Abstract
The human microbiome is a vast reservoir of microbial diversity and increasingly recognized to play a fundamental role in human health. In polymicrobial communities, the presence of one species can modulate the demography (growth and distribution) of other species. These demographic impacts generate feedbacks in multi-species interactions, which can be magnified in spatially structured populations (e.g., host-associated communities). Here we argue that demographic feedbacks between species are central to microbiome development, shaping whether and how potential metabolic interactions come to be realized between expanding lineages of bacteria. Understanding how demographic feedbacks tune metabolic interactions and in turn shape microbiome structure and function is now a key challenge to our abilities to better manage microbiome health.
Keywords: microbiome, multispecies communities, metabolic interactions, demography, spatial organization
The human microbiome: an ecological network of metabolic interactions
The human body is home to an extraordinary diversity of microbes, which are increasingly suggested to play pivotal roles in human health. Human microbiome (see Glossary) sequencing projects have revealed intriguing correlations between specific patterns of microbial diversity and multiple aspects of host health, including autoimmune disorders [1,2], diabetes [3], obesity [4,5], and even psychiatric conditions [6]. The establishment of microbial causal roles (particularly in obesity) is gathering pace thanks to experimental manipulations of germ-free mice (e.g., [7]), however the causal mechanisms frequently remain obscure.
A major challenge to unraveling the mechanisms of microbiome functioning is the necessity to combine molecular and ecological approaches to the study of highly complex assemblies of billions of interconnected bacterial cells. Systems biological approaches are beginning to make important headway by building and analyzing complex computational models of metabolic interactions within microbial communities [8-10], however these approaches typically make strongly simplifying assumptions on the spatio-temporal dynamics of constituent species, reducing their population biology to a simple ‘presence/absence’ dichotomy. This simplification (shared by ecological approaches to microbial community assembly [11]) allows a mapping of potential metabolic interactions among species, but fails to predict the extent to which any interaction will be realized. To address this issue, we propose a spatially-explicit population dynamic framework of microbiome development, to understand when and how potential metabolic interactions come to be realised via demographic feedbacks (reciprocal impacts on growth and distribution) between expanding lineages of bacteria.
Metabolic interactions and demographic feedbacks within a minimal microbiome
To develop a complete mechanistic understanding of a microbial community, it is key to understand how the presence of one species modulates the growth of each of the other species, and how these coupled demographies together shape the functional and spatial structuring or architecture of the community. Microbes constantly modify their environment through the secretion and excretion of both functional exo-products [12,13] and metabolic by-products [14,15] setting the stage for complex interspecific interactions. Of particular interest are ‘cross-feeding’ interactions, where species use metabolic by-products of others as energy or nutrient resources. Some cross-feeding relationships are characterized as mutualistic (enhancing both species’ growth rates [14,16]), however the exchange of metabolites can also promote exploitation where one species gains at the expense of another [17].
If we reduce the complexity of species interactions to a simple menu of discrete impacts on interacting species (positive +, negative -, or neutral 0), then for a two-species community there are six distinct patterns of potential ecological interaction [(0,0), (0,+), (0,-), (+,+), (+,-), (-,-)]. However, there is a combinatorial explosion in potential ecological complexity with increasing community diversity [18]. Given the high dimensionality of interactions within the human microbiome, there is a pressing need to create tractable model systems, as a necessary step towards understanding more complex multispecies dynamics. Here we suggest that in order to understand more diverse and complex microbial communities (such as the human microbiome), we first need to develop a thorough mechanistic understanding of coupled metabolic and demographic dynamics in defined ‘minimal’ microbiomes.
Previous studies on defined two-species communities highlight that these are capable of considerable ecological complexity [19-22]. Recent theoretical work has suggested that a single mechanism of interspecific metabolic exchange among two species can generate a diverse array of ecological relationships, spanning mutualism, competition, and exploitation; and that such diversity can arise by simply changing the properties of the metabolite that is exchanged (Figure 1A; [19]). A relevant empirical example of this diversity of outcomes is the interaction between the human oral commensal bacterium Streptococcus gordonii (Sg) and the pathogenic oral bacterium Aggregatibacter actinomycetemcomitans (Aa). Co-culture experiments of Sg and Aa in well-mixed liquid cultures have highlighted that the Sg–Aa interaction (mediated by Sg metabolic by-products lactate and H2O2) can readily move between mutualism and competition, depending on environmental conditions (Figure 1B) [23]. In aerobic conditions, Aa consumes lactate and relieves Sg of H2O2 toxicity, generating a marginally mutualistic relationship. However, under anaerobic conditions Aa cannot grow on lactate, and therefore competitive interactions (mediated by shared consumption of glucose) dominate (Figure 1B). In contrast, when grown in a structured in vivo model infection system, Aa gains significantly from the association while Sg neither benefits nor is harmed [23]. A key outstanding question is whether this stronger benefit to Aa is due solely to the many biochemical differences between the in vitro and in vivo growth environment, or whether there is a significant contribution from the effect of growing in a spatially structured environment.
Figure 1.
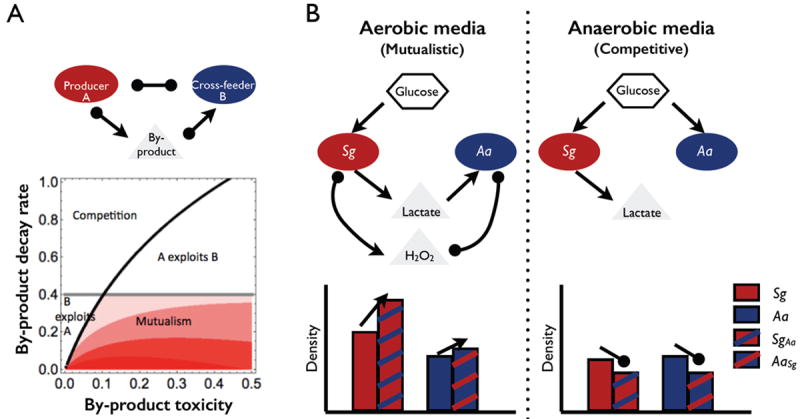
Metabolic interactions within a two-species community. (A) A single mechanism of metabolic exchange can generate diverse functional relationships in liquid (well-mixed) cultures. Figure adapted from [19] © 2012 by The University of Chicago. Mutualism: density of A in co-culture (AB) is larger than when alone (A), and B in co-culture (BA) is larger than alone (i.e., AB>A & BA>B). Red-scale indicates strength of mutualism (AB + BA − (A + B)). Competition: AB<A & BA< B. A exploits B: AB>A & BA<B. B exploits A: AB<A & BA>B. (B) Streptococcus gordonii (Sg) and Aggregatibacter actinomycetemcomitans (Aa) engage in multiple forms of metabolic interactions. Schematic model of Sg–Aa metabolic exchange under aerobic (left) and anaerobic (right) conditions [63]. Open arrows represent a positive effect, whereas oval arrows represent a negative effect upon the population or metabolite they are pointing toward. Sg and Aa form mutualistic or competitive interactions in liquid culture, dependent on oxygenation. Redrawn from [23].
Demography matters in spatially structured communities
Empirical work on microbial cross-feeding has shown that spatial structure plays an important role in shaping species interactions (e.g., [21,22,24-26]. From a modeling perspective, studying the role of metabolic interactions and demographic feedbacks in shaping the dynamics of spatially structured microbial communities is a challenging task, in part due to the computational challenge of studying mechanistically explicit models over space and time. In the past few years, there has been a rising interest in developing in silico models of microbial communities (for recent reviews see [27,28]). For instance, population-level models have been extended to study the dynamics and stability of the gut microbiota [29,30]. Metagenomic data combined with metabolic network analysis has recently been used to provide new insights into the correlation between species co-occurrence and predicted potential metabolic interactions in the gut microbiome [9]. Multispecies stoichiometric metabolic models [31-33] have also proven useful to predict potential species interactions. While these models provide valuable information, they do not explicitly consider spatial structure nor the effect of the chemico-physical environment.
Novel approaches, however, are emerging to address this gap. Recently, Harcombe et al. developed a novel computational approach that incorporates spatial structure into stoichiometric models of multispecies communities [34]. Specifically, they showed that their computational framework is able to predict the spatio-temporal dynamics of two- and three-species cross-feeding engineered microbial communities from the genome-scale metabolic network information of each individual species. Moreover, in recent years there has been considerable effort to develop individual-based models (IBMs) that simulate the growth of spatially extended microbial communities, such as biofilms [35,36]. Spatially explicit IBMs provide an excellent framework to investigate the demographic consequences of species interactions [20,21,26,37]. Recent computational studies on two metabolically interacting species have illustrated that when species provide mutual benefits (growth in a mutualism media, Figure 2), specific patterns of lineage mixing are promoted and the resulting pattern of species mixing is highly robust in the face of variation in initial demographic conditions (initial species frequency and spatial distribution) [20,21,26,34]. In contrast, when competitive interactions dominate, lineages tend to segregate and system behavior is highly contingent on initial conditions (Figure 2, [20,21,26]). Together, these studies support the idea that demographic feedbacks between neighbouring lineages are magnified in spatially structured populations.
Figure 2.
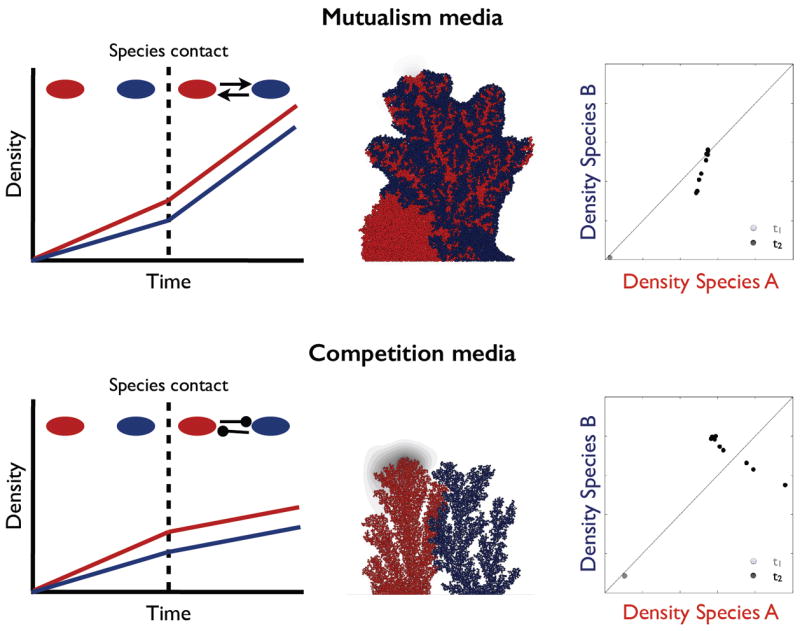
Distinct metabolic interactions produce distinct emergent functional and spatial relationships. Top panel, ‘mutualism medium’- the red species produces and blue species consumes a by-product toxic to the red species. The two species tend to mix and we observe a positive correlation between the two species densities across replicate communities (right plot, n =9, t1 and t2 represent initial and final timepoints). Bottom panel, ‘competition medium’- no metabolic exchange and competition for nutrients and space. The two species segregate and we observe a negative correlation between the two species densities (right plot). Both simulations were initiated with an identical segregated inoculum. Biofilm images and scatter plots are adapted from [20].
In a microbiome context, in addition to interacting with other microbial species, microbes also interact with their host. This raises many questions, such as what is the effect of the host in shaping these interactions, and how the host manipulates its own microbiota and selects for beneficial symbionts instead of harmful symbionts. Using an IBM of a host–gut microbiome, Schluter and Foster showed that host epithelial secretions of nutrients and antimicrobials play a critical role in modulating the microbiota [38]. They suggest that the selective effects imposed by the host at the epithelial surface are stronger than the selective effects of nutrients or antibiotics in the lumen contents, because of basic demographic asymmetries; cells favoured at the epithelial surface are more likely to replicate and found lineages growing out towards the lumen, whereas cells favoured in the lumen are much less likely to be founder cells for future generations, due to the elevated risk of being sloughed off [38].
Metabolically mediated demographic interactions and host health
Previous theoretical and experimental work have focused on the links between microbial metabolic interactions, spatial patterning, and ecological functioning among constituent microbial species [20-22,37,39,40]. However these microbe–microbe interactions are in turn central to the ability of microbial communities to perform services for the host, both positive, such as the production of nutrients [14] and protection against pathogens [41], and negative, such as enhanced virulence [23,42,43]. Here we discuss how a better understanding of the coupled effect of demographic and metabolic feedbacks can foster our understanding of services by the microbiome and how this can lead to potential new treatments.
Phenotypic antibiotic resistance through spatial structuring and partner shading
In a microbiome context, of particular interest is to understand the emergent resistance properties of microbial communities to antibiotic assault, and how antibiotics will affect microbiome services to the host. Generally, the success of antibiotic-resistant pathogens is due to genetic mutations that confer protection against the antibiotic. However, resistance can also be achieved without mutations, known as non-inherited [44] or phenotypic [45] antibiotic resistance. Unlike genetic resistance, phenotypic resistance is transient and environment-dependent, and includes mechanisms such as the dormancy of persister cells in biofilms [46], or reduced viral adsorption rate in bacterial resistance to phage [45]. Recently, another potential mechanism of phenotypic antibiotic resistance has been suggested. Connell et al. provide evidence for the key role played by the spatial relationships of species in conferring cross-species phenotypic antibiotic resistance [47]. Using a two-species microbial community constituted of genetically resistant beta-lactamase producing Pseudomonas aeruginosa (Pa), and a susceptible Staphylococcus aureus (Sa), they showed that Pa protected the susceptible Sa from beta-lactam antibiotics, and this protection was significantly enhanced due to their pre-defined spatial arrangement (Sa was confined within a shell of Pa) [47]. Although in this study the species spatial arrangement was imposed a priori, this raises the interesting question of whether antibiotic protection could arise via spatial structuring of emergent species. And if so, can we manipulate microbe–microbe functional and spatial relationships to favourably influence microbiome services to the host? Our prediction is that mutualistic biofilms, with a high degree of species mixing, are more resistant to narrow-spectrum antimicrobial clearance due to partner shading. The rational for this prediction is the following: under competitive interactions the two species segregate (Figure 2), rendering the susceptible strain isolated and vulnerable to clearance. In contrast, under mutualistic interactions, the two species interdigitate (Figure 2) and thus increase the average distance between the bulk fluid (maximal antimicrobial density) and target cells.
In addition to cross-species phenotypic resistance via partner shading, other mechanisms of cross-species antibiotic resistance have been suggested (see [48] for a recent review). For example, it was recently shown that co-culture of Salmonella enterica serovar Typhimurium with Escherichia coli enhances S. typhimurium tolerance to antibiotics [49]. When grown in co-culture, S. typhimurium can sense indole, a metabolite produced by E. coli but not produced by S. typhimurium, and this induces S. typhimurium antibiotic tolerance [49]. Enhanced tolerance of S. typhimurium to antibiotics when grown with E. coli had also been observed previously, but this time, tolerance was due to the exploitation of beta-lactamase producing E. coli [50].
Another important phenotypic mechanism of antibiotic resistance is the ‘inoculum effect’, described as an enhanced antibiotic resistance (i.e., higher minimum inhibitory concentration or MIC) with increasing inoculum density [51]. This density-dependent antibiotic resistance can have important implications for community-mediated resistance [48]. Our prediction here is that the inoculum effect will also contribute to enhanced resistance in mutualistic communities (as these are by definition more productive and achieve higher densities than competitive communities, see legend Figure 1A). Furthermore, a synergy between the inoculum effect and the partner shading effect could further increase resistance to narrow-spectrum antimicrobial clearance as increasing density and mixing combine to limit control. Hence, integrating structural and functional relationships into the broader theme of community-mediated resistance could shed light into the mechanisms underlying drug resistance.
Promoting microbiome health by managing polymicrobial interactions
Theoretical and experimental work on spatially explicit ‘minimal’ microbiomes can shed light on the causal mechanisms linking environmental inputs (e.g., nutrients or drugs) with microbiome structure and function as well as host health. Measuring host health (and disease) as a function of defined multispecies interactions can be approached experimentally using in vivo model systems [23,42,52], but also using simpler in vitro proxies, such as community productivity, antibiotic resistance, and robustness (i.e., invasion resistance). The main goal is then to identify the metabolic and demographic consequences of varying key environmental drivers (such as nutrients, drugs, flow rates, and initial mixing) for the structure and functioning of two-species communities (Figure 3). Knowing how the community responds to these environmental changes would present us with novel points of intervention, potentially allowing the tuning of species numbers (densities and frequencies), behaviour, and emergent structuring of communities.
Figure 3.
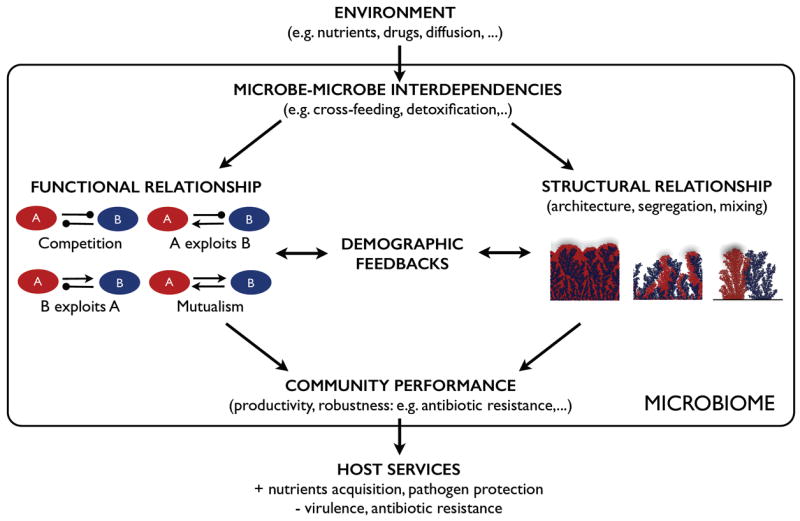
Schematic illustrating an integrative approach that aims at bridging the gap between cell and community properties to build an integrated mechanistic account of the functioning of the microbiome. Demographic feedbacks create links between structural and functional relationships. For example, mutualistic functional relationships will tend to increase lineage mixing (Figure 2), which in turn will enhance mutualistic functional relationships (Figure 2). Conversely, competitive functional relationships promote lineage segregation, which in turn attenuates competition (see [20]).
If we can influence human microbiome health via manipulation of their nutrient, drug, and mixing parameters, then what are the goals? For some microbial communities, the medical priority will be to prevent the establishment of a known pathogenic species (as in the case of Sg and Aa interactions where the priority is to prevent Aa establishment, [23]), but for other communities, the objective may be to encourage the growth of specific species or sets of species that are associated with human health (e.g., Bifidobacteria [53,54] or Bacteroides thetaiotomicron [55], suggested to reduce the occurrence of metabolic disorders associated with obesity and/or diabetes).
Moreover, virulence often emerges as a property of multi-species interactions, in particular mutualistic ones. For example, it has been demonstrated theoretically that mutualistic interactions among pathogens (e.g., HIV and tuberculosis [56]) are particularly dangerous due to the compounding effect of within-host demographic feedback [57], highlighting the importance of identifying effective control strategies to reduce mutualistic interdependency involving pathogens.
More generally, the existence of strong demographic feedbacks between species implies that treatment strategies cannot always focus on the behaviour of target species in isolation, but rather on managing the properties of polymicrobial interactions. These interactions open the door towards potential control strategies aiming to maximise (when beneficial – top row, Figure 4) or minimise (when harmful – bottom row, Figure 4) positive metabolic interactions between species.
Figure 4.
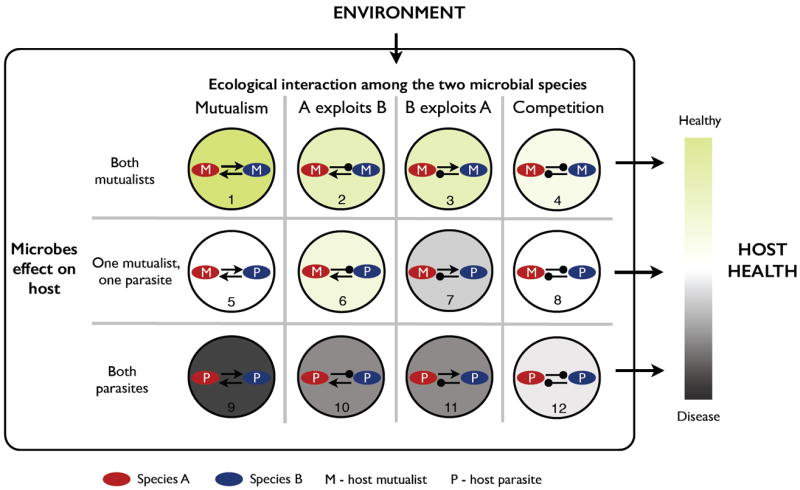
Schematic illustrating the diversity of potential ecological scenarios occurring between a host and a ‘minimal’ two-species community. To promote health and prevent disease, we aim at maximizing mutualism between host mutualists (1) and minimizing mutualism between host parasites (9). What we want to maximize crucially depends on the species that are present (i.e., mutualist or parasite) and the nature of their interaction (mutualism, exploitation, or competition), which is contingent on their environment. For instance, let’s focus on our minimal Sg–Aa microbiome model. When under anaerobic conditions, Sg and Aa are engaged in a competitive interaction. Sg is a better competitor, so it outcompetes Aa and thus protects the host. When under aerobic conditions, Sg and Aa are engaged in a mutualistic interaction. Although Sg is usually a commensal to the host, the harm to the host by Aa reduces the host fitness, thus this potential scenario should be minimized. For simplicity, the color shading illustrates potential symbiont effects on host health, assuming symmetric symbiont interactions, but the outcome will depend on the strength and symmetry of the interaction.
Concluding remarks: challenges and opportunities for progress
Microbiomes are immensely diverse and complex. While focusing on a two-species ‘minimal’ microbiome model is a vast simplification of real systems, a ‘minimal’ microbiome already gives rise to a diverse and complex network of potential ecological scenarios of microbe–microbe (Figure 1) and microbiome–host interactions (Figure 4). Here we suggest that a thorough mechanistic understanding of a two-species spatially-extended microbiome is therefore a necessary step towards understanding more diverse microbial communities and developing a better management of the human microbiome health. Furthermore, this approach is more easily testable experimentally (both in vitro and in vivo). A major challenge, however, is to integrate the findings of a ‘minimal’ microbiome approach into our understanding of diverse, natural microbiomes. A potential approach to this challenge is to group microbial species by functional traits rather than by phylogenetic similarity [58,59]. Grounded in ecological theory, the idea is that species with similar metabolisms and pathways would be grouped into the same functional group [59]. Using this functional classification approach provides a powerful complexity-reducing filter, and could allow testing the predictions of a ‘minimal’ microbiome model in natural microbial communities.
Another major challenge to furthering our understanding of microbiome dynamics is the ability to link scales from molecules to cells and communities (Box 1). But this is an exciting time to work at this interface, as new technologies are emerging that allow us to unravel the spatio-temporal dynamics of microbial communities [60]. For example, it is now possible to confine microbial cells in diffusive pico-litre scale traps to study their social behaviour [61], visualize single microbial cells in space and in real time [e.g., confocal laser scanning microscopy and fluorescence in situ hybridization (FISH)], as well as spatially map the molecular environment of microbial communities (e.g., scanning electrochemical microscopy [62,63] and nano-scale secondary-ion mass spectrometry (NanoSIMS) [64]). Major progress has been made by combining some of these techniques. For example, the coupling of FISH with SIMS or NanoSIMS (FISH-SIMS or FISH-NanoSIMS, respectively) has bridged the gap between species identity and metabolic function [65,66]. Using FISH-NanoSIMS, a recent study revealed the important role played by methanotrophic archaea in mediating the spatial distribution and extent of nitrogen fixation in methane seep sediments [67]. These approaches are particularly relevant when one wants to unravel the potential roles played by behavioural and regulatory interactions in the spatial and functional interactions of species. For instance, recent empirical work in a mouse model suggests that Sg–Aa synergistic interaction is enhanced by Aa’s ability to sense H2O2 and respond by modulating its own spatial positioning via dispersal. This ensures that the benefits of Aa cross-feeding on lactate are maximized while the costs incurred by the toxic effect of H2O2 are minimized [43]. This example illustrates the important role played by bacterial regulation (phenotypic plasticity) in shaping species spatial organization and enhancing bacterial virulence.
Box 1. Outstanding questions.
How do different mechanisms of metabolic interaction translate into functional relationships among species (competition, exploitation, and mutualism)?
How do functional relationships among species dictate patterns of spatial mixing?
Can we reverse-engineer functional and metabolic relationships from an analysis of spatial mixing patterns?
What are the implications of polymicrobial spatial and functional relationships for community performance (e.g., productivity, robustness to environmental perturbations)?
How do metabolic interactions and demographic feedbacks between species shape community-mediated resistance to antibiotics?
How do hosts tune metabolic and demographic interactions within host-associated communities to promote microbiome health?
Can we promote microbiome beneficial services to their host through interventions targeting microbe–microbe metabolic and demographic interactions?
From a modeling perspective, IBMs of biofilms have been of particular relevance to investigate questions at this interface as IBMs use a bottom-up approach where the community structure and dynamics arise as an emergent property of the interactions between individual cells [68]. In particular, these models have emphasized the importance of looking at structuring of surface attached communities as an emergent property of collective bacterial behavior and demography [20,21,26,37,69].
Coupling experimental techniques with spatially explicit IBMs of microbe–microbe interactions or host–microbe interactions is a challenging but promising research direction. But, in a time where multi-’omics’ approaches can produce a vast amount of data at an unprecedented pace, integrating experiments with theory can be essential to effectively exploit and interpret this information.
Highlights.
Metabolic interactions drive demographic feedbacks between species.
Demographic feedbacks strengthen or attenuate underlying metabolic interactions.
Demography is a key determinant of microbiome development and functioning.
Treatment strategies must consider the dynamical properties of species interactions.
Acknowledgments
We thank Ben Kerr, Luke McNally, Roman Popat, and two anonymous referees for comments on the manuscript. We thank the Human Frontier Science Program (HFSP RGP0011/2014, S.P.B. and M.W.), the National Institutes of Health (Grant 1RO1DE020100, M.W.), and FCT (SFRH/BD/33856/2009, S.E.) for funding, and PDBC-IGC for support (S.E.).
Glossary
- Competition
ecological interaction where all species are negatively affected by association due to, for example, consumption of shared and limiting resources.
- Cross-feeding
act of using the extracellular metabolic by-products of other organisms for growth.
- Demography
the study of population structure, emerging from patterns of births, deaths, migration, and development.
- Demographic feedback
an interaction where one species influences the demographic processes (births, deaths, and movement) of another species, and vice-versa.
- Exo-product
molecule secreted extracellularly and usually costly to produce (e.g., exopolysaccharide or exoenzyme).
- Exploitation
ecological interaction where one species benefits from association at the expense of the other species.
- Individual-based models (IBMs)
computational models tracking the dynamics of individual ‘agents’, often used to explore emergent properties of the population aggregate behavior.
- Microbiome
the set of microorganisms sharing a particular habitat, e.g. the human lower intestine.
- Mutualism
mutually beneficial ecological interaction between species.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nature Reviews Rheumatology. 2011;7:569–578. doi: 10.1038/nrrheum.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 4.Ley RE, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster JA, McVey Neufeld K-A. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 2013;36:305–312. doi: 10.1016/j.tins.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341 doi: 10.1126/science.1241214. 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenblum S, et al. Towards a predictive systems-level model of the human microbiome: progress, challenges, and opportunities. Current Opinion in Biotechnology. 2013;24:810–820. doi: 10.1016/j.copbio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy R, Borenstein E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc Natl Acad Sci U S A. 2013;110:12804–12809. doi: 10.1073/pnas.1300926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freilich S, et al. Competitive and cooperative metabolic interactions in bacterial communities. Nature Communications. 2011;2:589. doi: 10.1038/ncomms1597. [DOI] [PubMed] [Google Scholar]
- 11.Costello EK, et al. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nogueira T, et al. Horizontal gene transfer of the secretome drives the evolution of bacterial cooperation and virulence. Curr Biol. 2009;19:1683–1691. doi: 10.1016/j.cub.2009.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNally L, et al. Cooperative secretions facilitate host range expansion in bacteria. Nature Communications. 2014;5:4594. doi: 10.1038/ncomms5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahowald MA, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence D, et al. Species Interactions Alter Evolutionary Responses to a Novel Environment. Plos Biol. 2012;10:e1001330. doi: 10.1371/journal.pbio.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jagmann N, et al. Parasitic growth of Pseudomonas aeruginosa in co-culture with the chitinolytic bacterium Aeromonas hydrophila. Environmental Microbiology. 2010;12:1787–1802. doi: 10.1111/j.1462-2920.2010.02271.x. [DOI] [PubMed] [Google Scholar]
- 18.Großkopf T, Soyer OS. Synthetic microbial communities. Current Opinion in Microbiology. 2014;18:72–77. doi: 10.1016/j.mib.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estrela S, et al. From Metabolism to Ecology: Cross-Feeding Interactions Shape the Balance between Polymicrobial Conflict and Mutualism. Am Nat. 2012;180:566–576. doi: 10.1086/667887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estrela S, Brown SP. Metabolic and demographic feedbacks shape the emergent spatial structure and function of microbial communities. PLoS Comput Biol. 2013;9:e1003398. doi: 10.1371/journal.pcbi.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Momeni B, et al. Strong inter-population cooperation leads to partner intermixing in microbial communities. elife. 2013;2:e00230. doi: 10.7554/eLife.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller MJI, et al. Genetic drift opposes mutualism during spatial population expansion. Proc Natl Acad Sci U S A. 2014;111:1037–1042. doi: 10.1073/pnas.1313285111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramsey MM, et al. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011;7:e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harcombe W. Novel cooperation experimentally evolved between species. Evolution. 2010;64:2166–2172. doi: 10.1111/j.1558-5646.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- 25.Hillesland KL, Stahl DA. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Natl Acad Sci U S A. 2010;107:2124–2129. doi: 10.1073/pnas.0908456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Momeni B, et al. Spatial self-organization favors heterotypic cooperation over cheating. elife. 2013;2:e00960. doi: 10.7554/eLife.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bucci V, Xavier JB. Towards Predictive Models of the Human Gut Microbiome. Journal of Molecular Biology. 2014 doi: 10.1016/j.jmb.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manor O, et al. Mapping the Inner Workings of the Microbiome: Genomic- and Metagenomic-Based Study of Metabolism and Metabolic Interactions in the Human Microbiome. Cell Metab. 2014 doi: 10.1016/j.cmet.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bucci V, et al. Social interaction, noise and antibiotic-mediated switches in the intestinal microbiota. PLoS Comput Biol. 2012;8:e1002497. doi: 10.1371/journal.pcbi.1002497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stein RR, et al. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput Biol. 2013;9:e1003388. doi: 10.1371/journal.pcbi.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wintermute EH, Silver PA. Emergent cooperation in microbial metabolism. Molecular Systems Biology. 2010;6:1–7. doi: 10.1038/msb.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sergey S, et al. Metabolic modeling of a mutualistic microbial community. Molecular Systems Biology. 2007;3:92. doi: 10.1038/msb4100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klitgord N, Segrè D. Environments that induce synthetic microbial ecosystems. PLoS Comput Biol. 2010;6:e1001002. doi: 10.1371/journal.pcbi.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harcombe WR, et al. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Reports. 2014;7:1104–1115. doi: 10.1016/j.celrep.2014.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xavier JB, et al. A framework for multidimensional modelling of activity and structure of multispecies biofilms. Environmental Microbiology. 2005;7:1085–1103. doi: 10.1111/j.1462-2920.2005.00787.x. [DOI] [PubMed] [Google Scholar]
- 36.Lardon LA, et al. iDynoMiCS: next-generation individual-based modelling of biofilms. Environmental Microbiology. 2011;13:2416–2434. doi: 10.1111/j.1462-2920.2011.02414.x. [DOI] [PubMed] [Google Scholar]
- 37.Mitri S, et al. Social evolution in multispecies biofilms. Proc Natl Acad Sci U S A. 2011;108:10839–10846. doi: 10.1073/pnas.1100292108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schluter J, Foster KR. The evolution of mutualism in gut microbiota via host epithelial selection. Plos Biol. 2012;10:e1001424. doi: 10.1371/journal.pbio.1001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christensen BB, et al. Metabolic commensalism and competition in a two-species microbial consortium. Applied and Environmental Microbiology. 2002;68:2495–2502. doi: 10.1128/AEM.68.5.2495-2502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen SK, et al. Evolution of species interactions in a biofilm community. Nature. 2007;445:533–536. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 41.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 42.Korgaonkar A, et al. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A. 2013;110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stacy A, et al. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci U S A. 2014;111:7819–7824. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levin BR, Rozen DE. Non-inherited antibiotic resistance. Nature Reviews Microbiology. 2006;4:556–562. doi: 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- 45.Bull JJ, et al. Phenotypic Resistance and the Dynamics of Bacterial Escape from Phage Control. PLoS ONE. 2014;9:e94690. doi: 10.1371/journal.pone.0094690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis K. Persister cells and the riddle of biofilm survival. Biochemistry-Moscow. 2005;70:267–274. doi: 10.1007/s10541-005-0111-6. [DOI] [PubMed] [Google Scholar]
- 47.Connell JL, et al. 3D printing of microscopic bacterial communities. Proc Natl Acad Sci U S A. 2013;110:18380–18385. doi: 10.1073/pnas.1309729110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vega NM, Gore J. Collective antibiotic resistance: mechanisms and implications. Current Opinion in Microbiology. 2014;21:28–34. doi: 10.1016/j.mib.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vega NM, et al. Salmonella Typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc Natl Acad Sci U S A. 2013;110:14420–14425. doi: 10.1073/pnas.1308085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perlin MH, et al. Protection of Salmonella by ampicillin-resistant Escherichia coli in the presence of otherwise lethal drug concentrations. Proc Biol Sci. 2009;276:3759–3768. doi: 10.1098/rspb.2009.0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brook I. Inoculum effect. Review of Infectious Diseases. 1989 doi: 10.1093/clinids/11.3.361. [DOI] [PubMed] [Google Scholar]
- 52.Ramsey MM, Whiteley M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc Natl Acad Sci U S A. 2009;106:1578–1583. doi: 10.1073/pnas.0809533106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cani PD, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 54.Cani PD, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Falony G, et al. Coculture Fermentations of Bifidobacterium Species and Bacteroides thetaiotaomicron Reveal a Mechanistic Insight into the Prebiotic Effect of Inulin-Type Fructans. Applied and Environmental Microbiology. 2009;75:2312–2319. doi: 10.1128/AEM.02649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pawlowski A, et al. Tuberculosis and HIV co-infection. PLoS Pathog. 2012;8:e1002464. doi: 10.1371/journal.ppat.1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eswarappa SM, et al. Within-Host Dynamics of Multi-Species Infections: Facilitation, Competition and Virulence. PLoS ONE. 2012;7:e38730. doi: 10.1371/journal.pone.0038730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burke C, et al. Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci U S A. 2011;108:14288–14293. doi: 10.1073/pnas.1101591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shafquat A, et al. Functional and phylogenetic assembly of microbial communities in the human microbiome. Trends Microbiol. 2014;22:261–266. doi: 10.1016/j.tim.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wessel AK, et al. Going local: technologies for exploring bacterial microenvironments. Nature Publishing Group. 2013;11:337–348. doi: 10.1038/nrmicro3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Connell JL, et al. Probing Prokaryotic Social Behaviors with Bacterial “Lobster Traps”. mBio. 2010;1:e00202–10. doi: 10.1128/mBio.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koley D, et al. Discovery of a biofilm electrocline using real-time 3D metabolite analysis. Proc Natl Acad Sci U S A. 2011;108:19996–20001. doi: 10.1073/pnas.1117298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu X, et al. Real-time mapping of a hydrogen peroxide concentration profile across a polymicrobial bacterial biofilm using scanning electrochemical microscopy. Proc Natl Acad Sci U S A. 2011;108:2668–2673. doi: 10.1073/pnas.1018391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dekas AE, et al. Deep-Sea Archaea Fix and Share Nitrogen in Methane-Consuming Microbial Consortia. Science. 2009;326:422–426. doi: 10.1126/science.1178223. [DOI] [PubMed] [Google Scholar]
- 65.Pernthaler A, et al. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci U S A. 2008;105:7052–7057. doi: 10.1073/pnas.0711303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marlow JJ, et al. Carbonate-hosted methanotrophy represents an unrecognized methane sink in the deep sea. Nature Communications. 2014;5:5094. doi: 10.1038/ncomms6094. [DOI] [PubMed] [Google Scholar]
- 67.Dekas AE, et al. Spatial distribution of nitrogen fixation in methane seep sediment and the role of the ANME archaea. Environmental Microbiology. 2014;16:3012–3029. doi: 10.1111/1462-2920.12247. [DOI] [PubMed] [Google Scholar]
- 68.Kreft JU, et al. Individual-based modelling of biofilms. Microbiol-Uk. 2001;147:2897–2912. doi: 10.1099/00221287-147-11-2897. [DOI] [PubMed] [Google Scholar]
- 69.Nadell CD, et al. Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput Biol. 2010;6:e1000716. doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]


