Abstract
The emergence and spread of extended-spectrum beta-lactamases and carbapenemases among common bacterial pathogens are threatening our ability to treat routine hospital- and community-acquired infections. With the pipeline for new antibiotics virtually empty, there is an urgent need to develop novel therapeutics. Bacteria require iron to establish infection, and specialized pathogen-associated iron acquisition systems like yersiniabactin, common among pathogenic species in the family Enterobacteriaceae, including multidrug-resistant Klebsiella pneumoniae and pathogenic Escherichia coli, represent potentially novel therapeutic targets. Although the yersiniabactin system was recently identified as a vaccine target for uropathogenic E. coli (UPEC)-mediated urinary tract infection (UTI), its contribution to UPEC pathogenesis is unknown. Using an E. coli mutant (strain 536ΔfyuA) unable to acquire yersiniabactin during infection, we established the yersiniabactin receptor as a UPEC virulence factor during cystitis and pyelonephritis, a fitness factor during bacteremia, and a surface-accessible target of the experimental FyuA vaccine. In addition, we determined through transcriptome sequencing (RNA-seq) analyses of RNA from E. coli causing cystitis in women that iron acquisition systems, including the yersiniabactin system, are highly expressed by bacteria during natural uncomplicated UTI. Given that yersiniabactin contributes to the virulence of several pathogenic species in the family Enterobacteriaceae, including UPEC, and is frequently associated with multidrug-resistant strains, it represents a promising novel target to combat antibiotic-resistant infections.
INTRODUCTION
Widespread and increasing antibiotic resistance among bacterial pathogens that cause some of our most common health care-associated and community-acquired infections is jeopardizing our ability to prevent and treat routine infectious diseases (1). Two pathogens of particular concern are uropathogenic Escherichia coli (UPEC), which causes the majority (80%) of uncomplicated urinary tract infections (UTI) (2), and Klebsiella pneumoniae, a frequent cause of hospital-acquired pneumonia and UTI (3). Without adequate treatment, both UPEC and K. pneumoniae can breach epithelial and endothelial barriers to gain access to the bloodstream, causing life-threatening bacteremia (4). E. coli rates of resistance to fluoroquinolones and third-generation cephalosporins now exceed 50% in 5 of 6 global regions, and similar resistance rates were reported for K. pneumoniae worldwide (5). Unfortunately, the treatment of severe infections caused by these species must rely on carbapenems, the last resort to treat severe community- and hospital-acquired infections (6). Not only are these antibacterial compounds more expensive and less available in resource-constrained settings, but their extended use contributes to the spread of carbapenem-resistant Enterobacteriaceae (CRE), a serious global public health concern (7).
Increasing rates of antimicrobial resistance and limited new therapeutics in the development pipeline have created a critical need for new antibiotics with novel mechanisms of action (8). We hypothesized that targeting nutrient acquisition in pathogenic bacteria, specifically systems to acquire iron, could provide a novel mechanism to prevent or treat infection. Iron is an essential cofactor for normal cell physiology, and bacteria require a source of iron to establish infection (9). Most tissues in the body limit iron availability to microorganisms, sequestering it in storage and carrier molecules such as transferrin, lactoferrin, and ferritin, or binding it to heme in hemoglobin and hemopexin (10). During infection, additional iron sequestration occurs as epithelial cells and neutrophils secrete lipocalin-2, a competitor for bacterial iron-scavenging siderophores, and iron absorption and recycling pathways are repressed (11). Collectively, these antimicrobial mechanisms are characterized as “nutritional immunity” (12), and the ability to circumvent these barriers is a hallmark of successful pathogens.
Many pathogenic species in the family Enterobacteriaceae have multiple and often-redundant iron acquisition systems to facilitate infection (13). The genome of UPEC strain 536, for example, encodes two heme receptors (Hma, ChuA), three siderophore systems (enterobactin, salmochelin, and yersiniabactin), and receptors for two fungal siderophores (FhuA, FhuE), siderophores that UPEC does not synthesize but can import (14). Of particular interest is the yersiniabactin system, which is often pathogen associated (15) and encoded by the high-pathogenicity island, a horizontally acquired 30-kb chromosomal region common among highly pathogenic strains of Yersinia pestis, Yersinia enterocolitica, Yersinia pseudotuberculosis, K. pneumoniae, Klebsiella oxytoca, Salmonella enterica, and E. coli (16). Animal studies confirm that yersiniabactin contributes to the virulence of K. pneumoniae during respiratory infection (17) and to that of Y. pestis during bubonic and pneumonic plague (18). Recently, we identified the receptor for yersiniabactin, FyuA, as a protective vaccine target against E. coli-mediated pyelonephritis in a murine model of UTI (19). Although the yersiniabactin system is often pathogen associated, a protective vaccine target in the urinary tract, and more prevalent among UPEC isolates that cause pyelonephritis (94%) and cystitis (87%) than among commensal E. coli strains (59%), the contribution of the yersiniabactin system to E. coli pathogenesis during UTI is unknown (20).
The purpose of this study was to determine if yersiniabactin contributes to UPEC pathogenesis during UTI and whether yersiniabactin-mediated virulence in the kidney is different from that in the bladder, which would clarify the kidney-specific protection of the experimental FyuA vaccine (19). Understanding yersiniabactin-mediated pathogenesis has the potential to provide a new therapeutic target for a number of highly pathogenic bacterial species that cause some of our most common community- and hospital-acquired infections as well as to guide UTI vaccine design against a highly prevalent vaccine target.
Here we describe the use of a yersiniabactin receptor mutant (ΔfyuA mutant) to establish the yersiniabactin system as a UPEC virulence factor during cystitis and pyelonephritis, a fitness factor during bacteremia, and the surface-accessible target of the FyuA vaccine. In addition, we demonstrate through transcriptome sequencing (RNA-seq) analysis of RNA, isolated directly from E. coli in urine from women with cystitis, that iron acquisition systems, including the yersiniabactin system, are highly expressed by bacteria during natural uncomplicated UTI.
MATERIALS AND METHODS
Ethical statement.
Protocols involving human subjects were approved by the Institutional Review Board of the University of Michigan Medical School (HUM00029910). Mouse experimental procedures were conducted in accordance to protocols approved by the University Committee on Use and Care of Animals at the University of Michigan.
E. coli gene expression during human infection.
Urine samples collected from women with cystitis at the University Health Services Clinic were immediately stabilized with RNAprotect (Qiagen) to preserve bacterial RNA transcriptional profiles. Bacteria were pelleted by centrifugation and treated with proteinase K (0.06 mAU/μl), and RNA was extracted with the RNeasy minikit (Qiagen). DNA was removed by Turbo DNase (Ambion) treatment and RNA integrity assessed by the Bioanalyzer system (Agilent). Isolated E. coli strains were also cultured statically at 37°C in pooled, filter-sterilized human urine and LB to mid-log phase and processed according to the same protocol. Cystitis RNA samples were depleted of human RNA using the MicrobEnrich kit (Ambion). Sequencing libraries were generated using the Ovation Prokaryotic RNA-seq system (NuGen) and the Encore next generation sequencing library system (NuGen).
Libraries were sequenced using an Illumina HiSeq2000 at the Institute for Genome Sciences at the University of Maryland, Baltimore. Illumina reads were analyzed using an automated pipeline (21), and Bowtie (22) was used to align the reads to the sequenced reference genome of the isolate (23). Gene expression was calculated as reads per kilobase of a gene per million mapped reads (RPKM) (24). Differences in gene expression exhibited by each E. coli strain in two samples (normal urine sample and UTI urine sample compared to LB) were determined by calculating the log2-fold change (FC) of the RPKM values. A comprehensive report of this clinical study is presented by Subashchandrabose and colleagues (25). A heat map of differential gene expression was produced in R version 2.15.1 (R Development Core Team) (26) with the package “pheatmap” (27), and histograms of transcript abundance were generated with Artemis (Wellcome Trust Sanger Institute) (28).
Bacterial strains and culture conditions.
E. coli strains were cultured in lysogeny broth (LB; 10 g/liter tryptone, 5 g/liter yeast extract, 0.5 g/liter NaCl) at 37°C with aeration. E. coli 536 was isolated from the urine of a patient with UTI (14), and E. coli HM27, HM46, HM65, and HM69 were individually cultured from the urine of women with cystitis at the University of Michigan University Health Services Clinic, Ann Arbor. The isogenic mutant 536ΔfyuA::kan was generated in E. coli 536 using the λ Red recombinase system (29) as described previously (13).
Murine model of ascending UTI.
Six- to 8-week-old female CBA/J mice (Harlan Laboratories) were inoculated transurethrally with 50 μl of 1 × 108 CFU total bacteria/mouse as described previously (30), using a modification of the Hagberg protocol (31). In competition (cochallenge) experiments, mice were inoculated with 50 μl of a 1:1 mixture of strains 536ΔfyuA::kan and 536 for a total of 1 × 108 CFU of bacteria/mouse. Total CFU for each inoculum was quantified by plating dilutions onto LB agar with and without kanamycin (25 μg/ml). After 48 h, mice were euthanized and organs were removed and homogenized in 3 ml phosphate-buffered saline (PBS) (128 mM NaCl, 2.7 mM KCl, pH 7.4) using an Omni GLH homogenizer (Omni International). Homogenized organs were plated onto LB agar plates using an Autoplate 4000 spiral plater (Spiral Biotech), and total CFU/g tissue was determined (output) for each inoculating strain. For cochallenge experiments with two inoculated strains, competitive indices (C.I.) were calculated by dividing the ratio of the mutant to the wild type in the output by the ratio of the mutant to the wild type in the inoculum [(CFUΔfyuA/CFU536)output/(CFUΔfyuA/CFU536)inoculum]. Coinoculated strains that colonize to a similar level would have a C.I. of 1.
Murine model of bacteremia.
Six- to eight-week-old female CBA/J mice (Harlan Laboratories) were inoculated via tail vein injection with 100 μl of a 1:1 mixture of E. coli strains 536 and 536ΔfyuA::kan for a total of 1 × 107 CFU/mouse. After 21 h, mice were euthanized and kidneys and spleen were removed. Organs were homogenized and plated, and competitive indices were calculated as described above.
Vaccine antigen preparation.
The gene encoding the yersiniabactin receptor, fyuA, was PCR amplified from E. coli 536 genomic DNA and cloned into the expression vector pBAD-myc-HisA (Invitrogen). Recombinant FyuA expression was induced in E. coli TOP10 cells cultured in Terrific broth (12 g/liter tryptone, 24 g/liter yeast extract, 4 ml/liter glycerol, 100 ml/liter filter-sterilized 0.17 M KH2PO4 and 0.72 M K2HPO4) at 37°C with aeration to an optical density at 600 nm (OD600) of 0.8 by the addition of 1 mM l-arabinose. After 4 h, the induced cultures were harvested by centrifugation (8,000 × g, 10 min, 4°C) and resuspended in 10 mM HEPES, pH 7, and 100 U Benzonase nuclease (Sigma-Aldrich). Bacteria were lysed by passage through a French pressure cell (20,000 lb/in2), and the lysate was cleared by centrifugation. Bacterial membranes were pelleted by ultracentrifugation (112,000 × g, 30 min, 4°C) and solubilized in 5 ml of 100 mM NaH2PO4, 10 mM Tris-HCl, and 8 M urea, pH 8.0. His6-tagged FyuA was purified on nickel-nitrilotriacetic acid-agarose columns (Qiagen) under denaturing conditions according to the manufacturer's instructions. Eluted FyuA-His6 was renatured by four successive dialysis steps at 4°C into a final solution of 0.05% Zwittergent in PBS, pH 7.4, and quantified by the bicinchoninic acid (BCA) protein assay (Pierce).
Vaccination.
Recombinant FyuA was chemically cross-linked to cholera toxin (CT; Sigma-Aldrich) at a ratio of 10:1 using N-succinimidyl 3-(2-pyridyldithio)propionate (Pierce) according to the manufacturer's recommendations. Cross-linked FyuA-CT was administered to six- to eight-week-old female CBA/J mice (Harlan Laboratories) intranasally at 20 μl/mouse (10 μl/nare). A primary dose of 100 μg FyuA cross-linked to 10 μg CT was administered on day 0, and two booster doses of 25 μg FyuA cross-linked to 2.5 μg CT were administered on days 7 and 14. On day 21, mice were challenged with E. coli as described above.
MAb generation.
A female BALB/c mouse aged 6 to 8 weeks was immunized intraperitoneally with 20 μg of recombinant FyuA emulsified in complete Freund's adjuvant (Sigma-Aldrich) on day 0, followed by booster injections of 20 μg of FyuA with incomplete Freund's adjuvant (Sigma-Aldrich) on days 14, 42, and 56. The induction of FyuA-specific antibodies in serum was monitored by enzyme-linked immunosorbent assay (ELISA), and on day 60 the mouse was euthanized and the spleen removed. Harvested splenocytes were fused to the murine cell line P3X63-Ag8.653 (32) using polyethylene glycol and conventional somatic cell hybridization techniques (33, 34), and hybridoma clones were screened for antibody production by ELISA, Western blotting, and flow cytometry. The monoclonal antibody (MAb) selected for the highest affinity for FyuA (5E7.22) was isotyped as IgG2bκ using a Pierce Rapid ELISA Mouse mAB Isotyping kit (ThermoFisher).
Flow cytometric analysis of yersiniabactin receptor surface accessibility.
Iron receptor expression was induced in freshly diluted bacterial cultures by adding 200 μM 2,2′-dipyridyl (Sigma-Aldrich) and incubating the mixture at 37°C with aeration for 6 h. Cultures were pelleted, washed with PBS, and incubated in supernatant from hybridoma clone 5E7.22 for 30 min at room temperature. FyuA-specific antibody binding was quantified by staining with secondary antibody fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (H+L) (Jackson ImmunoResearch) and analyzed by FACSCanto (Becton Dickinson). Histograms were produced with FlowJo software (Tree Star Inc.).
Statistical analysis.
Significance was assessed using a two-tailed Mann-Whitney test or a Wilcoxon signed-rank test with a theoretical median of zero, when appropriate. All P values are two tailed at a 95% confidence interval. Analyses were performed using GraphPad Prism, version 6.0d (GraphPad Software, San Diego, CA).
RESULTS
The yersiniabactin system is highly expressed during uncomplicated UTI.
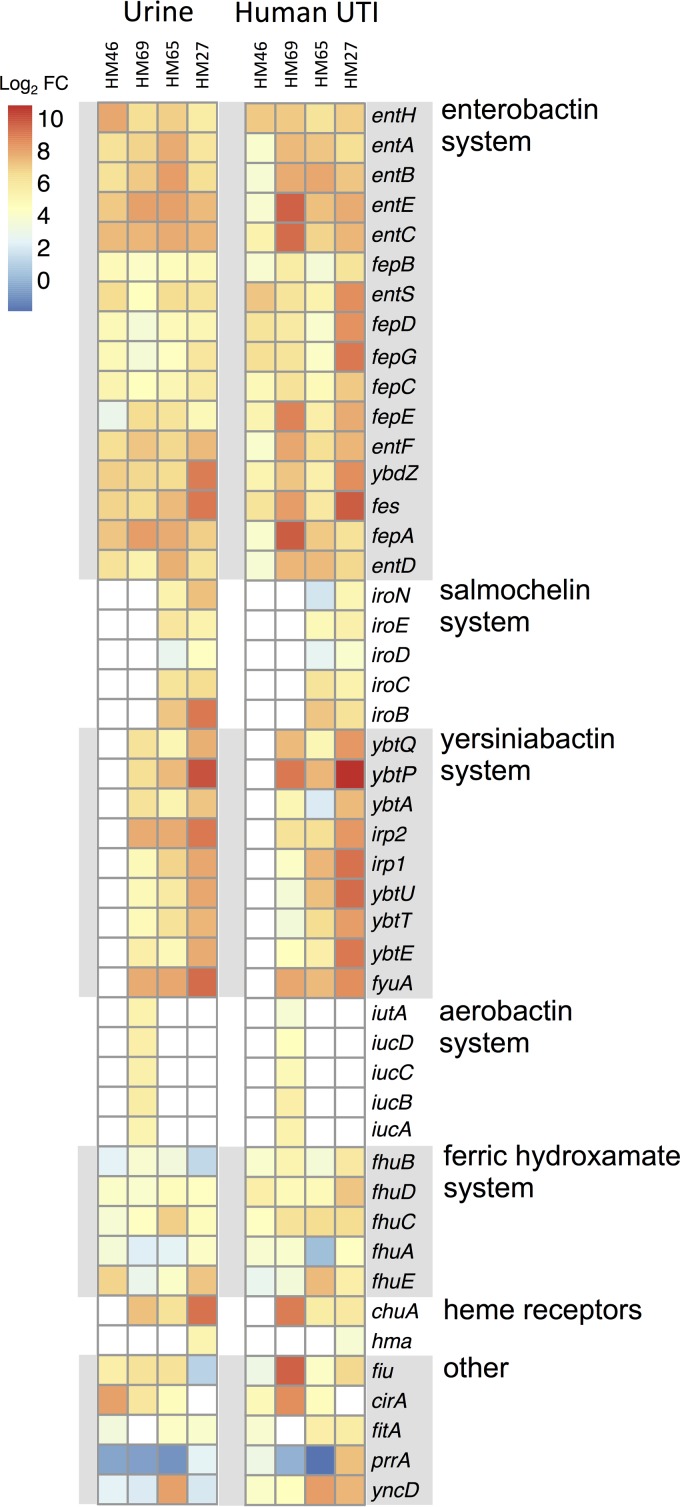
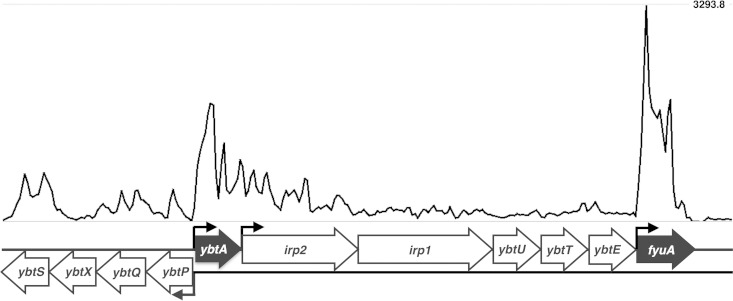
To determine if bacterial iron acquisition systems are expressed during natural, uncomplicated UTI, urine was collected and immediately stabilized with RNAprotect from women seeking treatment for cystitis at the University of Michigan University Health Services Clinic. Urine samples contained both planktonic bacteria infecting the bladder and bacteria adherent to or within exfoliated uroepithelial cells. Of the 86 urine samples collected, 42 were positive for bacteria by culture, and of those, 38 (90%) contained E. coli at ≥105 CFU/ml. An additional four species were isolated: Citrobacter freundii, Citrobacter koseri, Enterobacter aerogenes, and Proteus mirabilis. Bacterial RNA transcript levels from five E. coli samples were quantified by RNA-seq analysis, and four mapped sufficiently to their respective genomes for quantification (23). To provide comparison, RNA-seq analyses were also performed for RNA isolated from the same four E. coli strains cultured in LB (an iron-replete medium) and filter-sterilized human urine, which is naturally iron limited. All four UTI isolates from natural bladder infection had measurable RNA transcript levels for bacterial genes involved in iron acquisition, mirroring levels observed during culture in human urine, in contrast to levels in LB (Fig. 1). Of the three E. coli siderophore systems that are pathogen associated (salmochelin, aerobactin, and yersiniabactin), the yersiniabactin system was the most prevalent, found in three of the four isolates (Fig. 1). Our observations support previous molecular epidemiological data on UPEC virulence factors, which found that 87% of cystitis E. coli isolate genomes carried fyuA, compared to 74% and 35% for iroN (salmochelin) and iutA (aerobactin), respectively (20). Transcript levels for the gene fyuA were among the most abundant, as all three yersiniabactin-encoding isolates had fyuA in the top 15% of genes expressed during cystitis, which was not the case for growth in LB (HM27, 11.1%; HM69,10.4%; HM65,14.4%). Transcriptional start sites in the yersiniabactin operon could also be predicted by a histogram of transcript abundance, suggesting strong induction of the promoter upstream of ybtA, which encodes an AraC-like transcriptional activator of fyuA (35), and the promoter directly upstream of fyuA (Fig. 2).
FIG 1.
UPEC iron acquisition gene expression by E. coli strains during cystitis in women and culture in urine. Fresh urine samples from four women with uncomplicated E. coli UTI were stabilized immediately and processed for bacterial RNA (HM46, HM69, HM65, HM27). RNA transcript levels were quantified by RNA-seq and compared to transcript levels in the same strains during culture in normal human urine and LB. Differential transcript abundance between UTI urine samples or normal urine samples compared to LB for each gene is presented as the log2-fold change (FC) and visualized on a heat map. Genes with more abundant RNA transcripts during cystitis or cultured growth in urine than in growth in LB are in darker red. Genes not present in the genome of the isolate are in white.
FIG 2.
RNA-seq read coverage of the yersiniabactin operon in E. coli cultured in human urine. Histogram displaying sequence coverage per nucleotide for reads mapped to the yersiniabactin operon of the E. coli HM27 genome from RNA-seq data of E. coli HM27 cultured in urine. Open reading frames are illustrated below the histogram, with known promoter regions identified. The genes ybtA and fyuA, which encode an AraC-like transcriptional activator of fyuA and the yersiniabactin receptor FyuA, respectively, are highlighted. Genes irp2, irp1, ybtU, ybtT, ybtE, and ybtS encode proteins involved in yersiniabactin synthesis, ybtP and ybtQ encode proteins that facilitate yersiniabactin transport across the inner membrane, and ybtX encodes a protein that contributes to yersiniabactin-mediated zinc acquisition (52, 53).
Blocking yersiniabactin import attenuates UPEC during UTI.
The yersiniabactin receptor FyuA is an effective vaccine target to prevent pyelonephritis but not cystitis (19). Given that gene expression studies from experimental infections in mice (36) and now natural uncomplicated infections in women indicate that iron acquisition systems are highly expressed by bacteria infecting the bladder, the lack of bladder protection by an FyuA-based vaccine is surprising. One possible mechanism for this organ-specific immunity is that the yersiniabactin system mediates infection of the kidneys but not the bladder. To test whether the yersiniabactin system contributes to E. coli pathogenesis in the urinary tract, we evaluated the virulence of an E. coli mutant unable to acquire iron by yersiniabactin during infection. Mice inoculated transurethrally with wild-type E. coli 536 had significantly higher bacterial colonization in the bladder (2.6-fold) and the kidneys (6.4-fold) than did mice inoculated with an isogenic mutant deficient in the yersiniabactin receptor (ΔfyuA mutant) (Fig. 3). No growth defect was observed for the ΔfyuA mutant during cultured growth in vitro under iron-replete and iron-limiting conditions (see Fig. S1 in the supplemental material). These data support yersiniabactin as a significant E. coli virulence factor in the bladder and kidneys and suggest that the failure of an FyuA vaccine to prevent cystitis may be due to immunological differences, such as antibody concentration, between the bladder and the kidneys rather than differences in E. coli pathogenesis.
FIG 3.
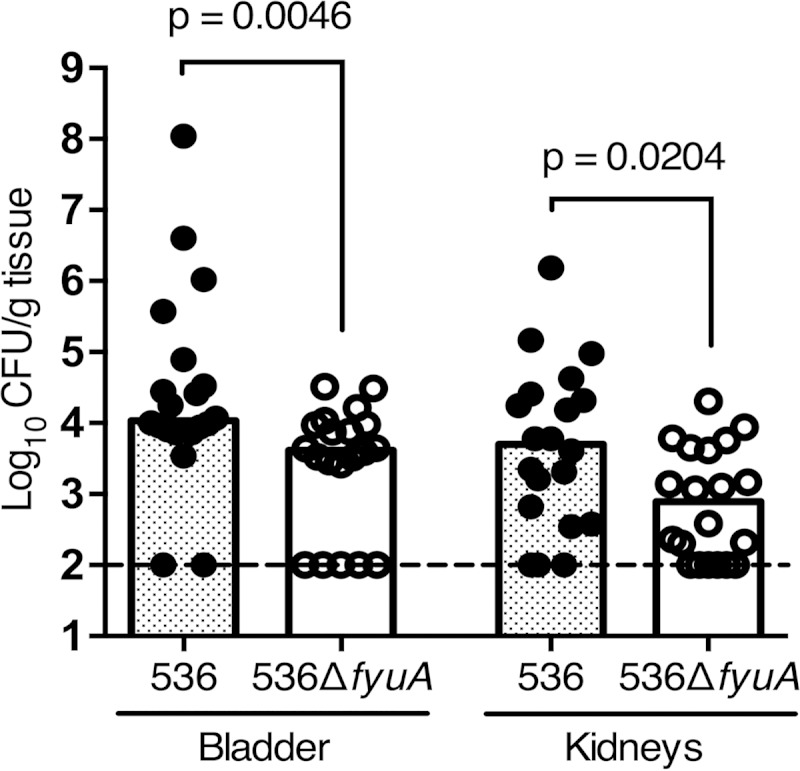
Blocking yersiniabactin import attenuates E. coli during UTI. Female CBA/J mice were transurethrally inoculated with 1 × 108 CFU of E. coli 536 or E. coli 536ΔfyuA in independent challenges, and colonization was measured 48 h postinoculation. Data from two independent experiments are presented, with the total number of animals per group (n) being 20 for strain 536 and 20 for strain 536ΔfyuA. Symbols represent CFU/g tissue from an individual mouse, and bars indicate median values. The dashed line shows the limit of detection for the assay, 100 CFU/g. Significance was determined using a two-tailed Mann-Whitney test.
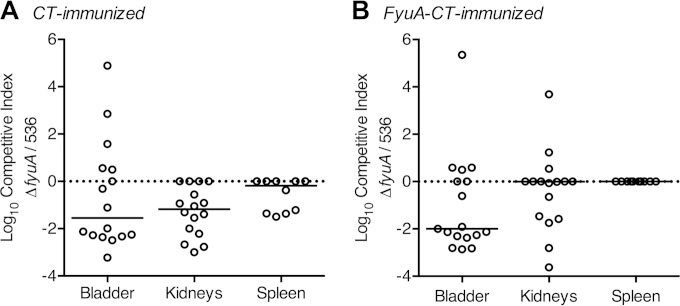
The recombinant FyuA vaccine is target specific.
The yersiniabactin iron acquisition system facilitates E. coli infection of the kidneys and bladder, but immunization with an experimental FyuA vaccine protected against only pyelonephritis, not cystitis (19). To confirm that the protective target of the FyuA vaccine is indeed the yersiniabactin receptor, mice were immunized and then cochallenged with a mixture of two isogenic E. coli strains: wild-type 536, which produces FyuA, and 536ΔfyuA::kan, which does not. E. coli isolates expressing FyuA have a substantial fitness advantage in the urinary tract (Fig. 3), but in the presence of an FyuA-specific immune response, we hypothesized that the advantage would be reduced or lost. Female CBA/J mice, intranasally immunized with FyuA-CT, were transurethrally coinoculated with a 1:1 mixture of 536 and 536ΔfyuA::kan. Forty-eight hours after inoculation, mice were euthanized and organs were removed to quantify the level of bacterial infection from each strain. As expected, wild-type E. coli outcompeted the ΔfyuA mutant when infecting the bladder and kidneys of control mice immunized with only adjuvant (Fig. 4A), reinforcing the observation that FyuA acts as a UPEC virulence factor during UTI (Fig. 3). Furthermore, in the five mice that had their infection reach the spleen, wild-type E. coli was found exclusively, suggesting a role for yersiniabactin during systemic infection (Fig. 4A). However, when mice were immunized with FyuA-CT prior to experimental infection, the fitness advantage of wild-type E. coli over the ΔfyuA mutant was lost exclusively in the kidneys (Fig. 4B). Kidney-specific attenuation of E. coli 536 in FyuA-immunized mice is consistent with the kidney-specific protection of the FyuA vaccine (19) and confirms the yersiniabactin receptor, FyuA, as the protective target of the FyuA vaccine.
FIG 4.
The FyuA vaccine is specific for the yersiniabactin receptor. Female CBA/J mice immunized with CT (A) or FyuA-CT (B) were transurethrally coinoculated with a mixture of E. coli strains 536 and 536ΔfyuA::kan. Colonization was measured 48 h postinfection by plating for CFU/g tissue, and competitive indices (C.I.) were calculated by dividing the fraction of the ΔfyuA mutant in the output (CFU/g tissue) by the fraction of the ΔfyuA mutant in the input (CFU/ml inoculum). Solid lines represent the median value for each group, and the dotted line indicates a theoretical median of zero. Symbols represent the C.I. of an organ from a single mouse. Data were validated by two independent experiments for a total of 16 mice per group, with the exception of the spleen, which had only 10 mice per group. Significance was determined by the Wilcoxon signed-rank test with a theoretical median of zero. P values for each group, from left to right, are 0.2293, 0.0005, and 0.0625 (A) and 0.0245, 0.2402, and not applicable (all spleens from the FyuA-CT-immunized mice had bacterial counts that were below the limit of detection) (B).
Yersiniabactin contributes to UPEC pathogenesis during systemic infection.
During severe infection, bacteria infecting the kidneys can breach epithelial and endothelial barriers to gain access to the bloodstream, causing systemic and life-threatening disease (4). Since blocking yersiniabactin attenuates E. coli in the urinary tract and FyuA-immunized mice are protected from systemic infection (37), we hypothesized that the yersiniabactin system may contribute to E. coli pathogenesis during bacteremia. To test this, mice were challenged systemically by intravenous injection with a 1:1 mixture of wild-type E. coli 536 and an isogenic mutant unable to import yersiniabactin. Twenty-one hours after inoculation, the E. coli mutant, unable to acquire iron by the yersiniabactin system, was significantly outcompeted by wild-type E. coli in the spleen and kidneys (Fig. 5), indicating that iron acquisition by the yersiniabactin system contributes significantly to UPEC fitness during bacteremia.
FIG 5.
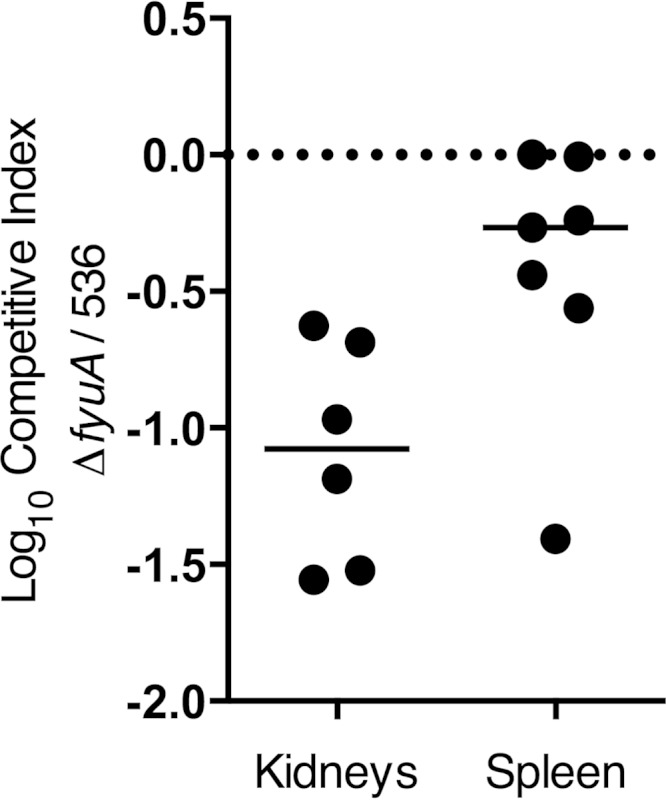
Blocking yersiniabactin import attenuates E. coli during systemic infection. Ten female CBA/J mice were intravenously coinoculated with an equal mixture of wild-type E. coli 536 and E. coli 536ΔfyuA::kan, and organ colonization was measured 21 h postinoculation. Competitive indices were calculated by dividing the fraction of the mutant in the output (CFU/g tissue) by the fraction of the mutant in the input (CFU/g inoculum). The median value for each group is represented as a solid line, and the dotted line indicates a theoretical median of zero, which would occur if both wild-type and mutant E. coli strains had equal representation in both the input and the output. Three mice succumbed to the infection before being euthanized and were excluded from the analysis. Significance was determined by the Wilcoxon signed-rank test with a theoretical median of zero, and the P value for each group was 0.0313 for the kidneys and 0.0313 for the spleen.
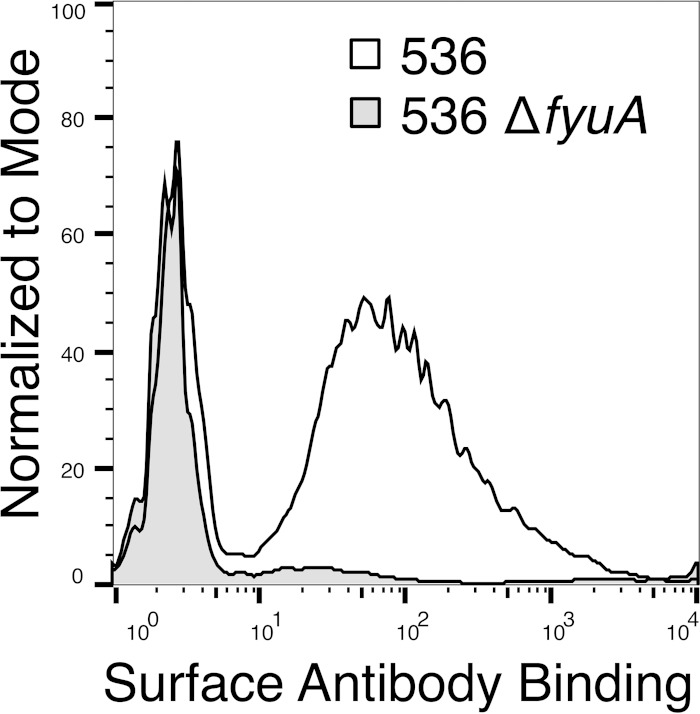
The yersiniabactin receptor is accessible on the bacterial cell surface.
Preventing iron acquisition by yersiniabactin attenuates UPEC in the urinary tract and bloodstream. Therapeutically blocking the yersiniabactin system, either with vaccine-induced antibodies or a pharmaceutical agent, would likely be simplest by targeting the siderophore receptor, FyuA, on the bacterial outer membrane. To confirm that FyuA is exposed and accessible on the cell surface, despite the presence of capsule and surface polysaccharides, we developed an FyuA-specific monoclonal antibody and quantified cell surface binding by flow cytometry. E. coli 536 cultured under iron limitation, mimicking conditions during infection, had a dramatic increase in median fluorescence intensity in comparison to an FyuA-deficient isogenic mutant (536ΔfyuA), indicating that a portion of FyuA is accessible to antibody binding on the bacterial cell surface (Fig. 6).
FIG 6.
The yersiniabactin receptor is surface exposed and accessible to antibodies. Histogram overlay of FyuA surface expression and antibody binding as assessed by flow cytometry. UPEC strains 536 and 536ΔfyuA were cultured under iron limitation and incubated with an FyuA-specific MAb (5E7.22), followed by a secondary antibody conjugated to FITC. The shaded area represents a control E. coli strain unable to express FyuA (536ΔfyuA), and the unshaded area denotes antibody binding to wild-type E. coli.
DISCUSSION
Here we demonstrate the yersiniabactin receptor to be a virulence factor for UPEC during cystitis and pyelonephritis, a fitness factor during bacteremia, and the specific, surface-accessible target of the experimental FyuA vaccine. Furthermore, we demonstrate that genes encoding bacterial systems for iron acquisition, including the yersiniabactin system, are highly expressed during natural, uncomplicated cystitis in women. Overall, our data support the yersiniabactin system as a therapeutic target to prevent or treat UPEC-mediated UTI and reinforce the potential of the yersiniabactin system as a common target for several, increasingly multidrug-resistant, Gram-negative enteric pathogens (38, 39).
Given that UPEC strains frequently encode numerous, often-redundant iron acquisition systems, the ability to attenuate infection by targeting just a single system is surprising. While individual iron uptake mechanisms uniquely contribute to bacterial pathogenesis (13, 40), it seems reasonable to assume that their similar function would allow for compensation. Of the four siderophores that UPEC isolates produce, three are so-called “stealth” siderophores (salmochelin, yersiniabactin, aerobactin) for their ability to avoid sequestration by lipocalin-2 (40, 41). Isolating UPEC strains with all three stealth siderophore systems is uncommon, possibly due to their outer membrane receptors being the frequent target of bacteriophages and bacteriocins (42). UPEC strain 536 synthesizes two of these stealth siderophores, i.e., salmochelin and yersiniabactin, but not aerobactin. It is possible that the less common aerobactin system may compensate for the loss of yersiniabactin during E. coli UTI, but this remains to be tested.
Although the protective mechanism of the kidney-specific FyuA vaccine is unknown, fyuA expression and significant FyuA-mediated virulence during cystitis suggest that the absence of vaccine protection in the bladder (19) is not due to reduced yersiniabactin expression or pathogenesis in this organ. Thus, immunological differences, such as lower antibody concentrations, between the bladder and kidneys may explain the disparity in vaccine efficacy between tissues. Indeed, despite being prone to recurrent infection, the immunological networks of the bladder are distinct from those of the kidney and remain poorly defined (43). Therefore, it may be possible to extend the protection of the FyuA vaccine to the bladder by modifying the immunization route, formula, adjuvant, or timeline to improve the adaptive immune response in the bladder.
It is important to note that UTI vaccine development has been difficult, in part, due to heterogeneity of UPEC strains and because no core set of virulence factors required for UPEC to cause infection has been identified (44). A vaccine against the diverse UPEC population will likely need to be multivalent, targeting multiple, commonly expressed UPEC virulence factors. Therefore, the yersiniabactin receptor, whose gene is highly represented among the genomes of pyelonephritis (94%) and cystitis (87%) strains (20), is a strong candidate antigen for inclusion in a multivalent UTI vaccine. Whether a UTI vaccine targeting FyuA would be broadly effective in all patient subpopulations remains to be determined, as yersiniabactin gene expression in UPEC strains from atypical UTI patient cohorts may be less uniform. A recent study evaluating UPEC gene expression in elderly men and women with UTI found yersiniabactin system genes to be expressed in only 13 of 21 UPEC cystitis isolates (45). In contrast, a microarray-based study evaluating UPEC gene expression from women at a hospital urology clinic found fyuA to be highly expressed in seven of eight UPEC cystitis isolates (46). Potential variability in UPEC yersiniabactin system gene expression between UTI patient subpopulations will likely need to be taken into account during UPEC vaccine design.
Lastly, since yersiniabactin is predominantly associated with highly pathogenic strains and is a major virulence factor for several pathogenic bacteria in the family Enterobacteriaceae, targeting FyuA may provide a mechanism to specifically eliminate bacterial pathogens without harming the beneficial bacteria constituting the microbiome, which is a common side effect of conventional antibiotics (47). In addition, yersiniabactin is disproportionately associated with antibiotic-resistant strains, including >90% of strains of the E. coli sequence type 131 (ST131), an alarming pandemic multidrug-resistant (MDR) clonal group (48–51), and >60% of respiratory carbapenemase-producing K. pneumoniae strains, compared to 10% of susceptible K. pneumoniae strains (41). Novel antimicrobial agents or vaccines that target yersiniabactin iron transport or other pathogen-associated nutrient acquisition systems may provide an effective strategy to combat the rising surge of antibiotic-resistant common infections.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Elizabeth Smith from the University of Michigan Hybridoma Core for help in generating an FyuA monoclonal antibody, Basel Abuaita for training assistance with the BD FACSCanto, Robert D. Ernst and the University of Michigan University Health Services Laboratory for support with UTI patient sample collection, and members of the Mobley lab for their helpful comments, critiques, and support.
This work was supported by Public Health Service Grants from the National Institutes of Health under award numbers R01 DK094777 and R56 A1043363.
Potential conflicts of interest: H.L.T.M. has a collaboration with Novartis International AG.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02904-14.
REFERENCES
- 1.Howard SJ, Hopwood S, Davies SC. 2014. Antimicrobial resistance: a global challenge. Sci Transl Med:6(236):236ed210. doi: 10.1126/scitranslmed.3009315. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Foxman B. 2003. Molecular epidemiology of Escherichia coli mediated urinary tract infections. Front Biosci 8:e235–e244. doi: 10.2741/1007. [DOI] [PubMed] [Google Scholar]
- 3.Jones RN. 2010. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis 51(Suppl 1):S81–S87. doi: 10.1086/653053. [DOI] [PubMed] [Google Scholar]
- 4.Miajlovic H, Smith SG. 2014. Bacterial self-defence: how Escherichia coli evades serum killing. FEMS Microbiol Lett 354:1–9. doi: 10.1111/1574-6968.12419. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance, p 256 World Health Organization, Geneva, Switzerland. [Google Scholar]
- 6.Schito GC, Naber KG, Botto H, Palou J, Mazzei T, Gualco L, Marchese A. 2009. The ARESC study: an international survey on the antimicrobial resistance of pathogens involved in uncomplicated urinary tract infections. Int J Antimicrob Agents 34:407–413. doi: 10.1016/j.ijantimicag.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Moayednia R, Shokri D, Mobasherizadeh S, Baradaran A, Fatemi SM, Merrikhi A. 2014. Frequency assessment of beta-lactamase enzymes in Escherichia coli and Klebsiella isolates in patients with urinary tract infection. J Res Med Sci 19(Suppl 1):S41–S45. [PMC free article] [PubMed] [Google Scholar]
- 8.Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards J Jr, Infectious Diseases Society of America . 2008. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis 46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 9.Miethke M, Marahiel MA. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nairz M, Schroll A, Sonnweber T, Weiss G. 2010. The struggle for iron-metal at the host-pathogen interface. Cell Microbiol 12:1691–1702. doi: 10.1111/j.1462-5822.2010.01529.x. [DOI] [PubMed] [Google Scholar]
- 11.Drakesmith H, Prentice AM. 2012. Hepcidin and the iron-infection axis. Science 338:768–772. doi: 10.1126/science.1224577. [DOI] [PubMed] [Google Scholar]
- 12.Skaar EP. 2010. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog 6:e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia EC, Brumbaugh AR, Mobley HL. 2011. Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect Immun 79:1225–1235. doi: 10.1128/IAI.01222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger H, Hacker J, Juarez A, Hughes C, Goebel W. 1982. Cloning of the chromosomal determinants encoding hemolysin production and mannose-resistant hemagglutination in Escherichia coli. J Bacteriol 152:1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spurbeck RR, Dinh PC Jr, Walk ST, Stapleton AE, Hooton TM, Nolan LK, Kim KS, Johnson JR, Mobley HL. 2012. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect Immun 80:4115–4122. doi: 10.1128/IAI.00752-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schubert S, Cuenca S, Fischer D, Heesemann J. 2000. High-pathogenicity island of Yersinia pestis in enterobacteriaceae isolated from blood cultures and urine samples: prevalence and functional expression. J Infect Dis 182:1268–1271. doi: 10.1086/315831. [DOI] [PubMed] [Google Scholar]
- 17.Lawlor MS, O'Connor C, Miller VL. 2007. Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect Immun 75:1463–1472. doi: 10.1128/IAI.00372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry RD, Fetherston JD. 2011. Yersiniabactin iron uptake: mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect 13:808–817. doi: 10.1016/j.micinf.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brumbaugh AR, Smith SN, Mobley HL. 2013. Immunization with the yersiniabactin receptor, FyuA, protects against pyelonephritis in a murine model of urinary tract infection. Infect Immun 81:3309–3316. doi: 10.1128/IAI.00470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson JR, Kuskowski MA, Gajewski A, Soto S, Horcajada JP, Jimenez de Anta MT, Vila J. 2005. Extended virulence genotypes and phylogenetic background of Escherichia coli isolates from patients with cystitis, pyelonephritis, or prostatitis. J Infect Dis 191:46–50. doi: 10.1086/426450. [DOI] [PubMed] [Google Scholar]
- 21.Orvis J, Crabtree J, Galens K, Gussman A, Inman JM, Lee E, Nampally S, Riley D, Sundaram JP, Felix V, Whitty B, Mahurkar A, Wortman J, White O, Angiuoli SV. 2010. Ergatis: a web interface and scalable software system for bioinformatics workflows. Bioinformatics 26:1488–1492. doi: 10.1093/bioinformatics/btq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subashchandrabose S, Hazen TH, Rasko DA, Mobley HL. 2013. Draft genome sequences of five recent human uropathogenic Escherichia coli isolates. Pathog Dis doi: 10.1111/2049-632X.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 25.Subashchandrabose S, Hazen TH, Brumbaugh AR, Himpsl SD, Smith SN, Ernst RD, Rasko DA, Mobley HL. 2014. Host-specific induction of Escherichia coli fitness genes during human urinary tract infection. Proc Natl Acad Sci U S A 111:18327–18332. doi: 10.1073/pnas.1415959112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Development Core Team. 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 27.Kolde R. 2013. Pretty heatmaps. R package version 0.7.7. http://cran.r-project.org/web/packages/pheatmap/index.html. [Google Scholar]
- 28.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 29.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson DE, Lockatell CV, Hall-Craigs M, Mobley HL, Warren JW. 1987. Uropathogenicity in rats and mice of Providencia stuartii from long-term catheterized patients. J Urol 138:632–635. [DOI] [PubMed] [Google Scholar]
- 31.Hagberg L, Engberg I, Freter R, Lam J, Olling S, Svanborg Eden C. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect Immun 40:273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kearney JF, Radbruch A, Liesegang B, Rajewsky K. 1979. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J Immunol 123:1548–1550. [PubMed] [Google Scholar]
- 33.Gefter ML, Margulies DH, Scharff MD. 1977. A simple method for polyethylene glycol-promoted hybridization of mouse myeloma cells. Somatic Cell Genet 3:231–236. doi: 10.1007/BF01551818. [DOI] [PubMed] [Google Scholar]
- 34.Claflin JL, Hudak S, Maddalena A. 1981. Anti-phosphocholine hybridoma antibodies. I. Direct evidence for three distinct families of antibodies in the murine response. J Exp Med 153:352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fetherston JD, Bearden SW, Perry RD. 1996. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol 22:315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- 36.Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, Welch RA, Mobley HL. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect Immun 72:6373–6381. doi: 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durant L, Metais A, Soulama-Mouze C, Genevard JM, Nassif X, Escaich S. 2007. Identification of candidates for a subunit vaccine against extraintestinal pathogenic Escherichia coli. Infect Immun 75:1916–1925. doi: 10.1128/IAI.01269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukacik P, Barnard TJ, Keller PW, Chaturvedi KS, Seddiki N, Fairman JW, Noinaj N, Kirby TL, Henderson JP, Steven AC, Hinnebusch BJ, Buchanan SK. 2012. Structural engineering of a phage lysin that targets gram-negative pathogens. Proc Natl Acad Sci U S A 109:9857–9862. doi: 10.1073/pnas.1203472109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Branger CG, Sun W, Torres-Escobar A, Perry R, Roland KL, Fetherston J, Curtiss R III. 2010. Evaluation of Psn, HmuR and a modified LcrV protein delivered to mice by live attenuated Salmonella as a vaccine against bubonic and pneumonic Yersinia pestis challenge. Vaccine 29:274–282. doi: 10.1016/j.vaccine.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachman MA, Miller VL, Weiser JN. 2009. Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS Pathog 5:e1000622. doi: 10.1371/journal.ppat.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bachman MA, Oyler JE, Burns SH, Caza M, Lepine F, Dozois CM, Weiser JN. 2011. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun 79:3309–3316. doi: 10.1128/IAI.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neilands JB. 1979. The ironic function of bacteriophage receptors. Trends Biochem Sci 4:115–118. doi: 10.1016/0968-0004(79)90396-7. [DOI] [Google Scholar]
- 43.Chan CY, St John AL, Abraham SN. 2013. Mast cell interleukin-10 drives localized tolerance in chronic bladder infection. Immunity 38:349–359. doi: 10.1016/j.immuni.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brumbaugh AR, Mobley HL. 2012. Preventing urinary tract infection: progress toward an effective Escherichia coli vaccine. Expert Rev Vaccines 11:663–676. doi: 10.1586/erv.12.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bielecki P, Muthukumarasamy U, Eckweiler D, Bielecka A, Pohl S, Schanz A, Niemeyer U, Oumeraci T, von Neuhoff N, Ghigo JM, Haussler S. 2014. In vivo mRNA profiling of uropathogenic Escherichia coli from diverse phylogroups reveals common and group-specific gene expression profiles. mBio 5:e01075–14. doi: 10.1128/mBio.01075-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hagan EC, Lloyd AL, Rasko DA, Faerber GJ, Mobley HL. 2010. Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog 6:e1001187. doi: 10.1371/journal.ppat.1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keeney KM, Yurist-Doutsch S, Arrieta MC, Finlay BB. 2014. Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol 68:217–235. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- 48.Olesen B, Hansen DS, Nilsson F, Frimodt-Moller J, Leihof RF, Struve C, Scheutz F, Johnston B, Krogfelt KA, Johnson JR. 2013. Prevalence and characteristics of the epidemic multiresistant Escherichia coli ST131 clonal group among extended-spectrum beta-lactamase-producing E. coli isolates in Copenhagen, Denmark. J Clin Microbiol 51:1779–1785. doi: 10.1128/JCM.00346-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oteo J, Navarro C, Cercenado E, Delgado-Iribarren A, Wilhelmi I, Orden B, Garcia C, Miguelanez S, Perez-Vazquez M, Garcia-Cobos S, Aracil B, Bautista V, Campos J. 2006. Spread of Escherichia coli strains with high-level cefotaxime and ceftazidime resistance between the community, long-term care facilities, and hospital institutions. J Clin Microbiol 44:2359–2366. doi: 10.1128/JCM.00447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 51.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Bano J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rakin A, Schneider L, Podladchikova O. 2012. Hunger for iron: the alternative siderophore iron scavenging systems in highly virulent Yersinia. Front Cell Infect Microbiol 2:151. doi: 10.3389/fcimb.2012.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bobrov AG, Kirillina O, Fetherston JD, Miller MC, Burlison JA, Perry RD. 2014. The Yersinia pestis siderophore, yersiniabactin, and the ZnuABC system both contribute to zinc acquisition and the development of lethal septicaemic plague in mice. Mol Microbiol 93:759–775. doi: 10.1111/mmi.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.