Abstract
Periplasmic localization of recombinant proteins offers advantages over cytoplasmic protein expression. In this study signal sequence of amicyanin, which is encoded by the mauC gene of Paracoccus denitrificans, was used to express the light chain variable domain of the human κIO8/O18 germline antibody in the periplasm of E. coli. The expressed protein was purified in good yield (70 mg/L of culture) in one step from the periplasmic fraction by affinity chromatography using an engineered hexahistidine tag. Circular dichroism spectroscopy was used to determine if the secondary and tertiary structures of the protein and its thermal stability corresponded to those of the native folded protein. The expressed and purified protein was indeed properly folded and exhibited a reasonable thermal transition temperature of 53°C. These results indicate that the amicyanin signal sequence may be particularly useful for prokaryotic expression of proteins which are prone to mis-folding, aggregation or formation of inclusion bodies, all of which were circumvented in this study.
Introduction
Escherichia coli is widely used in the production of recombinant proteins, including approximately 30% of therapeutic proteins that have been approved by the FDA [1]. Problems associated with protein expression in E. coli include degradation by intracellular proteases and the correct formation of disulfide bonds in the cytoplasm. The correct formation of disulfide bonds is especially important in biological therapeutics, where inefficient disulfide formation has been shown to limit the yield of the expressed protein [2]. High levels of cytoplasmic expression of recombinant proteins can also lead to formation of inclusion bodies and mis-folding of the proteins. This is especially prevalent during the expression of recombinant immunoglobulin domains [3–6]. These problems can be circumvented by translocation of the recombinant proteins into the periplasm of E. coli. The periplasm is an oxidizing environment compared to the cytoplasm. This facilitates formation of disulfide bonds that are often required for correct protein folding. The periplasm has a lower concentration of proteolytic enzymes and host cell proteins in general [7]. Thus, the periplasm is enriched in the relative concentration of the recombinant protein which facilitates its purification. For the above reasons the formation of inclusion bodies in the periplasm is much less likely than in the cytoplasm. Recombinant proteins are translocated to the periplasm via membrane-associated secretion systems that recognize an N-terminal signal sequence. The cleavage of the signal peptide during export of the recombinant protein to the periplasm yields a protein with its correct N-terminal residue. A variety of signal peptides have been used for this purpose. Some of the most widely used signal peptides are pelB from E. carotovora and ompA, DsbA, and TolB from E. coli [8–10]
Amicyanin [11] is a periplasmic type I copper protein, referred to as a cupredoxin [12], which is encoded by the mauC gene [13] of Paracoccus denitrificans. Recombinant amicyanin was previously expressed in E. coli at high levels [14]. In this study, we describe the use of the N-terminal signal sequence of amicyanin to express the light chain variable domain (VL) of a human antibody in the periplasm of E. coli. The κI O8/O18 germline antibody VL was chosen for expression as it is a well characterized member of this family [15]. Correct folding of this protein requires formation of a disulfide bond and as with many antibodies and single chain fragments, aggregation and incorrect folding of these recombinant proteins has been problematic [16]. The folding and stability of this VL has previously been characterized by circular dichroism (CD) and thermal stability studies [15]. Thus, it was possible to assess the integrity of the recombinant protein that was expressed in this study by comparison to those results. The results of this study demonstrate the utility of the amicyanin signal sequence as an alternative for recombinant protein expression in E. coli. It should also be noted that an increasing number of recombinant human single chain fragments are being developed for therapeutics and so there may be additional applications of the results which are presented.
Materials and Methods
Protein expression
DNA encoding the κI O8/O18 germline VL with the N-terminal signal sequence of mauC from P. denitrificans and a C-terminal hexahistidine tag was codon-optimized for expression in E. coli, and synthesized by GenScript (Piscataway, NJ). The sequence of the DNA and the amino acid sequence of the protein that it encodes are shown in Table 1. The synthetic gene was cloned into a pET11a expression plasmid at the Nde1 and BamH1 restriction sites. E. coli BL21(DE3) cells were transformed with this plasmid, and cells were cultured in LB media at 37° C in the presence of 100 μg/ml ampicillin. When the A600 of the culture reached 0.6, 0.4 mM IPTG was added to the culture to induce expression and incubation was continued at 30° C for 12 hours.
Table 1.
Sequences of the synthetic gene used to express κI O18/O8 VL and the protein that is encoded by the gene.
|
ATGATTTCCGCTACCAAAATCCGCTCATGCCTCGCGGCCTGTGTCTTGGCTGCCT TTGGAGCCACCGGAGCCCTTGCCTCGACATTCAAATGACTCAATCCCCGTCATCC CTGTCAGCGAGTGTCGGTGATCGCGTCACGATCACGTGCCAGGCGTCTCAAGACA TTAGCAACTACCTGAATTGGTACCAGCAGAAACCAGGTAAGGCCCCGAAACTCTTG ATCTACGACGCGTCCAATTTGGAAACAGGCGTGCCGAGTCGCTTTAGCGGTAGCG GAAGCGGCACCGATTTCACCTTCACCATCAGTTCCCTTCAGCCGGAAGACATCGC CACCTACTATTGTCAACAGTATGACAATCTGCCATATACGTTTGGCCAGGGCACCA AACTGGAAATCAAGCACCATCATCATCATCATTAG |
|
M I S A T K I R S C L A A C V L A A F G A T G A L A D I Q M T Q S P S S L S A S V G D R V T I T C Q A S Q D I S N Y L N W Y Q Q K P G K A P K L L I Y D A S N L E T G V P S R F S G S G S G T D F T F T I S S L Q P E D I A T Y Y C Q Q Y D N L P Y T F G Q G T K L E I K H H H H H H |
(Top) The DNA sequence of the gene encoding κIO18/O8 VL with the N-terminal mauC signal sequence and the sequence encoding the C-terminal hexahistidine tag underlined.
(Bottom) The amino acid sequence encoded from this gene is shown. The N-terminal signal sequence which is cleaved during expression and hexahisidine tag that is used for affinity purification are underlined.
Protein purification
The recombinant κI O8/O18 VL was purified from the periplasmic fraction of the E. coli cells. The periplasmic fraction was obtained using a lysozyme/osmotic shock method [11, 17]. The harvested cells were resuspended in 10 mM Tris-HCl buffer, pH 8.0, at a ratio of 6 mL/g of wet cell weight. The buffer also contained 20% w/v sucrose, 0.7 mM EDTA, 2 mg/mL of lysozyme, 1 mM MgCl2, 0.01 mg/mL of DNase and 200 µM phenylmethylsulfonyl fluoride. After incubation for 20 min at 30° C with shaking, an equal volume of H2O was added and incubation continued for a total of one hour. The spheroplasts were then removed by centrifugation and the periplasmic fraction was in the supernatant. The His-tagged κI O8/O18 VL was purified from the periplasmic fraction by affinity chromatography using a Ni-NTA column (Qiagen). Protein concentration was determined from the absorbance of the pure protein at 280 nm using an extinction coefficient of 16,055 M−1 cm−1 that was calculated from the amino acid composition of the protein using the Expasy ProtParam tool (http://web.expasy.org/protparam/) [18].
Protein characterization
Size exclusion chromatography was performed with a HiPrep 16/60 Sephacryl S-300 HR column (GE Healthcare). The equilibration and elution buffer was 10 mM potassium phosphate, pH 7.5, with 150 mM NaCl. The flow rate was 1.0 mL/min. Methylamine dehydrogenase, cytochrome c-553, cytochrome c-551i, and amicyanin with molecular weights of 124, 30, 22, and 11.5 kDa, respectively, were used as standards.
CD spectra were recorded using a J-810 spectropolarimeter equipped with a Peltier temperature controller (Jasco corp., Tokyo, Japan). Samples contained 3 µM protein in 10 mM potassium phosphate buffer, pH 7.5. For temperature-dependence studies, samples were equilibrated at each temperature for 2 min before CD measurements were recorded. The measured ellipticity, θmeas, was converted to mean residue molar ellipticity [θ], through the formula [θ] = θmeas/nlc, where n = 113 is the number of amino acid residues in the protein, l is the optical pathlength in mm, and c is the molar concentration of the protein. Thermal unfolding of the protein secondary structure was determined from the increase in the negative CD signal at 205 nm which results from an increase in the fraction of unordered structure [19]. The value of the thermal transition temperature (Tm) was determined as the inflection point of the sigmoidal temperature-dependence curves of the corresponding normalized signal intensity, which was identified as the extremum of the first derivative with respect to the temperature. The fraction of folded protein at a given temperature T was determined using eq 1 where [θ] is the mean residue molar ellipticity at 205 nm and the subscript indicates the temperature. Values of [θ] at 40° C and 70° C were deemed to correspond to 100% and 0% folded state, respectively.
| (1) |
Results
Protein expression and purification
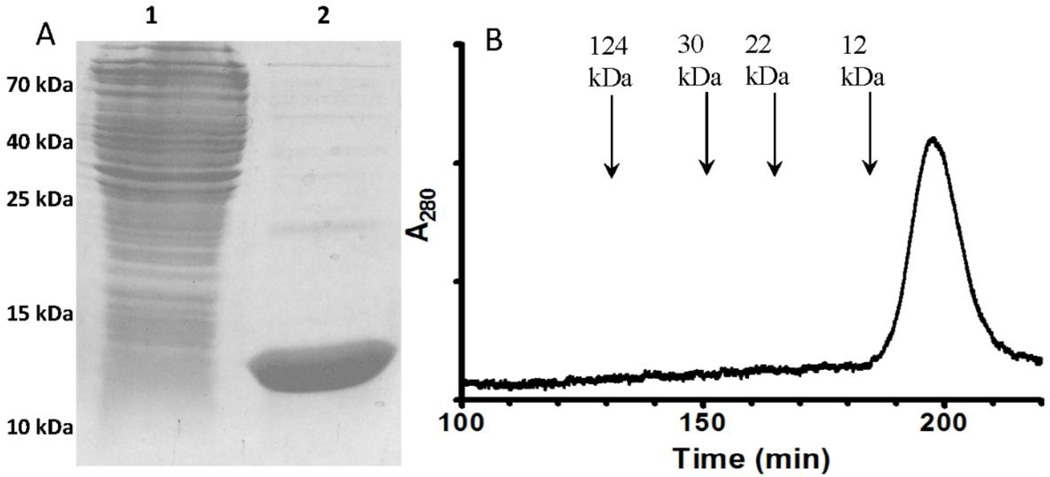
The recombinant κI O18/O8 germline VL was successfully expressed in E. coli and purified. The yield of pure protein was 70 mg/L of cell culture. The purified protein ran as a single band on SDS-PAGE (Figure 1A). When the protein was subjected to size exclusion chromatography it eluted as a single peak consistent with it being present exclusively as a monomer (Figure 1B). Comparison of the elution volume of the VL to other protein standards indicated that it eluted at a position corresponding to a molecular mass of about 9 kDa which is lower than that determined by SDS-PAGE and calculated from the amino acid sequence of 12.8 kDa. However, this observation is consistent with anomalously lower values for molecular mass determined by size exclusion chromatography that have been previously observed with VL domains [20–22]. Since the expressed protein is an antibody domain and not an enzyme, it does not have an activity to assay. Evidence for the structural and functional integrity of the expressed protein was provided by the demonstration that it binds to protein L resin, which is specific for antibody kappa light chains, and is often used for affinity purification of immunoglobulins [23]. In this study we chose to add a His-tag for purification because the Ni-NTA superflow resin used for purification has a much higher capacity for bound protein than the protein L resin. It was also confirmed by Western blot that the purified protein reacted with an anti-His-tag antibody. These results show that use of the amicyanin signal sequence allowed for expression of high levels of monomeric VL in the periplasm, which could be easily purified in a single step with no evidence of aggregation or inclusion bodies.
Figure 1.
SDS-PAGE and size exclusion chromatography of the expressed and purified κIO18/O8 VL. A. SDS-PAGE was performed using a 12.5% gel. Lane 1 is a whole cell extract of the E. coli cells from which the recombinant VL was purified. Lane 2 is the protein that was purified from the periplasmic fraction of the cells. The positions of MW markers which were also run on the gel are indicated. B. The elution profile obtained when the purified recombinant VL was subjected to size exclusion chromatography is shown with the positions of elution of other proteins of known molecular weight indicated.
Protein structure and stability
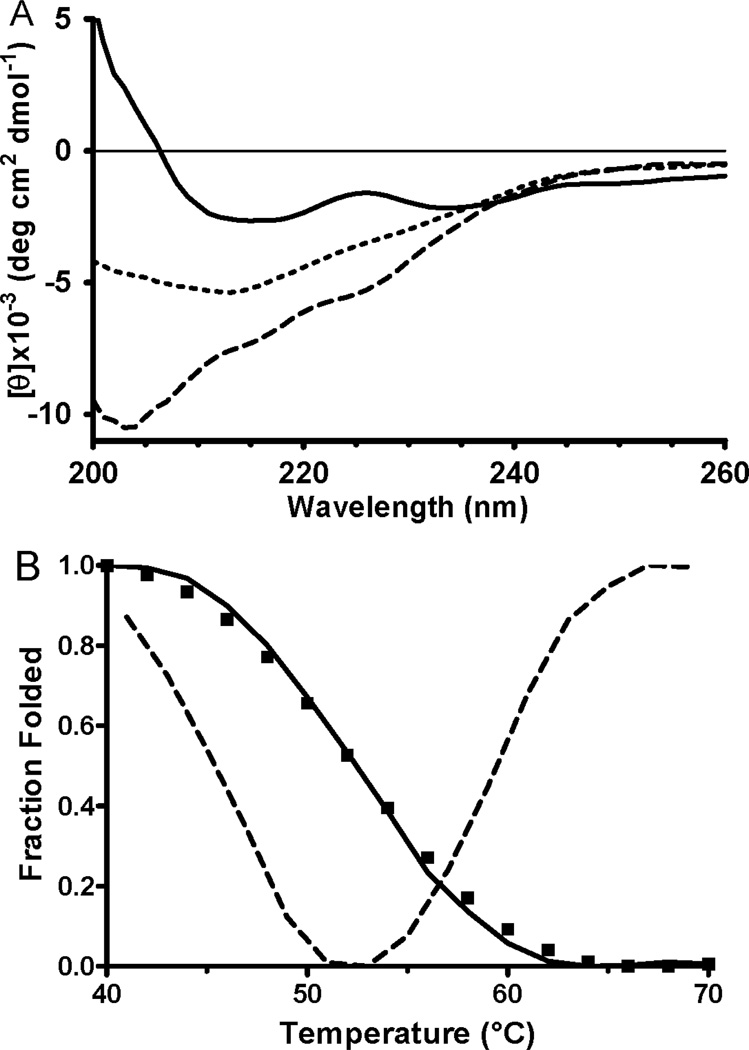
The secondary structure of the κI O18/O8 VL has previously been characterized by CD spectroscopy [15]. As such, the CD spectrum of the purified recombinant protein was obtained and compared with the published data. The spectrum shown in Figure 2A (solid line) is essentially identical to that previously reported for κIO18/O8 VL. It has a β-sheet secondary structure as indicated by the minimum around 216 nm. The spectrum also exhibits a second minimum at 232 nm, which can be assigned to type I β-turn structure [24]. The minimum between 200–205 nm that appears at higher temperatures (Figure 2A, dashed line) is generated by unordered structure [19]. These features of the CD spectrum are essentially identical to those reported for the native protein and are consistent with the x-ray crystal structure of the germline Vk1 O18/O8 light chain variable domain [15] (PDB entry 2Q20).
Figure 2.
CD spectra of purified recombinant κI O18/O8 VL and determination of the Tm for the protein. CD spectra of the protein were recorded at a range of temperatures up to 70°C. A. For clarity only three spectra are shown which were recorded at 40°C (solid line), 53°C (short dashed line) and 70°C (long dashed line). B. The values of normalized mean residue molar ellipticity at 205 nm as a function of temperature are shown (solid line) as well as the 1st derivative of that line which was used to determine the Tm (dashed line).
These results indicate that the recombinant VL protein that was expressed in the periplasm using the amicyanin signal sequence is properly folded with regard to secondary and tertiary structures. It is also important to assess the thermal stability of this recombinant protein. CD spectra were recorded at temperatures from 40° C to 70° C. In Figure 2A the spectra of the native protein at 40° C, completely denatured protein at 70° C, and the protein at the Tm are shown. The Tm of the protein, as determined from the temperature-dependent perturbations of the CD signal at 205 nm was 53° C (Figure 2B). This value is in agreement with the previously reported value of 56° C for κI O18/O8 VL 15].
Discussion
As stated earlier, there are certain advantages to expressing recombinant proteins in the periplasm of E. coli rather than in the cytoplasm. The amicyanin signal sequence is shown to be an efficient tool for periplasmic recombinant protein expression. It may also be particularly useful for expression of proteins which are prone to mis-folding or formation of inclusion bodies. The mauC gene encodes amicyanin, a Type I copper protein that is localized in the periplasm [11]. It was shown that incorporation of the very tightly bound copper occurs during the folding of amicyanin in the periplasm [25]. In cases such as this, the timing of the translocation of the protein is important to avoid mis-folding and aggregation that can result from unavailability of copper at the critical point in protein folding. Recombinant expression of VL proteins and immunoglobulins has also been problematic due to the tendency to form inclusion bodies. For expression of some proteins such amicyanin and κI O18/O8 VL, as well as many others, the level of recombinant protein expression may not necessarily be as important as the timing and rate at which the protein is being expressed and co-translationally translocated into the periplasm. The amicyanin signal sequence may be a useful alternative to other commonly used signal sequences for expression of proteins in E. coli and other prokaryotes.
Acknowledgments
The authors thank Yu Tang for providing technical assistance. This research was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R37GM41574 (VLD).
Abbreviations
- VL
light chain variable domain
- CD
circular dichroism
- Tm
thermal transition temperature
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huang CJ, Lin H, Yang X. Industrial production of recombinant therapeutics in Escherichia coli and its recent advancements. J Ind Microbiol Biotechnol. 2012;39:383–399. doi: 10.1007/s10295-011-1082-9. [DOI] [PubMed] [Google Scholar]
- 2.Proba K, Ge L, Pluckthun A. Functional antibody single-chain fragments from the cytoplasm of Escherichia coli: influence of thioredoxin reductase (TrxB) Gene. 1995;159:203–207. doi: 10.1016/0378-1119(95)00018-2. [DOI] [PubMed] [Google Scholar]
- 3.Tsumoto K, Shinoki K, Kondo H, Uchikawa M, Juji T, Kumagai I. Highly efficient recovery of functional single-chain Fv fragments from inclusion bodies overexpressed in Escherichia coli by controlled introduction of oxidizing reagent--application to a human single-chain Fv fragment. J Immunol Methods. 1998;219:119–129. doi: 10.1016/s0022-1759(98)00127-6. [DOI] [PubMed] [Google Scholar]
- 4.Sinacola JR, Robinson AS. Rapid refolding and polishing of single-chain antibodies from Escherichia coli inclusion bodies. Protein Expr Purif. 2002;26:301–308. doi: 10.1016/s1046-5928(02)00538-7. [DOI] [PubMed] [Google Scholar]
- 5.Kurucz I, Titus JA, Jost CR, Segal DM. Correct disulfide pairing and efficient refolding of detergent-solubilized single-chain Fv proteins from bacterial inclusion bodies. Mol Immunol. 1995;32:1443–1452. doi: 10.1016/0161-5890(95)00105-0. [DOI] [PubMed] [Google Scholar]
- 6.Fink AL. Protein aggregation: folding aggregates, inclusion bodies and amyloid. Fold Des. 1998;3:R9–R23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 7.Swamy KH, Goldberg AL. Subcellular distribution of various proteases in Escherichia coli. J Bacteriol. 1982;149:1027–1033. doi: 10.1128/jb.149.3.1027-1033.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei SP, Lin HC, Wang SS, Callaway J, Wilcox G. Characterization of the Erwinia carotovora pelB gene and its product pectate lyase. J Bacteriol. 1987;169:4379–4383. doi: 10.1128/jb.169.9.4379-4383.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Movva NR, Nakamura K, Inouye M. Amino acid sequence of the signal peptide of ompA protein, a major outer membrane protein of Escherichia coli. Biol Chem. 1980;255:27–29. [PubMed] [Google Scholar]
- 10.Schierle CF, Berkmen M, Huber D, Kumamoto C, Boyd D, Beckwith J. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J Bacteriol. 2003;185:5706–5713. doi: 10.1128/JB.185.19.5706-5713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Husain M, Davidson VL. An inducible periplasmic blue copper protein from Paracoccus denitrificans. Purification, properties, and physiological role. J Biol Chem. 1985;260:14626–14629. [PubMed] [Google Scholar]
- 12.Choi M, Davidson VL. Cupredoxins--a study of how proteins may evolve to use metals for bioenergetic processes. Metallomics. 2011;3:140–151. doi: 10.1039/c0mt00061b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chistoserdov AY, Boyd J, Mathews FS, Lidstrom ME. The genetic organization of the mau gene cluster of the facultative autotroph Paracoccus denitrificans. Biochem Biophys Res Commun. 1992;184:1181–1189. doi: 10.1016/s0006-291x(05)80007-5. [DOI] [PubMed] [Google Scholar]
- 14.Davidson VL, Jones LH, Graichen ME, Mathews FS, Hosler JP. Factors which stabilize the methylamine dehydrogenase-amicyanin electron transfer protein complex revealed by site-directed mutagenesis. Biochemistry. 1997;36:12733–12738. doi: 10.1021/bi971353m. [DOI] [PubMed] [Google Scholar]
- 15.Baden EM, Owen BA, Peterson FC, Volkman BF, Ramirez-Alvarado M, Thompson JR. Altered dimer interface decreases stability in an amyloidogenic protein. J Biol Chem. 2008;283:15853–15860. doi: 10.1074/jbc.M705347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee CC, Perchiacca JM, Tessier PM. Toward aggregation-resistant antibodies by design. Trends Biotechnol. 2013;31:612–620. doi: 10.1016/j.tibtech.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Davidson VL, Sun D. Lysozyme-osmotic shock methods for localization of periplasmic redox proteins in bacteria. Methods Enzymol. 2002;353:121–130. doi: 10.1016/s0076-6879(02)53042-1. [DOI] [PubMed] [Google Scholar]
- 18.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sreerama N, Woody RW. Circular dichroism of peptides and proteins. In: Berova N, Nakanishi K, Woody RW, editors. Circular Dichroism: Principles and Applications. New York: John Wiley & Sons; 2000. pp. 601–620. [Google Scholar]
- 20.Dudgeon K, Rouet R, Kokmeijer I, Schofield P, Stolp J, Langley D, Stock D, Christ D. General strategy for the generation of human antibody variable domains with increased aggregation resistance. Proc Natl Acad Sci U S A. 2012;109:10879–10884. doi: 10.1073/pnas.1202866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ewert S, Huber T, Honegger A, Pluckthun A. Biophysical properties of human antibody variable domains. J Mol Biol. 2003;325:531–553. doi: 10.1016/s0022-2836(02)01237-8. [DOI] [PubMed] [Google Scholar]
- 22.Jespers L, Schon O, Famm K, Winter G. Aggregation-resistant domain antibodies selected on phage by heat denaturation. Nat Biotechnol. 2004;22:1161–1165. doi: 10.1038/nbt1000. [DOI] [PubMed] [Google Scholar]
- 23.Nilson BHK, Logdberg L, Kastern W, Bjorck L, Akerstrom B. Purification of antibodies using protein-L-binding framework structures in the light-chain variable domain. J Immunol Methods. 1993;164:33–40. doi: 10.1016/0022-1759(93)90273-a. [DOI] [PubMed] [Google Scholar]
- 24.Woody RW. In: Circular Dichroism and the Conformational Analysis of Biomolecules. Fasman GD, editor. New York: Plenum Press; 1996. pp. 25–67. [Google Scholar]
- 25.Ma JK, Lee S, Choi M, Bishop GR, Hosler JP, Davidson VL. The axial ligand and extent of protein folding determine whether Zn or Cu binds to amicyanin. J Inorg Biochem. 2008;102:342–346. doi: 10.1016/j.jinorgbio.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]