Abstract
There is a long-standing unmet clinical need for biomarkers with high specificity for distributed stem cells (DSCs) in tissues, or for use in diagnostic and therapeutic cell preparations (e.g., bone marrow). Although DSCs are essential for tissue maintenance and repair, accurate determination of their numbers for medical applications has been problematic. Previous searches for biomarkers expressed specifically in DSCs were hampered by difficulty obtaining pure DSCs and by the challenges in mining complex molecular expression data. To identify DSC such useful and specific biomarkers, we combined a novel sparse feature selection method with combinatorial molecular expression data focused on asymmetric self-renewal, a conspicuous property of DSCs. The analysis identified reduced expression of the histone H2A variant H2A.Z as a superior molecular discriminator for DSC asymmetric self-renewal. Subsequent molecular expression studies showed H2A.Z to be a novel “pattern-specific biomarker” for asymmetrically self-renewing cells with sufficient specificity to count asymmetrically self-renewing DSCs in vitro and potentially in situ.
Keywords: adult stem cells, asymmetric stem cell division, cell microarray analysis, cDNA microarrays, computational biology, biomarkers, sparse feature selection
Introduction
Despite the important role of distributed stem cell (DSCs) in homeostatic tissue cell renewal and tissue repair, and their use in currently available cell replacement therapies (e.g., hematopoietic stem cell transplantation), there are no effective means for quantifying their number. This unmet research and clinical need is due to the lack of biomarkers with sufficient specificity for distinguishing DSCs from other tissue cell types. Previous searches for these illusive biomarkers relied on global gene expression profiles for DSCs that were based on comparisons of genes expressed in embryonic stem cells (ESCs) and genes expressed in cell populations enriched for adult stem cells (ASCs) (Ivanova et al., 2002; Ramalho-Santos et al., 2002; Fortunel et al., 2003), a sub-category of DSCs (Sherley, 2008; Noh et al., 2011). These ASC-enriched populations also contained a significant fraction of non-stem committed progenitors and differentiating progeny cells that limited their utility for identifying genes whose expression was unique to DSCs, i.e., “stemness genes” (Ivanova et al., 2002; Ramalho-Santos et al., 2002; Fortunel et al., 2003, Burns and Zon, 2002). In addition, gene expression profiles based on specific expression in both ESCs and DSC-enriched populations necessarily exclude genes whose expression is specific to either of these distinctive stem cell types. One essential distinction is that ESCs do not undergo asymmetric self-renewal, which is a universal defining property of DSCs (Noh et al., 2011; Loeffler and Potten, 1997; Sherley, 2002; Sherley, 2005).
We applied a different strategy to identify DSC-specific genes based on targeting deterministic asymmetric self-renewal, which is one of the two forms of asymmetric self-renewal, the other being stochastic asymmetric self-renewal (See Discussion later). Mammalian DSCs self-renew asymmetrically to replenish cells in tissues that undergo cell turnover but maintain a constant cell mass (Loeffler and Potten, 1997; Sherley, 2002; Sherley, 2005). Each deterministic asymmetric self-renewal division by DSCs yields a new DSC and a non-stem cell sister. The non-stem cell sister is a committed progenitor of differentiated cells responsible for mature tissue functions (Loeffler and Potten, 1997; Sherley, 2002; Sherley, 2005). Because asymmetric self-renewal is unique to DSCs, some genes whose expression is highly associated with asymmetric self-renewal may also identify DSCs (Noh et al., 2011).
Here, we describe the application of this strategy using genetically engineered cell lines that adopt deterministic asymmetric self-renewal conditionally. Restoration of normal wild-type p53 protein expression induces these lines to undergo asymmetric self-renewal in a similar way to DSCs (Noh et al., 2011; Sherley et al., 1995a; Liu et al., 1998a; Rambhatla et al., 2001; Rambhatla et al., 2005). When p53 expression is reduced, the cells switch to symmetric self-renewal. In vivo, symmetric self-renewal by DSCs is regulated to increase tissue mass during adult maturation growth and to repair injured tissues (Loeffler and Potten, 1997; Sherley, 2002; Sherley, 2005). When controls that constrain DSCs to asymmetric self-renewal are disrupted (e.g., by p53 mutations), the risk of proliferative disorders like cancer increases (Sherley, 2002; Sherley et al., 1995a; Rambhatla et al., 2005). In vitro, the experimental capture of this functional feature of DSCs served as a surrogate for natural occurring DSC properties that are otherwise inaccessible. The novelty in this work is manifold but derives primarily from the combination these experimental models with a highly effective but unconventional gene microarray bioinformatics approach to identify a small number of genes predicted to have a high degree of biomarker specificity for DSCs.
Materials And Methods
Analysis of cell division kinetics and quality control of cell cultures
All studies involving mice were conducted at the Boston Biomedical Research Institute (BBRI), the previous host institution for the Adult Stem Cell Technology Center. All procedures were approved by the BBRI Institutional Animal Care and Use Committee. Euthanasia was conducted according to the guidelines of the U.S. National Institutes of Health Office of Laboratory Animal Welfare.
During asymmetric self-renewal in culture, cells give rise to two different progeny with each cell division, dividing and non-dividing. PDCasym was calculated by PDCasym = (Nt − N0)/(Fd × N0) where N0 and Nt are the number of cells present in culture at time 0 h and time t, respectively (Merok et al., 2002; Sherley et al., 1995) [14,15]. Fd is the fraction of new sister cells that divide in a population. The PDC value of symmetrically dividing cells (PDCsym) was calculated with the equation, PDCsym = ln(Nt/N0)/ln2, derived from the exponential cell growth equation, Nt = N0ekt. (Merok et al., 2002; Sherley et al., 1995b). To assess whether the cell division kinetics were close to ideal asymmetric self-renewal for transcription profiling, the PDCsym/PDCasym ratio was calculated at the time when cells were harvested (See Supplementary Information). The generation time (GT) for each asymmetric cell growth model was determined from time-lapse microscopy observations, or from the average doubling time of the symmetrically dividing counterpart (Rambhatla et al., 2001; Sherley et al., 1995b). We used 0.5 for Fd to calculate the cell kinetics parameters for all experiments and generate the ideal ratio curves for asymmetric self-renewal (Supplementary Fig. S1; Supplementary Table S1).
Cell culture
The Ind-8, tC-2, and 1h-3 cells for asymmetric self-renewal and Con-3, tI-3, and 1g-1 cells for respective control symmetric self-renewal were maintained as described (Sherley et al., 1995a; Liu et al., 1998a; Rambhatla et al., 2001; Rambhatla et al., 2005; Merok et al., 2002; Liu et al., 1998b). For the analyses of the self-renewal kinetics of Ind-8 and Con-3 cells, cells were grown over a 3-day period to about 50% confluency, trypsinized, and replated in Zn-free medium (DMEM, 10% dialyzed fetal bovine serum [DFBS], 5 μg/ml puromycin) at a cell:plating area:medium volume of 100,000:75 cm2:20 ml. This ratio was held constant for all experiments, unless specified otherwise. Sixteen to 24 hours later, the culture medium was replaced with the same volume of Zn-free medium, or medium supplemented to the specified concentration of ZnCl2. This time was designated as 0-hour in the analyses. For the 1h-3 and 1g-1 cell growth, cells were grown to 50% confluency at 37°C and replenished with fresh growth medium (DMEM, 10% DFBS, 1 mg/ml G418 sulfate). Sixteen hours later, the cells were trypsinized and replated at either 37°C or 32.5°C. This time was 0-hour in the analyses. In case of the growth analyses of tI-3 and tC-2 cells, cells were grown and re-plated in the same way as for Ind-8 and Con-3 cells except for the medium (DMEM, 10% DFBS, 1 mg/ml G418 sulfate plus 5 μg/ml puromycin). Cells were harvested by trypsin treatment and counted with a Model ZM Coulter Counter to confirm the induction of asymmetric self-renewal in culture by PDC ratio analysis.
Human hSATs (strain 18Ubic) and HFSCs (strain 3C5) were cultured and maintained as previously described (Huh et al., 2011; Homma et al., 2012). To suppress the asymmetric self-renewal and H2A.Z asymmetry of hSATs, their culture medium was supplemented with 1.5 mM xanthine as described for HFSCs (Huh et al., 2011). Mouse p53 KO MEFs were kindly provided by L. Donehower. Mouse p53 wt/cdkn1A KO fibroblasts were kindly provided by T. Jacks. They were subsequently immortalized on a standard 3T3 cell schedule (Todaro and Green, 1963).
cDNA micro-array data analysis
Cells for total RNA extraction were harvested at the time estimated for PDC = 2 (36 hours for Ind-8 and Con-3, 48 hours for tC-2, tI-3, 1h-3, and 1g-1) and were selected for microarray experiments based on the actual PDC ratio curves derived from replicate cell cultures grown in parallel. The same PDC ratio values were maintained to obtain similar fractions of cycling stem-like cells and non-cycling differentiating cells for all self-renewal pattern comparison models. Total RNA was extracted with the Trizol reagent (Invitrogen, Carlsbad, CA) and impurities were removed with the Qiagen RNeasy kit (Qiagen, Valencia, CA). Fifty to seventy μg of total RNA was used for cDNA syntheses. Arabidopsis thaliana mRNAs (Stratagene, La Jolla, CA) were introduced as internal probe standards into reverse transcription reactions to normalize data between different arrays. Cy3- or Cy5-fluorescently labeled cDNAs were hybridized onto the National Institute for Aging 15K mouse cDNA prefabricated arrays (Tanaka et al., 2000) [20], supplied by the Massachusetts Institute of Technology (MIT)-BioMicro Center, using the procedure provided by the MIT-BioMicro Center. Hybridized microarrays were scanned with the arrayWoRxeTM Biochip Reader (Applied Precision LLC, Northwest Issaquah, WA). The fluorescence intensity of each spot was analyzed from the scanned tiff images by using the DigitalGenome™ software (MolecularWare, Inc. Cambridge, MA). The Cy3 and Cy5 fluorescence intensities were normalized by calculating the normalization factor from total intensity normalization (Quackenbush, 2001). Analyses for each self-renewal pattern comparison were performed as duplicate independent experiments. For each comparison, we performed two chip hybridizations with reciprocally labeled Cy3 or Cy5 target cDNAs to each biological sample. The entire analysis incorporated data from 16 independent chips, which comprised two dye-swap technical replicate arrays for each of the four asymmetric-symmetric comparisons.
A gene was selected for data analyses only if the mean value of foreground pixels of the spot was greater than the sum of the mean and two standard deviations of the background pixels. For individual gene probe spots, the expression intensities of Cy5 and Cy3 channels were estimated by subtracting mean backgrounds from mean foregrounds. The ratios of the final gene expression intensities for the asymmetrically self-renewing states to the respective symmetrically self-renewing states were calculated. These ratio values were used for sparse feature selection. The ratio data were deposited for public access in National Center for Biotechnology Information Gene Expression Omnibus Database the under accession number GSE40183.
Sparse feature selection
The EM algorithm was applied to the cDNA array data provided. The data were aggregated so that all asymmetric cell division array data were given a dependent variable class label of -1 and all symmetric cell division array data were given a class label of +1. The different culture treatments used to promote symmetric or asymmetric division were not modeled separately in the computational experiments. All symmetrically self-renewing cells were assigned to the symmetric class, and all those self-renewing asymmetrically were assigned to the asymmetric class, regardless of how the symmetric was controlled experimentally. This was to avoid artifacts caused by the different methods of inducing symmetry or asymmetry of division.
The cDNA micro-array dataset (GEO Accession number GSE40183) was screened to remove missing or zero expression values. We subsequently removed genes whose expression across replicates was less than the mean expression of the entire array dataset plus two standard deviations of the expression of the entire array data. This filter removed genes whose expression was not significantly different than the array background noise fluctuation at the 95% confidence limit. This processing resulted in 1,648 genes available for EM algorithm analysis (see Supplementary Information for mathematical details of the method). The selection of genes was found to be quite robust, with very similar subsets of genes being selected for varied filtering models with varying degrees of imposed sparsity. After the filters were applied, the EM algorithm reduced the pool of candidate genes to 4-7 genes at the higher levels of sparsity control applied. These genes were able to classify the self-renewal division pattern with very high efficacy, with r2 values exceeding 0.99. Most of the selected genes made negative contributions to the model, implying they would be down regulated in cells self-renewing asymmetrically compared to those self-renewing symmetrically. Conversely, genes making positive contributions to the model would be up regulated in cells self-renewing asymmetrically compared to those self-renewing symmetrically. The signs and contributions of selected genes to the models were also consistent across multiple calculations using varied filtering criteria, suggesting our novel gene selection algorithm was quite robust and reproducible.
Phosphorylated histone H3-H2A.Z asymmetry detection
For mitotic cell analysis of H2A.Z with phosphorylated histone H3 (pH3; phospho-S10), cells grown on glass chamber slides were washed with ice cold PBS and immediately fixed for 15 minutes at room temperature in PBS containing 3.7% formaldehyde. Thereafter, dual indirect in situ immunofluorescence (ISIF) analyses were performed for H2A.Z and pH3 as described for mitotic cell analysis of H2A.Z and α-tubulin (Huh and Sherley, 2011). In brief, after blocking, cells were incubated overnight at 4°C in a humidified chamber with an anti-rabbit H2A.Z polyclonal antibody (Cell Signaling Technology, Inc., Danvers, MA) diluted 1:200 in PBS containing 2% goat serum. After five rinses in PBS containing 0.5% BSA, the cells were incubated for 1 hour at room temperature with Alexa Fluor® 488-conjugated goat anti-rabbit IgG (Invitrogen, Inc., Carlsbad, CA), diluted 1:300 in the blocking solution. Next, the cells were washed five times with PBS containing 0.5% BSA. Cells stained for H2A.Z detection were subsequently incubated in the same manner with anti-mouse anti-pH3 monoclonal antibody (Abcam, Inc., Cambridge, MA) at a 1:1,000 dilution followed by Alexa Fluor® 568-conjugated goat anti-mouse IgG (Invitrogen, Inc., Carlsbad, CA) diluted 1:500. After washing, the cells were mounted with 4’-6-diamido-2-phenylindole (DAPI)-containing VectaShield® mounting media (Vector Laboratories, Inc., Burlingame, CA). Epifluorescence images were captured with a Leica DMR microscope and Leica DC300F digital camera system. Pair-wise control analyses that omitted anti-H2A.Z and anti-pH3 antibodies separately and together were evaluated to ensure that all detected fluorescence required the specific antibodies.
For tissue analyses, sections (10 μm) of paraffin-embedded skin from adult FVB mice (Taconic Farms, Inc., Hudson, NY; Institutional Animal Care and Use Committee-approved procedures) that included hair follicles were cut on a Leica RM2255 microtome (Leica Microsystems, Inc., Bannockburn, IL) and picked up on Superfrost/Plus glass slides (Fisher Scientific, Inc., Pittsburgh, PA). For ISIF, sections were deparaffinized in xylene and rehydrated with a descending series of ethanol solutions (100%, 95%, 90%, 80%, 70%). Sections were then washed three times with PBS and incubated with phosphate buffered sodium citrate solution (0.01M, pH 7.4) for 20 minutes in a food steamer (Oster) to expose epitopes blocked during fixation and embedding. After washing in PBS, the sections were permeablized with 0.2% Triton X-100 (v/v in PBS) at room temperature for 10 minutes and afterwards washed once with PBS. Thereafter, sections were blocked with 10% goat serum for 1 hour at room temperature and dual ISIF analyses were performed for H2A.Z and pH3 as described above for cultured cells.
Results
An orthogonal-intersection strategy for identification of asymmetric self-renewal associated genes
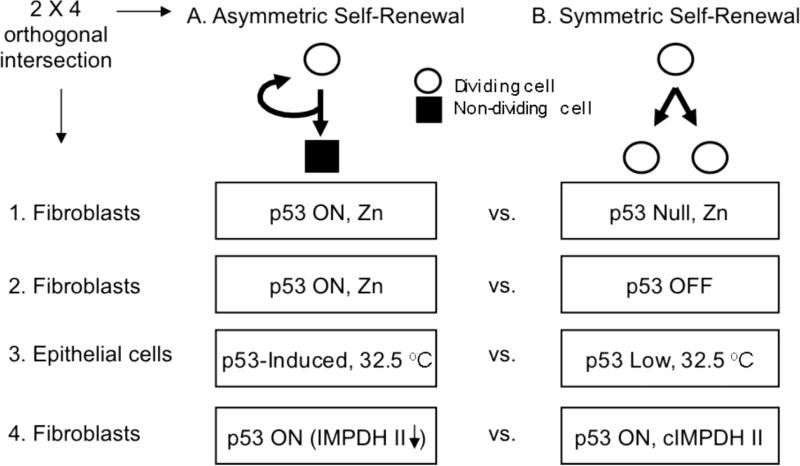
Previously, we used non-tumorigenic, immortalized mouse mammary epithelial C127 cells and mouse embryo fibroblasts (MEFs) to derive lines with conditional self-renewal patterns. The self-renewal pattern of these cells can be reversibly switched between symmetric and asymmetric by varying either culture temperature or Zn concentration, respectively, as a consequence of controlling p53 expression from respectively responsive promoters (Noh et al., 2011; Sherley et al., 1995a; Liu et al., 1998a; Rambhatla et al., 2001; Rambhatla et al., 2005; Liu et al., 1998b). These and related cell lines were used to design a 2 × 4 orthogonal-intersection microarray analysis for the purpose of identifying genes whose expression consistently showed the same pattern of change between asymmetric self-renewal versus symmetric self-renewal (See Fig. 1). Four different pair-wise comparisons were developed in which a state of asymmetric self-renewal was compared to a congruent state of symmetric self-renewal. The first 3 comparisons were based on a difference in p53 expression, but each had a different biological context. For asymmetric versus symmetric, respectively, these comparisons were:
-
1)
Zn-responsive p53-inducible MEFs (Ind-8 cells) versus p53-null control MEFs (Con-3 cells), both cultured in Zn-supplemented medium;
-
2)
Zn-responsive p53-inducible MEFs in Zn-supplemented medium versus in Zn-free medium (Noh et al., 2011; Liu et al., 1998a; Rambhatla et al., 2001; Rambhatla et al., 2005; Liu et al., 1998b); and
-
3)
low temperature (32.5°C)-dependent p53-inducible C127 cells (1h-3 cells) versus vector-control C127 cells (i.e., no p53 transgene; 1g-1 cells), both cultured at 32.5°C (Sherley et al., 1995a; Rambhatla et al., 2005; Liu et al., 1998b).
Figure 1. A 2 × 4 orthogonal-intersection microarray analysis to detect genes associated with asymmetric self-renewal.
cDNA micro-arrays were developed as described in Materials and Methods for four distinct comparisons of cells in congruent states of asymmetric self-renewal (A) versus symmetric self-renewal (B). In this strategy, asymmetric self-renewal associated (ASRA) genes [5] are defined as those whose expression consistently changes by a significant degree between asymmetric self-renewal versus symmetric self-renewal states in all four self-renewal pattern comparison models. Comparison 1 paired asymmetrically self-renewing, p53-induced Ind-8 MEFs to symmetrically self-renewing, p53-null Con-3 MEFs, both with ZnCl2-supplemention (Rambhatla et al., 2001; Rambhatla et al., 2005). Comparison 2 paired asymmetrically self-renewing, p53-induced Ind-8 cells (ZnCl2-supplemented) to symmetrically self-renewing, non-induced Ind-8 cells (ZnCl2-free). Comparison 3 paired asymmetrically self-renewing, p53-induced 1h-3 mouse mammary epithelial cells to symmetrically self-renewing, p53-expressing 1g-1 cells, both grown at 32.5°C (Sherley et al., 1995a; Rambhatla et al, 2001; Rambhatla et al, 2005). Culture of 1h-3 cells at 32.5°C causes a 1.5-fold elevation in p53 protein expression and a shift to asymmetric self-renewal due to a stably transfected low temperature-inducible p53 mini-gene. Line 1g-1 cells are congenic control cells that have subnormal p53 expression, lack the inducible p53 cDNA sequences, and maintain symmetric self-renewal at 32.5°C. Comparison 4-paired two stably transfected derivatives of Ind-8 cells under conditions of ZnCl2-supplementation. Line tC-2, which was derived by transfection with only gene expression regulatory elements, retains asymmetric self-renewal like the parental Ind-8 cells. In contrast, tI-3 cells, which were derived with synonymous expression constructs that contained a hamster type II inosine-5’-monophosphate dehydrogenase (IMPDH II) cDNA, no longer shift to asymmetric self-renewal with ZnCl2-supplemention (Liu et al., 1998a; Rambhatla et al., 2005). In tC-2 cells, IMPDH II protein levels are reduced as a result of p53 expression. In tI-3 cells, although the endogenous IMPDH II is reduced by p53 expression as well, the overall cellular level of IMPDH II is maintained by the constitutive expression the IMPDH II transgene (cIMPDH II) (Liu et al., 1998a).
The fourth comparison had a special purpose. It provided a comparison of asymmetric versus symmetric self-renewal that was not due to a difference in p53 expression (Noh et al., 2011). Two previously described derivatives of the Zn-responsive p53-inducible MEFs were used to make this comparison. One line (tI-3 cells) is stably transfected with a constitutively expressed type II inosine monophosphate dehydrogenase (IMPDH II) mini-gene (Noh et al., 2011; Liu et al., 1998a; Rambhatla et al., 2005). IMPDH II is the rate-limiting enzyme for guanine ribonucleotide biosynthesis. Its down-regulation by p53 is required for asymmetric self-renewal (Liu et al., 1998a; Rambhatla et al., 2005). Therefore, even in Zn-supplemented medium, which induces normal p53 expression, cells derived with a stably expressed IMPDH II transgene continue to undergo symmetric self-renewal (Liu et al., 1998a; Rambhatla et al., 2005). This abrogation of p53 effects on self-renewal pattern occurs even though other p53-dependent responses remain intact (Liu et al., 1998a). Under the same conditions, control vector-only transfectants (tC-2 cells) continue to exhibit asymmetric self-renewal (Liu et al., 1998a; Rambhatla et al., 2005). Thus, this fourth comparison could be used to exclude genes whose change in expression was primarily due to changes in p53 expression and not specifically transitions in self-renewal pattern as well (Noh et al., 2011).
We identified genes whose expression varied in the same manner for all 4 orthogonal comparisons of cells in states of asymmetric self-renewal versus symmetric self-renewal. In order for a gene to be considered, its average expression ratio – based on the two dye-swapped technical replicates – had to lie consistently among the top 5% of all average gene expression ratios determined (i.e., up-regulated) or among the bottom 5% of all average gene expression ratios (i.e., down-regulated). Moreover, this requirement had to be met for all 4 compared biological contexts.
We developed a new quality control metric called the population division cycle ratio (PDC ratio) to ensure consistent degrees of asymmetric self-renewal and symmetric self-renewal across all 4 orthogonal comparisons (Merok et al., 2002) (See Supplementary Information; Supplementary Fig. S1; Supplementary Table S1). Prefabricated cDNA micro-arrays constructed with the National Institute for Aging mouse 15K mouse clone set were used for the analysis (Tanaka et al., 2000; Kargul et al., 2001). As detailed in Materials and Methods, for each of the 4 experimental comparisons, we isolated two independent samples of RNA; and each of the RNA samples was labeled independently with Cy5 and Cy3 fluorescent dyes. Each independent set of fluorescent RNA samples was used to develop two reciprocal cohybridizations for each experimental comparison. Thus, data from 4 microarrays, representing 2 independent experiments, were available for each of the 4 orthogonal contexts for asymmetric versus symmetric self-renewal.
A sparse feature approach to identification of an asymmetric self-renewal biomarker
Conventional analysis of the microarray data identified 21 genes that were consistently up regulated in all 4 contexts of deterministic asymmetric self-renewal and 31 genes that were consistently down-regulated based on comparisons to symmetric self-renewal (See e.g. Supplementary Table S2) (Noh, 2006). Even a set of genes of this limited number poses a significant challenge to traditional methods for identifying DSC biomarkers. Therefore, we reevaluated the entire microarray dataset (GEO Accession number GSE40183) using the expectation maximization (EM) algorithm method, a previously described sparse feature approach (Burden and Winkler, 2009a,b). This very sparse feature selection method is ideally suited to select small sets of relevant genes from very large numbers of candidates, in a context-dependent manner.
Figeuiredo reported an EM algorithm using a sparse (non-informative) prior that provided parameter-free adaptive sparseness methodology (Figeuiredo, 2003). This algorithm iteratively and progressively sets irrelevant parameters exactly to zero. Figeuiredo's approach does not involve any (hyper)parameters to be adjusted or estimated. This is achieved by the adoption of a Jeffreys’ non-informative hyperprior. Figeuiredo showed that this approach yields state-of-theart performance that outperforms support vector machines, and performs competitively with the best alternative techniques. The EM method is set out very clearly in Figeuiredo's paper (Figeuiredo, 2003) and ours (Burden and Winkler, 2009b) [27]. (See Material and Methods and Supplementary Information [Fig. S2] for details of the method). In order to control manually the sparsity of the model, we added additional hyperparameters χ and ξ (Burden and Winkler, 2009b) [27].
The analysis identified a very small set of genes whose expression patterns were significantly related to changes in cell state between asymmetric self-renewal and symmetric self-renewal (Table 1). One gene that was down regulated during asymmetric self-renewal with respect to symmetric self-renewal had clearly superior significance as a discriminator of the two self-renewal states. This gene encoded H2A.Z (also known as H2afz), a variant of the nucleosomal core histone H2A.
Table 1.
Top discriminators of symmetric self-renewal versus asymmetric self-renewal identified by sparse feature analysis.
| Contribution Factor ± SEa | |||
|---|---|---|---|
| cDNA probe ID | Gene Name | pb | |
| H3009F03 | H2A.Z | −18.4 ± 1.1 | 7.0 × 10−6 |
| H3056B11 | None to date | −15.8 ± 4.2 | < 0.007 |
| H3132G04 | DNAjb11 | 11 ± 3.2 | < 0.01 |
| H3085E10 | TMEM39B | 12.1 ± 4.5 | 0.02 |
| H3114D11 | BTG1 | 8.7 ± 3.7 | 0.03 |
The mean contribution, in descending order of statistical significance, that a gene's expression change makes to the underlying model discriminating symmetric versus asymmetric self-renewal induced by any change in culture conditions. Positive values indicate up-regulation when cells shift to asymmetric self-renewal; and negative values indicate down-regulation when cells shift to asymmetric self-renewal. SE, the standard error.
Student's t-test confidence level for the gene's expression contribution to the discrimination model.
H2A.Z DSC biomarker properties
A priori, H2A.Z seems an impractical candidate as a DSC biomarker, because in the engineered cell lines, both H2A.Z mRNA (Noh, 2006) [25] and protein (Supplementary Fig. S3) are down regulated during asymmetric self-renewal. H2A.Z protein is also reduced when natural DSCs undergo asymmetric self-renewal (3C5 and 5B8 mouse hair follicle stem cells [HFSCs]; Supplementary Fig. S3). However, H2A.Z expression is not only reduced during asymmetric self-renewal, but the protein also takes on a unique molecular character. During asymmetric self-renewal, DSCs adopt a non-random pattern of chromosome segregation. With each asymmetric division, the DSC retains the set of sister chromatids with the older template DNA strands, which are called immortal DNA strands (Sherley, 2008; Cairns, 1975). Although H2A.Z is readily detected on immortal chromosomes by specific antibodies, on the corresponding mortal chromosomes it is masked from detection (Huh and Sherley, 2011). This unique molecular character, “H2A.Z asymmetry,” represents a potentially highly specific biomarker for DSCs, because non-random chromosome segregation has not been described in any other cell type to date, including committed progenitors, whose detection compromises the specificity of previously reported DSC indicators.
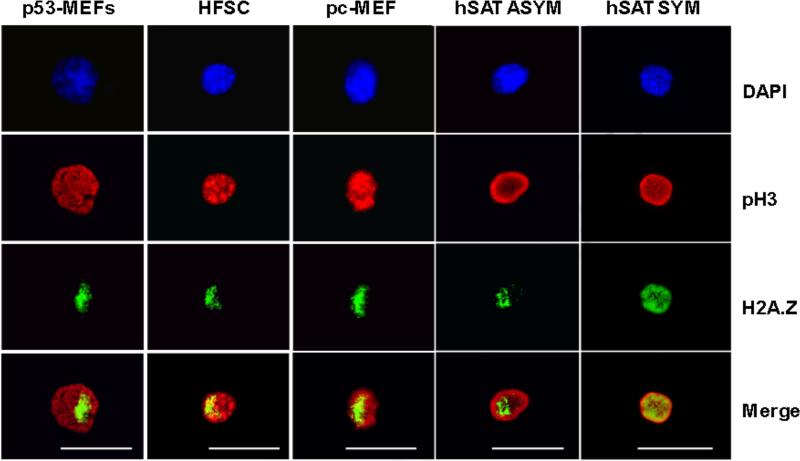
We adapted detection of H2A.Z asymmetry to cultures of expanded DSCs, cultures of pre-crisis mouse embryo fibroblast (pc-MEFs), and tissue sections containing adult mouse hair follicles. We paired the mitosis-specific biomarker phosphorylated histone H3 (pH3) with H2A.Z to develop a highly sensitive and specific assay for detecting rare mitotic tissue cells with H2A.Z asymmetry. Fig. 2 shows examples of the images obtained in evaluations of cultured cells. Ninety-three percent of prophase, symmetrically self-renewing, engineered cells that were pH3-positive had H2A.Z immunofluorescence that was symmetrically distributed with respect to pH3 immunofluorescence (e.g., See Fig. 2, hSAT SYM as an example; Table 2). In contrast, 32% and 20% of pH3-positive prophase cells in cultures of engineered cells and expanded mouse HFSCs (Huh et al., 2011), respectively, under conditions that promote asymmetric self-renewal, had a smaller, asymmetric distribution of H2A.Z immunofluorescence (Fig. 2; Table 2).
Figure 2. H2A.Z asymmetry is detected in prophase nuclei, identified by phosphorylated histone H3, for a variety of cultured cell types.
p53-MEFs, example of H2A.Z asymmetry in genetically engineered mouse embryo fibroblasts (line Ind-8) (Rambhatla et al., 2001; Rambhatla et al., 2005; Liu et al., 1998b) under conditions that promote asymmetric self-renewal. HFSC, example of H2A.Z asymmetry in mouse hair follicle stem cells (strain 3C5) (Huh et al., 2011; Huh and Sherley, 2011) under conditions that promote asymmetric self-renewal. pc-MEF, example of cell with H2A.Z asymmetry detected in cultures of pre-crises MEFs (See also Fig. 3). In cultures enriched for human skeletal muscle satellite stem cells: hSAT ASYM, example of cell with H2A.Z asymmetry, and hSAT SYM, example of cell with symmetric H2A.Z. DAPI, nuclear DNA fluorescence. pH3, indirect in situ immunofluorescence (ISIF) with antibodies specific for phosphorylated histone H3. H2A.Z, ISIF with antibodies specific for H2A.Z. ISIF was performed simultaneously for pH3 and H2A.Z. Merge, overlaid anti-pH3 and anti-H2A.Z images. Scale bar = 25 microns.
Table 2.
Percent H2A.Z asymmetry with respect to the self-renewal pattern regulation of diverse cultured cell types.
| Cell Type | Promoted Self-Renewal Patterna | pb | |||
|---|---|---|---|---|---|
| (basis) | ASYM | (basis) | SYM | ||
| p53-MEFsc | (p53 wt) | 32 | (p53 null) | 7 | < 0.024 |
| pc-MEFsd | (p53 wt) | 7 | (p53 KO) | <1 | < 0.009 |
| immMEFse | n/af | n/a | (immort) | <1 | n/a |
| HFSCs | (−Xng) | 20 | Ndh | nd | n/a |
| hSATs | (−Xn) | 12 | (+Xn) | 4 | < 0.004 |
Listed are the observed percentages of pH3-positive prophase nuclei showing H2A.Z asymmetry for the indicated cell types under conditions that promote either asymmetric self-renewal and non-random chromosome segregation (ASYM) or symmetric self-renewal and random segregation (SYM).
Statistical confidence, determined by the two-tailed Fisher's exact test, for the differences in percentage between states promoting asymmetric self-renewal versus symmetric self-renewal.
Wild-type p53-inducible MEFs and p53-null MEFs grown in ZnCl2-supplemented medium, corresponding to micro-array Comparison 1 (See also Fig. 1).
Pre-crises mouse embryonic fibroblasts from mice with a wild-type p53 genotype were compared to analogous cells derived from mice with a p53 gene knockout (KO) after 17 and 12 population doublings in culture, respectively. (The p53 wt cells had a transgenic KO of the cdkn1A gene.)
The same strain of p53 wt/cdkn1A KO MEFs was evaluated after spontaneous immortalization and culture for a total of 50 population doublings.
n/a = not applicable
Xn = xanthine
nd = not done
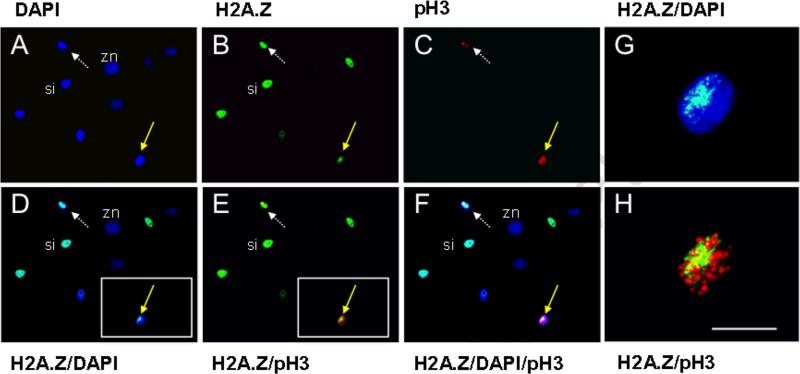
Early passage pre-crisis cultures are closer in cell make-up to primary dissociated tissue cell preparations than are immortalized cell lines. In analyses with cultures of early passage pc-MEFs (17 population doublings), 7% of prophase cells displayed H2A.Z asymmetry (Table 2; Fig. 3; also Fig. 2, pc-MEF). In contrast, in later immortalized cultures (50 population doublings), none of 110 examined prophase cells showed H2A.Z asymmetry (< 0.9%). Similarly, none of 123 examined prophases in early passage cultures from p53 gene knockout mice showed H2A.Z asymmetry (12 population doublings; < 0.8%). This significant difference in detection (p < 0.009) is consistent with our proposal that immortalization, which is highly associated with p53 gene mutation, manifests a permanent conversion of long-lived DSCs from asymmetric self-renewal to symmetric self-renewal, with loss of non-random segregation (Rambhatla et al., 2001; Rambhatla et al., 2005; Merok et al., 2002).
Figure 3. H2A.Z asymmetry in prophase nuclei detected by phosphorylated histone H3 in cultures of wild-type, pre-crises MEFs.
DAPI, nuclear DNA fluorescence. H2A.Z, ISIF with antibodies specific for H2A.Z. pH3, ISIF with antibodies specific for phosphorylated histone H3. ISIF was performed simultaneously for pH3 and H2A.Z. A-C, independent fluorescence images for the indicated markers. D-F, merged combinations of A-C images as denoted. Yellow solid arrows, example of a prophase nucleus with H2A.Z asymmetry. White dotted arrows, example of a prophase nucleus with symmetric H2A.Z. si, example of interphase nucleus (i.e., pH3-negative) with symmetric H2A.Z localization. zn, H2A.Z-negative interphase nucleus. G and H, 10X magnification of nuclear images in the inset boxes of D and E, respectively. Scale bar = 25 microns.
We used the pH3-H2A.Z asymmetry assay to provide evidence for non-random segregation in normal human DSCs. Earlier human studies evaluated cancer stem cells (Pine et al., 2010; Hari et al., 2011; Xin et al., 2012). Pre-senescent cultures containing human skeletal muscle satellite stem cell cultures were evaluated (Fig. 2, hSAT) (Homma et al., 2012). Non-random segregation by mouse skeletal muscle satellite stem cells was reported earlier (Shinin et al., 2006; Conboy et al., 2007). When grown under routine culture conditions, 12% of mitotic cells in hSAT cultures exhibited H2A.Z asymmetry (Fig. 2, hSAT ASYM; Table 2). Asymmetry was detected throughout mitosis. Consistent with indicating non-random segregation, the frequency of H2A.Z asymmetric mitotic cells in hSAT cultures was reduced to 4% by supplementation with 1.5 mM xanthine (p < 0.004; Fig. 2, hSAT, SYM; Table 2), as occurs for mouse HFSCs (Huh et al., 2011).
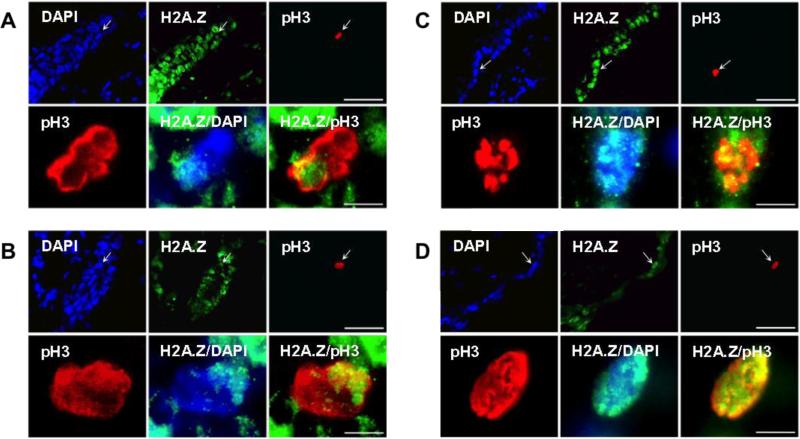
Recently, we showed that Lgr5-expressing DSCs expanded from mouse hair follicles undergo non-random segregation with H2A.Z asymmetry (Huh et al., 2011; Huh and Sherley, 2011) (Also Fig. 2, HFSC). To evaluate whether cells with H2A.Z asymmetry were also found in tissues, we applied the pH3-H2A.Z asymmetry assay to evaluate 10-micron thick sections of adult mouse skin that contained hair follicles. Cells positive for pH3 were rare in these sections, as expected for mitotic cells. However, we found that 22% of the identified pH3-positive cells (i.e., 14 of 64 detected in 183 sectioned hair follicles) showed H2A.Z asymmetry as shown in the examples given in Fig. 4 (p < 0.0001; two-tailed Fisher's exact test with respect to no detection). The mitotic cells with asymmetric H2A.Z were consistently found at the base of the hair shaft in the secondary germ region, the location in which Lgr5-expressing HFSCs have also been shown to reside (Jaks et al., 2008), whereas mitotic cells with symmetric H2A.Z localization were found both in hair follicles and epidermal regions.
Figure 4. Detection of mitotic cells with H2A.Z asymmetry in mouse hair follicles.
Ten-micron thick, paraffin-embedded sections of adult mouse skin were evaluated by ISIF with antibodies specific for H2A.Z and phosphorylated histone H3 (pH3), a biomarker for mitotic cells. DAPI, total nuclear DNA fluorescence. Note: In A images, the hair shaft emerges through the lower left corner. A and B, examples of mitotic hair follicle cells with H2A.Z asymmetry. C and D, examples of epidermal mitotic cells with symmetric H2A.Z. Top rows, low magnification images; scale bars = 50 microns. Bottom rows, 10X magnification images of the cell identified with arrows in upper images; scale bars = 5 microns. H2A.Z/DAPI, merged images of H2A.Z and DAPI fluorescence. H2A.Z/pH3, merged images of H2A.Z and phosphorylated histone H3 epifluorescence.
Discussion
Our results show the potential of H2A.Z to provide a highly specific biomarker for identifying cells in tissues that are undergoing two tightly linked processes, deterministic asymmetric self-renewal and non-random segregation (Rambhatla et al., 2005; Huh et al., 2011), which specify DSCs. The key evidence for this conclusion is based on the cell culture data presented with expanded HFSCs. We have shown previously that these expanded cell strains exhibit many of the unique properties of DSCs (Huh et al., 2011; Huh and Sherley, 2011). These include long-term symmetric self-renewal; asymmetric self-renewal with production of multi-lineage differentiated cells based on loss of Lgr5 expression and expression of K5, K10, or filaggrin; expression of Lgr5, which is asymmetrically limited to the stem cell sister during asymmetric self-renewal divisions; and non-random chromosome segregation (Huh et al., 2011; Huh and Sherley, 2011). In addition, we show several additional examples of H2A.Z asymmetry detection in murine and human cell types, both in cell culture and in tissue sections, which are consistent with the specific detection of asymmetrically self-renewing DSCs.
Supporting a general functional role for H2A.Z in DSC function in many human tissues, we found that in human cancer cell lines of diverse tissue origin higher expression of H2A.Z mRNA was significantly correlated with higher expression of IMPDH II mRNA (Supplementary Fig. S4). Up-regulation of IMPDH II is predicted to shift tissue DSCs from asymmetric self-renewal to symmetric self-renewal, with loss of non-random segregation, constituting an important carcinogenic mechanism (Rambhatla et al., 2005; Huh et al., 2011).
Whereas H2A.Z is predicted to be a highly specific biomarker for DSCs, its sensitivity for detecting DSCs may vary from tissue to tissue because of differences in DSC self-renewal kinetics programs. DSCs undergoing symmetric self-renewal with random sister chromatid segregation would not be detected. Such self-renewal pattern excursions by deterministic asymmetrically self-renewing DSCs are predicted to be limited to periods of tissue mass expansion (e.g., during adult maturation) and repair of injuries. Quiescent DSCs might also go undetected, if they arrested without H2A.Z asymmetry (Li and Clevers, 2010). This possibility might be evaluated in cell culture studies.
A third possible cause of lower sensitivity awaits more extensive tissue investigations with the biomarker. An ongoing discussion in tissue stem cell biology is the balance of deterministic asymmetric self-renewal programs, reported here, compared to stochastic asymmetric self-renewal programs employed by DSCs in mammalian tissues (Loeffler and Potten, 1997; Enver et al., 1998; Klein and Simons, 2011)). In stochastic programs for asymmetric self-renewal by DSCs, tissue renewal is accomplished with nearly equivalent frequencies of symmetric self-renewal divisions and divisions that give rise to one or two lineage-committed cells. If tissues were renewing primarily by stochastic asymmetric self-renewal programs, H2A.Z asymmetry might have low sensitivity for detecting DSCs. However, this detection shortcoming would still serve to advance knowledge of the cell kinetics properties of DSCs in diverse mammalian tissues.
The identification of H2A.Z, a down regulated gene, by the sparse feature approach underlines how this method can detect genes of unique biological significance that would often be overlooked. A down-regulated protein would hardly be considered as a practical expression biomarker candidate in the absence of any other distinguishing properties. Conceptually, the quantitative extraction of H2A.Z from the complex 16-dimensional data set by the sparse feature analysis reflects the detection of a consistent relationship of H2A.Z mRNA expression level and expression variance to two biologically linked cellular phenotypes (i.e., asymmetric self-renewal versus symmetric self-renewal) over an experimentally devised varying cellular landscape. Although conventional bioinformatics methods also identified H2A.Z, it was a relatively low priority member of a much longer list of candidate genes. Of the 930 genes identified as being up regulated or down regulated with respect to self-renewal pattern in the previous work (Noh et al., 2011), H2A.Z appeared towards the bottom of the list sorted by expression fold ratio and would normally be overlooked. In contrast, the sparse feature selection method identified just a handful of candidate genes among which H2A.Z was the most prominent. This capability will undoubtedly be very powerful in many other types of investigations in which gene expression is evaluated with respect to specific physiological conditions of biomedical interest. As an additional proof of concept, we very recently used the same sparse feature selection approach to identify genes implicated in a novel mechanism for the strontium-induced differentiation of mesenchymal stem cells down the osteogenic pathway (Autefage et al., 2015).
Currently, because of the lack of biomarkers that identify DSCs specifically, the only way to monitor relative changes in their number and quality is by functional assays. In animal model research, changes in DSCs are monitored by the ability of transplanted donor cells to reconstitute recipient tissues after ablation of host DSCs. Similarly, the measure of human hematopoietic stem cells is successful reconstitution of patients whose hematopoietic stem cells were either deficient due to disease or iatrogenic ablation. These function-based approaches are not quantitative. They cannot discern differences in stem cell number versus stem cell quality; and they evaluate stem cells under conditions of physiological stress (e.g., transplantation). The research tools required to address these important deficiencies, specific and universal biomarkers for DSCs, have not be described previously. The availability of such biomarkers would greatly accelerate tissue stem cell research and advances in cell replacement medicine. Stem cell research would join the ranks of other quantitative biological sciences; and transplant physicians would have a first quantitative metric for predicting transplant efficacy. This report of an H2A.Z asymmetry biomarker for DSCs is the beginning of this new era of DSC biology and biomedicine.
Conclusions
Our paper contains several sources of novelty. Primarily, our results show how an unconventional computational feature selection method can be applied effectively to large microarray gene expression datasets to identify sparse sets of a few genes that have a high degree of functional significance for a biological process under study. In this work the small set of genes were implicated in the symmetry of cell division. These genes were only some of many that would have been selected using standard expression level/fold ratio bioinformatics criteria. The efficacy of this sparse feature approach has important advantages in functional genomics research.
Amongst our set of candidate markers was a down-regulated gene encoding the histone H2A variant H2A.Z. We specifically chose to validate experimentally H2A.Z because it was identified by the sparse feature selection properties of the EM algorithm to have the highest Bayesian relevance. We showed that it was a specific indicator of asymmetric self-renewal in cell lines genetically engineered to model DSC asymmetric self-renewal in several experimental contexts. Cellular analyses further revealed that down-regulated H2A.Z gene expression was associated with asymmetric detection of the H2A.Z protein on only one set of segregating mitotic chromosomes in DSCs undergoing non-random segregation, which is functionally linked to asymmetric self-renewal. Using expanded natural hair follicle tissue stem cell lines, we confirmed that this H2A.Z asymmetry was a specific biomarker for DSCs. Studies with presenescent mouse cell populations, human muscle satellite stem cell-enriched cultures, and mouse hair follicle tissue sections showed the potential of H2A.Z asymmetry to be a universal, specific biomarker for asymmetrically self-renewing mammalian DSCs with the capability of quantifying DSCs for the first time. Our work identifies the potential for sparse feature selection bioinformatics methods to uncover novel crucial insights into previously impenetrable biological systems and processes.
We also presented new stem cell methods and data: -
A new H2A.Z/phospho histone H3 assay for detecting H2A.Z asymmetry, prospectively, before a cell divides. We applied this new assay in the studies of in vitro cultured hair follicle stem cells.
We also applied this assay to show that H2A.Z asymmetry can also be detected in cultures containing human adult tissue stem cells, and that it is regulated by xanthine – evidence that the asymmetry is occurring in asymmetrically cycling human muscle tissue stem cells.
In primary MEF cultures, we showed that H2A.Z is regulated by p53 expression – additional evidence of its efficacy in identifying asymmetrically self-renewing primary tissue stem cells.
We extended our prior in vitro studies on H2A.Z asymmetry in asymmetrically self-renewing cultured hair follicle stem cells to the in vivo situation, showing the presence of cells with H2A.Z asymmetry in a region known to contain hair follicle tissue stem cells.
The showed the utility of the PDC ratio for evaluating the cell kinetics of cultures containing asymmetrically self-renewing cells
These new data support our hypothesis that H2A.Z, identified by the EM algorithm, has potential to be a general highly-specific marker of asymmetrically self-renewing tissue stem cells both in mouse tissues and human tissues. No biomarker described to date as the ability to identify tissue stem cells exclusively.
Supplementary Material
Highlights.
There is a long standing unmet need for biomarkers for distributed stem cells (DSC)
We used engineered cells that deterministically self-renew in response to multiple triggers
Analysis of gene expression data from experiments used unconventional, sparse gene selection
Identified H2A.Z as a novel, pattern-specific biomarker for asymmetrically self-renewing DSCs
This specificity of this biomarker was validated in subsequent experiments
Acknowledgments
This work was supported by NIH-NHGRI grant #PSO HG 003170, NIH-NIEHS grant #689471, and NIH-NIGMS Director's Pioneer Award #5DP1OD000805. JCC was supported by the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health under Award Number U54HD060848. We thank Shirley A. Bohn and Hetal Parekh for technical assistance in deriving immortalized p53 wt/cdkn1A KO MEFs. DAW also acknowledges support from the CSIRO Newton Turner Award for Exceptional Senior Scientists.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Autefage H, Gentleman E, Littmann E, Hedegaard M, Von Erlach T, O'Donnell M, Burden FR, Winkler DA, Stevens MM. Novel sparse feature selection methods identify unexpected global cellular response to strontium-containing materials. Proc. Natl. Acad. Sci. 2015 doi: 10.1073/pnas.1419799112. USA under revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden FR, Winkler DA. An optimal self-pruning neural network that performs nonlinear descriptor selection for QSAR. QSAR Comb. Sci. 2009a;28:1092–1097. [Google Scholar]
- Burden FR, Winkler DA. Optimum QSAR feature selection using sparse Bayesian methods. QSAR Comb. Sci. 2009b;28:645–653. [Google Scholar]
- Burns CE, Zon LI. Portrait of a stem cell. Dev. Cell. 2002;3:612–613. doi: 10.1016/s1534-5807(02)00329-5. [DOI] [PubMed] [Google Scholar]
- Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- Cawley GC, Talbot NL. Gene selection in cancer classification using sparse logistic regression with Bayesian regularization. Bioinfo. 2006;22:2348–2355. doi: 10.1093/bioinformatics/btl386. [DOI] [PubMed] [Google Scholar]
- Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:1120–1126. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enver T, Heyworth CM, Dexter TM. Do stem cells play dice? Blood. 1998;92:348–351. [PubMed] [Google Scholar]
- Figueiredo MAT. Adaptive sparseness for supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 2003;25:1150–1159. [Google Scholar]
- Fortunel NO, Out HH, Ng HH, Chen J, Mu X, Chevassut T, Li X, Joseph M, Bailey C, Hatzfeld C, et al. Comment on “’Stemness: transcriptional profiling of embryonic and adult stem cells” and “A stem cell molecular signature”. Science. 2003;302:393. doi: 10.1126/science.1086384. [DOI] [PubMed] [Google Scholar]
- Hari D, Xin HW, Jaiswal K, Wiegand G, Kim BK, Ambe C, Burka D, Koizumi T, Ray S, Garfield S, et al. Isolation of live label-retaining cells and cells undergoing asymmetric cell division via nonrandom chromosomal cosegregation from human cancers. Stem Cells Dev. 2011;20:1649–1658. doi: 10.1089/scd.2010.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma S, Chen JC, Rahimov F, Beermann ML, Hanger K, Bibat GM, Wagner KR, Kunkel LM, Emerson CP, Jr, Miller JB. A unique library of myogenic cells from facioscapulohumeral muscular dystrophy subjects and unaffected relatives: family, disease and cell function. Eur. J. Hum. Genet. 2012;20:404–410. doi: 10.1038/ejhg.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh YH, King J, Cohen J, Sherley JL. SACK-expanded hair follicle stem cells display asymmetric nuclear Lgr5 expression with non-random sister chromatid segregation. Sci Rep. 2011;1:176. doi: 10.1038/srep00176. doi:10.1038/srep00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh YH, Sherley JL. Molecular cloaking of H2A.Z on mortal DNA chromosomes during non-random segregation. Stem Cells. 2011;29:1620–1627. doi: 10.1002/stem.707. [DOI] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgård R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nature Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- Kargul GJ, Dudekula BB, Qian Y, Lim MK, Jaradat SA, Tanaka TS, Carter MG, Ko MSH. Verification and initial annotation of the NIA mouse 15K cDNA clone set. Nature Genet. 2001;28:17–18. doi: 10.1038/ng0501-17. [DOI] [PubMed] [Google Scholar]
- Kiiveri HT. A general approach to simultaneous model fitting and variable elimination in response models for biological data with many more variables than observations. BMC Bioinfo. 2008;9:195. doi: 10.1186/1471-2105-9-195. doi:10.1186/1471-2105-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Simons BD. Universal patterns of stem cell fate in cycling adult tissues. Devel. 2011;138:3103–3111. doi: 10.1242/dev.060103. [DOI] [PubMed] [Google Scholar]
- Krishnapuram B, Hartemink AJ, Carin L, Figueiredo MA. Bayesian approach to joint feature selection and classifier design. IEEE Trans. Pattern. Anal. Mach. Intell. 2004;26:1105–1111. doi: 10.1109/TPAMI.2004.55. [DOI] [PubMed] [Google Scholar]
- Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bohn SA, Sherley JL. Inosine-5’-monophosphate dehydrogenase is a rate-limiting factor for p53-dependent growth regulation. Mol. Biol. Cell. 1998a;9:15–28. doi: 10.1091/mbc.9.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Riley LB, Bohn SA, Boice JA, Stadler PB, Sherley JL. Comparison of bax, waf1, and IMP dehydrogenase regulation in response to wild-type p53 expression under normal growth conditions. J. Cell. Physiol. 1998b;177:364–376. doi: 10.1002/(SICI)1097-4652(199811)177:2<364::AID-JCP18>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Loeffler M, Potten CS. Stem cells and cellular pedigrees – a conceptual introduction. In: Potten CS, editor. Stem Cells. Academic Press; London: 1997. pp. 1–27. [Google Scholar]
- Merok JR, Lansita JA, Tunstead JR, Sherley JL. Cosegregation of chromosomes containing immortal DNA strands in cells that cycle with asymmetric stem cell kinetics. Cancer Res. 2002;62:6791–6795. [PubMed] [Google Scholar]
- Noh M, Smith JL, Huh YH, Sherley JL. A resource for discovering specific and universal biomarkers for distributed stem cells. Plos ONE. 2011;6(7):e22077. doi: 10.1371/journal.pone.0022077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh M. Doctoral thesis. Massachusetts Institute of Technology; 2006. https://dspace.mit.edu/handle/1721.1/37955. [Google Scholar]
- Pine SR, Ryan BM, Varticovski L, Robles AI, Harris CC. Microenvironmental modulation of asymmetric cell division in human lung cancer cells. Proc. Natl. Acad. Sci. USA. 2010;107:2195–2200. doi: 10.1073/pnas.0909390107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J. Computational analysis of microarray data. Nature Rev. Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. Stemness: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Rambhatla L, Bohn SA, Stadler PB, Boyd JT, Coss RA, Sherley JL. Cellular senescence: ex vivo p53-dependent asymmetric cell kinetics. J. Biomed. Biotech. 2001;1:28–37. doi: 10.1155/S1110724301000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambhatla L, Ram-Mohan S, Cheng JJ, Sherley JL. Immortal DNA strand co-segregation requires p53/IMPDH-dependent asymmetric self-renewal associated with adult stem cells. Cancer Res. 2005;65:3155–3161. doi: 10.1158/0008-5472.CAN-04-3161. [DOI] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nature Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- Sherley JL. A new mechanism for aging: chemical ‘age spots’ in immortal DNA strands in distributed stem cells. Breast Dis. 2008;29:37–46. doi: 10.3233/bd-2008-29105. [DOI] [PubMed] [Google Scholar]
- Sherley JL. Asymmetric cell kinetics genes: The key to expansion of adult stem cells in culture. Stem Cells. 2002;20:561–572. doi: 10.1634/stemcells.20-6-561. [DOI] [PubMed] [Google Scholar]
- Sherley JL. Asymmetric self-renewal: the mark of the adult stem cell. In: Habib NA, Gordon MY, Levicar N, Jiao L, Thomas-Black G, editors. Stem Cell Repair and Regeneration. Imperial College Press; London: 2005. pp. 21–28. [Google Scholar]
- Sherley JL, Stadler PB, Johnson DR. Expression of the wild-type p53 anti-oncogene induces guanine nucleotide-dependent stem cell division kinetics. Proc. Natl. Acad. Sci. USA. 1995a;92:136–140. doi: 10.1073/pnas.92.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherley JL, Stadler PB, Stadler JS. A quantitative method for the analysis of mammalian cell proliferation in culture in terms of dividing and non-dividing cells. Cell Prolif. 1995b;28:137–144. doi: 10.1111/j.1365-2184.1995.tb00062.x. [DOI] [PubMed] [Google Scholar]
- Shinin V, Gayraud-Morel B, Gomès D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nature Cell. Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, et al. Genome-wide expression profiling of mid-gestation placental and embryo using 15k mouse developmental cDNA microarray. Proc. Natl. Acad. Sci USA. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell. Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toni T, Stumpf MP. Parameter inference and model selection in signaling pathway models. Methods Mol. Biol. 2010;673:283–295. doi: 10.1007/978-1-60761-842-3_18. [DOI] [PubMed] [Google Scholar]
- Xin HW, Hari DM, Mullinax JE, Ambe CM, Koizumi T, Ray S, Anderson AJ, Wiegand GW, Garfield SH, Thorgeirsson SS, et al. Tumor-initiating label-retaining cancer cells in human gastrointestinal cancers undergo asymmetric cell division. Stem Cells. 2012;30:591–598. doi: 10.1002/stem.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.