HR, Williams DH. Identification of 1,25-dihydroxycholecalciferol, a new kidney hormone controlling calcium metabolism. Nature. 1971;230:228–230. doi: 10.1038/230228a0. [DOI] [PubMed] [Google Scholar]
- 34.Lin CH, Su CH, Tseng DY, Ding FC, Hwang PP. Action of vitamin D and the receptor, VDRa, in calcium handling in zebrafish (Danio rerio) PLoS One. 2012;7:e45650. doi: 10.1371/journal.pone.0045650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu N, Nguyen L, Chun RF, Lagishetty V, Ren S, et al. Altered endocrine and autocrine metabolism of vitamin D in a mouse model of gastrointestinal inflammation. Endocrinology. 2008;149:4799–4808. doi: 10.1210/en.2008-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu NQ, Kaplan AT, Lagishetty V, Ouyang YB, Ouyang Y, et al. Vitamin D and the regulation of placental inflammation. J Immunol. 2011;186:5968–5974. doi: 10.4049/jimmunol.1003332. [DOI] [PubMed] [Google Scholar]
- 37.Malloy PJ, Feldman D. Genetic disorders and defects in vitamin d action. Endocrinol Metab Clin North Am. 2010;39:333–346. doi: 10.1016/j.ecl.2010.02.004. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martineau AR. Old wine in new bottles: vitamin D in the treatment and prevention of tuberculosis. Proc Nutr Soc. 2012;71:84–89. doi: 10.1017/S0029665111003326. [DOI] [PubMed] [Google Scholar]
- 39.McCurley AT, Callard GV. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol Biol. 2008;9:102. doi: 10.1186/1471-2199-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nebert DW, Gonzalez FJ. P450 genes: structure, evolution, and regulation. Annu Rev Biochem. 1987;56:945–993. doi: 10.1146/annurev.bi.56.070187.004501. [DOI] [PubMed] [Google Scholar]
- 41.Nelson DR, Goldstone JV, Stegeman JJ. The cytochrome P450 genesis locus: the origin and evolution of animal cytochrome P450s. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120474. doi: 10.1098/rstb.2012.0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ooi JH, McDaniel KL, Weaver V, Cantorna MT. Murine CD8+ T cells but not macrophages express the vitamin D 1alpha-hydroxylase. J Nutr Biochem. 2014;25:58–65. doi: 10.1016/j.jnutbio.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panda DK, Miao D, Tremblay ML, Sirois J, Farookhi R, et al. Targeted ablation of the 25-hydroxyvitamin D 1alpha -hydroxylase enzyme: evidence for skeletal, reproductive, and immune dysfunction. Proc Natl Acad Sci U S A. 2001;98:7498–7503. doi: 10.1073/pnas.131029498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao DS, Raghuramulu N. Vitamin D metabolism in tilapia (Oreochromis mossambicus) Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1998;120:145–149. doi: 10.1016/s0742-8413(98)00035-8. [DOI] [PubMed] [Google Scholar]
- 45.Shinki T, Shimada H, Wakino S, Anazawa H, Hayashi M, et al. Cloning and expression of rat 25-hydroxyvitamin D3-1alpha-hydroxylase cDNA. Proc Natl Acad Sci U S A. 1997;94:12920–12925. doi: 10.1073/pnas.94.24.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sundh H, Larsson D, Sundell K. Environmental salinity regulates the in vitro production of [3H]-1,25-dihydroxyvitamin D3 and [3H]-24,25 dihydroxyvitamin D3 in rainbow trout (Oncorhynchus mykiss) Gen Comp Endocrinol. 2007;152:252–258. doi: 10.1016/j.ygcen.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 47.Takaki K, Davis JM, Winglee K, Ramakrishnan L. Evaluation of the pathogenesis and treatment of Mycobacterium marinum infection in zebrafish. Nat Protoc. 2013;8:1114–1124. doi: 10.1038/nprot.2013.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tobin DM, Ramakrishnan L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol. 2008;10:1027–1039. doi: 10.1111/j.1462-5822.2008.01133.x. [DOI] [PubMed] [Google Scholar]
- 50.Tobin DM, Vary JC, Jr., Ray JP, Walsh GS, Dunstan SJ, et al. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell. 2010;140:717–730. doi: 10.1016/j.cell.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tobin DM, Roca FJ, Oh SF, McFarland R, Vickery TW, et al. Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell. 2012;148:434–446. doi: 10.1016/j.cell.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Townsend K, Evans KN, Campbell MJ, Colston KW, Adams JS, et al. Biological actions of extra-renal 25-hydroxyvitamin D-1alpha-hydroxylase and implications for chemoprevention and treatment. J Steroid Biochem Mol Biol. 2005;97:103–109. doi: 10.1016/j.jsbmb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 53.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, et al. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ustianowski A, Shaffer R, Collin S, Wilkinson RJ, Davidson RN. Prevalence and associations of vitamin D deficiency in foreign-born persons with tuberculosis in London. J Infect. 2005;50:432–437. doi: 10.1016/j.jinf.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 55.Vanhooke JL, Prahl JM, Kimmel-Jehan C, Mendelsohn M, Danielson EW, et al. CYP27B1 null mice with LacZreporter gene display no 25-hydroxyvitamin D3-1 alpha-hydroxylase promoter activity in the skin. Proc Natl Acad Sci U S A. 2006;103:75–80. doi: 10.1073/pnas.0509734103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Volkman HE, Pozos TC, Zheng J, Davis JM, Rawls JF, et al. Tuberculous granuloma induction via interaction of a bacterial secreted protein with host epithelium. Science. 2010;327:466–469. doi: 10.1126/science.1179663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wakino S, Meguro M, Suzuki H, Saruta T, Ogishima T, et al. Evidence for 54-kD protein in chicken kidney as a cytochrome P450 with a high molecular activity of 25-hydroxyvitamin D3 1 alpha-hydroxylase. Gerontology. 1996;42(Suppl 1):67–77. doi: 10.1159/000213826. [DOI] [PubMed] [Google Scholar]
- 58.Wallis JW, Aerts J, Groenen MA, Crooijmans RP, Layman D, et al. A physical map of the chicken genome. Nature. 2004;432:761–764. doi: 10.1038/nature03030. [DOI] [PubMed] [Google Scholar]
- 59.Yamamoto K, Uchida E, Urushino N, Sakaki T, Kagawa N, et al. Identification of the amino acid residue of CYP27B1 responsible for binding of 25-hydroxyvitamin D3 whose mutation causes vitamin D-dependent rickets type 1. J Biol Chem. 2005;280:30511–30516. doi: 10.1074/jbc.M505244200. [DOI] [PubMed] [Google Scholar]
- 60.Yu S, Bruce D, Froicu M, Weaver V, Cantorna MT. Failure of T cell homing, reduced CD4/CD8alphaalpha intraepithelial lymphocytes, and inflammation in the gut of vitamin D receptor KO mice. Proc Natl Acad Sci U S A. 2008;105:20834–20839. doi: 10.1073/pnas.0808700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zehnder D, Bland R, Walker EA, Bradwell AR, Howie AJ, et al. Expression of 25-hydroxyvitamin D3-1alpha-hydroxylase in the human kidney. J Am Soc Nephrol. 1999;10:2465–2473. doi: 10.1681/ASN.V10122465. [DOI] [PubMed] [Google Scholar]
- 62.Zhu JG, Ochalek JT, Kaufmann M, Jones G, Deluca HF. CYP2R1 is a major, but not exclusive, contributor to 25-hydroxyvitamin D production in vivo. Proc Natl Acad Sci U S A. 2013;110:15650–15655. doi: 10.1073/pnas.1315006110. [DOI] [PMC free article] [PubMed] [Google Scholar]