Abstract
Rickettsia rickettsii is an obligate intracellular pathogen that is the causative agent of Rocky Mountain spotted fever. Strains of R. rickettsii differ dramatically in virulence. In a guinea pig model of infection, the severity of disease as assessed by fever response varies from the most virulent, Sheila Smith, to Iowa, which causes no fever. To identify potential determinants of virulence in R. rickettsii, the genomes of two additional strains were sequenced for comparison to known sequences (comparative genome sequencing [CGS]). R. rickettsii Morgan and R strains were compared to the avirulent R. rickettsii Iowa and virulent R. rickettsii Sheila Smith strains. The Montana strains Sheila Smith and R were found to be highly similar while the eastern strains Iowa and Morgan were most similar to each other. A major surface antigen, rickettsial outer membrane protein A (rOmpA), is severely truncated in the Iowa strain. The region of ompA containing 13 tandem repeats was sequenced, revealing only seven shared SNPs (four nonsynonymous) for R and Morgan strains compared to Sheila Smith, with an additional 17 SNPs identified in Morgan. Another major surface antigen and autotransporter, rOmpB, exhibits a defect in processing in the Iowa strain such that the beta fragment is not cleaved. Sequence analysis of ompB reveals identical sequences between Iowa and Morgan strains and between the R and Sheila Smith strains. The number of SNPs and insertions/deletions between sequences of the two Montana strains and the two eastern strains is low, thus narrowing the field of possible virulence factors.
INTRODUCTION
Rickettsia rickettsii is the tick-borne, etiologic agent of Rocky Mountain spotted fever (RMSF). Strains of R. rickettsii have been known to differ dramatically in virulence since the earliest recognition of the disease. Indeed, Rocky Mountain spotted fever in the Bitterroot Valley of western Montana had a case fatality rate of over 80% in the era before antibiotics versus a case fatality rate of less than 5% in nearby Idaho (1). It is now recognized that the highest incidence of the disease is not in the Rocky Mountain region but in the south-central United States. Comparisons of virulence of selected “eastern” versus “western” strains in animal model systems suggested a less severe disease caused by eastern strains (2–6) although there is significant variation in virulence even within relatively small geographic areas (1, 7). The molecular bases for these differences in virulence are unknown.
With limited genetic systems, it has been difficult to definitively identify virulence factors in R. rickettsii. R. rickettsii contains a small, reduced genome of approximately 1.27 Mbp with ∼1,350 predicted genes (8). We have taken a comparative genomics approach in an attempt to identify genomic distinctions between closely related R. rickettsii strains that exhibit differences in virulence. A number of genomic differences have been identified between the virulent R. rickettsii Sheila Smith strain and the avirulent R. rickettsii Iowa strain, including the absence of rickettsial outer membrane protein A (rOmpA) from the avirulent R. rickettsii Iowa strain (8). While this comparison revealed important distinctions between R. rickettsii Sheila Smith and R. rickettsii Iowa, the number of polymorphisms makes it difficult to ascertain which are responsible for the variation in virulence between the two strains. Here, we have extended these analyses to compare multiple genomes of R. rickettsii which differ in virulence to identify unique differences which may be involved in the pathogenesis of R. rickettsii.
MATERIALS AND METHODS
Rickettsiae.
R. rickettsii strains R, Sheila Smith, Iowa, Sao Paulo, Morgan, and HLP7421 were propagated in Vero cells using M199 medium and were purified by Renografin density gradient centrifugation (9) (Table 1).
TABLE 1.
R. rickettsii strain history of isolates used in this study
| Strain | Isolate location | Year | Source | Passage historya |
|---|---|---|---|---|
| Sheila Smith | Missoula, MT | 1946 | Human | 5 YS, 4 V, 1 B, 10 V |
| Morgan | Kannapolis, NC | 1975 | Human | 4 YS, 7 V, 1 B, 14 V |
| Sao Paulo | Sao Paulo, Brazil | 1933 | Amblyomma cajennense | 8 YS, 4 V, 1 B, 5 V |
| R | Bitterroot Valley, MT | 1949 | Dermacentor andersoni | 10 YS, 3 V, 1B, 12 V |
| HLP7421 | Bitterroot Valley, MT | 1961 | Haemaphysalis leporispalustris | 7 YS, 6 V, 1 B, 5 V |
| Iowa | Iowa | 1938 | Dermacentor variabilis | 271 YS, 1 GP, 9 V, 1 B, 13 V |
Number of passages in yolk sac (YS), Vero cells (V), guinea pig (GP), and BALB/c mouse (B).
Genomic DNA purification.
To isolate R. rickettsii genomic DNA, approximately 1 × 1010 purified R. rickettsii organisms were lysed by incubation in 50 mM Tris-HCl (pH 8.0), 50 mM EDTA, 1% sodium dodecyl sulfate, 10 mM dithiothreitol, and 0.1 mg/ml proteinase K for 2 h at 60°C. After 2 h, 1 volume of chloroform-isoamyl alcohol was added, and the mixture was centrifuged for 3 min at 20,000 × g. The aqueous phase was removed and subjected to another round of chloroform-isoamyl alcohol extraction. DNA was precipitated with 0.1 volume of 3 M sodium acetate (pH 5.0) plus 0.6 volume of isopropanol and resuspended in Tris-EDTA (pH 8.0). A typical yield was approximately 30 μg of DNA.
Comparative genome sequencing, alignment, and annotation.
Approximately 20 μg of genomic DNA was provided to NimbleGen for comparative genome sequencing. DNA samples from R and Morgan strains were compared to DNA from the reference strain Iowa (GenBank accession number CP000766), and DNA from the R strain was compared to that of Sheila Smith (GenBank accession number CP000848). In addition, the R and Morgan strains were sequenced using a SOLiD next-generation sequencing system (Applied Biosystems) by the Rocky Mountain Laboratories (RML) Genomics Lab. Sequence coverage was approximately 200× for the R strain and 235× for the Morgan strain. Referenced assembly was performed with Corona Lite (version 0.4r2.0) software from Applied Biosystems and ZOOM (version 1.0.5) from Bioinformatics Solutions, Inc. Sequence-verified single nucleotide polymorphisms (SNPs) and indels were included in final genomes uploaded to the NCBI pipeline for annotation.
Guinea pig inoculations.
Female Hartley strain guinea pigs (400 g) were purchased from Charles River Laboratories, Massachusetts, and housed in an animal biosafety level 3 laboratory under a protocol approved by the Rocky Mountain Laboratories Animal Care and Use Committee. R. rickettsii strains Sheila Smith, Iowa, HLP7421, R, Morgan, and Sao Paulo and an equivalent amount of formaldehyde-fixed Sheila Smith or diluent control were inoculated intradermally with 100 PFU. Temperatures were monitored rectally for 14 days after infection. Animals were sacrificed on day 30 after sera were collected via heart puncture under deep anesthesia.
Plaque cloning.
Vero cells were seeded at 3 × 105 cells/ml (3 ml/well) into Falcon six-well plates and allowed to adhere overnight. The cell monolayers were infected with serial dilutions of R. rickettsii in brain heart infusion (BHI) broth for 30 min in a humidified 34°C chamber. Each well was then overlaid with 5 ml of M199 medium containing 5% fetal bovine serum and 0.5% agarose (GenePure ME; ISC Bioexpress) (10). All strains were grown for 9 days before cells were stained with tetrazolium bromide (3 mg/ml; 0.5 ml/well).
Transposon sequencing.
The transposon insertion kit EZ-Tn5 (DHFR-1) (Epicentre) was used according to the manufacturer's instructions to sequence and arrange the rOmpA series of repeats for both R. rickettsii Morgan and R. This method generated randomly inserted primer binding sites that allowed for bilateral extended sequence reads and assembly of the repeats using DNAStar Lasergene SeqMan Pro and Geneious, version 4.7.
Western blotting.
Purified R. rickettsii strains (2 × 108 particles) were resuspended in 100 μl of Laemmli buffer. Protein from equal volumes of solubilized rickettsiae was separated by electrophoresis on a 12% sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel for 1 h at 150 V. Proteins were transferred at 100 V for 1 h to a nitrocellulose membrane, and rOmpA was detected using anti-rOmpA monoclonal antibody 13-3 (11). rOmpB proteins were detected with a rabbit anti-rOmpB 120-kDa antigen polyclonal antibody (12).
Multilocus sequence alignment.
DNA sequences were chosen from eight regions consisting of the conserved genes recA and gltA, outer membrane genes sca0, sca1, sca2, and sca5, and two major deletions contained only in the Sheila Smith and R strains, ATPase and a 10-kb deletion region. The sequences were concatenated and collected from the following R. rickettsii genomes (GenBank accession numbers are indicated in parentheses): R. rickettsii Iowa (CP000766.3), Hino (CP003309.1) Hauke (CP003318.1), Arizona (CP003307.1), Brazil (CP003305.1), Colombia (CP003306.1), Sheila Smith (CP000848.1), Hlp#2 (CP003311.1), Morgan (CP006010.1), and R (CP006009.1).
Nucleotide sequence accession numbers.
Genome assembly contig sequence and annotation files have been deposited in the GenBank under accession numbers CP006009.1 for R. rickettsii R and CP006010.1 for R. rickettsii Morgan.
RESULTS
Virulence of R. rickettsii strains in a guinea pig model system.
Groups of five guinea pigs were infected intradermally with 100 PFU of R. rickettsii strains Sheila Smith, R, Morgan, Sao Paulo, HLP7421, and Iowa, and temperatures were monitored for 14 days (Fig. 1). As previously shown (2), a typical eastern strain, Morgan, produced a reduced fever response compared to the more virulent Sheila Smith strain. The Sao Paulo strain produced a fever response almost identical to that of the Morgan strain. The R strain, which was isolated from a Rocky Mountain wood tick, Dermacentor andersoni, from western Montana, produced an even more attenuated fever curve. The HLP7421 strain of R. rickettsii produced only a very minimal fever response, as is typical for this strain (2, 13), and the Iowa strain produced no fever response. Absence of fever with R. rickettsii Iowa infection is characteristic of this strain (8, 14). Although animals infected with the Iowa strain did not develop fever, as previously shown (8), all seroconverted, indicating that replication of the rickettsiae had occurred. No seroconversion was observed in animals given the equivalent mass of formalin-killed organisms (data not shown).
FIG 1.
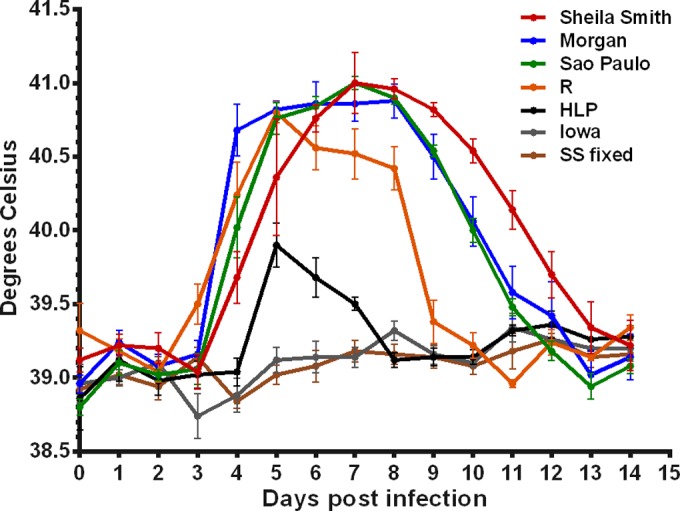
Fever curves for R. rickettsii strains. Female Hartley strain guinea pigs were inoculated intradermally with 100 PFU of R. rickettsii strains Sheila Smith, R, Morgan, Sao Paulo, HLP7421, and Iowa. Controls received an equivalent mass of formalin-killed Sheila Smith (SSfixed) in K36 diluent. Temperatures were monitored for 14 days.
Protein profiles of R. rickettsii strains.
Lysates of purified R. rickettsii strains Sheila Smith, R, Sao Paulo, Morgan, Iowa, and HLP7421 were subjected to SDS-PAGE and immunoblotting against anti-rOmpA and anti-rOmpB antisera. As previously shown, the Iowa strain is deficient in rOmpA and does not efficiently process the rOmpB autotransporter (8, 15) (Fig. 2). Protein profiles of the remaining strains were very similar.
FIG 2.
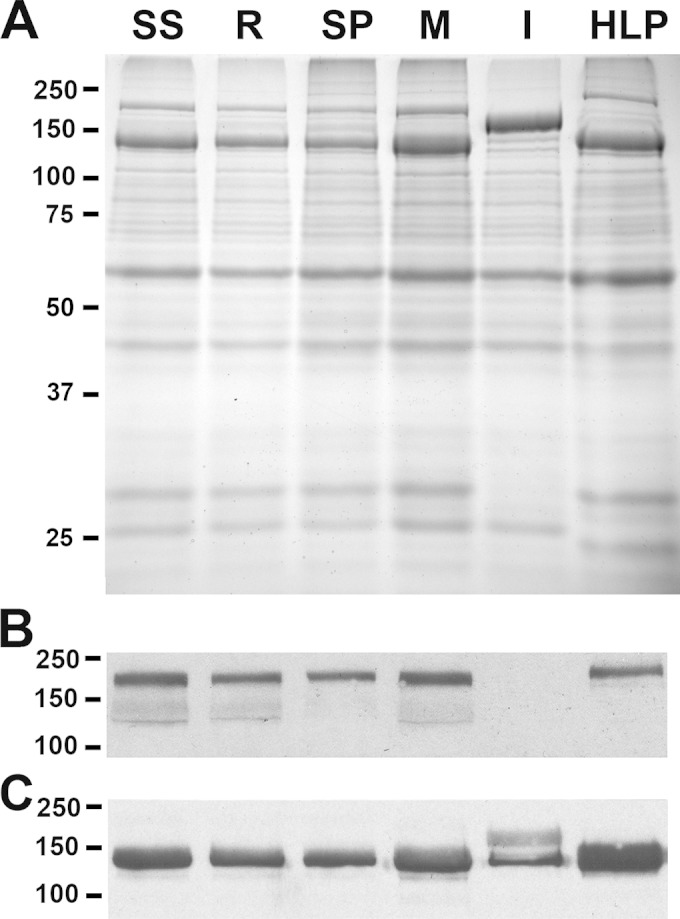
SDS-PAGE and Western blotting of major outer membrane proteins. All strains examined in the animal model were separated by SDS-PAGE. SS, Sheila Smith; SP, Sao Paulo; M, Morgan; I, Iowa; HLP, HLP7421. (A) Coomassie staining showing the presence of rOmpA in all strains except Iowa and the 120-kDa rOmpB protein in which the larger unprocessed form is abundant in the Iowa strain. (B) Immunoblot of rOmpA with 13-3 monoclonal antibody. (C) Immunoblot of the 120-kDa rOmpB protein using a rabbit polyclonal anti-R. rickettsii rOmpB antiserum. In the Iowa strain, both the processed and larger, unprocessed forms are detected.
Genome sequence of R. rickettsii R and Morgan.
Previous comparisons of a highly virulent strain of R. rickettsii, Sheila Smith, to the avirulent Iowa strain identified 143 insertions/deletions and 492 single nucleotide polymorphisms (SNPs) between the genomes (8). To refine the number of potential virulence determinants in R. rickettsii, two additional strains displaying intermediate levels of virulence, Morgan and R, were subjected to comparative genome sequencing. A complete list of verified SNPs, insertions, and deletions between these four strains of R. rickettsii are provided in Data Sets S1 and S2 in the supplemental material.
Sequence analysis of rOmpA.
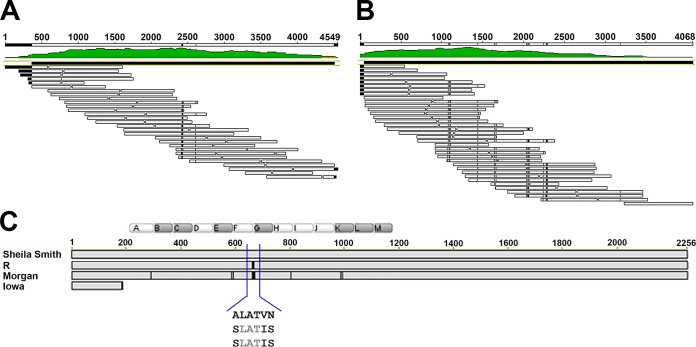
The autotransporter rOmpA is a large, surface-exposed antigenic protein of approximately 190 kDa (16). rOmpA contains a repeat region of 13 copies of near-identical repeat units of 72 to 75 amino acids (aa). The region of the gene ompA encoding the repeats encompasses approximately 3 kbp, which due to the identity of the repeat sequences, is very difficult to directly read through in sequencing. The repeat regions of ompA from R. rickettsii R and Morgan were cloned, and a transposon library was created for sequencing. Four nonsynonymous SNPs resulting in three amino acid changes (two SNPs in one codon) were identified in the R strain relative to the sequence of Sheila Smith. These same SNPs were also identified in the Morgan strain, as were nine additional nonsynonymous SNPs (Fig. 3).
FIG 3.
Sequencing of the rOmpA repeat region and alignment of rOmpA. The transposon insertion kit EZ-Tn5 (DHFR-1) (Epicentre) was used to sequence and arrange the rOmpA series of repeats. This method generated randomly inserted primer binding sites that allowed for extended sequence reads and assembly of the repeats. (A and B) Sequence coverage from E. coli clones for the R (A) and Morgan (B) strains. Transposon insertion sites from which sequencing was primed are shown as gaps (H) in the sequence. (C) Sequence alignment of R and Morgan strains to Sheila Smith reveals seven shared SNPs, resulting in three nonsynonymous amino acid changes at the beginning of the type II G repeat. Clear boxed letters represent type I repeats, and shaded boxes represent type II. The area of shared changes is expanded to show hydrophobic A-to-hydrophilic S residue substitution. Morgan also contains 17 other SNPs, 9 of which result in 8 amino acid changes (two SNPs in one codon).
Genomic distinction of virulent from avirulent R. rickettsii strains.
Although all four strains were very similar, the eastern and western strains clearly were distinguished. The two western strains, Sheila Smith and R, showed a high degree of identity while the two eastern strains (Morgan and Iowa) were most similar to each other. However, the two western strains were almost as different from the virulent Morgan strain as they were from the avirulent Iowa strain. Notably, both western strains displayed the 10-kb deletion that overlaps remnants of a predicted integral membrane protein which is intact in other rickettsial species (8). Exclusive of this large deletion, there was approximately 99% identity among the four strains of R. rickettsii. The similarities between the two western strains and the two eastern strains, each of which differed in virulence levels, simplified comparisons. Only 12 nonsynonymous SNPs (four in ompA), six synonymous SNPs, and eight SNPs in intergenic regions distinguished Sheila Smith from the R strain (Table 2). A single in-frame insertion of 588 bp into a hypothetical ankyrin repeat-containing protein of Sheila Smith also distinguished these two strains (Table 3). Similarly, only eight unique nonsynonymous SNPs specifically distinguished the Morgan and the Iowa strains (sequence differences that were seen in only the Iowa strain and none of the virulent strains are addressed below). In addition, 8 synonymous SNPs and 15 SNPs in intergenic regions uniquely differentiated the Iowa and Morgan strains (Table 4). Eight insertions/deletions, five in noncoding regions and three in coding regions, completed the distinctions between Iowa and Morgan (Table 5). Interestingly, the same 588-bp sequence in an ankyrin domain-containing protein which is present in the Sheila Smith strain sequence is present in Morgan but absent from the R and Iowa strains.
TABLE 2.
Single nucleotide polymorphisms between Sheila Smith and R strains
| SNP type and coordinate (Iowa) | Iowa locus or region | Product | Nucleotide in: |
Amino acid change in: |
|||
|---|---|---|---|---|---|---|---|
| Iowa | Sheila Smith | R | Sheila Smith | R | |||
| Nonsynonymous | |||||||
| 24960 | RrIowa_0029 | Cell surface antigen Sca1 | C | T | R1592C | ||
| 245938 | RrIowa_0287 | Chaperone protein DnaJ | C | T | D150N | ||
| 471604 | RrIowa_0564 | Outer membrane assembly protein | G | T | P818T | ||
| 485033 | RrIowa_0580 | O antigen polymerase | G | A | T139I | ||
| 1019673 | RrIowa_1296 | Guanosine polyphosphate pyrophosphohydrolase | G | A | H118Y | ||
| 1033960 | RrIowa_1312 | Proline/betaine transporter | T | C | K13R | ||
| 1106406 | RrIowa_1417 | Hypothetical protein | A | T | K30N | ||
| 1160493 | RrIowa_1466 | Glutathione-regulated potassium-efflux system protein | C | T | D6N | ||
| 1180414 | RrIowa_1493 | Outer membrane protein A | C | T | S669N | ||
| 1180416 | RrIowa_1493 | Outer membrane protein A | T | C | I668V | ||
| 1180418 | RrIowa_1493 | Outer membrane protein A | T | C | I668V | ||
| 1180430 | RrIowa_1493 | Outer membrane protein A | A | C | S664A | ||
| Synonymous | |||||||
| 429835a | RrIowa_0512 | Type I protein secretion ATP-binding protein | A | G | |||
| 814439 | RrIowa_1018 | Hypothetical protein | C | G | |||
| 814460 | RrIowa_1018 | Hypothetical protein | A | G | |||
| 863111 | RrIowa_1085 | Transcription-repair coupling factor | A | G | |||
| 1180419 | RrIowa_1493 | Outer membrane protein A | C | T | |||
| 1180422 | RrIowa_1493 | Outer membrane protein A | G | T | |||
| Intergenic | |||||||
| 75616 | Intergenic | G | T | ||||
| 75736 | Intergenic | A | G | ||||
| 254168 | Intergenic | C | T | ||||
| 547584 | Intergenic | T | C | ||||
| 552680 | Intergenic | C | T | ||||
| 942266 | Intergenic | C | T | ||||
| 1082862 | Intergenic | T | C | ||||
| 1090438 | Intergenic | T | C | ||||
NCBI Sheila Smith reference sequence alignment. This insertion was not observed by sequencing of the Sheila Smith strain in the Rocky Mountain Laboratories collection.
TABLE 3.
Insertions distinguishing the Sheila Smith strain from the R strain
| Coordinate (Iowa) | Iowa locus | Product | Length of insertion in Sheila Smith (bp) |
|---|---|---|---|
| 1152185a | Intergenic | 154 | |
| 563473a | RrIowa_0677 | Acriflavin resistance plasma membrane protein | 3 |
| 881389 | RrIowa_1113 | Hypothetical protein (ankyrin repeat) | 588b |
NCBI Sheila Smith reference sequence alignment. These insertions were not observed by sequencing of the Sheila Smith strain in the Rocky Mountain Laboratories collection.
In-frame insertion of 196 aa. This sequence is present also in the Morgan strain but absent from the Iowa and R strains.
TABLE 4.
Single nucleotide polymorphisms specifically distinguishing the Iowa and Morgan strains
| SNP type and coordinate (Iowa) | Iowa locus or regiona | Product | Nucleotide in: |
Amino acid change in Morgan | |
|---|---|---|---|---|---|
| Iowa | Morgan | ||||
| Nonsynonymous | |||||
| 41166 | RrIowa_0056 | Hypothetical protein | G | T | P44T |
| 73049 | RrIowa_0101 | Hypothetical protein | G | A | P118S |
| 462800 | RrIowa_0554 | Hypothetical protein | C | T | A218T |
| 592024 | RrIowa_0716 | Hypothetical protein/AbrB family transcriptional regulator | C | T | V16I |
| 977093 | RrIowa_1246 | DNase, TatD family | G | T | H175N |
| 1037601 | RrIowa_1322 | Hypothetical protein | T | G | Y12S |
| 1045924 | RrIowa_1330 | Malonyl-CoA-acyl-carrier protein transacylaseb | A | G | I274V |
| 1180401 | RrIowa_1493 | Outer membrane protein A | C | T | D673G |
| Synonymous | |||||
| 163376 | RrIowa_0203 | Hypothetical protein/predicted nucleoside-diphosphate-sugar epim | G | A | |
| 424451 | RrIowa_0506 | TolA | G | A | |
| 425483 | RrIowa_0509 | Hypothetical protein | C | T | |
| 431074 | RrIowa_0512 | Type I protein secretion ATP-binding protein | T | C | |
| 645866 | RrIowa_0788 | Hypothetical protein | G | A | |
| 664744 | RrIowa_0808 | Pyruvate phosphate dikinase | G | A | |
| 852969 | RrIowa_1071 | ATP-dependent DNA helicase | C | T | |
| 1178379 | RrIowa_1493 | Outer membrane protein A | G | A | |
| Intergenic | |||||
| 227151 | Intergenic | T | G | ||
| 401017 | Intergenic | A | C | ||
| 425483 | Intergenic | C | T | ||
| 564723 | Intergenic | C | A | ||
| 713118 | Intergenic | G | A | ||
| 725479 | Intergenic | A | T | ||
| 825946 | Intergenic | C | T | ||
| 840609 | Intergenic | A | T | ||
| 841977 | Intergenic | G | A | ||
| 873138 | Intergenic | C | T | ||
| 967190 | Intergenic | G | A | ||
| 1002812 | Intergenic | G | A | ||
| 1067848 | Intergenic | G | A | ||
| 1162554 | Intergenic | G | A | ||
| 1204185 | Intergenic | T | C | ||
NCBI.
CoA, coenzyme A.
TABLE 5.
Insertions and deletions distinguishing the Iowa strain from the Morgan strain
| Coordinate (Iowa) | Iowa region or locus | Product | Length of indel in Morgan (bp) | Comment |
|---|---|---|---|---|
| 58901 | Intergenic | 1 | Deletion | |
| 332908 | Intergenic | 7 | Deletion | |
| 704722 | Intergenic | 106 | Insertion | |
| 796549 | Intergenic | 2 | Deletion | |
| 898566 | Intergenic | 1 | Deletion | |
| 972349 | RrIowa_1237 | Hypothetical protein | 1 | Insertion |
| 1020010 | RrIowa_1296 | Guanosine polyphosphate | 13 | Deletion |
| 1091293 | RrIowa_1396 | Hypothetical protein | 1 | Insertion |
Particular attention was paid to genomic differences in the Iowa strain that were not observed in any of the other virulent strains. Only 10 nonsynonymous SNPs are unique to the Iowa strain as are 8 synonymous SNPs and 11 SNPs in noncoding regions (Table 6). As previously described (8), a single nucleotide deletion from the ompA gene shifts the open reading frame of Iowa and prevents rOmpA expression (Table 7). The virulent strains also contain an additional downstream insertion of 891 bp in ompA which differentiates them from the Iowa strain. An ankyrin repeat-containing protein (RrIowa_290) is truncated in the Iowa strain, but an A-to-G transition restores the open reading frame in each of the virulent strains. An 11-bp insertion which truncates a hypothetical protein (RrIowa_450) at 129 amino acids and an in-frame deletion of 3 bp in RrIowa_1432, another hypothetical protein, also distinguish Iowa from each of the virulent strains. Three additional insertions/deletions in noncoding regions are unique to the Iowa strain.
TABLE 6.
Single nucleotide polymorphisms unique to the Iowa strain
| SNP type and coordinate (Iowa) | Iowa region or locus | Product | Nucleotide in: |
Amino acid change in: |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Iowa | Morgan | Sheila Smith | R | Morgan | Sheila Smith | R | |||
| Nonsynonymous | |||||||||
| 65860 | RrIowa_0091 | Succinate dehydrogenase iron-sulfur subunit | A | G | G | G | S260G | S260G | S260G |
| 194500 | RrIowa_0228 | Hypothetical protein | A | G | G | G | K13E | K13E | K13E |
| 229483 | RrIowa_0265 | Prolyl endopeptidase | A | C | C | C | L6W | L6W | L6W |
| 250900 | RrIowa_0290 | Hypothetical protein/ankyrin | A | G | G | G | *68Wa | *68Wa | *68Wa |
| 857393 | RrIowa_1080 | Arp2/3 complex activation protein/RickA | A | G | G | G | M257T | M257T | M257T |
| 882319 | RrIowa_1113 | Hypothetical protein/ankyrin repeat | T | G | G | G | D27A | D27A | D27A |
| 1036868 | RrIowa_1321 | RNase H | T | C | C | C | K99E | K99E | K99E |
| 1090471 | RrIowa_1396 | Hypothetical protein/endonuclease subunit | A | G | G | G | I4 M | I4 M | I4 M |
| 1127496 | RrIowa_1431 | Methyltransferase | A | G | G | G | K208R | K208R | K208R |
| 1252300 | RrIowa_1582 | Type I restriction-modification system methylation subunit | A | G | G | G | F25S | F25S | F25S |
| Synonymous | |||||||||
| 140280 | RrIowa_0180 | Channel protein | A | G | G | G | |||
| 221550 | RrIowa_0258 | 16S rRNA methyltransferase RsmE | A | G | G | G | |||
| 329851 | RrIowa_0392 | Hypothetical protein | T | C | C | C | |||
| 631283 | RrIowa_0773 | Protein U | T | C | C | C | |||
| 654808 | RrIowa_0797 | Antigenic heat-stable 120-kDa protein/Sca4 | A | G | G | G | |||
| 758713 | RrIowa_0944 | Hypothetical protein | T | C | C | C | |||
| 1063307 | RrIowa_1363 | Hypothetical protein/PD-(D/E)XK nuclease family transposase | T | C | C | C | |||
| 1204862 | RrIowa_1526 | Hypothetical protein | C | T | T | T | |||
| Intergenic | |||||||||
| 246462 | Intergenic | G | T | T | T | ||||
| 246467 | Intergenic | G | A | A | A | ||||
| 250900 | Intergenic | A | G | G | G | ||||
| 261290 | Intergenic | T | G | G | G | ||||
| 375768 | Intergenic | A | C | C | C | ||||
| 469765 | Intergenic | T | C | C | C | ||||
| 816598 | Intergenic | T | G | G | G | ||||
| 1189812 | Intergenic | A | G | G | G | ||||
| 1202096 | Intergenic | A | G | G | G | ||||
| 1252416 | Intergenic | A | G | G | G | ||||
| 1264129 | Intergenic | T | C | C | C | ||||
In the Iowa strain, RrIowa_0290 is prematurely terminated at amino acid 68.
TABLE 7.
Insertions and deletions unique to the Iowa strain
| Iowa coordinate | Iowa region or locus | Product | Length of indel (bp) |
Comment | ||
|---|---|---|---|---|---|---|
| Morgan | Sheila Smith | R | ||||
| 64628 | Intergenic | 26 | 26 | 26 | Insertion | |
| 376310 | RrIowa_0450 | Hypothetical protein | 11 | 11 | 11 | Insertion; truncates a hypothetical protein at aa 129 |
| 767329 | Intergenic | 9 | 9 | 9 | Insertion | |
| 956246 | Intergenic | 1 | 1 | 1 | Deletion | |
| 1128779 | RrIowa_1432 | Putative transcriptional regulator | 3 | 3 | 3 | In-frame deletion |
| 1180402 | RrIowa_1493 | Outer membrane protein A | 891 | 891 | 891 | In-frame insertion of 297 aa |
| 1181871 | RrIowa_1494 | Outer membrane protein A | 1 | 1 | 1 | 1-bp insertion; restores open reading frame for rOmpA |
Sequence analysis of rOmpB.
R. rickettsia Iowa was described as defective in the processing of rOmpB. In all other species of Rickettsia examined, the 32-kDa autotransporter domain of rOmpB is cleaved although the 120-kDa passenger domain apparently remains associated with the B fragment since they coimmunoprecipitate. Sequence near the cleavage site was identical to that from R. rickettsii R (15). Comparison of the Iowa ompB gene to that of Sheila Smith and R identified only four nonsynonymous SNPs, and these are identical to those found in the Morgan strain (Fig. 4). The defect in processing of rOmpB in the Iowa strain thus does not appear to be due to a defect in the protein itself.
FIG 4.
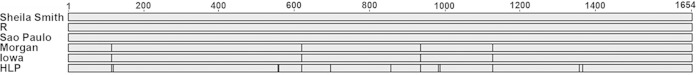
Comparison of rOmpB sequences. Alignment of each strain's sequenced rOmpB to Sheila Smith shows that the R and Sao Paulo sequences are identical to the reference sequence. Morgan and Iowa are identical to each other, with 4 amino acid substitutions compared to the sequence of Sheila Smith. HLP7421 has these same changes plus 9 more compared to the Sheila Smith sequence.
Phylogenetic analysis.
A multilocus sequence alignment confirmed the close relationship of the Sheila Smith and R strains and their divergence from the Iowa and Morgan strains (Fig. 5).
FIG 5.
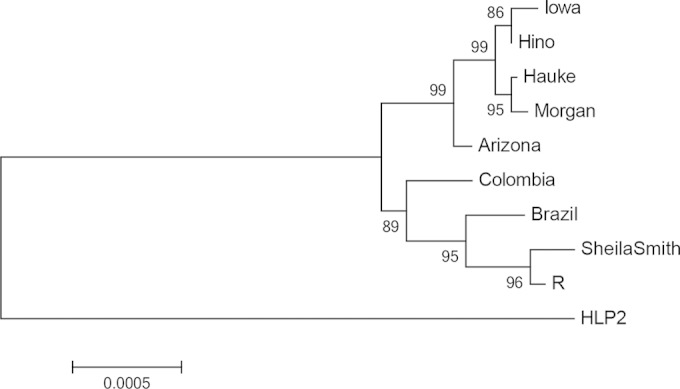
Phylogenetic analysis of closely related Rickettsia rickettsii strains. The evolutionary history was inferred by using a maximum likelihood method based on the Tamura-Nei model (45). The tree with the highest log likelihood (−58,350.3501) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The initial tree(s) for the heuristic search was obtained automatically by applying neighbor joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood approach and then selecting the topology with the superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (five categories [+G, parameter = 0.0500]). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 10-nucleotide sequences. Codon positions included were 1st, 2nd, 3rd, and noncoding. There were a total of 42,243 positions in the final data set. Evolutionary analyses were conducted in MEGA, version 5 (46).
DISCUSSION
Strains of R. rickettsii differ dramatically in their abilities to cause disease. This can be seen in differences in case fatality rates (1, 17) and is supported by animal model systems (2, 17). The genetic basis for these strain-dependent differences in virulence is unknown; however, R. rickettsii's reduced genome, high genetic homology, and heterogeneous interstrain virulence allow the use of a direct bioinformatics approach to identify potential virulence factors. A previous genomic comparison of a highly virulent strain isolated from an RMSF patient in the Bitterroot Valley and an avirulent strain isolated from a tick in Iowa showed over 99% identity. However, with 492 SNPs and 143 deletions between them (8), it was difficult to experimentally assess each for a role in virulence. Here, we have completed two additional genomes from R. rickettsii strains displaying intermediate degrees of virulence in a guinea pig model of infection. Interestingly, the two western Montana strains, Sheila Smith and R, were more similar to each other than to the two eastern strains, while the eastern strains Morgan and Iowa were very similar to each other.
Major distinctions included the absence of the major surface antigen, rOmpA, from the Iowa strain. Indeed, Iowa is alone among all strains of R. rickettsii with published genomes that contains a single nucleotide deletion 660 bp poststart of ompA, leading to protein truncation. Interestingly, rOmpA is also absent from a closely related, avirulent spotted fever group rickettsia, Rickettsia peacockii (18). rOmpA is a large autotransporter and protective immunogen (19) that has also been implicated as a possible adhesin (20). The passenger domain of rOmpA contains 13 nearly identical 75-aa repeat units (16) and has evolved under strong positive selection, consistent with antigenic variation (21, 22). The number and order of repeat units can differ between rickettsial species. Here, we observed only minor differences in sequences between strains of R. rickettsii.
The most distinct difference between the R. rickettsii strains used in this analysis is an approximately 10-kbp deletion from the Sheila Smith and R strains. The significance of this deletion is unclear as the region is retained in both Iowa and Morgan as well as in other strains of R. rickettsii, including Hino, Hauke, Arizona, Colombia, and Hlp#2. The only other sequenced strain of R. rickettsii which exhibits this deletion is the Brazil strain. The presence or absence of this region does not seem to directly influence virulence since it is present in the virulent Morgan strain. However, many of the genes in this region appear to be fragmented in R. rickettsii, including a predicted integral membrane protein which is intact in some other species of spotted fever group rickettsiae (8). The high degree of pseudogenicity and interstrain disparity in this region may be an intermediate step in genomic reduction (8). A comparative analysis of various rickettsial species demonstrated that genome decay in rickettsiae positively correlated with increased virulence (23), a phenomena observed in other bacterial pathogens (24–26). This phenomenon may be due to deregulation of genes involved in intracellular reproduction, virulence, and metabolism. Therefore, a comparative genomics approach must take into account both absent and intact genes unique to avirulent strains such as Iowa.
Another major surface antigen, rOmpB, shows defects in proteolytic processing in the Iowa strain (8, 15). Four nonsynonymous SNPs not associated with the cleavage site were identified in rOmpB of the Iowa strain (8); however, these four SNPs were also found in the closely related Morgan strain, which is processed normally. Therefore, differences in the rOmpB protein itself do not appear to be responsible for the defect in processing. Because there are very few mutations that are unique to the Iowa strain alone, it may be possible to analyze those genes to identify a potential peptidase cleaving the beta fragment from autotransporters.
R. peacockii is an endosymbiont of D. andersoni ticks and is not known to infect mammals (27). Its closest pathogenic relative is R. rickettsii (23). While there is a high degree of sequence similarity between many of their shared genes, there are also some important differences. Notably, R. peacockii contains 42 copies of the ISRpe1 transposon which is absent from R. rickettsii. Recombination between the multiple copies is believed to have led to genomic rearrangements and an overall lack of synteny between the two genomes. Nevertheless, there is sufficient similarity that several genes were identified that may contribute to the lack of pathogenic potential in R. peacockii, including an ankyrin repeat-containing protein, dsbA, rickA, ompA, sca1, protease II, and a putative phosphoethanolamine transferase (23).
Surprisingly, the R. rickettsii Sheila Smith genome contains 14 genes expressing proteins annotated as ankyrin and/or containing ankyrin repeat domains (see Data Set S3 in the supplemental material). Only two of these, however, were uniquely different in the Iowa strain. The ankyrin repeat-containing protein (RrIowa_1113) had previously been associated with the avirulence of R. rickettsii Iowa (8). Interestingly, the 588-bp deletion in RrIowa_1113 is also observed in the R strain, which is the less virulent of the two western strains of R. rickettsii compared here. The ortholog of RrIowa_1113 in Rickettsia typhi has recently been annotated as Rickettsia ankyrin repeat protein-2 (RARP-2; RT0600) (28). Also annotated as an ankyrin-containing protein is A1G_01345 in Sheila Smith. In the Iowa strain, this protein (RrIowa_0290) is prematurely terminated at amino acid 68. A role of any of these ankyrin repeat-containing proteins in pathogenesis has not been established. Although the sites and mechanisms of action are unknown, the possibility of redundancy in function must be considered.
Early studies suggested that eastern stains of R. rickettsii were generally less virulent than western strains based upon severity of fever, fatalities, scrotal pathology, and length of incubation period in a guinea pig model of infection (3–6). In the west, R. rickettsii is transmitted by the Rocky Mountain wood tick, Dermacentor andersoni (29), while in the eastern United States, it is spread by the dog tick, Dermacentor variabilis (4); thus, adaptations to the distinct tick vectors were considered a possible basis for this generalization. Subsequent isolation of highly virulent strains from D. variabilis ticks in the east (30, 31) and low-virulence strains from D. andersoni ticks in the west (32) largely discounted the idea of a broad distinction between eastern and western strains (33). However, these studies were conducted before the development of plaque assay procedures to determine inoculum size. Indeed, many of these studies did not define the infectious dose. With the development of improved methods for enumeration of viable rickettsiae (10, 34–36), Anacker et al. (2) were able to distinguish a less severe disease caused by selected eastern strains than by Bitterroot Valley strains in an animal model system. More recently, a phylogenetic analysis of specimens from fatal cases of Rocky Mountain spotted fever revealed that the clades associated with infections in the eastern, central, and southern United States were distinct from those from the western and northwestern United States (37). Although the question of distinctions between eastern and western strains remains, it is clear that there is significant variation in virulence even within relatively small geographic areas, irrespective of the tick vector (1, 7, 17). The molecular basis for these differences in virulence have not been determined.
Although there are relatively few differences in the coding regions between the western and eastern pairs, there are many more SNPs and indels occurring in regions not predicted to encode polypeptides. Regulatory RNAs have not yet been described in rickettsiae but have been described in many pathogenic bacteria (38, 39), including other obligate intracellular bacteria such as chlamydiae (40, 41). The possibility of alterations to regulatory elements cannot be discounted.
Although this screen was intended to aid in the identification of rickettsial determinants of virulence, not all may be apparent in this comparison. For example, Sca2, has four nonsynonymous SNPs and three separate deletions of 3, 18, and 72 bp that are shared by Sheila Smith and R but do not demonstrably alter actin tail formation in those virulent strains. The sca2 gene of the Morgan strain is identical to Iowa's. However, Sca2 is clearly a virulence determinant in R. rickettsii as knockout of Sca2 abolishes not only actin-based motility but also the fever response in guinea pigs (42). Actin-based motility is therefore required for full virulence but is not in itself sufficient since the Iowa strain, which forms normal actin tails, is avirulent. Similarly, phospholipase D was shown to be required for virulence of the etiologic agent of epidemic typhus, Rickettsia prowazekii (43), although no defects in pld were observed in the avirulent Iowa strain examined here. Genomic screens thus represent only one means of identification of determinants and must be supplemented by alternative means of identifying potential virulence determinants, such as bioinformatic searches, unbiased forward screens such as transposon mutagenesis, or development of isogenic mutants. Clearly, virulence in spotted fever group rickettsiae is multifactorial. Indeed, transmission from arthropods to mammals is a critical aspect of rickettsial disease that would not be addressed in the experiments described here.
The availability of genetic tools for the study of rickettsiae has rapidly increased over the past decade (44). The continued improvements in means to modify, complement, and delete specific genes offer unprecedented opportunities to definitively identify the molecular basis for intracellular parasitism and pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the NIAID, NIH.
We thank Stacy Ricklefs and Dan Bruno of the Rocky Mountain Laboratories Genomics Unit, Research Technologies Branch, for DNA sequencing.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.03140-14.
REFERENCES
- 1.Ricketts HT. 1909. Some aspects of Rocky Mountain spotted fever as shown by recent investigation. Med Rec 76:843–855. [DOI] [PubMed] [Google Scholar]
- 2.Anacker RL, Philip RN, Williams JC, List RH, Mann RE. 1984. Biochemical and immunochemical analysis of Rickettsia rickettsii strains of various degrees of virulence. Infect Immun 44:559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badger LF, Dyer RE, Rumreich A. 1931. An infection of the Rocky Mountain spotted fever type. Public Health Rep 46:463–469. doi: 10.2307/4579958.19315284 [DOI] [Google Scholar]
- 4.Dyer RE. 1931. Rocky Mountain spotted fever (Eastern type): transmission by the American dog tick (Dermacentor variabilis). Public Health Rep 46:1403–1412. doi: 10.2307/4580068. [DOI] [Google Scholar]
- 5.Lillie RD. 1931. Pathology of the Eastern type of Rocky Mountain spotted fever. Public Health Rep 46:2840–2859. doi: 10.2307/4580261. [DOI] [Google Scholar]
- 6.Lillie RD, Dyer RE. 1936. Brain reaction in Guinea pigs infected with endemic typhus, epidemic (European) typhus, and Rocky Mountain spotted fever, Eastern and Western types. Public Health Rep 51:1298–1307. [Google Scholar]
- 7.Harden VA. 1990. Rocky Mountain spotted fever: history of a twentieth century disease. Johns Hopkins University Press, Baltimore, MD. [Google Scholar]
- 8.Ellison DW, Clark TR, Sturdevant DE, Virtaneva K, Porcella SF, Hackstadt T. 2008. Genomic comparison of virulent Rickettsia rickettsii Shiela Smith and avirulent Rickettsia rickettsii Iowa. Infect Immun 76:542–550. doi: 10.1128/IAI.00952-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss E, Coolbaugh JC, Williams JC. 1975. Separation of viable Rickettsia typhi from yolk sac and L cell host components by renografin density gradient centrifugation. Appl Microbiol 30:456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cory J, Yunker CE, Ormsbee RA, Peacock MG, Meibos H, Tallent G. 1974. Plaque assay of rickettsiae in a mammalian cell line. Appl Microbiol 27:1157–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anacker RL, Mann RE, Gonzales C. 1987. Reactivity of monoclonal antibodies to Rickettsia rickettsii with spotted fever and typhus group rickettsiae. J Clin Microbiol 25:167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilmore RD Jr, Cieplak W, Policastro PF, Hackstadt T. 1991. The 120 kilodalton outer membrane protein (rOmp B) of Rickettsia rickettsii is encoded by an unusually long open reading frame: evidence for protein processing from a large precursor. Mol Microbiol 5:2361–2370. doi: 10.1111/j.1365-2958.1991.tb02082.x. [DOI] [PubMed] [Google Scholar]
- 13.Parker RR, Pickens EG, Lackman DB, Bell EJ, Thrailkill FB. 1951. Isolation and characterization of Rocky Mountain spotted fever rickettsiae from the rabbit tick Haemaphysalis leporis-paulstris Packard. Public Health Rep 66:455–463. doi: 10.2307/4587691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox HR. 1941. Cultivation of Rickettsiae of the Rocky Mountain spotted fever, typhus and Q fever groups in the embryonic tissues of developing chicks. Science 94:399–403. doi: 10.1126/science.94.2444.399. [DOI] [PubMed] [Google Scholar]
- 15.Hackstadt T, Messer R, Cieplak W, Peacock MG. 1992. Evidence for the proteolytic cleavage of the 120-kilodalton outer membrane protein of rickettsiae: identification of an avirulent mutant deficient in processing. Infect Immun 60:159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson BE, McDonald GA, Jones DC, Regnery RL. 1990. A protective protein antigen of Rickettsia rickettsii has tandemly repeated, near-identical sequences. Infect Immun 58:2760–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price WH. 1953. The epidemiology of Rocky Mountain spotted fever. I. The characterization of strain virulence of Rickettsia rickettsii. Am J Hyg 58:248–268. [DOI] [PubMed] [Google Scholar]
- 18.Baldridge GD, Burkhardt NY, Simser JA, Kurtti TJ, Munderloh UG. 2004. Sequence and expression analysis of the ompA gene of Rickettsia peacockii, an endosymbiont of the Rocky Mountain wood tick, Dermacentor andersoni. Appl Environ Microbiol 70:6628–6636. doi: 10.1128/AEM.70.11.6628-6636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald GA, Anacker RL, Garjian K. 1987. Cloned gene of Rickettsia rickettsii surface antigen: candidate vaccine for Rocky Mountain spotted fever. Science 235:83–85. doi: 10.1126/science.3099387. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Walker DH. 1998. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb Pathog 24:289–298. doi: 10.1006/mpat.1997.0197. [DOI] [PubMed] [Google Scholar]
- 21.Blanc G, Ngwamidiba M, Ogata H, Fournier PE, Claverie JM, Raoult D. 2005. Molecular evolution of Rickettsia surface antigens: evidence of positive selection. Mol Biol Evol 22:2073–2083. doi: 10.1093/molbev/msi199. [DOI] [PubMed] [Google Scholar]
- 22.Jiggins FM. 2006. Adaptive evolution and recombination of Rickettsia antigens. J Mol Evol 62:99–110. doi: 10.1007/s00239-005-0080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felsheim RF, Kurtti TJ, Munderloh UG. 2009. Genome sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: identification of virulence factors. PLoS One 4:e8361. doi: 10.1371/journal.pone.0008361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lescot M, Audic S, Robert C, Nguyen TT, Blanc G, Cutler SJ, Wincker P, Couloux A, Claverie JM, Raoult D, Drancourt M. 2008. The genome of Borrelia recurrentis, the agent of deadly louse-borne relapsing fever, is a degraded subset of tick-borne Borrelia duttonii. PLoS Genet 4:e1000185. doi: 10.1371/journal.pgen.1000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parish T, Smith DA, Kendall S, Casali N, Bancroft GJ, Stoker NG. 2003. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect Immun 71:1134–1140. doi: 10.1128/IAI.71.3.1134-1140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D, Zheng X, Sun W, Karaolis DK. 2003. The Vibrio pathogenicity island-encoded mop protein modulates the pathogenesis and reactogenicity of epidemic Vibrio cholerae. Infect Immun 71:510–515. doi: 10.1128/IAI.71.1.510-515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niebylski ML, Schrumpf ME, Burgdorfer W, Fischer ER, Gage KL, Schwan TG. 1997. Rickettsia peacockii sp. nov., a new species infecting wood ticks, Dermacentor andersoni, in western Montana. Int J Syst Bacteriol 47:446–452. doi: 10.1099/00207713-47-2-446. [DOI] [PubMed] [Google Scholar]
- 28.Gillespie JJ, Kaur SJ, Rahman MS, Rennoll-Bankert K, Sears KT, Beier-Sexton M, Azad AF. 4 December 2014. Secretome of obligate intracellular Rickettsia. FEMS Microbiol Rev doi: 10.1111/1574-6976.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricketts HT. 1907. Further experiments with the woodtick in relation to Rocky Mountain spotted fever. JAMA 49:1278–1281. [Google Scholar]
- 30.Brigham GD, Watt J. 1940. Highly virulent strains of Rocky Mountain spotted fever virus isolated from ticks (D. variabilis) in Georgia. Public Health Rep 55:2125–2126. doi: 10.2307/4583511. [DOI] [Google Scholar]
- 31.Topping NH. 1940. A highly virulent strain of Rocky Mountain spotted fever virus isolated in the eastern United States. Public Health Rep 55:728–731. doi: 10.2307/4583268. [DOI] [Google Scholar]
- 32.Topping NH. 1941. A strain of Rocky Mountain spotted fever virus of low virulence isolated in the western United States. Public Health Rep 56:2041–2043. doi: 10.2307/4583896. [DOI] [Google Scholar]
- 33.Topping NH. 1941. Rocky Mountain spotted fever: a note on some aspects of its epidemiology. Public Health Rep 56:1699–1703. doi: 10.2307/4583839. [DOI] [Google Scholar]
- 34.McDade JE, Stakebake JR, Gerone PJ. 1969. Plaque assay system for several species of Rickettsia. J Bacteriol 99:910–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ormsbee R, Peacock M, Gerloff R, Tallent G, Wike D. 1978. Limits of rickettsial infectivity. Infect Immun 19:239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinberg EH, Stakebake JR, Gerone PJ. 1969. Plaque assay for Rickettsia rickettsii. J Bacteriol 98:398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paddock CD, Denison AM, Lash RR, Lui L, Bollweg BC, Dahlgren FS, Kanamura CT, Angerami RN, dos Santos P, Martines RB, Karpathy SE. 2014. Phylogeograpy of Rickettsia rickettsii genotypes associated with fatal Rocky Mountain spotted fever. Am J Trop Med Hyg 91:589–597. doi: 10.4269/ajtmh.14-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papenfort K, Vogel J. 2010. Regulatory RNA in bacterial pathogens. Cell Host Microbe 8:116–127. doi: 10.1016/j.chom.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Waters LS, Storz G. 2009. Regulatory RNA in bacteria. Cell 136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grieshaber NA, Grieshaber SS, Fischer ER, Hackstadt T. 2006. A small RNA inhibits translation of the histone-like protein Hc1 in Chlamydia trachomatis. Mol Microbiol 59:541–550. doi: 10.1111/j.1365-2958.2005.04949.x. [DOI] [PubMed] [Google Scholar]
- 41.Abdelrahman YM, Rose LA, Belland RJ. 2011. Developmental expression of non-coding RNAs in Chlamydia trachomatis during normal and persistent growth. Nucleic Acids Res 39:1843–1854. doi: 10.1093/nar/gkq1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleba B, Clark TR, Lutter EI, Ellison DW, Hackstadt T. 2010. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect Immun 78:2240–2247. doi: 10.1128/IAI.00100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Driskell LO, Yu XJ, Zhang L, Liu Y, Popov VL, Walker DH, Tucker AM, Wood DO. 2009. Directed mutagenesis of the Rickettsia prowazekii pld gene encoding phospholipase D. Infect Immun 77:3244–3248. doi: 10.1128/IAI.00395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood DO, Wood RR, Tucker AM. 2014. Genetic systems for studying obligate intracellular pathogens: an update. Curr Opin Microbiol 17:11–16. doi: 10.1016/j.mib.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, P D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.