Summary
Fever-associated syndromic epilepsies ranging from febrile seizures plus (FS+) to Dravet syndrome have a significant genetic component. However, apart from SCN1A mutations in over 80% of patients with Dravet syndrome, the genetic underpinnings of these epilepsies remain largely unknown. Therefore, we performed a genome-wide screening for copy number variations (CNVs) in 36 patients with SCN1A-negative fever-associated syndromic epilepsies. Phenotypes included Dravet syndrome (n=23; 64%), GEFS+/FS+ (n=11; 31%) and unclassified fever-associated epilepsies (n=2; 6%). Array CGH was performed using Agilent 4×180K arrays. We identified 13 rare CNVs in 8/36 (22%) individuals. These included known pathogenic CNVs in 4/36 (11%) patients: a 1q21.1 duplication in a proband with Dravet syndrome, a 14q23.3 deletion in a proband with FS+ and two deletions at 16p11.2 and 1q44 in two individuals with fever-associated epilepsy with concomitant autism and/or intellectual disability. In addition, a 3q13.11 duplication in a patient with FS+ and two de novo duplications at 7p14.2 and 18q12.2 in a patient with atypical Dravet syndrome were classified as likely pathogenic. Six CNVs were of unknown significance. The identified genomic aberrations overlap with known neurodevelopmental disorders, suggesting that fever-associated epilepsy syndromes may be a recurrent clinical presentation of known microdeletion syndromes.
Keywords: copy number variation, fever-associated syndromic epilepsy, febrile seizures plus, Dravet syndrome, SCN1A-negative
Introduction
Febrile seizures (FS) are a major manifestation of fever-associated syndromic epilepsies such as febrile seizures plus (FS+), genetic epilepsy with febrile seizures plus (GEFS+) and Dravet syndrome. As opposed to simple FS that usually occur as isolated events, fever-associated syndromic epilepsies present with recurrent FS, FS persisting beyond the age of six years, additional afebrile seizures, additional features such as intellectual disability (ID), autism or congenital anomalies (CGA) and in cases of GEFS+ with family members also affected with FS or fever-associated epilepsy.
Although fever acts as the precipitating factor, the etiology of fever-associated syndromic epilepsies shows a significant genetic contribution. Investigation of classic FS did not reveal any major causative genes, but progress was made examining patients with fever-associated syndromic epilepsies. Known genes include SCN1A, SCN1B, SCN2A and GABRG21.
Sequence mutations in SCN1A are the most common genetic variant in Dravet syndrome, accounting for at least 70% of cases2. In addition, deletions or duplications of SCN1A can be detected in 2–3% of patients with Dravet syndrome3. Rare copy number variants (CNVs) identified in 1/19 Dravet4 and 2/15 SIGEI5 patients and 17q12 duplications in 0.45% (1/222) of patients with familial FS/GEFS+6 suggest CNVs play a role, but the overall significance of CNVs remains unclear. CNVs are deletions and duplications larger than 1 kb and rare CNVs have been established as an important source of genetic morbidity in a range of neurodevelopmental phenotypes including ID, schizophrenia and autism7–9. Rare recurrent microdeletions in 15q13.3, 15q11.2 and 16p13.11 are important risk factors in 3% of patients with idiopathic/genetic generalized epilepsy (IGE/GGE)5. Rare structural genomic variations have been identified as a frequent cause of epileptic encephalopathies4.
In this study, we performed a genome-wide CNV analysis in patients with fever-associated syndromic epilepsy and identified pathogenic CNVs in 11% of patients.
Methods
Patient selection
This study has been approved by the ethics committee of the Christian-Albrechts-University of Kiel, the Antwerp University Hospital and the University of Washington.
36 patients with fever-associated syndromic epilepsies were selected from the biobanks of the Department of Neuropediatrics at the Christian-Albrechts-University of Kiel and the VIB Department of Molecular Genetics at the University of Antwerp. All 36 samples were negative for sequence mutations in SCN1A; 18/36 (50%) were negative for SCN1A deletions and duplications using MLPA or MAQ.
All patients had fever-related seizures or status epilepticus as the most prominent clinical features in the first years of life. Patient phenotypes were classified in three large groups (Table 1). The first group consisted of 13 patients with typical and 10 with atypical Dravet syndrome (n=23; 64%). Most patients with atypical features were classified as having severe idiopathic generalized epilepsy of infancy (SIGEI) as described by Doose10, with predominant generalized tonic-clonic seizures (GTCS), but without the alternating hemiclonic seizures of typical Dravet syndrome.
Table 1.
Phenotypes of patients included in the current study
| Epilepsy phenotype | Number of patients | Number of patients with | ||||
|---|---|---|---|---|---|---|
| ID with onset | autism | CGA | ||||
| prior to epilepsy | after epilepsy | unknown | ||||
| SCN1A-negative Dravet syndrome | 23 (64%) | 2 | 18 | 3 | 4 | 4 |
| typical Dravet syndrome | 13 | |||||
| atypical Dravet syndrome/ SIGEI | 10 | |||||
| GEFS+/FS+ | 11 (31%) | 1a | 0 | 0 | 0 | 0 |
| FS+ afebrile | 4 | |||||
| FS+ prolonged | 4 | |||||
| FS+ absence | 1 | |||||
| FS+ other | 2 | |||||
| unclassified fever-associated epilepsy | 2 (6%) | 2 | 0 | 0 | 1 | 2 |
| Total | 36 | 5 (14%) | 18 (50%) | 3 (8%) | 5 (14%) | 6 (17%) |
CGA, congenital anomalies; FS+, febrile seizures plus; GEFS+, genetic epilepsy with febrile seizures plus; ID, intellectual disability; SIGEI, severe idiopathic generalized epilepsy of infancy;
patient with borderline ID (HAWIK-IV full scale IQ: 75).
According to the presentation, patients were classified as “FS+ afebrile” if additional afebrile GTCS occurred, “FS+ prolonged” if FS persisted beyond the age of six years, “FS+ absence” if additional absence seizures were noted, “FS+ other” if other seizure types were found. In the two patients of this category, astatic and tonic seizures were observed.
The second group comprised 11 patients (n=11; 31%) with GEFS+/FS+. All patients had simple FS with additional seizure types or FS persisting beyond the usual age of remission. The third group included two patients (n=2; 6%) with “unclassified” epilepsy who had additional syndromic features including profound ID, autism, short stature, dysmorphic features, macrocephaly or hypotonia, though a diagnosis of Dravet/SIGEI or FS+ would be justifiable from the epileptology point of view. These patients were included since the clinical features of fever-induced epilepsy were prominent.
Among all patients, a diagnosis of ID was present in 26/36 (72%) patients, autism was diagnosed in 5/36 (14%) individuals and 6/36 (17%) patients had CGA. 18/26 (69%) of the patients with ID had normal development before seizure onset and cognitive impairment started with epileptic activity in terms of an epileptic encephalopathy.
Array CGH
Genomic DNA of all patients was screened for CNVs using array comparative genomic hybridization (array CGH). We applied Agilent 4×180K arrays with an overall median probe spacing of 13 kb.
Labeling and hybridization were performed following standard protocols. Microarray slides were scanned on a G2505C DNA Microarray Scanner (Agilent Technologies) using Agilent Scan Control (version A.8.4.1) with the preset Agilent G3 CGH/CNV/ChIP scan profile (resolution 3μm), then processed by the Agilent Feature Extraction software (version 10.7.3.1) using the default CGH_107_Sep09 protocol. The Derivative Log Ratio Spread (DLRS) was used for quality control. All arrays had a DLRS ≤ 0.3 and 26 of 36 arrays had a DLRS < 0.2.
CNV analysis
Further data processing and analysis were performed using Agilent Genomic Workbench (standard edition, version 5.0.14). Log2 ratios for each array were normalized (centralization algorithm with bin size 10, centralization threshold 6.0). CNVs were called applying the ADM-2 algorithm (threshold 6.0). Additionally, arrays were visually inspected for CNVs. Only CNVs containing at least 5 probes with a minimum average absolute log ratio of 0.5 were reported. Variants were filtered as previously described4: CNVs were eliminated from analysis if they did not overlap any RefSeq genes, lay entirely within segmental duplications or had a >50% overlap with a CNV detected in 4519 published controls11, 12. The last criterion was not applied to established hotspot CNVs known to rarely occur in healthy controls. All filtered CNVs were assessed by visual analysis in Genomic Workbench. Where available, parental DNA was investigated to determine whether the CNV had arisen de novo. Patients with pathogenic and likely pathogenic variants underwent detailed phenotyping including review of available clinical documentation, imaging and electroencephalography.
Evaluation of pathogenicity
Pathogenicity of rare CNVs was evaluated based on guidelines for the interpretation of clinical array CGH results in patients with ID, autism or multiple CGA13 and as previously performed4. In brief, CNVs were classified as pathogenic if they were de novo deletions, were previously described in neurodevelopmental phenotypes or included known epilepsy genes. CNVs were evaluated as likely pathogenic if they were de novo duplications or CNVs > 1 Mb regardless of inheritance. All remaining CNVs were classified as variants of unknown significance.
Deletion breakpoint mapping, microsatellites
Deletion breakpoints were refined in patient #2 using long-range PCR as described in the Supporting Information.
Microsatellite analysis (patient #5, PowerPlex S5 System, Promega) or CNV segregation studies (patient #2) were used to confirm maternity and paternity.
Results
CNV screening
We identified eight patients with a total of 13 rare CNVs (Table 2) including four patients with a single, three patients with two and one patient with three CNVs. To determine the inheritance pattern of rare CNVs, parental DNA was available for both parents in two patients, for one parent in three patients and unavailable in three patients. However, two of these patients carried hotspot CNVs at 1q21.1 and 16p11.2, which were considered pathogenic regardless of inheritance. In total, we identified three de novo, three maternally inherited and one paternally inherited variant. Six CNVs were of unknown inheritance.
Table 2.
Rare CNVs identified in 36 patients with SCN1A-negative fever-associated syndromic epilepsies
| Pat. | Epilepsy phenotype | ID, autism, CGA | CNV, chr location | Coordinates (Build 36, Mb) | Size (kb) | RefSeq genes included | Inheritance | Evaluation of pathogenicity |
|---|---|---|---|---|---|---|---|---|
| #1 | FS+ afebrile | - | del, 4q28.3 | 139.29–139.49 | 197 | SLC7A11 | not in mother | of unknown significance |
| del, 14q23.3 | 66.18–66.27 | 85 | GPHN | M | pathogenic | |||
| #2 | unclassified fever-associated epilepsy | ID, short stature, dysmorphic features | del, 1q44 | 243.11–244.41a | 1301a | HNRNPU a, EFCAB2, KIF26B, SMYD3 | de novo | pathogenic |
| dup, 6q23.3 | 135.89–136.07 | 184 | MIR548H4, C6orf217 | M | of unknown significance | |||
| del, 7p22.1 | 4.88–4.97 | 85 | RADIL, MMD2 | F | of unknown significance | |||
| #3 | unclassified fever-associated epilepsy | ID, autism, generalized muscular hypotonia, macrocephaly | del, 16p11.2 | 28.73–28.95 | 218 | ATXN2L, TUFM, SH2B1, ATP2A1, RABEP2, CD19, NFATC2IP, SPNS1, LAT | unknown | pathogenic |
| #4 | FS+ prolonged | borderline ID b | dup, 3q13.11 | 106.11–107.73 | 1623 | ALCAM, CBLB | not in mother | likely pathogenic |
| #5 | SCN1A-negative atypical Dravet syndrome | ID | dup, 7p14.2 | 36.93–37.12 | 191 | ELMO1 | de novo | likely pathogenic |
| dup, 18q12.2 | 31.02–31.24 | 217 | ZNF397, ZNF397OS, ZNF271, ZNF24, ZNF396 | de novo | likely pathogenic | |||
| #6 | SCN1A-negative Dravet syndrome | ID, autism | dup, 11p15.1 | 21.30–21.39 | 92 | NELL1 | M | of unknown significance |
| #7 | SCN1A-negative Dravet syndrome | ID, microcephaly | dup, 1p22.3 | 87.17–87.28 | 115 | HS2ST1 | unknown | of unknown significance |
| dup, 4p15.31 | 21.87–22.73 | 855 | GPR125, GBA3 | unknown | of unknown significance | |||
| #8 | SCN1A-negative Dravet syndrome | ID | dup, 1q21.1 | 144.53–146.27 | 1744 | GPR89A, GPR89C, PDZK1P1, LOC200030, NBPF11, LOC728989, PRKAB2, PDIA3P, FMO5, CHD1L, BCL9, ACP6, GJA5, GJA8, GPR89B | unknown | pathogenic |
CGA, congenital anomalies; F, inherited from father; FS+, febrile seizures plus; ID, intellectual disability; M, inherited from mother;
HNRNPU was shown to be interrupted by the deletion by refining the breakpoints with long-range PCR and gel electrophoresis to chr1: 243090300-243093300 and chr1: 244408023-244411023 (NCBI Build 36),
HAWIK-IV full scale IQ: 75
All CNVs were then classified according to pathogenicity. Four patients carried pathogenic CNVs (Figure 1), two patients carried likely pathogenic duplications (Supporting Information, Figure S1) and in two patients CNVs were of unknown significance. All pathogenic CNVs in our study had additional supportive data for pathogenicity as some assessment criteria suggest caution when interpreting CNVs as pathogenic due to a single de novo deletion.
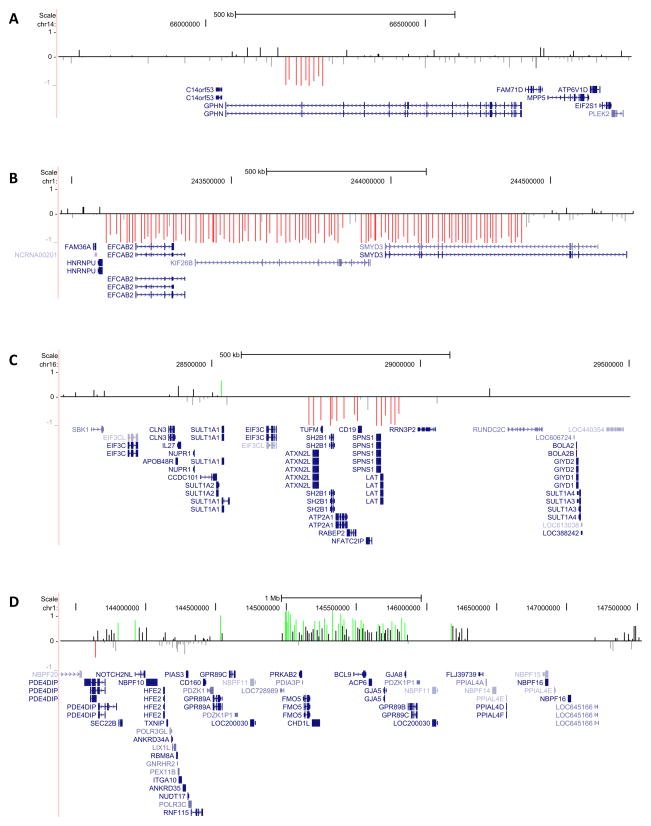
Figure 1.
Array CGH results are shown for all pathogenic CNVs detected in 36 patients: patient #1, del 14q23.3 (A); patient #2, del 1q44 (B); patient #3, distal del 16p11.2 (C); patient #8, dup 1q21.1 (D). Probe log2 ratios are plotted against the loci of probes and depicted by vertical lines: between −0.5 and 0 colored gray, between 0 and 0.5 colored black, < −0.5 colored red, > 0.5 colored green. RefSeq genes are shown in blue.
All pathogenic variants have been previously described in neurodevelopmental disorders. They comprised two rare recurrent CNVs or hotspot variants of unknown inheritance, namely a 1q21.1 duplication in patient #8 with Dravet syndrome and a distal 16p11.2 deletion in patient #3 with unclassified fever-associated epilepsy, ID, autism, generalized muscular hypotonia and macrocephaly. Furthermore, a de novo 1q44 deletion was identified in patient #2 with unclassified fever-associated epilepsy, ID, dysmorphic features and short stature. This patient also carried two additional rare inherited CNVs of unknown significance, including a duplication of 6q23.3 and a deletion of 7p22.1. Patient #1 with FS+ presented with a maternally inherited pathogenic 14q23.3 deletion interrupting GPHN and a 4q28.3 deletion of unknown significance. Overall, three likely pathogenic CNVs were discovered. Patient #4 with FS+ carried a 1.6 Mb likely pathogenic duplication at 3q13.11 including ALCAM and CBLB. Patient #5 carried two likely pathogenic de novo CNVs, a 191 kb duplication at 7p14.2 including the ELMO1 gene and a 217 kb duplication at 18q12.2 comprising five zinc finger genes. Three additional CNVs in two patients were duplications of unknown significance in the chromosomal regions 11p15.1, 1p22.3 and 4p15.31.
The breakpoints of the 1q44 deletion in patient #2 were narrowed down to chr1: 243090300-243093300 and chr1: 244408023-244411023 (NCBI Build 36), interrupting HNRNPU by deleting the first exon of the gene.
Microsatellite and CNV segregation analysis confirmed maternity and paternity for the parents of patient #5 and #2.
Patient phenotypes
Phenotypes of patients carrying pathogenic or likely pathogenic CNVs are described in the Supporting Information.
Discussion
We performed genome-wide array CGH in 36 patients with SCN1A-negative fever-associated syndromic epilepsies and identified 13 rare CNVs in eight individuals including pathogenic CNVs in 11% (4/36) of patients and likely pathogenic CNVs in 6% (2/36) of patients. These CNVs and the corresponding phenotypes are discussed in the Supporting Information.
The investigated cohort included several patients with ID, autism and CGA in addition to fever-associated epilepsy. As CNVs are found more frequently in patients with epilepsy and additional neurodevelopmental phenotypes than in patients with epilepsy alone, a diagnosis of ID, autism or CGA might upwardly bias the diagnostic yield in our study. Spreiz et al. identified rare CNVs in 9/43 (21%) patients with epilepsy, ID and minor dysmorphisms including causative CNVs in 3/43 (7%) individuals14. Mullen et al. detected rare CNVs in 17/60 (28%) patients with GGE and ID15. These findings are consistent with 8/36 (22%) patients from our study carrying rare and 4/36 (11%) patients carrying pathogenic CNVs. However, most of our patients with ID (18/26; 69%) had normal development before seizure onset and cognitive impairment occurred in terms of an epileptic encephalopathy. Considering prior studies, the diagnostic yield of chomosomal micorarrays in epileptic encephalopathies is lower than in GGE with ID (8% vs. 28%)4, 15 and about equals the diagnostic yield in GGE alone (8% vs. 9%)4, 5 indicating that ID linked to epileptic encephalopathies does not relevantly increase the amount of CNV findings. Comparing patients with fever-associated epilepsy and ID present before epilepsy onset or with unknown starting point, autism or CGA to patients with fever-associated epilepsy without these additional features there is no significant difference in the amount of rare CNVs (5/14 and 3/22, p=0.22, Fisher’s exact test). However, as we investigated a small cohort replication of our findings in a larger cohort will help to provide a more accurate estimate of diagnostic yields.
The pathogenic CNVs detected in this study were not specific for fever-associated syndromic epilepsies, but are known to occur in patients with different neurodevelopmental disorders.
Collectively, our findings indicate that several known microdeletion syndromes can present as fever-associated epilepsy syndromes, either in isolation or in association with other neurodevelopmental features. In conclusion, our study suggests that array CGH represents a useful diagnostic tool to identify the genetic basis in fever-associated syndromic epilepsies negative for prime candidate genes such as SCN1A even though we cannot know from our sample whether the frequency of pathogenic CNVs is higher than in other epilepsy cohorts. This conclusion cannot be generalized to individuals with simple FS as these were not investigated in this study. In a clinical setting, array CGH analysis may be the next step of the diagnostic work-up in patients with fever-associated syndromic epilepsies and overlapping neurodevelopmental phenotypes once common genetic causes have been excluded.
Supplementary Material
Table S1. Deletion breakpoint mapping: Primer pairs and length of PCR fragments
Figure S1. Array CGH results for all likely pathogenic CNVs detected in 36 patients
Acknowledgments
We thank all the patients and their families for participating in this study. We are also grateful to Angelika Ackerhans and Klaus Moldenhauer (both Department of Neuropediatrics, Kiel) for database management.
I.H. was supported by intramural funds of the University of Kiel and by a grant from the German Research Foundation (HE5415/3-1) within the EuroEPINOMICS framework of the European Science Foundation. The project also received infrastructural support through the Institute of Clinical Molecular Biology in Kiel, supported in part by DFG Cluster of Excellence “Inflammation at Interfaces” and “Future Ocean”. The project was also supported by the popgen 2.0 network (P2N) through a grant from the German Ministry for Education and Research (01EY1103).
Part of the research was supported by the EUROCORES program EuroEPINOMICS of the European Science Foundation from the Fund for Scientific Research Flanders, and the University of Antwerp (P.D.J.). H.C.M. is supported by NIH (NINDS R01NS06960) and a Burroughs Wellcome Fund Career Award for Medical Scientists. A.S. is a postdoctoral fellow and G.B. a Senior Clinical Investigator of the Research Foundation – Flanders (FWO Vlaanderen).
Footnotes
Disclosure of Conflicts of Interest
None of the authors has any conflict of interest to disclose.
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Additional Supporting Information may be found in the online version of this article:
Deletion breakpoint mapping
Phenotypes of patients with pathogenic and likely pathogenic CNVs
Discussion of pathogenic and likely pathogenic CNVs
References
- 1.Baulac S, Gourfinkel-An I, Nabbout R, et al. Fever, genes, and epilepsy. Lancet Neurol. 2004;3:421–430. doi: 10.1016/S1474-4422(04)00808-7. [DOI] [PubMed] [Google Scholar]
- 2.Marini C, Scheffer IE, Nabbout R, et al. The genetics of Dravet syndrome. Epilepsia. 2011;52 (Suppl 2):24–29. doi: 10.1111/j.1528-1167.2011.02997.x. [DOI] [PubMed] [Google Scholar]
- 3.Marini C, Scheffer IE, Nabbout R, et al. SCN1A duplications and deletions detected in Dravet syndrome: implications for molecular diagnosis. Epilepsia. 2009;50:1670–1678. doi: 10.1111/j.1528-1167.2009.02013.x. [DOI] [PubMed] [Google Scholar]
- 4.Mefford HC, Yendle SC, Hsu C, et al. Rare copy number variants are an important cause of epileptic encephalopathies. Ann Neurol. 2011;70:974–985. doi: 10.1002/ana.22645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mefford HC, Muhle H, Ostertag P, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardies K, Weckhuysen S, Peeters E, et al. Duplications of 17q12 can cause familial fever-related epilepsy syndromes. Neurology. 2013;81:1434–1440. doi: 10.1212/WNL.0b013e3182a84163. [DOI] [PubMed] [Google Scholar]
- 7.de Vries BB, Pfundt R, Leisink M, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doose H, Lunau H, Castiglione E, et al. Severe idiopathic generalized epilepsy of infancy with generalized tonic-clonic seizures. Neuropediatrics. 1998;29:229–238. doi: 10.1055/s-2007-973567. [DOI] [PubMed] [Google Scholar]
- 11.Itsara A, Cooper GM, Baker C, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaikh TH, Gai X, Perin JC, et al. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spreiz A, Haberlandt E, Baumann M, et al. Chromosomal microaberrations in patients with epilepsy, intellectual disability, and congenital anomalies. Clin Genet. 2013 doi: 10.1111/cge.12288. [DOI] [PubMed] [Google Scholar]
- 15.Mullen SA, Carvill GL, Bellows S, et al. Copy number variants are frequent in genetic generalized epilepsy with intellectual disability. Neurology. 2013;81:1507–1514. doi: 10.1212/WNL.0b013e3182a95829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Deletion breakpoint mapping: Primer pairs and length of PCR fragments
Figure S1. Array CGH results for all likely pathogenic CNVs detected in 36 patients