Abstract
3-Hydroxypropionic acid (3-HP) is a commercially valuable platform chemical from which an array of C3 compounds can be generated. Klebsiella pneumoniae has been considered a promising species for biological production of 3-HP. Despite a wealth of reports related to 3-HP biosynthesis in K. pneumoniae, its commercialization is still in infancy. The major hurdle hindering 3-HP overproduction lies in the poor understanding of glycerol dissimilation in K. pneumoniae. To surmount this problem, this review aims to portray a picture of 3-HP biosynthesis, involving 3-HP-synthesizing strains, biochemical attributes, metabolic pathways and key enzymes. Inspired by the state-of-the-art advances in metabolic engineering and synthetic biology, here we advocate protocols for overproducing 3-HP in K. pneumoniae. These protocols range from cofactor regeneration, alleviation of metabolite toxicity, genome editing, remodeling of transport system, to carbon flux partition via logic gate. The feasibility for these protocols was also discussed.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-015-0513-0) contains supplementary material, which is available to authorized users.
Keywords: 3-Hydroxypropionic acid, Klebsiella pneumoniae, Cofactor regeneration, Genome editing, Logic gate, Synthetic biology
Introduction
Biosynthesis has emerged as an alternative to conventional chemical synthesis owing to fast consumption of petroleum resources. 3-Hydroxypropionic acid (3-HP) is one of the 12 top-added platform compounds suggested by US DOE [1]. As a versatile precursor, 3-HP can be readily converted into a series of economically important chemicals, including 1,3-propandiol (1,3-PDO), 3-hydroxypropionaldehyde (3-HPA), acrylic acid, and malonic acid [2]. Additionally, the hydroxyl and carboxyl in 3-HP molecule can be polymerized to form poly-3-hydroxypropionate [3], which is a novel polymer displaying higher mechanical strength and elongation at break compared with poly lactic acid (2-hydroxypropionic acid). Because of its broad applications, 3-HP has been fueled in recent years.
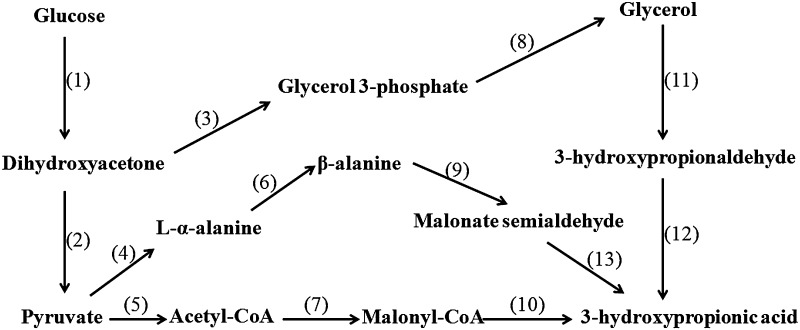
So far myriads of 3-HP biosynthesis pathways have been identified [4]. Based on carbon sources, 3-HP pathways can be divided into two types: glucose- and glycerol-based routes. In the glucose-based pathway, glucose is converted to acetyl-CoA which is catalyzed to propionyl-CoA by Acetyl-CoA carboxylase. Next, propionyl-CoA is catalyzed to 3-HP by malonyl-CoA reductase [5]. Except this, there exist other biosynthetic pathways where glucose is converted to glycerol and next to 3-HP [6]. In the glycerol pathway, glycerol dehydratase catalyzes glycerol into 3-hydroxypropionaldehyde (3-HPA), next, 3-HPA is converted to 3-HP by aldehyde dehydrogenase (AldH) [7], and to 1,3-propanediol by 1,3-propanediol oxidoreductase [8] (see Fig. 1).
Fig. 1.
Representative pathways of 3-HP production from glucose or glycerol. (1) and (2) indicate the enzymes in glycolytic pathway, (3) glycerol-3-phosphate dehydrogenase EC:1.1.1.8, (4) beta-alanine/pyruvate aminotransferase EC:2.6.1.18, (5) pyruvate oxidase EC:1.2.3.6, (6) alanine 2,3-aminomutase EC:5.4.3.8, (7) acetyl-CoA carboxylase EC:6.4.1.2 or biotin carboxylase EC:6.3.4.14, (8) glycerol 3-phosphatase EC:3.1.3.21, (9) beta -alanine/pyruvate aminotransferase EC:2.6.1.18, (10) malonyl-CoA reductase EC:1.2.1.75, (11) large subunit of glycerol dehydratase EC:4.2.1.30, (12) aldehyde dehydrogenase EC:1.2.1.4, (13) 3-hydroxypropionate dehydrogenase (NADP+) EC:1.1.1.298
As a main byproduct of biodiesel production, glycerol is a competitive substrate for 3-HP biosynthesis due to its decreasing price [9, 10]. In contrast to glucose pathway, glycerol pathway seems to be more applicable owing to its simple catalytic reactions and tractable manipulation. Apart from above mentioned two synthetic pathways, 3-HP can be derived from other carbon sources like sucrose and maltose [11]. Most of the carbon sources are converted to 3-HP via intermediate pyruvate or glycerol in vivo. In addition, recent research demonstrates that 3-HP can also be generated from CO2 through 3-HP cycle in Pyrococcus furiosus [12], which was previously reported as carbon dioxide fixation pathway [13].
Klebsiella pneumoniae and Glycerol Dissimilation
Among 3-HP-synthesizing strains, Klebsiella pneumoniae is of great attractiveness due to its biochemical attributes, including remarkable capacity to metabolize glycerol, active cell proliferation, and native capability to synthesize vitamin B12 [14]. Since vitamin B12 is the cofactor of glycerol dehydratase, K. pneumoniae has obvious advantage over E. coli which needs the addition of expensive vitamin B12 to fermentation medium [15]. Although Lactobacillus reuteri also naturally synthesizes 3-HPA and is benign to environment, its growth is significantly slow compared with K. pneumoniae, thus increasing production cost. Under anaerobic or microaerobic conditions, K. pneumoniae grow actively with glycerol as sole carbon source. An operon named dha governs the glycerol dissimilation which relates to glycerol oxidation and reduction pathways [16]. In glycerol oxidation pathway, glycerol dehydrogenase catalyzes glycerol into dihydroxyacetone (DHA), next, DHA is mutated into dihydroxyacetone phosphate (DHAP) by dihydroxyacetone kinase. In glycerol reduction pathway, glycerol is changed into 3-hydroxypropionaldehyde (3-HPA) by B12-dependent glycerol dehydratase (GDHt). Subsequently, 3-HPA is converted to 3-HP by aldehyde dehydrogenase (AldH), and to 1,3-propanediol (1,3-PDO) by 1,3-propanediol oxidoreductase [7, 17] (Fig. 1). It seems that dha regulon was tailored for the adaption to anaerobic or microaerobic environment. Of diverse 3-HP-synthesizing bacteria reported thus far, Klebsiella and Clostridium species harbor intact dha regulon [14]. These bacteria have developed a suite of mechanisms to adapt the changing milieus. For example, under anaerobic conditions glycerol dissimilation in K. pneumoniae is mediated by dha regulon. However, under aerobic conditions, glp instead of dha works because glp regulon is more efficient for glycerol consumption and cell respiration [16].
Despite K. pneumoniae can naturally synthesize 3-HP due to its innate aldehyde dehydrogenases (e.g., KPN_01468,KPN_01919) [15], the yield is far from commercialization. Namely, 3-HP pathway needs significant modification. Among all identified 3-HP pathways, the pathway native to K. pneumoniae has attracted most attention. Previous reviews have documented the research advances [11, 18], here we rethink the glycerol dissimilation in K. pneumoniae in the hope of addressing the problems arising from molecular breeding.
3-HP Production Relies Largely on the Activity of Aldehyde Dehydrogenase
As mentioned above, glycerol dehydratase catalyzes glycerol into 3-HPA, which is subsequently converted into 1,3-PDO by 1,3-propanediol oxidoreductase, or, in parallel, into 3-HP by aldehyde dehydrogenase [19]. Wild type K. pneumoniae strain produces more 1,3-PDO yet less 3-HP because 1,3-propanediol oxidoreductase shows higher activity than aldehyde dehydrogenase. The reasons behind include but are not limited to the following: (1) Compared with 3-HP, 1,3-PDO has lower polarity and does not cause significant pH fluctuation. Moreover, 1,3-PDO is an antifreeze that may enable K. pneumoniae to survive at low temperature. Thus, from the perspective of evolution, we hypothesize that K. pneumoniae prefers to synthesize 1,3-PDO rather than 3-HP. (2) High activity of aldehyde dehydrogenase resulted in the accumulation of 3-HP, which slowed down the cell growth [20]. In other words, low activity ALDH restricts the formation of 3-HP and benefits cell growth. (3) Biosynthesis of 3-HPA represents a self-protection mechanism for K. pneumoniae. This viewpoint is evidenced by the fact that some viruses are sensitive to aldehyde [21, 22], and intracellular 3-HPA may diffuse out of the cell to prevent virus spreading. Hence, low activity of ALDH enables the existence of 3-HPA, which is ready to cope with viral invasion. (4) 3-HPA acts as the core metabolite in glycerol dissimilation pathways, and its biosynthesis contributes to both rigidity and flexibility of dha operon. Presumably, when glycerol is ample, 3-HPA is converted to 3-HP or 1,3-PDO. Conversely, when glycerol is exhausted, 3-HPA as an intermediate blocks the conversion of glycerol to 3-HP and 1,3-PDO.
Aldehyde dehydrogenases are common in nearly all living organisms. The ubiquitous aldehyde dehydrogenases catalyze aldehydes to corresponding carboxylic acids [23]. In plants and bacteria, aldehyde dehydrogenases are expressed under extreme conditions to cope with hostile stimuli such as drought and high concentration of aldehyde [23]. Upon long term evolution, aldehyde dehydrogenase currently exhibits low activity because its overexpression collides with other functional aldehyde compounds. To overproduce 3-HP, high activity ALDH is imperative and the below are approaches toward this goal. One is screening strains from extreme environment, given that the adaptation to harsh conditions is usually correlated with high activity enzyme. One such example is the screened enzyme “KGSADH” that exhibited a high activity toward 3-HPA [15]. Another approach towards high ALDH activity is “directed evolution”. Through error-prone PCR or DNA shuffling and subsequent high throughput screening, high activity ALDH could be acquired. The third strategy is chemical synthesis of the enzyme gene based upon molecular simulation with emphasis on substrate specificity.
Insufficient Cofactor NAD+ Limits 3-HP Production
Cofactor availability is crucial for aldehyde dehydrogenase (ALDH) activity and 3-HP biosynthesis [24]. In glycerol reductive pathway, ALDH together with NAD+ catalyzes 3-HPA to 3-HP [25]. Since ALDH activity is lower than GDHt, plasmid-dependent ALDH overexpression is a common strategy for promoting 3-HPA to 3-HP. One drawback of this strategy is massive NAD+ consumption by the overexpression of ALDH, which leads to NAD+ exhaustion and the cessation of 3-HP formation. Another factor affecting ALDH activity is the formation of byproducts. Accompanied by 3-HP biosynthesis, lactate dehydrogenase and phosphotransacetylase catalyze the formation of lactic acid and acetic acid, respectively. Since phosphotransacetylase also utilizes NAD+ as a cofactor, there exists a competition for the limited amount of NAD+ between this enzyme and ALDH. Evidently, both ALDH activity and 3-HP biosynthesis are associated with the formation of lactic acid and acetic acid.
To boost 3-HP production, NAD+ regeneration system could be engineered in K. pneumoniae. Previously, NAD+ was regenerated mainly by overexpression of the enzymes using NADH as a cofactor. For example, both lactate dehydrogenase and glutamine dehydrogenase are NADH-dependent enzymes, and were expressed in host cell to regenerate NAD+ [26, 27]. However, plasmid-dependent cofactor regeneration overburdens the host cell and therefore hinders 3-HP productivity. To coordinate cofactor regeneration and cell growth, coproduction of two or more desired compounds was implemented. For example, 3-HP and 1,3-PDO were successfully coproduced, achieving titer up to 24.4 g/L of and 49.3 g/L, respectively [28]. The overall yield on glycerol is around 50–80 % [15]. So far although coenzyme regeneration systems have been engineered in K. pneumoniae for the production of 3-HP and 1,3-PDO, there existed an imbalance between oxidation and reduction pathways, because emphasis was preferentially placed on glycerol reduction pathway. Namely, the structural rigidity of dha regulon was ignored. Based on available conclusions, coproduction of 3-HP and 1,3-PDO seems to be feasible.
Excessive Lipopolysaccharide Attenuates the Carbon Flux Toward 3-HP
The third hurdle retarding 3-HP biosynthesis is lipopolysaccharide secreted from the cell wall of Gram-negative bacteria. As an endotoxin and the major ingredient of cell wall, lipopolysaccharide is generated when bacteria are subjected to lysozyme degradation or virus infection. Considering K. pneumoniae is a pathogenic bacterium, we reckon that it may produce more lipopolysaccharide than that of nonpathogenic bacteria. Since the biosynthesis of lipopolysaccharide consumes a large amount of carbon source, the conversion of glycerol to 3-HP or 1,3-PD is attenuated. Aside from this drawback, excess accumulation of lipopolysaccharide entangles the downstream separation and thereby increases the cost of waste disposal, particularly the quenching of pathogenicity or sterilization [29]. Considering lipopolysaccharide makes K. pneumoniae robust to survive under hostile environment, it is postulated that moderate rather than excessive lipopolysaccharide may benefits the formation of 3-HP and 1,3-PD. Park and his colleagues screened a new K. pneumoniae strain J2b, which produced less lipopolysaccharide yet more 1,3-PDO [30]. To reduce the formation of lipopolysaccharide, deletion or down-regulation of its synthetic genes is imperative.
3-HP Formation is Related to Cell Tolerance
3-HP cannot be overproduced because it is toxic to the host. Previously 3-HP was shown to have nematicidal activity. For example, some endophytic fungi secrete 3-HP to protect host plant from the invasion of nematode [31]. Also, some bacteria are sensitive to 3-HP [32, 33] because low concentration 3-HP is sufficient to inhibit bacteria. Beyond cytotoxicity, 3-HP overburdens the host strain K. pneumoniae, which in turn restricts its biosynthesis. One strategy to address this dilemma is the addition of a chemical into fermentation medium, which can specifically bind to 3-HP and therefore attenuate its toxicity. This strategy had been applied to the industrial production of 3-HPA, whereby achieving titer up to 108 g/L [34]. Another strategy is expression of a channel protein that timely pumps 3-HP out of cell once it is generated. The unknown protein may be discovered via computer stimulation. Although it seems difficult to uncover or engineer such a channel protein, this idea is a fascinating solution to this problem.
Attenuating the Metabolic Burden Caused by Plasmid
In microbial metabolic engineering, plasmid vectors are widely used for overexpression of the enzymes closely related to desired metabolite. Previous 3-HP production was achieved mainly by plasmid-dependent overexpression of glycerol dehydratase and aldehyde dehydrogenase in K. pneumoniae or E. coli [28, 35]. However, plasmid-dependent metabolic engineering shows the following drawbacks. First, plasmid replication overburdens the host K. pneumoniae, which retards cell growth and blocks 3-HP biosynthesis. Second, the addition of antibiotics and IPTG into fermentation medium increases the production cost. Third, the engineered strains show genetic instability mainly due to the plasmid loss. In fact, it is difficult to simultaneously express multiple enzymes in a single vector. Moreover, co-transformation of two or more vectors into host cell leads to metabolic imbalance which in turn hampers 3-HP production. Although 3-HP concentration was partially enhanced by plasmid-dependent method, the engineered strains are often fragile and unstable when they are fermented in a larger bioreactor, and, moreover, the productivity cannot maintain as high as that in 5 L bioreactor. Considering most industrialized products such as ethanol and 2,3-butanediol are achieved by using plasmid-free strategy [36, 37], plasmid-independent metabolic engineering is suggested. One such strategy is incorporation of 3-HP synthesis module into chromosome. Not only alleviating the burden to host cell, genome engineering also endows strain with genetic stability. Along this line, the engineered K. pneumoniae strain may divert ample carbon flux towards 3-HP.
Exploiting Synthetic Biology Approaches to 3-HP Production
Since glycerol dissimilation in K. pneumoniae involves parallel oxidation and reduction pathways, it is unlikely to preferentially intensify any branched pathway. Given the topological rigidity of dha operon, novel strategies are required to remodel 3-HP pathways. Recent years have been a watershed for microbial breeding due to the emergence of synthetic biology. The repertoire of synthetic biology is beyond imagination. For instance, a series of genome editing approaches have tapped into horizon, which are powerful in sculpting genome.
Rewiring 3-HP Pathway Via Genome Editing
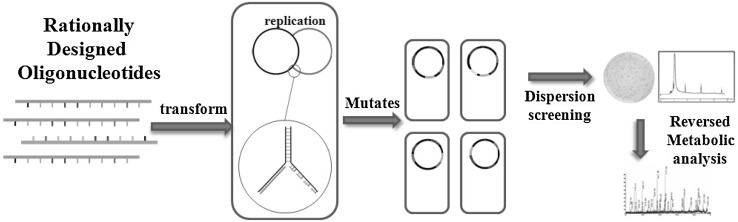
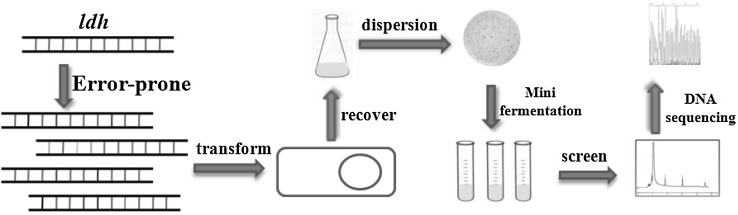
Early endeavors have revealed the involvement of multiple enzymes in 3-HP biosynthesis, indicating that 3-HP production depends on quantitative trait loci (QTL). In fact, overexpression or disruption of one or two specific genes has proven inefficient to significantly accumulate 3-HP. Hence, large-scale genome editing seems imperative. Recent years have seen great strides in the area of genome editing. Among the approaches, a creative technique named multiplex automated genomic engineering (MAGE) is eye-catching [38]. In light of Red recombination mechanism, artificially synthetic DNA fragments specific to target genes are continuously transferred into bacteria (see Fig. 2). Subsequently, high-yield strains are screened and analyzed for the mutation sites that affect 3-HP yield. The advantage of this method is two-fold. First, the chromosome is globally modified and the resulting clones are subjected to high throughput screening. Second, the strains are genetically stable because the genes are modified at the DNA level. For genome editing of K. pneumoniae via MAGE, the candidate genes may include those governing the formation of lipopolysaccharide, porin, lactic acid, and acetic acid. Also, the regulatory genes closely related to 3-HP synthesis are incorporated. By means of MAGE, the above mentioned genes could be deleted or mutated, and the biosynthesis pathways of byproducts could be determined. Apart from modification of multiple genes, one or two specific genes also deserve to be overexpressed or mutated. Briefly, the primers containing mutated gene sequence could be artificially designed, chemically synthesized, and electro-transformed into the host. The resulting positive clones display phenotype of the target genes (see Fig. 3). Aside from MAGE, other novel genome editing technologies also come to light, including CAGE [39], TALEN [40] and CRISPR-Cas9 System [41]. With the aid of these techniques, the K. pneumoniae genome can be thoroughly reshaped and 3-HP production may be significantly enhanced.
Fig. 2.
Protocols for enhancing 3-HP production via MAGE. Different oligonucleotides are chemically synthesized and transformed into K. pneumoniae, leading to insertion, mutation or deletion in 3-HP synthetic pathways. Metabolic flux is preferentially diverted into 3-HP pathway and high yield strains are acquired. DNA sequencing of the mutated nucleotides may reveal the limitations on 3-HP production, which facilitates further modification of the K. pneumoniae genome
Fig. 3.
Chromosome engineering of K. pneumoniae using double-strand DNA fragments. Primer design is based on reported sequence. Error-prone PCR is repeatedly performed. The mutated DNA fragments are transformed into host cell and subjected to homologous recombination. Positive clones are fermented for screening high-yielding strain. This method is free from selection marker and thus suitable for strain improvement
Switching Metabolic States Via On/Off Logic Gate
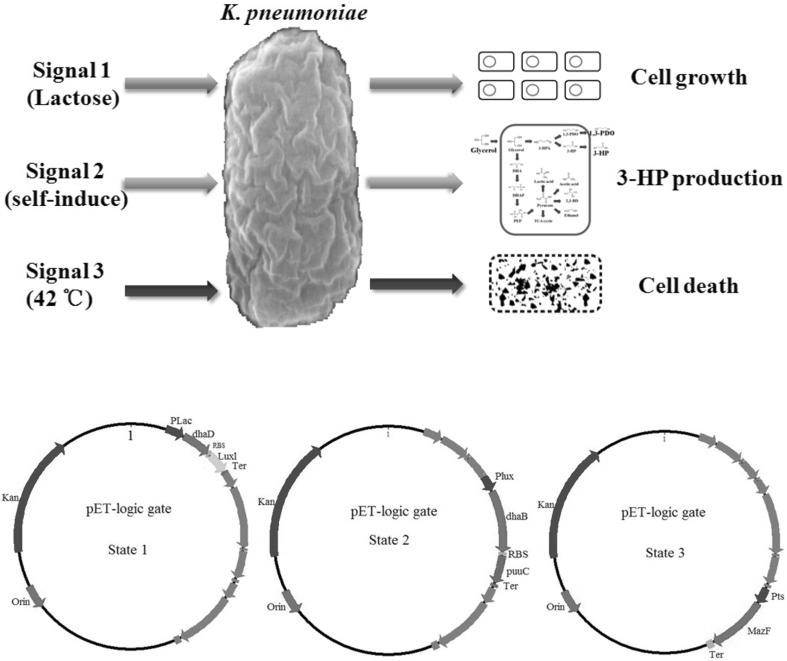
In K. pneumoniae, 3-HP production is positively correlated with biomass accumulation during log-phase. Interestingly, 3-HP in turn retards cell growth. To resolve this dilemma, two-phase fermentation strategy came into being. That is, biomass accumulation and 3-HP biosynthesis were implemented separately. Previously, biomass accumulation was achieved by using glucose as carbon source, and 3-HP biosynthesis was initiated by addition of glycerol into medium. To date, the burgeoning synthetic biology offers novel approach to two-phase fermentation. Based on bi-stability in bacteria [42, 43], an “on/off” logic gate could be engineered in K. pneumoniae, which executes the switch between two metabolic states: cell growth and 3-HP formation (see Fig. 4).
Fig. 4.
The entire 3-HP production is divided into three phases by logic gate. First, cell growth is facilitated by overexpression of dhaD to divert carbon flux into oxidative pathway. Meanwhile, Lux I is expressed to accumulate AHL, which is the inducer of next step. Second, when AHL concentration reaches a set threshold, dhaB and puuC are expressed for biosynthesis of 3-HP. This step is controlled by promoter Plux. Third, the gene MazF from E. coli is expressed for cell suicide and simplifying downstream separation process. This Pts-driven process is silenced at 37 °C but reactivated at 42 °C
Although glucose is the most efficient carbon source to bacterial growth, it impedes the consumption of other carbohydrates, which is so-called Crabtree effect [44]. Fortunately, K. pneumoniae grows actively using glycerol as the sole carbon. Hence, for glycerol-based 3-HP biosynthesis in K. pneumoniae, biomass accumulation is accomplished by the addition of glycerol into medium at the initial stage of fermentation. Once cell concentration reaches a set threshold, 3-HP biosynthesis is triggered by switching to another state (see Fig. 5). At this state, glycerol oxidation pathway is simultaneously running to coordinate the reduction and oxidation pathways. This proof-of-concept “on/off” strategy can alleviate feedback inhibition and attenuate 3-HP toxicity. The entire fermentation process is exactly controlled and the balance between oxidation and reduction is maintained.
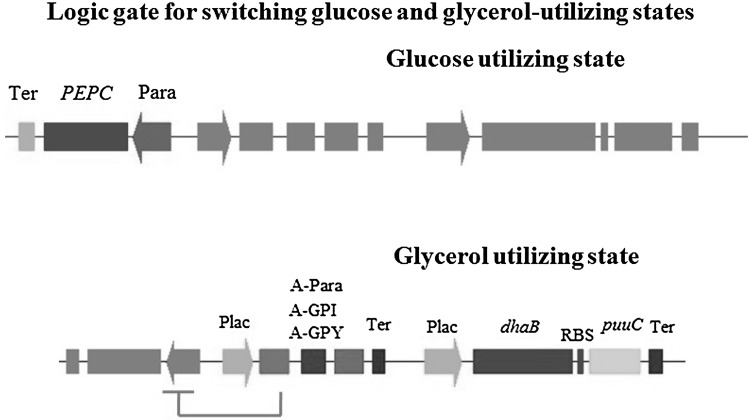
Fig. 5.
Logic gate for switching between glucose- and glycerol-utilizing states in K. pneumoniae. At the initial stage, cells grow using glucose instead of glycerol as carbon source, meanwhile, phosphoenolpyruvate carboxylase (PEPC) is expressed to benefit anaerobic respiration and cell proliferation. When glucose is exhausted, lactose is used to induce glycerol-utilizing state. Three antisense mRNA modules (A-Para, A-GPI, and A-GPY) are employed to turn off glucose metabolism, and Plac promoter drives the expression of dhaB and puuC for production of 3-HP. Bacteria prefer to metabolize glucose when it coexists with glycerol, therefore, when glycerol-utilizing state is turn on, the glucose-utilizing state must be turn off
Enhancing Enzymatic Activities Via Surface Display
3-HP biosynthesis depends largely on the activities of GDHt and ALDH. Previous metabolic engineering of K. pneumoniae centered upon in vivo biosynthesis but ignored in vitro catalysis. Surface display can enhance enzymatic activity due to enlarged specific surface area. In principle, scaffold proteins were expressed in vivo and assembled in vitro on cell surface. For 3-HP biosynthesis, the key enzymes GDHt and ALDH as ligands are transferred to cell surface with the aid of signal peptide and then specifically bind to the scaffold protein which acts as receptor. GDHt and ALDH could even be fused to form a “superbug” that continuously catalyzes glycerol to 3-HP. Due to enlarged specific surface area and spatial proximity, glycerol conversion rate may be enhanced (see Fig. 6). Although this strategy may encounter obstacles, it represents a novel strategy to implement sequential reactions. In fact, surface display has been applied to ethanol production from renewable carbon sources and CO2 fixation [45, 46]. Surface display is an important addition to the toolbox for improving enzymatic activity, attenuating toxicity, and alleviating the feedback inhibition arising from metabolite accumulation.
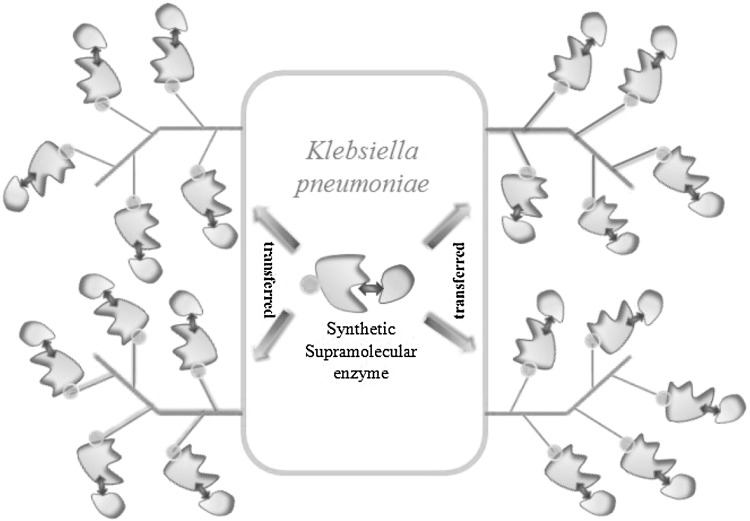
Fig. 6.
Diagram of scaffold protein for loading enzymes on the cell surface of K. pneumoniae. GDHt and ALDH are synthesized in vivo and tethered via a linker. A supramolecular enzyme catalyzing glycerol into 3-HP is generated and shipped to cell surface with the aid of signal peptide. The supermolecule is adhered to scaffold protein. The catalytic activity is enhanced due to enlarged surface area
Rational Design of 3-HP Pathways
Computational methods have been developed and applied to microbial metabolic engineering. The methods such as OptKnock [47], OptStrain [48], OptFlux [49] and Genetic Design through Local Search (GDLS) [50] are effective in the rational design of Escherichia coli and Saccharomyces cerevisiae [50, 51]. To date, K. pneumoniae genome has been sequenced and basic research has long been conducted because this bacterium is an important clinical microbe. In addition, K. pneumoniae is by far a model organism biochemically similar to E. coli. Thus, it is the time to rationally design the networks of K. pneumoniae for 3-HP biosynthesis purpose. Prior work has successfully anticipated the impacts of gene deletion on the biosynthesis of 1,3-PDO and other desired metabolites. The network BNICE [52] enables the precise prediction of 3-HP production from glucose. Despite this, 3-HP production cannot be further elevated simply by analysis of the experimental data. Computational methods in combination with experimental validations may be a wise solution to decipher 3-HP biosynthesis mechanism.
Concluding Remarks
Previously, metabolic engineering of K. pneumoniae for production of 3-HP was limited to the overexpression of enzymes from glycerol reduction pathway, or the deletion of genes from oxidation pathway. However, these approaches failed to overproduce 3-HP because dha regulon has topological rigidity. Although 3-HP cannot be highly accumulated in K. pneumoniae due to above mentioned bottlenecks, the burgeoning synthetic biology offers ways to overcome these limitations. Hence, in this review, the limitations on biological production of 3-HP were profoundly discussed and the strategies for refactoring K. pneumoniae were proposed. These strategies are multifaceted, involving cofactor regeneration, cytotoxicity attenuation, genome editing, recruitment of logic gate, alleviation of feedback inhibition, and so on. With the expending toolbox of microbial breeding, we believe that the remodeled K. pneumoniae strain will highly produce 3-HP, 1,3-PDO, or beyond.
Electronic supplementary material
Acknowledgments
This work was supported by National Basic Research Program of China (973 Program) (2012CB725200) and National Natural Science Foundation of China (Nos. 21276014, 21476011).
References
- 1.Werpy T, Petersen G. Top value added chemicals from biomass. Washington: U.S. DOE; 2004. [Google Scholar]
- 2.Southers PF, Cameron DC (2005) Production of 3-hydroxypropionic acid in recombinant organisms. United states patent US6852517
- 3.Andreeßen B, Steinbuchel A. Biosynthesis and biodegradation of 3-hydroxypropionate containing polyesters. Appl Environ Microbiol. 2010;76:4919–4925. doi: 10.1128/AEM.01015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang XL, Meng X, Xian M. Biosynthetic pathways for 3-hydroxypropionic acid production. Appl Microbiol Biotechnol. 2009;82:995–1003. doi: 10.1007/s00253-009-1898-7. [DOI] [PubMed] [Google Scholar]
- 5.Rathnasingh C, Raj SM, Lee Y, Catherinea C, Ashoka S, Park S. Production of 3-hydroxypropionic acid via malonyl-CoA pathway using recombinant Escherichia coli strains. J Biotechnol. 2012;157:633–640. doi: 10.1016/j.jbiotec.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Salles IM, Forchhammerb N, Croux C, Girbala L, Soucaille P. Evolution of a Saccharomyces cerevisiae metabolic pathway in Escherichia coli. Metab Eng. 2007;9:152–159. doi: 10.1016/j.ymben.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Forage RG, Foster MA. Glycerol fermentation in Klebsiella pneumoniae: functions of the coenzyme B12-dependent glycerol and diol dehydratases. J Bacteriol. 1982;149:413–419. doi: 10.1128/jb.149.2.413-419.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raj SM, Rathnasingh C, Jo JE, Park S. Production of 3-hydroxypropionic acid from glycerol by a novel recombinant Escherichia coli BL21 strain. Process Biochem. 2008;43:1440–1446. doi: 10.1016/j.procbio.2008.04.027. [DOI] [Google Scholar]
- 9.Kumar P, Mehariya S, Ray S, Mishra A, Kalia VC. Biodiesel industry waste: a potential source of bioenergy and biopolymers. Indian J Microbiol. 2015;55:1–7. doi: 10.1007/s12088-014-0509-1. [DOI] [Google Scholar]
- 10.Thompson JC, He BB. Characterization of crude glycerol from biodiesel production from multiple feedstocks. Appl Eng Agric. 2006;22:261–265. doi: 10.13031/2013.20272. [DOI] [Google Scholar]
- 11.Kumar V, Ashok S, Park S. Recent advances in biological production of 3-hydroxypropionic acid. Biotechnol Adv. 2013;31:945–961. doi: 10.1016/j.biotechadv.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Keller MW, Schut GJ, Lipscomb GL, Menon AL, Iwuchukwu IJ, Leuko TT, Thorgersen MP, Nixon WJ, Hawkins AS, Kelly RM, Adams MWW. Exploiting microbial hyperthermophilicity to produce an industrial chemical, using hydrogen and carbon dioxide. Proc Natl Acad Sci USA. 2013;110:5840–5845. doi: 10.1073/pnas.1222607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herter S, Farfsing J, Gad’On N, Rieder C, Eisenreich W, Bacher A, Fuchs G. Autotrophic CO2 fixation by Chloroflexus aurantiacus: study of glyoxylate formation and assimilation via the 3-hydroxypropionate cycle. J Bacteriol. 2001;183:4305–4316. doi: 10.1128/JB.183.14.4305-4316.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J, van den Heuvel J, Soucaille P, Qu YB, Zeng AP. Comparative genomic analysis of dha regulon and related genes for anaerobic glycerol metabolism in bacteria. Biotechnol Progr. 2003;19:263–272. doi: 10.1021/bp025739m. [DOI] [PubMed] [Google Scholar]
- 15.Ashok S, Raj SM, Rathnasingh C, Park S. Development of recombinant Klebsiella pneumoniae ΔdhaT strain for the co-production of 3-hydroxypropionic acid and 1,3-propanediol fromglycerol. Appl Microbiol Biotechnol. 2011;90:1253–1265. doi: 10.1007/s00253-011-3148-z. [DOI] [PubMed] [Google Scholar]
- 16.Forage RG, Lin ECC. Dha system mediating aerobic and anaerobic dissimilation of glycerol in Klebsiella pneumoniae NCIB 418. J Bacteriol. 1982;151:591–599. doi: 10.1128/jb.151.2.591-599.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Celinska E. Debottlenecking the 1,3-propanediol pathway by metabolic engineering. Biotechnol Adv. 2010;28:519–530. doi: 10.1016/j.biotechadv.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Valdehuesa KN, Liu H, Nisola GM, Chung WJ, Lee SH, Park SJ. Recent advances in the metabolic engineering of microorganisms for the production of 3-hydroxypropionic acid as C3 platform chemical. Appl Microbiol Biotechnol. 2013;97:3309–3321. doi: 10.1007/s00253-013-4802-4. [DOI] [PubMed] [Google Scholar]
- 19.Ko Y, Ashok S, Zhou S, Kumar V, Park S. Aldehyde dehydrogenase activity is important to the production of 3-hydroxypropionic acid from glycerol by recombinant Klebsiella pneumoniae. Process Biochem. 2012;47:1135–1143. doi: 10.1016/j.procbio.2012.04.007. [DOI] [Google Scholar]
- 20.Doleyres Y, Beck P, Vollenweider S, Lacroix C. Production of 3-hydroxypropionaldehyde using a two-step process with Lactobacillus reuteri. Appl Microbiol Biotechnol. 2005;68:467–474. doi: 10.1007/s00253-005-1895-4. [DOI] [PubMed] [Google Scholar]
- 21.Yin Z, Patel SJ, Wang WL, Chan WL, Rao KRR, Wang G, Ngew X, Patel V, Beer D, Knox JE, Ma NL, Ehrhardt C, Lim SP, Vasudevan SG, Keller TH. Peptide inhibitors of dengue virus NS3 protease. Part 2: SAR study of tetrapeptide aldehyde inhibitors. Bioorg Med Chem Lett. 2006;16:40–43. doi: 10.1016/j.bmcl.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z, Zhang Y, Ding XR, Chen SH, Yang J, Wang XJ, Jia GL, Chen HS, Bo XC, Wang SQ. Protocatechuic aldehyde inhibits hepatitis B virus replication both in vitro and in vivo. Antiviral Res. 2007;74:59–64. doi: 10.1016/j.antiviral.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Kirch HH, Bartels D, Wei YL, Schnable PS, Wood AJ. The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 2004;9:371–377. doi: 10.1016/j.tplants.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Su MY, Ge XZ, Tian PF. Enhanced aldehyde dehydrogenase activity by regenerating NAD+ in Klebsiella pneumoniae and implications for the glycerol dissimilation pathways. Biotechnol Lett. 2013;35:1609–1615. doi: 10.1007/s10529-013-1243-1. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez LB. Aldehyde dehydrogenase (CoA-Acetylating) and the mechanism of ethanol formation in the amitochondriate protist, Giardia lamblia. Arch Biochem Biophys. 1998;354:57–64. doi: 10.1006/abbi.1998.0664. [DOI] [PubMed] [Google Scholar]
- 26.Laval JM, Bourdillon C, Moiroux J. Enzymatic electrocatalysis: electrochemical regeneration of NAD+ with immobilized lactate dehydrogenase modified electrodes. J Am Chem Soc. 1984;106:4701–4706. doi: 10.1021/ja00329a011. [DOI] [Google Scholar]
- 27.Lee LG, Whitesides GM. Enzyme-catalyzed organic synthesis: a comparison of strategies for in situ regeneration of NAD from NADH. J Am Chem Soc. 1985;107:6999–7008. doi: 10.1021/ja00310a043. [DOI] [Google Scholar]
- 28.Huang Y, Li Z, Shimizu K, Ye Q. Co-production of 3-hydroxypropionic acid and 1,3-propanediol by Klebsiella pneumoniae expressing aldH under microaerobic conditions. Bioresour Technol. 2013;128:505–512. doi: 10.1016/j.biortech.2012.10.143. [DOI] [PubMed] [Google Scholar]
- 29.Cortes G, Borrell N, de Astorza B, Gomez C, Sauleda J, Albert S. Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect Immun. 2002;70:2583–2590. doi: 10.1128/IAI.70.5.2583-2590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arasu MV, Kumar V, Ashok S, Song H, Rathnasingh C, Lee HJ, Seung D, Park S. Isolation and characterization of the new Klebsiella pneumoniae J2B strain showing improved growth characteristics with reduced lipopolysaccharide formation. Biotechnol Bioprocess Eng. 2011;16:1134–1143. doi: 10.1007/s12257-011-0513-9. [DOI] [Google Scholar]
- 31.Schwarz M, Kopcke B, Weber RWS, Sterner O, Anke H. 3-Hydroxypropionic acid as a nematicidal principle in endophytic fungi. Phytochemistry. 2004;65:2239–2245. doi: 10.1016/j.phytochem.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 32.Warnecke TE, Lynch MD, Karimpour-Fard A, Lipscomb ML, Handke P, Mills T, Ramey CJ, Hoang T, Gill RT. Rapid dissection of a complex phenotype through genomic-scale mapping of fitness altering genes. Metab Eng. 2010;12:241–250. doi: 10.1016/j.ymben.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Warnecke TE, Lynch MD, Lipscomb ML, Gill RT. Identification of a 21 amino acid peptide conferring 3-hydroxypropionic acid stress-tolerance to Escherichia coli. Biotechnol Bioeng. 2012;109:1347–1352. doi: 10.1002/bit.24398. [DOI] [PubMed] [Google Scholar]
- 34.Krauter H, Willke T, Vorlop KD. Production of high amounts of 3-hydroxypropionaldehyde from glycerol by Lactobacillus reuteri with strongly increased biocatalyst lifetime and productivity. N Biotechnol. 2012;29:211–217. doi: 10.1016/j.nbt.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 35.Kumar V, Sankaranarayanan M, Jae KE, Durgapal M, Ashok S, Ko Y, Sarkar R, Park S. Co-production of 3-hydroxypropionic acid and 1,3-propanediol from glycerol using resting cells of recombinant Klebsiella pneumoniae J2B strain overexpressing aldehyde dehydrogenase. Appl Microbial Biotechnol. 2012;96:373–383. doi: 10.1007/s00253-012-4187-9. [DOI] [PubMed] [Google Scholar]
- 36.Jung MY, Ng CY, Song H, Lee J, Oh MK. Deletion of lactate dehydrogenase in Enterobacter aerogenes to enhance 2,3-butanediol production. Appl Microbiol Biotechnol. 2012;95:461–469. doi: 10.1007/s00253-012-3883-9. [DOI] [PubMed] [Google Scholar]
- 37.Mussatto SI, Dragone G, Guimaraes PMR, Silva JPA, Carneiro LM, Roberto IC, Vicente A, Domingues L, Teixeira JA. Technological trends, global market, and challenges of bio-ethanol production. Biotechnol Adv. 2010;28:817–830. doi: 10.1016/j.biotechadv.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, Bang D, Emig CJ, Hwang SJ, Jewett MC, Jacobson JM, Church GM. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaj T, Gersbach CA, Barbas CF., III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 43.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Bio. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodrussamee N, Lertwattanasakul N, Hirata K, Limtong S, Kosaka T. Growth and ethanol fermentation ability on hexose and pentose sugars and glucose effect under various conditions in thermotolerant yeast Kluyveromyces marxianus. Appl Microbiol Biotechnol. 2011;90:1573–1586. doi: 10.1007/s00253-011-3218-2. [DOI] [PubMed] [Google Scholar]
- 45.Fan LH, Liu N, Yu MR, Yang ST, Chen HL. Cell surface display of carbonic anhydrase on Escherichia coli using ice nucleation protein for CO2 sequestration. Biotechnol Bioeng. 2011;108:2853–2864. doi: 10.1002/bit.23251. [DOI] [PubMed] [Google Scholar]
- 46.Fan LH, Zhang ZJ, Yu XY, Xue YX, Tan TW. Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proc Natl Acad Sci USA. 2012;109:13260–13265. doi: 10.1073/pnas.1209856109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burgard AP, Pharkya P, Maranas CD. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng. 2003;84:647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- 48.Pharkya P, Burgard AP, Maranas CD. OptStrain: a computational framework for redesign of microbial production systems. Genome Res. 2004;14:2367–2376. doi: 10.1101/gr.2872004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rocha I, Maia P, Evangelista P, Vilaca P, Soares S, Pinto JP, Nielsen J, Patil KR, Ferreira EC, Rocha M. OptFlux: an open-source software platform for in silico metabolic engineering. BMC Syst Bio l. 2010;4:45. doi: 10.1186/1752-0509-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lun DS, Rockwell G, Guido NJ, Baym M, Kelner JA, Berger B, Galagan JE, Church GM. Large-scale identification of genetic design strategies using local search. Mol Syst Biol. 2009;5:296. doi: 10.1038/msb.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kennedy CJ, Boyle PM, Waks Z, Silver PA. Systems-level engineering of nonfermentative metabolism in yeast. Genetics. 2009;183:385–397. doi: 10.1534/genetics.109.105254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henry CS, Broadbelt LJ, Hatzimanikatis V. Discovery and analysis of novel metabolic pathways for the biosynthesis of industrial chemicals: 3-hydroxypropanoate. Biotechnol Bioeng. 2010;106:462–473. doi: 10.1002/bit.22673. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.