Abstract
Re-sequencing permits the mining of genome-wide variations on a large scale and provides excellent resources for the research community. To accelerate the development and application of molecular markers and identify the QTLs affecting the flowering time-related trait in pepper, a total of 1,038 pairs of InDel and 674 SSR primers from different sources were used for genetic mapping using the F2 population (n = 154) derived from a cross between BA3 (C. annuum) and YNXML (C. frutescens). Of these, a total of 224 simple PCR-based markers, including 129 InDels and 95 SSRs, were validated and integrated into a map, which was designated as the BY map. The BY map consisted of 13 linkage groups (LGs) and spanned a total genetic distance of 1,249.77 cM with an average marker distance of 5.60 cM. Comparative analysis of the genetic and physical map based on the anchored markers showed that the BY map covered nearly the whole pepper genome. Based on the BY map, one major and five minor QTLs affecting the number of leaves on the primary axis (Nle) were detected on chromosomes P2, P7, P10 and P11 in 2012. The major QTL on P2 was confirmed based on another subset of the same F2 population (n = 147) in 2014 with selective genotyping of markers from the BY map. With the accomplishment of pepper whole genome sequencing and annotations (release 2.0), 153 candidate genes were predicted to embed in the Nle2.2 region, of which 12 important flowering related genes were obtained. The InDel/SSR-based interspecific genetic map, QTLs and candidate genes obtained by the present study will be useful for the downstream isolation of flowering time-related gene and other genetic applications for pepper.
Introduction
Capsicum is a member of the Solanaceae family and consists of the following five most important cultivated species, C. annuum, C. chinense Jacq., C. baccatum, C. pubescens Ruiz & Pavon and C. frutescens [1]. Of which C. annuum is most widely cultivated for use as food, spice, ornaments, and medicine around the world. C. frutescens and C. chinense exhibit a relatively closer interspecific relationship with C. annuum and provide critical resources for the genetic improvement of pepper production [2,3]. Even though interspecific crosses suffer from low fertility and high segregation distortion, they benefit from higher level of polymorphism [4] and provide opportunities to introduce economically valuable traits into the cultivated species [5].
During the last few decades, genetic maps have become the basic tool necessarily for genetics and breeding such as genome assembly, QTL analysis, gene tagging and marker-assisted selection (MAS). Numerous genetic maps including integrated maps have been constructed for pepper [6,7] using either intraspecific [8–12] or interspecific populations [13–20]. In these studies, different marker systems such as Restriction Fragment Length Polymorphism (RFLP), Random Amplified Polymorphic DNA (RAPD), Amplified Fragment Length Polymorphism (AFLP), Simple Sequence Repeat (SSR) and Single Nucleotide Polymorphism (SNP) had been used. However, the number of simple PCR-based molecular markers for pepper remains to be increased [8,17]. Insertion/deletion (InDel) polymorphism, which is known as a user-friendly marker type, has high variability and co-dominant inheritance and is relatively abundant and uniformly distributed throughout the genome [21,22]. With the decreasing cost of next generation sequencing, InDels can be developed on a large scale through re-sequencing and are becoming a popular choice for plant and animal systems [21–25]. Unfortunately, InDel discovery efforts have lagged significantly behind SSR discovery efforts and relatively few InDels have been identified in pepper. In addition, to our knowledge, InDel markers have seldom been used for genetic mapping in spite of more intensive sequencing of pepper in recent years [26–29]. Fairly recently, we constructed an initial InDel-based genetic map (BB-InDel map) using an intraspecific population [30].
In flowering plants, the initiation of flower primordia indicates the start of the transition from the vegetative phase to the reproductive phase that will definitively reflect the flowering time, which is one of the most important economic traits in conventional pepper breeding [31]. Pepper is a member of the Solanaceae family and has a sympodial shoot structure [32]. The formation of flower primordia is controlled by the shoot apical meristem (SAM), which terminates in an inflorescence meristem (IM) that subsequently develops into a solitary flower along with the reproductive transition [33]. Up to now, several genes controlling the transition to flowering and shoot architecture were reported in pepper through EMS mutagenesis [33–37]. Of these, Ca-ANANTHA (Ca-AN), CaBLAND (CaBL), CaJOINTLESS (CaJ) and Capsicum annuum S (CaS) were all found to promote the early flower formation in pepper while FASCICULATE (FA) stimulated late flowering. In addition, both genes CaHAM and CaBL participated in the controlling of axillary meristem formation [38]. The relationships between these genes were also partly investigated. For example, CaJ showed epistasis over FA [36] and CaBL functions independently of FA in regulating sympodial growth, but is epistatic to FA in controlling axillary meristem formation [35]. Recent results also indicated that CaS is epistatic over other genes controlling the transition to flowering with respect to flower formation [33]. Even so, the molecular regulatory mechanism of pepper flowering primordia initiation is poorly understood. More importantly, the cause of wide natural variation in flowering time is still cryptic for pepper. In fact, pepper exhibits widespread natural variation in flowering time and the number of leaves on the primary axis (Nle) ranges from 1 to more than 20 in different species [39]. Classical quantitative genetic analysis and QTL mapping showed that Nle was commonly controlled by a few major genes with some minor factors, as well as the environment [12,31,39–44]. Additionally, so far QTLs affecting Nle had been identified on all pepper chromosomes with exception of P9 and P10 using different populations [31,40,44–46]. However, most of these studies were based on intraspecific populations.
In this study, a genetic linkage map was first constructed based on InDel and SSR markers using the F2 population derived from an interspecific cross between BA3 (C. annuum) and YNXML (C. frutescens). Subsequently, QTL analysis was performed to identify the genomic region associated with the flowering time-related trait (namely Nle) by using the two subsets of the same F2 population in 2012 and 2014, respectively. Finally, the candidate genes embed in the QTL region were discussed. The genetic map, QTLs and candidate genes therein this study will provide useful information for molecular assisted selection (MAS) breeding, and lay the foundation for the isolation of genes underlying the variation in flowering time in pepper.
Materials and Methods
Plant materials and trait evaluation
An F2 genetic mapping population was derived from the cross between the inbred lines BA3 (C. annuum) and YNXML (C. frutescens), both of which were re-sequenced [1]. BA3 is a cytoplasmic male sterility (CMS) line with Nle ranged from 8 to 12. YNXML, which is a pungent type with small size and erect fruit, was collected from Yunnan Province, China, and its Nle is approximately two times as BA3, which led directly to a flowering time that occurred 10~15 days later than BA3. The F2 population was divided into two sub-groups consisting of 154 and 147 individuals, respectively. Approved by the Office for Teaching & Research Bases, the two subsets, together with the parental lines and their hybrid population, were grown successively at the Zengcheng Experimental Station, South China Agricultural University (SCAU), Guangzhou, China (23° 083 N, 113mental) in 2012 and 2014, respectively. Genomic DNA was extracted from young leaves using the modified CTAB method [47]. The Nle were numbered successively from the node of the cotyledon to the first flower node on the main stem, as recommended by the IPGRI (The International Plant Genetic Resources Institute), for each individual plant after the formation of the first branch.
Sources and genotyping of InDel and SSR
Information on the InDel and SSR primers with different sources [17,48–50] that were used in the present study are summarized in. A total of 1000 pairs of InDel primers recently developed from re-sequencing [30] were selected and used to screen the parental lines (BA3 and YNXML) for polymorphism. An additional 38 InDels, which have been predicted between BA3 and YNXML using the same bioinformatics analysis pipeline, were selected to increase the marker density of P2. In addition, a total of 420 EST-SSRs from a public database (http://compbio.dfci.harvard.edu/tgi/plant.html) were previously identified by our group [48], and also used for polymorphism screening in the present study. 119 primer pairs of genomic SSR markers and 135 primer pairs of EST-SSR markers that were previously reported (Table 1) were also used. A PCR mixture contained 10 ng genomic DNA, 200 μM of each dNTP, 2 μM of each primer, 1 × reaction buffer, 37.5 μM of Mg2+, and 0.5 unit of Taq polymerase (Dsbio) in a final volume of 20 μL. The reaction was performed as follows: an initial cycle of 5 min at 94°C; 34 cycles of 45 s at 94°C, 45 s at 58°, and 1 min at 72°m, and a final 10 min at 72°m.
Table 1. Polymorphism screening of primers from different sources.
| Marker type | Sources | Prefixed | No. of primers | Successful amplification | Polymorphic | Sources | ||
|---|---|---|---|---|---|---|---|---|
| N | Percentage (%) | N | Percentage (%) | |||||
| InDel | Re-sequencing | CIDH_ | 1,000 | 976 | 97.60 | 129 | 12.90 | [30] |
| InDel | Re-sequencing | CIDHjw_ | 38 | 35 | 92.11 | 11 | 28.95 | In present study |
| SSR | EST | PSE_ | 420 | 403 | 95.95 | 64 | 15.24 | [48] |
| SSR | EST | EPMS_ | 135 | 130 | 96.30 | 22 | 16.30 | [50] |
| SSR | Genome | GPMS_, Hpms_, AF_ | 119 | 113 | 94.96 | 16 | 13.45 | [17,49] |
| Total | - | - | 1,712 | 1657 | 96.79 | 242 | 14.14 | - |
Genetic map construction and comparison with the physical map
A genetic map was constructed using JoinMap 4.0 software with a population type code, F2 [51]. Most of the InDel markers were grouped according to the physical mapping information [30]. The remaining InDel markers from P0 were assigned to the known groups using the Strongest Cross Link (SCL) information. Both the SCL information and BLAST tool [52] were used to map the SSR markers onto the pseudo chromosomes (groups) of the Zunla-1 reference genome (http://peppersequence.genomics.cn). Recombination values were converted to genetic distances using the Kosambi mapping function [53] and a comparative map was drawn using Mapchart 2.2 [54]. The segregation ratios of markers in the population were examined by Chi-square analysis. Markers with segregation ratios that differed from expected 1:2:1 or 3:1 at P <0.05 were classified as segregation distortion markers. Similar to the definition in previous study [55], a region with five or more adjacent skewed segregation markers was defined as a segregation distortion region (SDR) in present study.
QTL analysis of Nle
Both of the Inclusive Composite Interval Mapping (ICIM) [56] and Composite Interval Mapping (CIM) was initially applied to detect QTLs (LOD > 2.5) for Nle with the sub-population (n = 154) of 2012 using the QTL IciMapping 4.0 and Windows QTL Cartographer 2.5 [57], respectively. Further, a set of 111 markers from the above map, which were uniformly distributed on the whole physical map but with preference to the chromosome P2, was selected to genotype another sub-population (n = 147) of 2014 and subsequently independent QTL analysis was performed as above to evaluate the primary QTL results of 2012. On the other hand, Single Marker Analysis (SMA) was carried out on a dataset that normalized from the phenotypic data of two years (2012 and 2014) with the selected positive markers from the 2012 analyses as well.
Results and Discussion
Polymorphism screening
In order to construct an interspecific genetic map based purely on user-friendly PCR markers, InDel and SSR primers from different sources (Table 1) were used for polymorphism screening between BA3 (C. annuum) and YNXML (C. frutescens). A total of 1,000 pairs of InDel primers that were developed previously by our group [30] and an additional 38 predictably polymorphic InDels between BA3 and YNXML were selected and then subsequently used to screen for polymorphism between the parental lines (BA3 and YNXML). Finally, 140 out of 1,038 pairs of InDel primers were validated and used for further genetic mapping (Table 1 and S1 Table.). On the other hand, 64 polymorphic SSR loci (S2 Table.) between BA3 and YNXML were identified from the 420 pairs of EST-SSR primers that developed by our group [48]. None of these EST-SSR markers have been used for genetic mapping and their rate of polymorphism (15.24%) is similar to previous reports (16.30%), and slightly higher than that of genome-derived SSRs (13.45%, Table 1). In total, 1712 pairs of PCR-based primers with different sources were analyzed and 242 pairs of polymorphic primers (14.14%) were validated and then subsequently applied to the genotyping of the F2 individuals (Table 1).
Genetic map construction
The F2 progenies were genotyped with the 242 markers, including the 140 InDels and 102 SSRs (S1 and S2 Tables.). Of these, 4 SSR markers with inconsistent grouping results between SCL-based assigning and BLAST-based physical mapping were excluded before mapping (S2 Table.). Finally, a genetic map (Fig. 1 and S1 Fig.), designated as the BY map, was constructed with a total of 224 simple PCR-based markers (including 129 InDels and 95 SSRs), the 14 remaining markers (11 InDels and 3 SSRs) were not integrated because of insufficient linkage. The BY map consisted of 13 linkage groups (LGs) covering a total genetic distance of 1249.77 cM with an average density of one framework marker for every 5.60 cM (Table 2 and S3 Table.). The number of mapped markers per LG ranged from 5 to 28 with an average of 17.23 markers. The largest and smallest genetic distance between two markers was 30.45 cM and 0.10 cM, respectively. This interspecific map that purely based on InDel and SSR would be useful for both of the basic and applied research for pepper in the future.
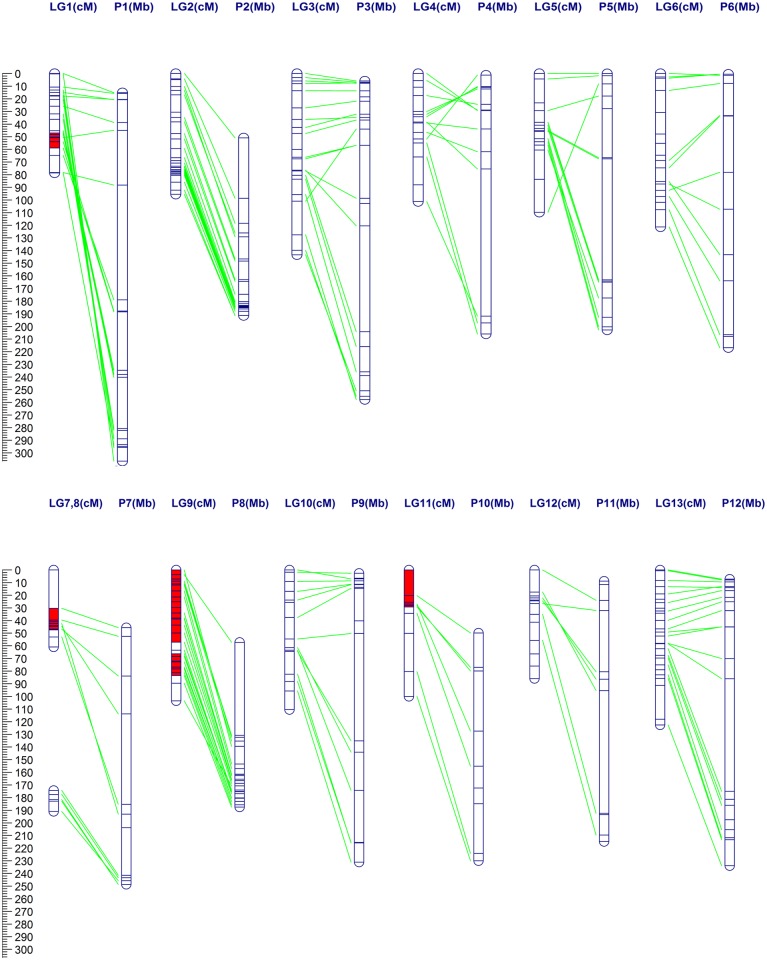
Fig 1. An interspecific genetic map of pepper based on 224 InDel and SSR markers, and the comparison with its physical map.
A total of 13 LGs (LG1 ~ LG13) were assigned to the corresponding chromosomes (P1 ~ P12) based on anchored markers. P7 was divided into LG7 and LG8 due to insufficient linkage. Green lines indicate the synteny between the genetic and physical maps. Five SDRs on LG1 (= P1), LG7 (= P7), LG9 (= P8) and LG11 (= P10) are filled with a red color.
Table 2. Statistics of the pepper BY map based on InDel and SSR markers.
| Linkage group | Chromosome | No. of markers | Marker distance (cM) | Map length | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| InDel | SSR | InDel and SSR | Distorted | Average | Min | Max | Genetic (cM) | Physical a (Mb) | ||
| LG1 | P1 | 11 | 10 | 21 | 11 | 3.93 | 0.22 | 13.59 | 78.57 | 291.27 |
| LG2 | P2 | 17 | 11 | 28 | 4 | 3.54 | 0.10 | 13.73 | 95.52 | 140.55 |
| LG3 | P3 | 12 | 9 | 21 | 10 | 7.16 | 0.57 | 26.58 | 143.26 | 251.91 |
| LG4 | P4 | 9 | 6 | 15 | 5 | 7.24 | 0.68 | 21.95 | 101.34 | 204.67 |
| LG5 | P5 | 9 | 6 | 15 | 2 | 7.83 | 0.67 | 26.00 | 109.66 | 202.72 |
| LG6 | P6 | 10 | 7 | 17 | 3 | 7.58 | 0.97 | 17.37 | 121.29 | 216.29 |
| LG7 | P7 | 7 | 4 | 11 | 8 | 6.09 | 0.60 | 30.45 | 60.91 | 147.49 |
| LG8 | P7 | 1 | 4 | 5 | 3 | 4.19 | 1.20 | 8.25 | 16.74 | 7.15 |
| LG9 | P8 | 13 | 14 | 27 | 25 | 3.98 | 0.58 | 13.86 | 103.47 | 130.30 |
| LG10 | P9 | 8 | 7 | 15 | 1 | 7.89 | 1.09 | 17.93 | 110.48 | 228.33 |
| LG11 | P10 | 9 | 1 | 10 | 8 | 11.11 | 1.12 | 30.13 | 100.03 | 180.06 |
| LG12 | P11 | 7 | 6 | 13 | 6 | 7.16 | 0.75 | 17.53 | 85.92 | 185.61 |
| LG13 | P12 | 16 | 10 | 26 | 5 | 4.90 | 0.33 | 26.61 | 122.56 | 226.55 |
| Total | - | 129 | 95 | 224 | 91 | 5.60 | - | - | 1249.77 | 2412.90 |
a Physical distance spanned by the BY map.
In this study, 91 out of 224 (40.63%) markers showed distorted segregation at an P<0.05 significance level (Table 2 and S3 Table.), which was considerably higher than that of several intraspecific populations [9,10,58] but similar to interspecific crossings [19,20]. Moreover, five segregation distortion regions (SDRs) were found on LG1, LG7, LG9 and LG11 (Fig. 1 and S3 Table.). Interestingly, all of the marker alleles within SDRs on LG7 (= P7) and LG11 (= P10) were skewed toward the female line BA3, whereas almost all were associated with the hybrid (F1) of the parental lines on LG1 (= P1) and LG9 (= P8). This indicates that there may be some segregation distorted factors in these regions [59]. For example, the incomplete chromosome pairing that resulted from reciprocal translocation between chromosome P1 and P8 [15] and the reduced recombination in this interspecific cross may be important determining factors in the selection of heterozygous genotypes in these areas.
Comparison of genetic and physical maps
Since the 12 pseudo chromosomes (nominated as P1~ P12 [15]) of the Zunla-1 reference genome were built previously [1], the 13 LGs of the present BY map were successfully assigned to the 12 corresponding chromosomes in the haploid pepper genome based on 184 anchored markers including the 116 InDels and 68 SSRs (S1 and S2 Tables.). Even though the density of one marker per 14.96 Mb was low relative to the Zunla-1 reference genome (3.35 Gb), the total physical distance spanned by the BY map was 2,412.90 Mb (Table 2), which accounted for 72.03% of the Zunla-1 reference genome. Based on the comparative analysis (Fig. 1), P7 was found to be divided into two LGs (LG7 and LG8) due to insufficient linkage between them. Additionally, the high degree of consistency between the genetic and physical positions on P2, P8, P10 and P12 indicated these chromosomes are relatively conserved between the C. annuum and C. frutescens genomes [15]. Meanwhile, many inversions, especially for P1, could account for the variations between C. frutescens and C. annuum. Regardless, the relatively high whole genome coverage suggests that the BY map can serve as a basic reference map for targeted saturation and genetic applications in the future.
Genetic analysis of Nle
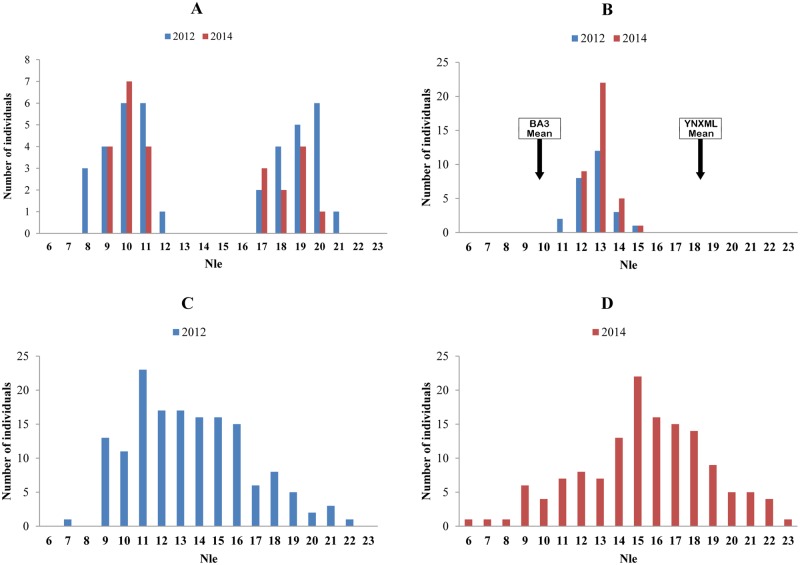
Nle is one of the component traits of the complex trait earliness, which is tightly correlated with flowering time [31,37]. There were significant differences (P < 0.01) in Nle between BA3 and YNXML (Table 3 and Fig. 2A), which led directly to a flowering time that occurred 10 ~ 15 days later in YNXML. The Nle that in the BA3 × YNXML hybrid (F1) is maternal biased (Fig. 2B), and it showed significantly continuous variation in the two subsets of F2 progenies with unimodal distribution (Figs. 2C and 2D). The heritability estimates of Nle were as high as 87.89% and 93.79% in 2012 and 2014, respectively (Table 3). These data indicate that Nle is suitable for artificial selection and is under polygenic control.
Table 3. Comparison of Nle in different generations and years.
| Years | BA3 | YNXML | BA3 × YNXML F1 | BA3 × YNXML F2 | Broad heritability (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Mean | SD | N | Mean | SD | N | Mean | SD | N | Mean | SD | ||
| 2012 | 20 | 9.90 | 1.17 | 18 | 19.00 | 1.14 | 26 | 12.73 | 0.92 | 154 | 13.52 | 3.10 | 87.89 |
| 2014 | 15 | 10.00 | 0.76 | 10 | 18.30 | 1.06 | 37 | 12.95 | 0.70 | 139 | 15.40 | 3.43 | 93.79 |
Fig 2. Frequency distribution of Nle for different populations in 2012 and 2014.
A: Parental lines (Left: BA3, Right: YNXML), B: F1 population, the mean of parental lines were shown with black arrows, C and D: F2 populations in 2012 and 2014, respectively.
Identification of the QTLs affecting Nle
By combining the new BY genetic map with the phenotypic value (S4 Table.) of Nle from the subset of the F2 population (n = 154) in 2012, a total of 6 QTLs, including one major (named Nle2.2) and 5 minor QTLs (Nle2.1, Nle7.1, Nle10.1 Nle10.2 and Nle11.1), were detected on P2, P7, P10 and P11 with both ICIM and CIM methods (Table 4). The results were consistent with the classic statistical genetic analysis in the present study as well as earlier studies [39]. Phenotypic variation explained by these QTLs varied between 2.09 and 51.63%. Except for the Nle11.1 identified by the CIM, the Nle-increasing alleles within the remaining 5 QTLs were all from the parent YNXML (Table 4).
Table 4. QTLs for Nle identified by two mapping methods in the present study.
| Year | Method | QTL a | Chromosome | Position | Interval b | LOD | PVE c (%) | Add | Dom |
|---|---|---|---|---|---|---|---|---|---|
| 2012 | ICIM | Nle2.1 | P2 | 10.00 | Hpms1_106—CIDH197 | 3.53 | 5.48 | 0.42 | -1.29 |
| Nle2.2 | P2 | 79.00 | EPMS677—CIDHjw1_24 | 22.79 | 48.32 | 3.16 | -0.46 | ||
| Nle7.1 | P7 | 42.00 | CIDH66—Hpms1_166 | 3.17 | 4.55 | 0.78 | -1.04 | ||
| Nle10.1 | P10 | 36.00 | CIDH607—CIDH619 | 2.90 | 5.19 | 1.02 | -0.12 | ||
| CIM | Nle2.1 | P2 | 10.20 | CIDH197—PSE342 | 4.24 | 7.59 | 0.52 | -1.36 | |
| Nle2.2 | P2 | 78.80 | EPMS677—CIDHjw1–24 | 19.80 | 51.63 | 2.98 | -0.84 | ||
| Nle10.2 | P10 | 27.00 | CIDH356—CIDH985 | 2.52 | 2.09 | 0.79 | 0.34 | ||
| Nle11.1 | P11 | 57.60 | CIDH799—CIDH146 | 2.54 | 3.32 | -1.02 | -0.21 | ||
| 2014 | ICIM | Nle2.2 | P2 | 82.00 | CIDHjw1_24—CIDHjw2_2 | 26.53 | 58.79 | 3.78 | 1.21 |
| CIM | Nle2.2 | P2 | 82.30 | CIDHjw2_2—CIDHjw2_6 | 29.68 | 31.14 | 3.87 | 1.42 |
a Two QTLs from the P10 were named Nle10.1 and Nle10.2, respectively, because the genetic distance between them was over 5 cM.
b The marker that was closer to the peak of LOD was unlined.
c PVE, phenotypic variation explained by the QTL.
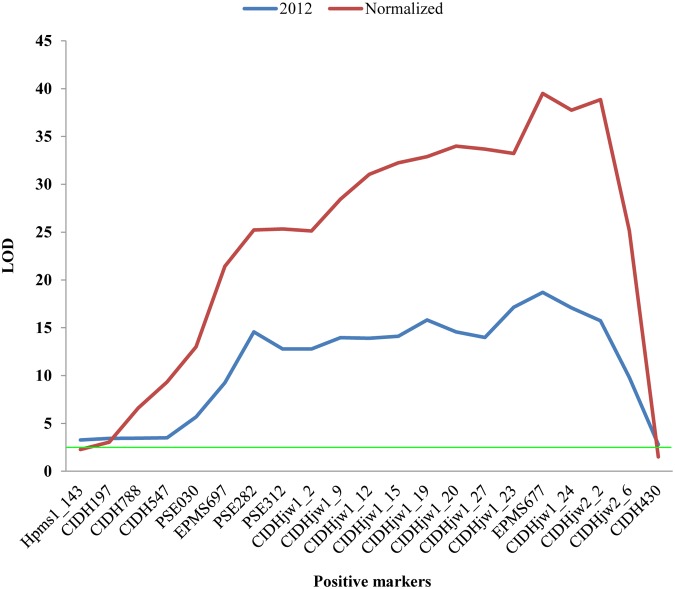
To evaluate the QTL results, another subset of the F2 population (n = 147) was genotyped using a set of selective markers (n = 111) from the BY map and another map consisted of 13 LGs with 94 markers was constructed (unpublished data). After collecting the Nle phenotypic data from 139 out of 147 individuals planted in 2014, the major QTL, which was assumed to be equal to Nle2.2 since the LOD peak markers EMPS677 and CIDHjw2_2 were tightly linked with physical distance of ~882.07 kb, was repeatedly identified using either ICIM or CIM method (Table 4). The results showed that the effect of Nle2.2 on controlling the Nle was less affected by the environment and were consistent with the characterization of high heritability of this trait (Table 3). In addition, single marker analysis (SMA) was also carried out on a dataset that normalized from the phenotypic data of two years (2012 and 2014) with 21 selected positive markers (total number of the positive makers is 28) from the 2012 analyses. Plot of LOD (Fig. 3) showed that both peaks of 2012 and normalized were agreement with the results of ICIM and CIM indicating again that the environment has little influence on the Nle.
Fig 3. Likelihood profile comparison of single marker analysis with phenotypic data from 2012 and normalized, respectively.
The threshold value of LOD = 2.5 was showed with green line.
Previously, QTL analysis for Nle was conducted using different populations [31,40,44–46], most of which were intraspecific populations (Table 5). Among these studies, the cross used for QTL detection that was most similar to the current study was “B9431 × H108”, which is also an interspecific cross between C. annuum and C. frutescens. In addition, a total of 3 QTLs were detected on LG1 (= P6), LG7 (= P5) and LG22 (= P2) [46]. Interestingly, the QTL with the highest PVE (12.6%) was on P2 and one of its flanking markers, the CAMS-327, was found approximately 477.15 kb and 1,359.00 kb downstream of our two LOD peak markers (CIDHjw2_2 and EMPS677). However, the relationship between the two major QTLs requires further research. In summary, the QTLs affecting Nle were detected on all chromosomes except for P9 in Capsicum so far (Table 5), and there is at least one major QTL underlying the significantly natural variation in Nle between C. annuum and C. frutescens on chromosome P2.
Table 5. Comparison of mapped QTLs for Nle among studies.
| Cross | Population type | Size | Number of QTLs | Chromosomes (LGs) | PVE(%) | References |
|---|---|---|---|---|---|---|
| BA3 × YNXML | Interspecific F2 | 154+139 | 6 | P2, P7, P10, P11 | 2.1 ~ 58.8 | This study |
| 83–58 × perennial | Intraspecific RIL | 122 | 3 | P1, P2, P6 | 7.2 ~ 21.2 | [45] |
| H3 × 83–60 | Intraspecific RIL | 100 | 8 | P1, P2, P3, P4, P7, P8, P12 | 18.5 ~ 55.1 | [45] |
| B9431× H108 | Interspecific F2 | 180 | 3 | LG1, LG7, LG22 | 6.4 ~ 12.6 | [46] |
| YW × CM334 | Intraspecific RIL | 149 | 2 | P3, P12 | - | [40] |
| CW × LS2341 | Intraspecific DH | 94 | 2 | P12, LG8 | 20.0 ~ 33.0 | [31] |
| YW × CM334 | Intraspecific RIL | 297 | 4 | P3, LG38 (= P11), LG45 (= P3), LG47 | 4.0 ~ 11.0 | [44] |
Candidate genes in the Nle2.2 region
Through integrating the QTL results from the two years (Table 4), we can delimit a loose candidate region for the major QTL Nle2.2 into an interval between the marker EPMS677 and CIDHjw2_6 with physical distance of ~2.76 Mb on chromosome P2. A total of 153 protein coding genes (S5 Table.), including 37 new genes without homologs in public database, were predicted to embed in this region based on the current annotations of the Zunla-1 reference genome (http://peppersequence.genomics.cn). Even though two of the six flowering related genes reported previously in pepper, CaS (KC414761, equal to Capana02g001854) and Ca-AN (FJ190669, equal to Capana02g002328), were both mapped/anchored on the chromosome P2 as well [33,34], they are not included in the Nle2.2 candidate region. On the other hand, because 12 out of the 153 genes that were found to be homologous to the flower/inflorescence development related proteins of Arabidopsis (Table 6), and consequently they were recommended as important candidate genes for the major QTL Nle2.2 of pepper. Especially for Capana02g003062, which is homologous to the Arabidopsis AP2 gene, as well as Capana02g003067 and Capana02g003070, both of which are the homologs of Arabidopsis CLF. More importantly, both of AP2 and CLF were important members of the flowering time pathway in Arabidopsis. For example, earlier studies reported that AP2 was involved in the specification of floral organ identity, establishment of floral meristem identity and suppression of floral meristem indeterminacy [60–62]. In addition, CLF was also found to have effects on the vegetative phase to reproductive phase transition of meristem [63]. Taken these together, we suppose that Nle2.2 is possibly a new member that participated in the flowering time regulation pathway of pepper and mainly controls the natural variation with respect to Nle in Capsicum population. The findings in present study would not only be useful for the isolation of genes controlling the initiation of flower primordia, but also provided insights into the molecular regulation of flowering time in pepper.
Table 6. List of 12 important candidate genes for the Nle2.2 of pepper and homologs of Arabidopsis.
| Gene ID | Position on chromosome P2 | Homologous gene symbol | Descriptions | |
|---|---|---|---|---|
| Start | Stop | |||
| Capana02g003062 | 154,819,441 | 154,822,016 | AP2 | Floral homeotic protein APETALA 2 |
| Capana02g003067 | 154,918,269 | 154,941,151 | CLF | Histone-lysine N-methyltransferase CLF |
| Capana02g003070 | 154,981,807 | 155,004,727 | CLF | Histone-lysine N-methyltransferase CLF |
| Capana02g003079 | 155,444,041 | 155,447,464 | SPT | Transcription factor SPATULA |
| Capana02g003089 | 155,587,450 | 155,588,112 | HEC2 | Transcription factor HEC2 |
| Capana02g003131 | 156,190,290 | 156,194,955 | ANP1 | Mitogen-activated protein kinase kinase kinase ANP1 |
| Capana02g003133 | 156,215,286 | 156,216,230 | LBD36 | LOB domain-containing protein 36 |
| Capana02g003199 | 157,107,846 | 157,110,095 | COL2 | Zinc finger protein CONSTANS-LIKE 2 |
| Capana02g003200 | 157,118,313 | 157,120,062 | COL2 | Zinc finger protein CONSTANS-LIKE 2 |
| Capana02g003201 | 157,124,787 | 157,126,407 | COL2 | Zinc finger protein CONSTANS-LIKE 2 |
| Capana02g003223 | 157,449,873 | 157,469,179 | BIG | Auxin transport protein BIG |
| Capana02g003224 | 157,469,217 | 157,471,764 | BIG | Auxin transport protein BIG |
Conclusions
An interspecific genetic map of pepper, comprising of a total of 224 purely anchored markers including InDel and SSR, was constructed. Assignment of the LGs to corresponding chromosomes indicated the relatively high coverage and confirmed the variations of Capsicum genomes. One major QTL (Nle2.2) influencing flowering time was identified on the chromosome P2 in 2012 and confirmed in 2014. Based on the annotations of Zunla-1 reference genome, 153 protein coding candidate genes were suggested. Hence, the map, QTLs and candidate genes obtained by the present study will be useful for future basic and applied research with respect to flowering time in Capsicum.
Supporting Information
A (red): homozygote as BA3, H (yellow): heterozygote as F1, B (dark blue): homozygote as YNXML, C (light blue): not A, D (brown): not B, U (gray), missing.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
This work was supported by the Guangdong Natural Science Foundation of China (S2011030001410), the National High Technology Research and Development Program ("863" Program) of China (2012AA100103), the National Natural Science Foundation of China (31372076), the Zunyi City Natural Science Foundation of China (No. 201201) and the Guizhou Province and Zunyi City Science and Technology Cooperation Project of China (No. 201307).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Guangdong Natural Science Foundation of China (S2011030001410), the National High Technology Research and Development Program (“863” Program) of China (2012AA100103), the National Natural Science Foundation of China (31372076), the Zunyi City Natural Science Foundation of China (No. 201201) and the Guizhou Province and Zunyi City Science and Technology Cooperation Project of China (No. 201307). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Qin C, Yu C, Shen Y, Fang X, Chen L, Min J, et al. (2014) Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proceedings of the National Academy of the Sciences of the United States of America 111: 5135–5140. 10.1073/pnas.1400975111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ibiza VP, Blanca J, Cañizares J, Nuez F (2012) Taxonomy and genetic diversity of domesticated Capsicum species in the Andean region. Genetic Resources and Crop Evolution 59: 1077–1088. [Google Scholar]
- 3. Nicolaï M, Cantet M, Lefebvre V, Sage-Palloix A- M, Palloix A (2013) Genotyping a large collection of pepper (Capsicum spp.) with SSR loci brings new evidence for the wild origin of cultivated C. annuum and the structuring of genetic diversity by human selection of cultivar types. Genetic Resources and Crop Evolution 60: 2375–2390. [Google Scholar]
- 4. Minamiyama Y, Tsuro M, Hirai M (2006) An SSR-based linkage map of Capsicum annuum . Molecular Breeding 18: 157–169. [Google Scholar]
- 5. Yoon JB, Yang DC, Do JW, Park HG (2006) Overcoming two post-fertilization genetic barriers in interspecific hybridization between Capsicum annuum and C. baccatum for introgression of Anthracnose resistance. Breeding Science 56: 31–38. [Google Scholar]
- 6. Lee HR, Bae IH, Park SW, Kim HJ, Min WK, Han JH, et al. (2009) Construction of an integrated pepper map using RFLP, SSR, CAPS, AFLP, WRKY, rRAMP, and BAC end sequences. Molecules and cells 27: 21–37. 10.1007/s10059-009-0002-6 [DOI] [PubMed] [Google Scholar]
- 7. Paran I, van der Voort JR, Lefebvre V, Jahn M, Landry L, van Schriek M, et al. (2004) An integrated genetic linkage map of pepper (Capsicum spp.). Molecular Breeding 13: 251–261. [Google Scholar]
- 8. Sugita T, Semi Y, Sawada H, Utoyama Y, Hosomi Y, Yoshimoto E, et al. (2013) Development of simple sequence repeat markers and construction of a high-density linkage map of Capsicum annuum . Molecular Breeding 31: 909–920. [Google Scholar]
- 9. Barchi L, Bonnet J, Boudet C, Signoret P, Nagy I, Lanteri S, et al. (2007) A high-resolution, intraspecific linkage map of pepper (Capsicum annuum L.) and selection of reduced recombinant inbred line subsets for fast mapping. Genome 50: 51–60. [DOI] [PubMed] [Google Scholar]
- 10. Sugita T, Kinoshita T, Kawano T, Yuji K, Yamaguchi K, Nagata R, et al. (2005) Rapid construction of a linkage map using high-efficiency genome scanning/AFLP and RAPD, based on an intraspecific, doubled-haploid population of Capsicum annuum . Breeding Science 55: 287–295. [Google Scholar]
- 11. Ogundiwin EA, Berke TF, Massoudi M, Black LL, Huestis G, Choi D, et al. (2005) Construction of 2 intraspecific linkage maps and identification of resistance QTLs for Phytophthora capsici root-rot and foliar-blight diseases of pepper (Capsicum annuum L.). Genome 48: 698–711. [DOI] [PubMed] [Google Scholar]
- 12. Ben-Chaim A, Paran I, Grube RC, Jahn M, van Wijk R, Peleman J (2001) QTL mapping of fruit-related traits in pepper (Capsicum annuum). Theoretical and Applied Genetics 102: 1016–1028. [Google Scholar]
- 13. Park SW, Jung JK, Choi EA, Kwon JK, Kang JH, Jahn M, et al. (2014) An EST-based linkage map reveals chromosomal translocation in Capsicum . Molecular Breeding 34: 963–975. [Google Scholar]
- 14. Yarnes SC, Ashrafi H, Reyes-Chin-Wo S, Hill TA, Stoffel KM, Van Deynze A (2013) Identification of QTLs for capsaicinoids, fruit quality, and plant architecture-related traits in an interspecific Capsicum RIL population. Genome 56: 61–74. 10.1139/gen-2012-0083 [DOI] [PubMed] [Google Scholar]
- 15. Wu F, Eannetta NT, Xu Y, Durrett R, Mazourek M, Jahn MM, et al. (2009) A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum . Theoretical and Applied Genetics 118: 1279–1293. 10.1007/s00122-009-0980-y [DOI] [PubMed] [Google Scholar]
- 16. Ben-Chaim A, Borovsky Y, Falise M, Mazourek M, Kang BC, Paran I, et al. (2006) QTL analysis for capsaicinoid content in Capsicum . Theoretical and Applied Genetics 113: 1481–1490. [DOI] [PubMed] [Google Scholar]
- 17. Lee JM, Nahm SH, Kim YM, Kim BD (2004) Characterization and molecular genetic mapping of microsatellite loci in pepper. Theoretical and Applied Genetics 108: 619–627. [DOI] [PubMed] [Google Scholar]
- 18. Rao GU, Ben Chaim A, Borovsky Y, Paran I (2003) Mapping of yield-related QTLs in pepper in an interspecific cross of Capsicum annuum and C. frutescens . Theoretical and Applied Genetics 106: 1457–1466. [DOI] [PubMed] [Google Scholar]
- 19. Kang BC, Nahm SH, Huh JH, Yoo HS, Yu JW, Lee MH, et al. (2001) An interspecific (Capsicum annuum ×C. chinese) F2 linkage map in pepper using RFLP and AFLP markers. Theoretical and Applied Genetics 102: 531–539. [Google Scholar]
- 20. Livingstone KD, Lackney VK, Blauth JR, van Wijk R, Jahn MK (1999) Genome mapping in Capsicum and the evolution of genome structure in the solanaceae. Genetics 152: 1183–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, et al. (2006) An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Research 16: 1182–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pacurar DI, Pacurar ML, Street N, Bussell JD, Pop TI, Gutierrez L, et al. (2012) A collection of INDEL markers for map-based cloning in seven Arabidopsis accessions. Journal of Experimental Botany 63: 2491–2501. 10.1093/jxb/err422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hou X, Li L, Peng Z, Wei B, Tang S, Ding M, et al. (2010) A platform of high-density INDEL/CAPS markers for map-based cloning in Arabidopsis. Plant Journal 63: 880–888. 10.1111/j.1365-313X.2010.04277.x [DOI] [PubMed] [Google Scholar]
- 24. Liu B, Wang Y, Zhai W, Deng J, Wang H, Cui Y, et al. (2013) Development of InDel markers for Brassica rapa based on whole-genome re-sequencing. Theoretical and Applied Genetics 126: 231–239. 10.1007/s00122-012-1976-6 [DOI] [PubMed] [Google Scholar]
- 25. Yan Y, Yi G, Sun C, Qu L, Yang N (2014) Genome-wide characterization of insertion and deletion variation in chicken using next generation sequencing. PLoS One 9: e104652 10.1371/journal.pone.0104652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martinez-Lopez LA, Ochoa-Alejo N, Martinez O (2014) Dynamics of the chili pepper transcriptome during fruit development. BMC Genomics 15: 143 10.1186/1471-2164-15-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu C, Ma N, Wang PY, Fu N, Shen HL (2013) Transcriptome sequencing and de novo analysis of a cytoplasmic male sterile line and its near-isogenic restorer line in chili pepper (Capsicum annuum L.). PLoS One 8: e65209 10.1371/journal.pone.0065209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ahn YK, Tripathi S, Kim JH, Cho YI, Lee HE, Kim DS, et al. (2014) Transcriptome analysis of Capsicum annuum varieties Mandarin and Blackcluster: assembly, annotation and molecular marker discovery. Gene 533: 494–499. 10.1016/j.gene.2013.09.095 [DOI] [PubMed] [Google Scholar]
- 29. Ahn Y- K, Tripathi S, Cho Y- I, Kim J- H, Lee H- E, Kim D- S, et al. (2013) De novo transcriptome assembly and novel microsatellite marker information in Capsicum annuum varieties Saengryeg 211 and Saengryeg 213. Botanical Studies 54: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li W, Cheng J, Wu Z, Qin C, Tan S, Tang X, et al. (2015) An InDel-based linkage map of hot pepper (Capsicum annuum). Molecular Breeding 35: 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mimura Y, Minamiyama Y, Sano H (2010) Mapping for axillary shooting, flowering date, primary axis length, and number of leaves in pepper (Capsicum annuum). Journal of the Japanese Society for Horticultural Science 79: 56–63. [Google Scholar]
- 32. Schmitz G, Theres K (1999) Genetic control of branching in Arabidopsis and tomato. Current opinion in plant biology 2: 51–55. [DOI] [PubMed] [Google Scholar]
- 33. Cohen O, Borovsky Y, David-Schwartz R, Paran I (2014) Capsicum annuum S (CaS) promotes reproductive transition and is required for flower formation in pepper (Capsicum annuum). New Phytologist 202: 1014–1023. 10.1111/nph.12711 [DOI] [PubMed] [Google Scholar]
- 34. Lippman ZB, Cohen O, Alvarez JP, Abu-Abied M, Pekker I, Paran I, et al. (2008) The making of a compound inflorescence in tomato and related nightshades. PLoS Biology 6: e288 10.1371/journal.pbio.0060288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jeifetz D, David-Schwartz R, Borovsky Y, Paran I (2011) CaBLIND regulates axillary meristem initiation and transition to flowering in pepper. Planta 234: 1227–1236. 10.1007/s00425-011-1479-8 [DOI] [PubMed] [Google Scholar]
- 36. Cohen O, Borovsky Y, David-Schwartz R, Paran I (2012) CaJOINTLESS is a MADS-box gene involved in suppression of vegetative growth in all shoot meristems in pepper. Journal of Experimental Botany 63: 4947–4957. 10.1093/jxb/ers172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Elitzur T, Nahum H, Borovsky Y, Pekker I, Eshed Y, Paran I (2009) Co-ordinated regulation of flowering time, plant architecture and growth by FASCICULATE: the pepper orthologue of SELF PRUNING . Journal of Experimental Botany 60: 869–880. 10.1093/jxb/ern334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. David-Schwartz R, Borovsky Y, Zemach H, Paran I (2013) CaHAM is autoregulated and regulates CaSTM expression and is required for shoot apical meristem organization in pepper. Plant Science 203: 8–16. 10.1016/j.plantsci.2012.12.011 [DOI] [PubMed] [Google Scholar]
- 39. Chen X, Chen J, Fang R, Cheng Z, Wang S (2006) Inheritance of the node for first flower in pepper (Capsicum annuum L.). Acta Horticulturae Sinica 33: 152–154 (in Chinese with English Abstract). 16529299 [Google Scholar]
- 40. Alimi NA, Bink MCAM, Dieleman JA, Nicolaï M, Wubs M, Heuvelink E, et al. (2013) Genetic and QTL analyses of yield and a set of physiological traits in pepper. Euphytica 190: 181–201. 10.1007/978-3-642-16037-0_12 [DOI] [PubMed] [Google Scholar]
- 41. Ben-Chaim A, Paran I (2000) Genetic analysis of quantitative traits in pepper (Capsicum annuum). Journal of the American Society for Horticultural Science 125: 66–70. [Google Scholar]
- 42. Marame F, Desalegne L, Fininsa C, Sigvald R (2009) Genetic analysis for some plant and fruit traits, and its implication for a breeding program of hot pepper (Capsicum annuum var. annuum L.). Hereditas 146: 131–140. 10.1111/j.1601-5223.2009.02101.x [DOI] [PubMed] [Google Scholar]
- 43. Shifriss C, Hakim Y (1977) Segregation for prebifurcation shooting, stem length and leaf number of main stem in two crosses of Capsicum annuum L. Euphytica 26: 491–495. [Google Scholar]
- 44. Barchi L, Lefebvre V, Sage-Palloix A- M, Lanteri S, Palloix A (2009) QTL analysis of plant development and fruit traits in pepper and performance of selective phenotyping. Theoretical and Applied Genetics 118: 1157–1171. 10.1007/s00122-009-0970-0 [DOI] [PubMed] [Google Scholar]
- 45.Duan MM (2014) Construction of Intraspecific Genetic Linkage Map and QTL Analysis of Phytological Traits and Phytophthora capsici Resistance in Pepper (Capsicum L.): Chinese Academy of Agricultural Sciences (in Chinese with English Abstract).
- 46.Zong HX (2013) Construction of a molecular linkage map and QTL analysis on fruit-related traits in pepper.: Jiangxi Agricultural University (in Chinese with English Abstract).
- 47. Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu Z, Liu W, Tang X, Cui J, Cheng J, Hu K (2012) Development and application of EST-SSR markers in pepper. Journal of South China Agricultural University 33: 171–174 (in Chinese with English Abstract). [Google Scholar]
- 49. Nagy I, Stágel A, Sasvári Z, Röder M, Ganal M (2007) Development, characterization, and transferability to other Solanaceae of microsatellite markers in pepper (Capsicum annuum L.). Genome 50: 668–688. [DOI] [PubMed] [Google Scholar]
- 50. Portis E, Nagy I, Sasvári Z, Stágel A, Barchi L, Lanteri S (2007) The design of Capsicum spp. SSR assays via analysis of in silico DNA sequence, and their potential utility for genetic mapping. Plant Science 172: 640–648. [Google Scholar]
- 51.Van Ooijen J (2006) JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations Kyazma BV, Wageningen, Netherlands.
- 52. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kosambi D (1943) The estimation of map distances from recombination values. Annals of Eugenics 12: 172–175. [Google Scholar]
- 54. Voorrips R (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. Journal of Heredity 93: 77–78. [DOI] [PubMed] [Google Scholar]
- 55. Ren Y, Zhao H, Kou Q, Jiang J, Guo S, Zhang H, et al. (2012) A high resolution genetic map anchoring scaffolds of the sequenced watermelon genome. PLoS One 7: e29453 10.1371/journal.pone.0029453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li H, Ye G, Wang J (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175: 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang S, Basten C, Zeng Z (2007) Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC.
- 58. Lefebvre V, Pflieger S, Thabuis A, Caranta C, Blattes A, Chauvet JC, et al. (2002) Towards the saturation of the pepper linkage map by alignment of three intraspecific maps including known-function genes. Genome 45: 839–854. [DOI] [PubMed] [Google Scholar]
- 59. Lu H, Romero-Severson J, Bernardo R (2002) Chromosomal regions associated with segregation distortion in maize. Theoretical and Applied Genetics 105: 622–628. [DOI] [PubMed] [Google Scholar]
- 60. Dinh TT, Girke T, Liu X, Yant L, Schmid M, Chen X (2012) The floral homeotic protein APETALA2 recognizes and acts through an AT-rich sequence element. Development 139: 1978–1986. 10.1242/dev.077073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wurschum T, Gross-Hardt R, Laux T (2006) APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell 18: 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang X, Gingrich DK, Deng Y, Hong Z (2012) A nucleostemin-like GTPase required for normal apical and floral meristem development in Arabidopsis. Molecular biology of the cell 23: 1446–1456. 10.1091/mbc.E11-09-0797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Doyle MR, Amasino RM (2009) A single amino acid change in the enhancer of zeste ortholog CURLY LEAF results in vernalization-independent, rapid flowering in Arabidopsis. Plant Physiol 151: 1688–1697. 10.1104/pp.109.145581 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A (red): homozygote as BA3, H (yellow): heterozygote as F1, B (dark blue): homozygote as YNXML, C (light blue): not A, D (brown): not B, U (gray), missing.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.