Abstract
Photobacterium damselae subsp. damselae is a marine bacterium that causes septicemia in marine animals and in humans. Previously, we had determined a major role of pPHDD1 plasmid-encoded Dly (damselysin) and HlyA (HlyApl) and the chromosome-encoded HlyA (HlyAch) hemolysins in virulence. However, the mechanisms by which these toxins are secreted remain unknown. In this study, we found that a mini-Tn10 transposon mutant in a plasmidless strain showing an impaired hemolytic phenotype contained an insertion in epsL, a component of a type II secretion system (T2SS). Reconstruction of the mutant by allelic exchange confirmed the specific involvement of epsL in HlyAch secretion. In addition, mutation of epsL in a pPHDD1-harboring strain caused an almost complete abolition of hemolytic activity against sheep erythrocytes, indicating that epsL plays a major role in secretion of the plasmid-encoded HlyApl and Dly. This was further demonstrated by analysis of different combinations of hemolysin gene mutants and by strain-strain complementation assays. We also found that mutation of the putative prepilin peptidase gene pilD severely affected hemolysis, which dropped at levels inferior to those of epsL mutants. Promoter expression analyses suggested that impairment of hemolysin secretion in epsL and pilD mutants might constitute a signal that affects hemolysin and T2SS gene expression at the transcriptional level. In addition, single epsL and pilD mutations caused a drastic decrease in virulence for mice, demonstrating a major role of T2SS and pilD in P. damselae subsp. damselae virulence.
INTRODUCTION
The marine bacterium Photobacterium damselae subsp. damselae is considered a primary pathogen of a wide range of marine animals, including wild and cultivated fish, causing losses of economical importance in marine aquaculture (1–3). In addition, this pathogen is of special concern for humans, since it can cause a highly severe necrotizing fasciitis that may lead to a fatal outcome (4, 5). The majority of the reported infections in humans have their primary origin in wounds exposed to seawater (6), inflicted during fish and fishing tool handling (7), while practicing aquatic sports (8), and occasionally after consumption of seafood (9).
Highly hemolytic strains of this pathogen harbor the virulence plasmid pPHDD1, which encodes the hemolysins damselysin (Dly) and HlyA (HlyApl) (10). In addition, all of the hemolytic P. damselae subsp. damselae strains encode a third hemolysin, chromosome-encoded HlyA (HlyAch), in chromosome I (11, 12). Dly is a potent phospholipase D cytotoxin with hemolytic activity that removes choline phosphate head groups from sphingomyelin (13, 14), whereas HlyApl and HlyAch are predicted to be pore-forming toxins (11). HlyAch and HlyApl show 92% identity in their amino acid sequences, but HlyApl is twice more active against sheep erythrocytes than HlyAch (11). When the two HlyA hemolysins are produced by a P. damselae subsp. damselae strain, their activities demonstrated to exert an additive effect in terms of hemolysis and virulence. The interaction of Dly with any of the two HlyA produces a synergistic effect against erythrocytes, and this synergy is responsible for maximum virulence for mice and fish (11). In addition, Dly enhances synergistically the hemolytic activity of toxins produced by different bacterial species, establishing CAMP (Christie, Atkins, and Munch-Petersen) reactions. CAMP reactions were originally defined as a synergistic hemolytic process produced by the interaction of a hemolysin (CAMP factor) of group B streptococci with the beta-toxin (a sphingomyelinase) of Staphylococcus aureus (15).
Although much has been ascertained on the molecular basis of hemolysis and virulence in P. damselae subsp. damselae, nothing is known about how the hemolysins are secreted. Proteins destined for the extracellular environment of Gram-negative bacteria have to be transported across the cytoplasmic membrane and the outer membrane (16). The type II secretion system (T2SS) belongs to a widespread superfamily of membrane nanomachines that promote specific transport of folded proteins across the outer membrane (17, 18). Exoproteins are synthesized with N-terminal signal peptides that target them for cytoplasmic membrane translocation through either the Sec or Tat complexes (19). After removal of the signal peptides, the exoproteins transiently reside in the periplasm prior to outer membrane translocation (20). Extracellular secretion of proteins is regarded as a major virulence mechanism in bacterial infection. Proteins secreted by the T2SS include proteases, cellulases, pectinases, phospholipases, lipases, hemolysins, and toxins in a broad sense. In general, these proteins are associated with destruction of tissues, which contributes to cell damage and disease. The species of Proteobacteria identified to date as harboring T2SS genes are mostly extracellular pathogens (16).
In this study, we identified, using a transposon-based approach, epsL, a gene of a predicted T2SS in P. damselae subsp. damselae. We demonstrated that epsL, but not the pPHDD1-carried tolC, is involved in the secretion of Dly, HlyApl, and HlyAch and that the putative prepilin peptidase gene pilD is essential for hemolysin secretion and pilus biogenesis. The major role of epsL and pilD in the virulence of P. damselae subsp. damselae for mice is also demonstrated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used here and those derived from the present study are listed in Table 1. P. damselae subsp. damselae cells were routinely grown at 25°C on tryptic soy agar (TSA) supplemented with 1% NaCl (TSA-1) and in Luria-Bertani (LB) broth and LB agar, supplemented with antibiotics when appropriate. Sheep blood agar plates (Oxoid) were used for hemolysis assays and for conjugative matings. For hemolysis assays on agar plates, a single colony of each strain grown on a TSA-1 plate was picked with the tip of a rounded wooden pick and seeded on the blood agar plate, and pictures were taken, unless otherwise stated, after 15 h of incubation at 25°C. Escherichia coli strains were routinely grown at 37°C in LB broth and LB agar supplemented with antibiotics when appropriate. Antibiotics were used at the following final concentrations: streptomycin (Sm) at 50 μg ml−1, kanamycin (Km) at 50 μg ml−1, ampicillin sodium salt at 50 μg ml−1, gentamicin at 15 μg ml−1, and rifampin at 50 μg ml−1.
TABLE 1.
Strains and plasmids used and constructed in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| P. damselae subsp. damselae | ||
| RM-71 | Wild type, isolated from turbot (Psetta maxima), strongly hemolytic, pPHDD1 | 1 |
| AR57 | RM-71 derivative, spontaneous rifampin-resistant mutant; Rfr | 10 |
| AR64 | AR57 with in-frame deletion of dly gene | 10 |
| AR78 | AR57 with in-frame deletion of dly and hlyApl genes | 10 |
| AR158 | AR57 with in-frame deletion of hlyApl and hlyAch genes | 11 |
| AR89 | AR57 with in-frame deletion of dly, hlyApl, and hlyAch genes | 11 |
| AR217 | AR57 with in-frame deletion of epsL gene | This study |
| AR222 | AR57 with in-frame deletion of dly and epsL genes | This study |
| AR223 | AR57 with in-frame deletion of hlyApl, hlyAch, and epsL genes | This study |
| AR235 | AR57 with in-frame deletion of tolC gene | This study |
| AR234 | AR57 with in-frame deletion of tolC, dly, and epsL genes | This study |
| AR227 | AR57 with in-frame deletion of hlyApl, hlyAch, tolC, and epsL genes | This study |
| AR251 | AR57 with in-frame deletion of hlyApl, dly, and epsL genes | This study |
| AR239 | AR57 with in-frame deletion of pilD gene | This study |
| AR252 | AR57 with in-frame deletion of pilD and tadV genes | This study |
| LD-07 | Wild type, isolated from seabream (Sparus aurata), weakly hemolytic | 1 |
| AR111 | LD-07 derivative, spontaneous rifampin-resistant mutant; Rfr | 11 |
| AR203 | AR111 with mini-Tn10 inserted in epsL gene; Kmr | This study |
| AR211 | AR111 with in-frame deletion of epsL gene | This study |
| AR240 | AR111 with in-frame deletion of pilD gene | This study |
| E. coli | ||
| DH5α | Cloning strain, recA | Laboratory stock |
| S17-1-λpir | recA thi pro ΔhsdR-hsdM RP4-2 Tc::Mu Km::Tn7 λpir; Tpr Smr | 21 |
| β-3914 | F− RP4-2-Tc::Mu ΔdapA::(erm-pir) gyrA462 zei-298::Tn10 (Kmr Emr Tcr) | 53 |
| AR201 | β-3914 donor for mini-Tn10 transposon; Kmr | This study |
| Plasmids | ||
| pUC118 | High-copy-number cloning vector; Apr | 54 |
| pWKS30 | Low-copy-number cloning vector; Apr | 55 |
| pNidkan | Suicide vector derived from pCVD442; Kmr | 22 |
| pHRP309 | lacZ reporter plasmid, mob; Gmr | 26 |
| pSEVA434 | Cloning and expression vector; Smr | 56 |
| pAJR45 | hlyAch::lacZ fusion in pHRP309 | 11 |
| pAJR51 | dly::lacZ fusion in pHRP309 | 11 |
| pAJR53 | hlyApl::lacZ fusion in pHRP309 | 11 |
| pAJR71 | epsC::lacZ fusion in pHRP309 | This study |
| pAJR76 | pHRP309 with pilD gene from strain RM-71 | This study |
| pAVL88 | pHRP309 with epsL gene from strain RM-71 | This study |
Rfr, rifampin resistance; Tcr, tetracycline resistance; Tpr, trimethoprim resistance; Smr, streptomycin resistance; Kmr, kanamycin resistance; Gmr, gentamicin resistance; Emr, erythromycin resistance; Apr, ampicillin resistance.
Mini-Tn10 mutagenesis based analysis.
Mini-Tn10 mutagenesis was carried out using a modification of the method developed by Herrero et al. with the suicide conjugative plasmid pLOFKm (21). Briefly, we introduced pLOFKm into the diaminopimelic acid-deficient strain Escherichia coli β-3914 (yielding strain AR201), which was subsequently mated with the plasmidless P. damselae subsp. damselae strain AR111. For conjugative matings, exponentially growing cells of donor and recipient strains were mixed, a drop (100 μl) was placed directly onto a sheep blood agar plate, and the plate was incubated at 25°C for 3 days. After incubation, cells were scraped off the plate and resuspended in TSB-1, and 100-μl aliquots of serial decimal dilutions were spread on LB plates containing kanamycin (marker for mini-Tn10 insertions) and 5% of sheep erythrocytes to select for P. damselae subsp. damselae colonies impaired in their hemolytic activity. Zones of hemolysis were evaluated by eye for both size and extent of translucency.
Cloning of the mini-Tn10 insertions into P. damselae subsp. damselae genome.
The regions surrounding the genes disrupted by the mini-Tn10 mutagenesis were cloned into pUC118 plasmid. Briefly, genomic DNA from the clones with impaired hemolysis of P. damselae subsp. damselae strain AR111 was purified using a genome DNA kit (Qbiogene), partially digested with BfuCI and then ligated using T4 DNA ligase into the calf intestinal alkaline phosphatase-treated and BamHI-digested site of pUC118. The ligated DNA was used to transform E. coli DH5α by electroporation. Transformants were selected on LB agar plates supplemented with kanamycin and ampicillin. The pUC118-cloned inserts were PCR amplified and subsequently sequenced with a CEQ DTCS118 quick start kit and a capillary DNA CEQ8000 sequencer (Beckman Coulter).
Mutant construction and gene complementation.
Nonpolar deletions were constructed by using PCR amplification of the amino- and carboxy-terminal fragments of each gene, which, when fused together, would result in an in-frame deletion of >90% of the coding sequence. Amplification was carried out using Hi-Fidelity Kapa Taq (Kapa). Allelic exchange was performed as previously described (22) using the Kmr suicide vector pNidKan containing the sacB gene, which confers sucrose sensitivity, and R6K ori, which requires the pir gene product for replication. The plasmid constructions containing the deleted alleles were transferred from E. coli S17-1-λpir into the rifampin-resistant derivatives AR57 and AR111, respectively. After conjugation on sheep blood agar plates (see above), cells were scraped off the plate and resuspended in LB medium, and 100-μl aliquots of serial decimal dilutions were spread on TSA-1 plates, selecting for kanamycin resistance for plasmid integration, and subsequently for sucrose resistance (15% [wt/vol]) for a second recombination event. This led to P. damselae subsp. damselae mutant strains listed in Table 1. The presence of the correct alleles was confirmed by PCR. Complementation plasmids for dly (pAJR39) and hlyApl genes (pAJR38) were constructed in a previous study and complementation of the original dly and hlyApl mutations was demonstrated (10). Plasmid pAJR55 constructed in a previous study (11) containing hlyAch was introduced into the respective P. damselae subsp. damselae hlyAch mutant derivatives to demonstrate functional complementation. For complementation of pilD mutants, pilD open reading frame (ORF), together with its respective promoter sequence, was PCR amplified with Hi-Fidelity Kapa Taq, cloned into pHRP309 vector, and mobilized from E. coli S17-1-λpir into the respective P. damselae subsp. damselae mutants. For complementation of epsL mutants, epsL ORF was PCR amplified with Hi-Fidelity Kapa Taq and cloned downstream of the Ptrc promoter into pSEVA434 vector. The epsL sequence fused to Ptrc promoter was PCR amplified, cloned into pHRP309 and mobilized from E. coli S17-1-λpir into the respective P. damselae subsp. damselae epsL mutants.
CAMP assays.
The CAMP reaction is a synergistic hemolytic process produced by the interaction of two different membrane-perturbing molecules and was initially described in group B streptococci (15). We used CAMP reactions to specifically dissect the effect of epsL and tolC deletions in Dly secretion. Due to the fact that Dly alone is weakly active against sheep erythrocytes (11), defects in Dly secretion in hlyA mutants might pass unperceived on sheep blood agar. For CAMP reaction assays, ΔhlyApl ΔhlyAch double mutant (AR158), ΔhlyApl ΔhlyAch ΔepsL triple mutant (AR223), and ΔhlyApl ΔhlyAch ΔepsL ΔtolC tetramutant (AR227) were cultured on sheep blood agar plates using the tip of a rounded wooden pick. After 3 days of growth (enabling Dly to be secreted and diffused through the agar plate), the indicator strain AR78 was radially streaked in close proximity to the rounded streaks of AR223, AR158 and AR227. AR78 is a Δdly ΔhlyApl mutant that yields a strongly synergistic halo of hemolysis in the presence of exogenous Dly molecules (11). Pictures were taken after 15 h of incubation.
Hemolytic assays with bacterial ECPs.
To obtain the extracellular products (ECPs), 100 μl of P. damselae subsp. damselae broth cultures adjusted to an optical density at 600 nm (OD600) of 1 were spread over cellophane covered TSA-1 plates with a sterile cotton swab as previously described (23). After 48 h of incubation at 25°C, cells were washed off the cellophane with a minimum volume of saline solution (0.85% [wt/vol] NaCl) and readjusted to an OD600 of 1. The cell suspensions were centrifuged at 15,000 × g for 5 min, and the supernatants were filtered through 0.22-μm-pore-size membranes and stored at −30°C until needed. For hemolytic liquid assays, a modification of the method of Bernheimer was used (24). In brief, sheep erythrocytes (Oxoid) were washed with phosphate-buffered saline (PBS) and centrifuged at 900 × g for 5 min at 4°C. Washes were repeated until the supernatant was visibly clear of hemoglobin. Different volumes of crude erythrocytes were washed and tested to assess the minimal volume necessary to get the maximal absorbance (the absorbance obtained by lysis of the erythrocytes with distilled water). Two hundred microliters of crude erythrocytes was enough to obtain the maximal absorbance and, consequently, the erythrocytes were adjusted with PBS to 0.5 ml per dilution tube assayed, and the 0.5 ml of serially diluted ECP samples was added. After 1 h of incubation at 25°C, the mixtures were centrifuged at 900 × g for 5 min at 4°C to remove undamaged erythrocytes. This assay is based on the measure of the released hemoglobin, whose concentration is estimated by reading the absorbance at 545 nm in a spectrophotometer and adjusted using nonlysed cells (0.5 ml of erythrocyte suspension with 0.5 ml of PBS) as a control.
One unit of hemolytic activity was expressed as the amount of ECPs that caused the release of 50% of the hemoglobin in the standardized erythrocyte suspension. All hemolytic assays were carried out in triplicate.
N-terminal sequencing of Dly protein and in silico prediction of signal peptides.
A search for N-terminal signal peptides within Dly, HlyApl, and HlyAch and their cleavage positions was conducted using the SignalP 4.1. program (25). To identify the N-terminal sequence of the presumed mature Dly, we purified the hemolytic principle from extracellular products of the AR158 strain (encoding Dly, but devoid of HlyApl and HlyAch). A protein with an apparent molecular mass of ∼60 kDa in SDS-PAGE eluted with a major peak from a mono-S cation-exchange column. Upon blotting onto a polyvinylidene difluoride (PVDF) membrane, the band was cut out and subjected to Edman degradation.
Construction of lacZ transcriptional fusions and β-galactosidase assays.
A DNA fragment corresponding to epsC presumptive promoter region extending from ∼1 kb upstream of the ATG start codon to ∼30 bp downstream of the start codon was PCR amplified and fused to a promoterless lacZ gene in the low-copy-number reporter plasmid pHRP309 (26). The resulting transcriptional fusion construct, epsC::lacZ (pAJR71), as well as the previously constructed dly::lacZ (pAJR51), hlyApl::lacZ (pAJR53), and hlyAch::lacZ (pAJR45) fusions (11), was mobilized by conjugation from E. coli S17-1-λpir into different P. damselae subsp. damselae parental and mutant strains. The P. damselae subsp. damselae strains carrying the promoter-lacZ fusion vectors were grown in standard LB broth (0.5% NaCl) and in LB broth supplemented with 3.5% NaCl. The β-galactosidase activities were measured by the method of Miller (27). The data shown correspond to three independent experiments.
TEM.
To visualize pili by transmission electron microscopy (TEM), bacterial cells were grown overnight in LB medium, fixed with 2.5% glutaraldehyde for 1 h at room temperature, centrifuged at 2,000 × g for 2 min, and washed once with PBS. Cells were negatively stained with 2% phosphotungstic acid on Parlodion-coated grids and examined with a JEOL JEM-2010 transmission electron microscope. For each strain tested, at least 100 cells on each grid were examined for the presence of pili.
Mouse virulence assays.
Virulence assays were carried out with BALB/c mice (age, 6 to 8 weeks; weight, 26 to 30 g) in three groups of five animals each per strain tested. The inoculum was prepared by suspending several colonies from a 24-h TSA-1 culture into saline solution to achieve the turbidity of a no. 2 McFarland standard. Mice were inoculated in the tail vein with 50 μl of a 2.5 μM hemoglobin solution (8 μg hemoglobin per mouse) 2 h before inoculation with the bacterial suspension, as previously described (28). Mice were inoculated intravenously in the tail vein with 0.1 ml of 10-fold serial dilutions of the bacterial suspensions. The actual number of injected CFU was determined by plate count on TSA-1. The final dose assayed corresponded to 2.1 × 106 bacterial cells per mouse. The mortalities were recorded daily for 3 days, and the results from the three independent groups of five animals were pooled and expressed as mortality percentages. All of the protocols of animal experimentation used in the present study have been reviewed and approved by the Bioethics Committee of the University of Santiago de Compostela.
Statistical analysis.
The statistical analysis of the hemolytic activity data to assess the robustness of the differences between pairs of strains was carried out with the Student t test. For the statistical analysis of the results of the mouse virulence experiments, differences among data for live/dead animals for each pooled group of 15 animals were compared using a chi-square test. The SPSS statistical software package (version 20; IBM SPSS, Inc., Chicago, IL) was used for statistical analyses. Adjusted P values of <0.05 were considered statistically significant.
RESULTS AND DISCUSSION
Mutation of the pPHDD1-encoded TolC component has no detectable effect in hemolysin secretion.
Although the role of the three P. damselae subsp. damselae hemolysins Dly (damselysin), HlyApl and HlyAch in virulence has been quite well described (11), nothing is known about how the cell secretes them. Pore-forming toxins can be secreted following different pathways in Gram-negative bacteria. E. coli HlyA is secreted by a Sec-independent pathway which requires TolC (29), whereas V. cholerae cytolysin VCC is secreted by a type II secretion pathway (T2SS) (30). In order to start ascertaining the mechanisms by which P. damselae subsp. damselae hemolysins get their way out of the cell, we first placed our attention on a cluster of putative secretion genes located immediately upstream of dly in pPHDD1 sequence, which includes a gene for the outer membrane protein TolC (Fig. 1A). We observed that deletion of tolC in the highly hemolytic strain AR57 (yielding AR235) did not cause any detectable effect in hemolysis on sheep blood agar plates (Fig. 2B). In addition, hemolytic activity assays with sheep erythrocyte suspensions and bacterial ECPs showed no significant differences in the hemolytic units (HU) produced by the parental strain AR57 and the ΔtolC mutant AR235 (Fig. 3).
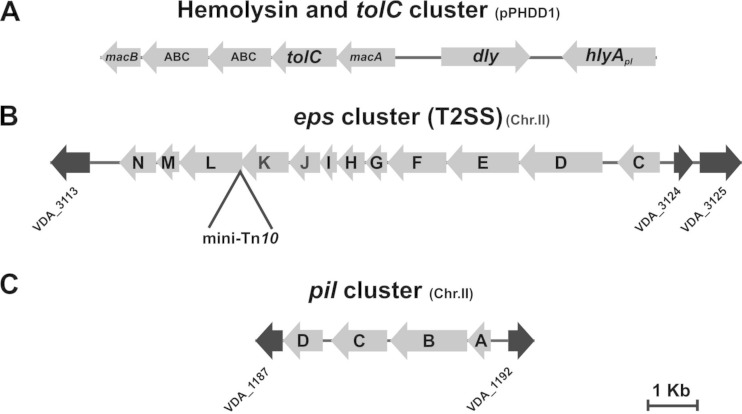
FIG 1.
Physical maps of three different P. damselae subsp. damselae genome regions. (A) Partial sequence region of pPHDD1 plasmid containing the genes encoding damselysin (dly) and HlyApl (hlyApl) hemolysins, as well as genes of a putative secretion system that includes the outer membrane protein TolC. (B) eps gene cluster encoding the T2SS. The point of a mini-Tn10 insertion in epsL is shown. (C) pil cluster containing the putative prepilin peptidase gene pilD. Note that eps and pil clusters are encoded within chromosome II. Flanking gene numbers (depicted as black arrows) refer to gene annotation of the P. damselae subsp. damselae type strain ATCC 33539 complete genome (GenBank accession no. ADBS00000000). epsM and epsI are not annotated in the ATCC 33539 genome.
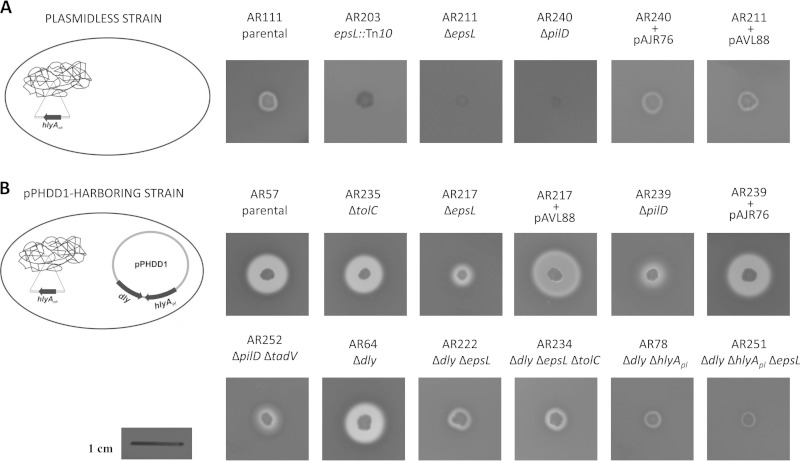
FIG 2.
Effects of tolC, epsL, pilD, and tadV deletions in the hemolytic haloes produced on sheep blood agar plates by different P. damselae subsp. damselae parental and hemolysin mutant strains. (A) Plasmidless strain AR111 (which only contains hlyAch gene in chromosome I) and its mutant derivatives. (B) pPHDD1-harboring strain AR57 (which contains the three hemolysin genes dly, hlyApl, and hlyAch) and its mutant derivatives. The genotypes are described below the strain names. epsL and pilD mutants complemented with epsL and pilD genes cloned into plasmids pAVL88 and pAJR76, respectively, are also indicated. Scale bar, 1 cm.
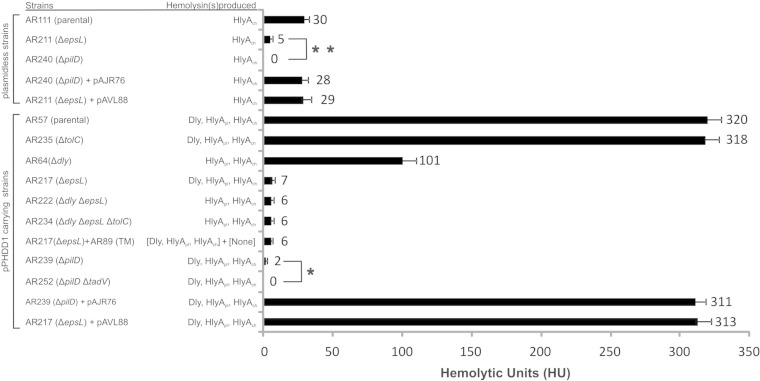
FIG 3.
Measurement of the hemolytic activity of the ECPs of P. damselae subsp. damselae strains on sheep erythrocytes. The release of hemoglobin was determined at A540. One hemolytic unit is defined as the amount of hemolysin that lyses 50% of sheep erythrocytes. All assays were carried out in triplicate and mean values with standard deviation are shown. “(TM)” refers to the triple mutant AR89 (Δdly ΔhlyApl ΔhlyAch). Asterisks denote statistically significant differences between certain pairs of strains using the Student t test. *, P < 0.05; **, P < 0.01.
Transposon-based mutagenesis identifies a T2SS in P. damselae subsp. damselae with a role in HlyAch secretion.
Since an in silico sequence analysis failed to reveal the presence of additional secretion systems encoded on pPHDD1, we hypothesized that the secretion system involved in P. damselae subsp. damselae hemolysins secretion might be encoded on the chromosomes. We therefore screened a library of mini-Tn10 mutants in the naturally pPHDD1-lacking strain AR111 and isolated two mutants displaying a nonhemolytic phenotype and one mutant severely affected in hemolysis. Further genetic characterization led us to determine that the two nonhemolytic mutants contained the transposon inserted in the hlyAch gene, the only hemolytic determinant in P. damselae subsp. damselae plasmidless strains (11). Interestingly, we found that the insertion in the mutant AR203 with a severely impaired hemolysis (Fig. 2A) took place in a gene whose deduced amino acid sequence was 41% identical to Vibrio cholerae EpsL (extracellular protein secretion L). EpsL, a component of the T2SS of Gram-negative bacteria, is a predicted inner-membrane protein that plays a crucial role in pseudopilus formation (31, 32). The T2SS is composed of 12 to 15 proteins and enables secretion of folded proteins to the outer medium (16). An in silico search in the draft genome of the P. damselae subsp. damselae type strain ATCC 33539 (GenBank accession no. ADBS00000000) revealed that epsL is the third-to-last gene of the 11.5-kb T2SS gene cluster that consists of 12 genes (from epsC to epsN) (Fig. 1B).
To confirm the association between the disruption of epsL and the severely impaired hemolytic phenotype, we constructed an in-frame deletion of this gene by allelic exchange, yielding strain AR211. As a result, we observed that targeted deletion of epsL caused the same reduction in the hemolytic halo that we previously observed in the transposon mutant (Fig. 2A). Similarly, deletion of epsL caused a decrease of the hemolytic activity from 30 HU produced by the parental (AR111) to 5 HU of the ΔepsL mutant (AR211) (Fig. 3). Complementation of the mutant with epsL cloned into pAVL88 plasmid restored the hemolytic activity values of the parental strain (Fig. 2A and Fig. 3). Since HlyAch is the only β-hemolysin produced by plasmidless P. damselae subsp. damselae strains (11, 12), these results clearly indicate that epsL plays a major role in HlyAch secretion in P. damselae subsp. damselae.
Deletion of epsL causes a defect in secretion of the pPHDD1-encoded hemolysins Dly and HlyApl.
In addition to HlyAch, P. damselae subsp. damselae strains carrying pPHDD1 produce Dly and HlyApl hemolysins (10). To test whether the T2SS is also involved in the secretion of the plasmid-encoded hemolysins, we deleted epsL in AR57. We observed that deletion of epsL (strain AR217) caused a strong reduction of the hemolytic halo (Fig. 2B), suggesting that the T2SS plays a major role in secreting the plasmid-encoded P. damselae subsp. damselae hemolysins.
We further investigated the contribution of T2SS to secretion of the two different types of hemolysins produced by P. damselae subsp. damselae: the two pore-forming HlyAs on the one hand and the phospholipase D cytotoxin Dly on the other hand. To assess the role in HlyA secretion, we analyzed the hemolytic haloes produced upon epsL mutation in the Δdly mutant (produces HlyApl and HlyAch) and in the Δdly ΔhlyApl double mutant (only produces HlyAch). Deletion of epsL in the Δdly mutant caused a severe decrease of the hemolytic halo (AR222) (Fig. 2B). In addition, the already reduced halo of the Δdly ΔhlyApl mutant (due to the rather low individual contribution of HlyAch to hemolysis) underwent a further reduction upon epsL deletion (AR251) (Fig. 2B). In a previous study, we found that production by some P. damselae subsp. damselae mutants of residual hemolytic haloes on sheep blood agar did not necessarily mean that the mutant retained the ability to cause the complete lysis of erythrocytes in liquid hemolytic assays (11). We thus carried out a quantitative assay using ECPs and erythrocyte suspensions (Fig. 3). Deletion of epsL caused a drastic decrease of the hemolytic activity from 320 HU produced by the parental (AR57) to 7 HU in the ΔepsL mutant (AR217). A strong drop in hemolytic activity was also measured in the double ΔepsL Δdly mutant (6 HU) with respect to the single Δdly mutant (101 HU). These results clearly demonstrate that the T2SS plays a major role in secretion of the P. damselae subsp. damselae hemolysins HlyApl and HlyAch.
To assess the effect in Dly secretion we deleted epsL in the ΔhlyApl ΔhlyAch double mutant (only produces Dly). It is important to note that Dly alone is weakly active against sheep erythrocytes (11), and thus the likely defect in Dly secretion in a ΔepsL mutant might pass unperceived on sheep blood agar. Hence, in order to demonstrate whether Dly is being secreted by the T2SS, we carried out CAMP assays (see Materials and Methods) using AR78 as an indicator strain to detect Dly secretion. We found that strain AR223 (ΔhlyApl ΔhlyAch ΔepsL) showed a strong impairment to produce CAMP reactions with strain AR78 (Fig. 4A) compared to the positive control AR158 (ΔhlyApl ΔhlyAch) (Fig. 4C), demonstrating that Dly is secreted by the T2SS. Complementation of the epsL mutant strain AR217 with epsL cloned into pAVL88 plasmid restored the hemolytic phenotype on sheep blood agar (Fig. 2B) and the hemolytic activity values (Fig. 3) observed with the parental strain. Notably, bacterial hemolysins with phospholipase D activity have been described in a limited number of species, and only a few studies have deciphered the molecular mechanisms of phospholipase D secretion (33). To the best of our knowledge, our study represents the first evidence of a bacterial phospholipase D with hemolytic activity which is secreted by a T2SS.
FIG 4.
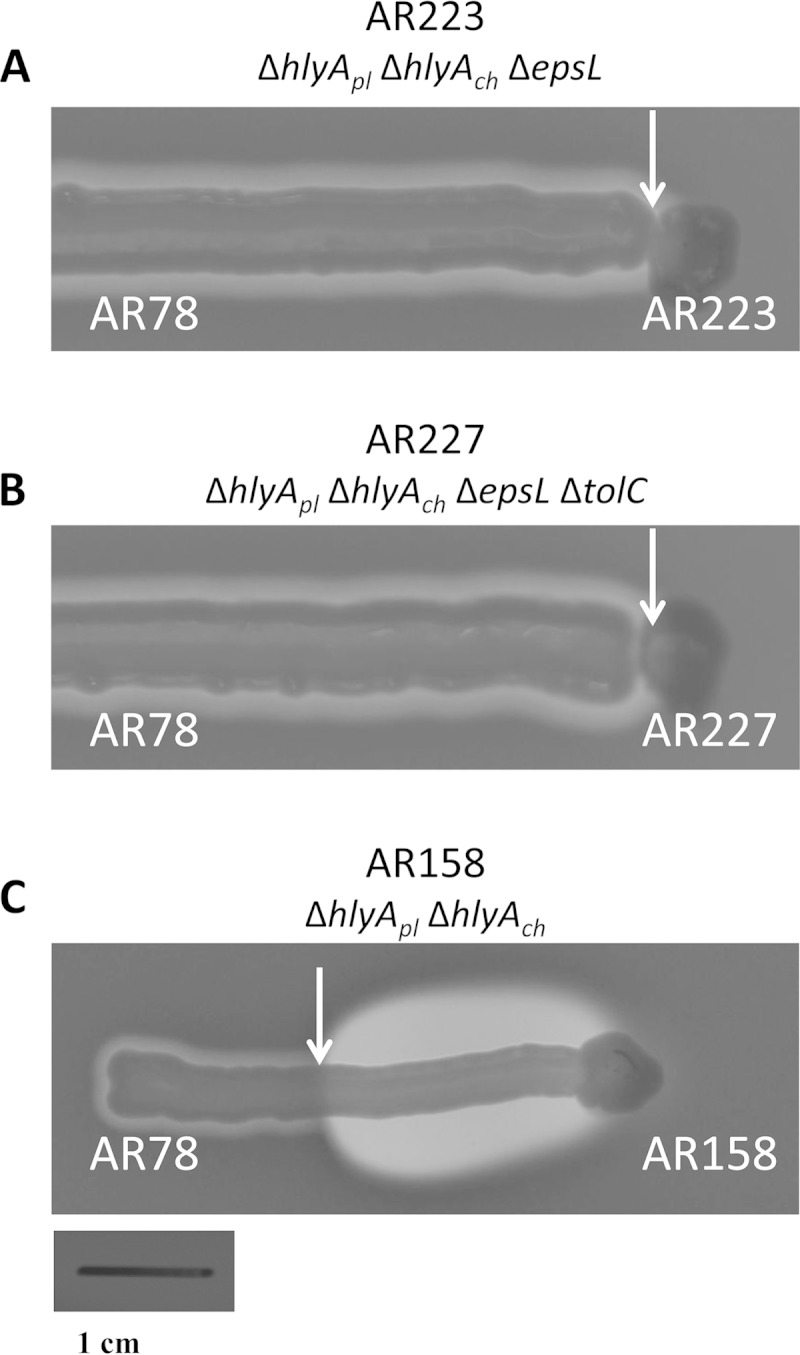
CAMP assays demonstrate that epsL plays a role in Dly secretion. (A) Mutation of epsL abolishes Dly secretion in strain AR223, which is unable to produce CAMP reactions with the indicator strain AR78 (see the lack of hemolytic halo in the zone indicated by a white arrow). (B) Further deletion of tolC in the AR223 mutant (rendering AR227) did not cause any detectable change. (C) Note that a strain producing Dly and with an intact epsL gene (AR158) is able to produce a large halo of hemolysis resultant from the synergistic action of Dly and HlyAch (white arrow depicts the extent of the synergistic halo, characteristic of a CAMP effect). Strains AR223 and AR227 and the positive control AR158 were cultured each in the center of a sheep agar plate with the tip of a rounded wooden pick and left to grow for 3 days. Subsequently, the indicator strain AR78 was streaked radially in a line-shape from the plate edge to the center. Pictures were taken after 15 h of incubation. The genotypes are described below the strain name. Scale bar, 1 cm.
Since the single epsL mutant AR217 still produces an observable hemolytic halo on sheep blood agar (Fig. 2B) and 7 HU in the liquid assay (Fig. 3), we wanted to make sure that this residual hemolysis is not due to a minor contribution of TolC that might have been underestimated in the single tolC mutant tested above. We thus deleted tolC in a double ΔepsL Δdly mutant, as well as in a ΔepsL ΔhlyApl ΔhlyAch triple mutant, in order to evaluate whether TolC is selectively involved in the residual secretion of either the pore-forming HlyA hemolysins or of Dly, respectively. As a result, we did not observe any further decrease of the hemolytic halo in the tolC mutant AR234 (Fig. 2B) or any variation in the CAMP reaction of mutant AR227 with respect to AR223 (Fig. 4B). In addition, the liquid hemolytic assays showed no significant effect of tolC deletion on the HU values of strain AR234 with respect to AR222 (Fig. 3). These observations, together with the results obtained with the single tolC mutant (see above), clearly indicate that the pPHDD1-encoded TolC is not involved in the secretion of P. damselae subsp. damselae hemolysins.
Although the majority of proteins released by T2SS are thought to be in their active forms (16), some proteins are secreted as proforms (34, 35). More pertaining to our study, V. cholerae VCC is secreted as an inactive protoxin that is matured after protease cleavage in the extracellular milieu (36, 37). It thus might be argued that the observed decrease in the hemolytic activity in epsL mutants could be due to a defect of epsL mutants to secrete a factor responsible for maturating the hemolysins, rather than to a defect in secreting the hemolysins themselves. In order to rule out this possibility, we preincubated ECPs from the single ΔepsL mutant AR217 with ECPs from the Δdly ΔhlyAch ΔhlyApl triple mutant AR89 and carried out liquid hemolytic assays. As a result, we did not observe any improvement in the hemolytic activity of the ΔepsL ECPs preincubated with AR89 ECPs (Fig. 3), confirming that the impaired hemolytic activity in ΔepsL mutants is indeed caused by a defect in hemolysin secretion rather than by the lack of an additional factor.
Dly, HlyApl, and HlyAch are predicted to contain N-terminal signal peptides characteristic of proteins released by Sec-dependent secretion.
Characteristic of the T2SS-secreted proteins is the presence of an N-terminal signal peptide (38) that targets them to either the Sec or Tat machinery for transport across the cytoplasmic membrane into the periplasm. Using the SignalP 4.1. program (25), we predicted the presence of N-terminal signal peptides within Dly, HlyApl, and HlyAch and their cleavage positions (Fig. 5). For nonsecretory proteins, all of the scores in the SignalP output should ideally be very low (close to the negative target value of 0.1). The D-score (discrimination score) is the score used to discriminate signal peptides from non-signal peptides. Dly, HlyApl, and HlyAch showed D-scores of 0.795, 0.829, and 0.832, respectively, with a D-score cutoff of 0.340, which clearly suggests that the three hemolysins possess a signal peptide. Confirming this, the SecretomeP program yielded for the three hemolysins a SecP score of 0.94, where scores >0.5 indicate possible secretion. Even though SecretomeP is trained to predict nonclassical secretion, it usually gives high score to proteins entering the classical secretory pathway (39). The predicted cleavage positions for each hemolysin are represented in Fig. 5.
FIG 5.
N-terminal signal peptides of Dly, HlyApl, and HlyAch predicted using the SignalP 4.1. program (25). The predicted cleavage positions for each hemolysin are represented as large uppercase letters in boldface. Leucine residues that suggest a Sec-dependent processing are indicated in boldface. The cleavage site position for Dly was corroborated by N-term Edman degradation.
Proteins to be secreted through the T2SS may cross the inner membrane barrier via either the Tat (twin-arginine) or the Sec systems (19). Sec-targeting signal peptides have leucine and alanine as the most abundant residues. On the other hand, the Tat signal peptides have much higher contents of glycine but less leucine (40). Dly, HlyApl, and HlyAch show signal peptides with leucine/glycine ratios of 2/0, 5/1, and 5/1, respectively. In addition, substrates of the Tat pathway are characterized by the presence of a twin-arginine motif pattern, Z-R-R-x-Φ-Φ, where “Z” represents any polar residue, and “Φ” represents hydrophobic residues (41). Dly, HlyApl, and HlyAch do not contain this motif in their signal peptides (Fig. 5), suggesting that the three P. damselae subsp. damselae hemolysins follow the Sec pathway.
V. cholerae cytolysin (VCC) is a T2SS-secreted toxin (30). The secreted protein needs to be proteolytically processed in order to become active (37). At the amino acid level, HlyA is 50% identical to VCC. Proteolytic cleavage of HlyA after secretion is expected to remove the signal peptide cleavage site, which will preclude its identification in mature toxin. In contrast, phospholipases are not generally thought to undergo proteolytic maturation. Hence, we chose to analyze the N terminus of extracellular Dly by Edman degradation. The resulting sequence (N terminus-F-T-Q-W-G-G-S-C terminus) matches positions 20 through 26 of the amino acid sequence obtained by in silico translation of dly and covers the predicted signal peptide cleavage site of Dly (Fig. 5). The corresponding mature Dly is predicted to comprise 554 amino acids and to have a molecular mass of 63 kDa, thus closely matching the molecular mass of native Dly in our preparation as estimated from SDS-PAGE.
The putative prepilin peptidase PilD plays a role in P. damselae subsp. damselae hemolysin secretion and pilus formation.
EpsG, EpsH, EpsI, and EpsJ are type IV pseudopilins that need to be processed by the prepilin peptidase EpsO to be translocated to the periplasm (17). Most eps operons contain an epsO homologue, although in V. cholerae and V. vulnificus this gene is not present in the T2SS cluster, and it is found (pilD) as part of a type IV pilus biogenesis cluster (42, 43). Since P. damselae subsp. damselae T2SS also lacks the epsO gene (Fig. 1B), we searched in the genome of the type strain ATCC 33539 and found a pilD homologue associated to three genes that constitute a putative pil cluster (pilABCD) (Fig. 1C). We therefore investigated the role that pilD plays in P. damselae subsp. damselae hemolysin secretion by deleting pilD in AR111 (plasmidless) and AR57 (pPHDD1-containing) backgrounds, resulting in the pilD mutants AR240 and AR239, respectively. Hemolytic activity was completely suppressed in the ΔpilD AR240 in blood agar plates (Fig. 2A) and in liquid hemolytic assays (Fig. 3), pointing out that PilD-mediated maturation of prepilin-like components is crucial for proper HlyAch secretion and suggesting that the hemolytic activity previously observed in the ΔepsL mutant AR211 might be due to residual activity of the T2SS secreting small amounts of HlyAch.
Regarding strains carrying pPHDD1, the hemolytic phenotype observed in the ΔpilD mutant AR239 in blood agar plates was similar to that observed in the ΔepsL mutant AR217, but it underwent even a further reduction of activity in liquid hemolytic assays, where it produced only two hemolytic units (Fig. 3). Complementation of AR239 and AR240 with pilD cloned into plasmid pAJR76 restored both the haloes (Fig. 2) and the hemolytic activity values (Fig. 3) of the parental strains.
In a previous work, we found that plasmid pPHDD1 contains a set of tad (tight adherence) genes (10). Homologues of these genes were found to provide adherence and biofilm formation functions by creating pili (44). In Actinobacillus actinomycetemcomitans a prepilin peptidase, TadV, is found as part of the tad cluster and is responsible for maturating tad pseudopilins (45). We found that deletion of tadV in the ΔpilD mutant AR239 (yielding AR252) produced an even less translucent and slightly smaller hemolytic halo on sheep blood agar (Fig. 2B) and led to a complete absence of hemolytic units in liquid assays (Fig. 3). This suggests that TadV might partially substitute for the putative prepilin peptidase function in pilD mutants.
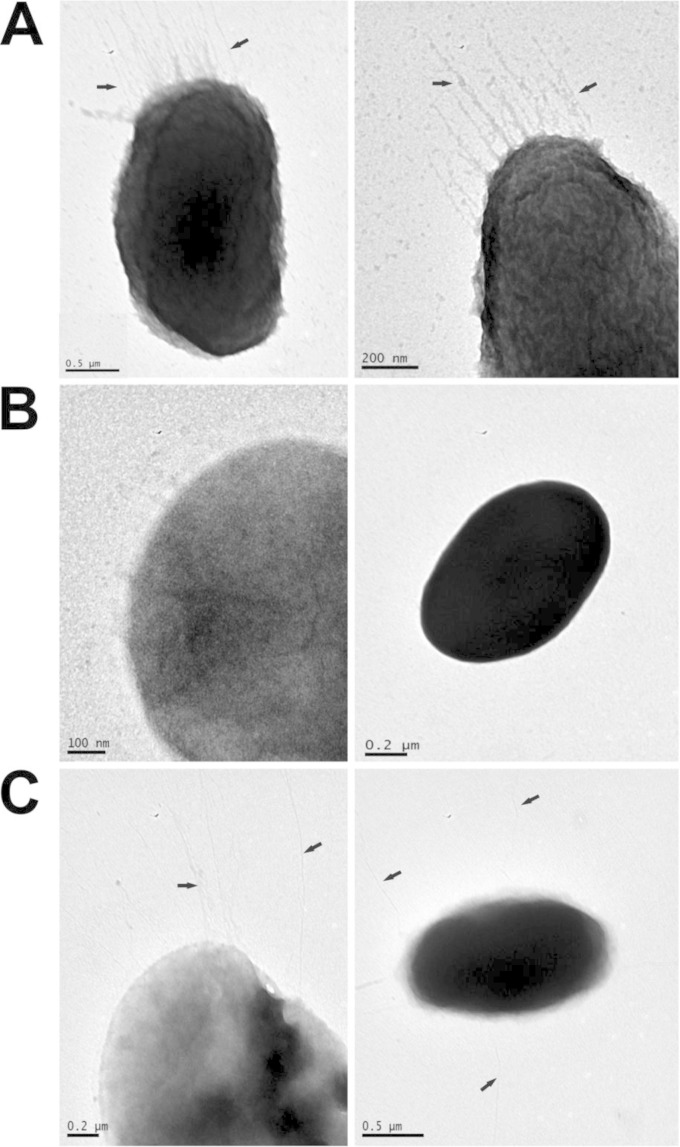
In different bacterial species it has been shown that upon pilD mutation, in addition to blocking T2SS-dependent secretion, the expression of pili on the cell surface is also prevented (43, 46). We investigated whether PilD is involved in P. damselae subsp. damselae pilus biogenesis. TEM revealed in the parental strain AR57 the presence of fine hair-like structures extending from the cell surface that likely corresponded to type IV pili (Fig. 6A). However, pili were not observed on the surface of the ΔpilD mutant strain AR239 (Fig. 6B). Complementation of the ΔpilD mutant with a plasmid containing the pilD gene restored the ability to produce pili (Fig. 6C). The reason for the lack of observable pili in pilD mutants of pPHDD1 strains that bear the tad cluster is unknown and might suggest that the pPHDD1-carried tad cluster does not lead to proper type IV pili formation in P. damselae subsp. damselae. Although V. cholerae pilD and epsL mutants display a severe growth defect and leaky outer-membrane phenotypes (42, 47), P. damselae subsp. damselae pilD and epsL mutants did not show any growth defect (data not shown).
FIG 6.
Transmission electron micrographs of the P. damselae subsp. damselae AR57 strain, pilD mutant derivatives, and complemented mutants, showing the presence or absence of surface pili (indicated by arrows). (A) Parental strain (AR57); (B) pilD mutant (AR239); (C) pilD mutant complemented with pilD gene (AR242). Cells were stained with 2% phosphotungstic acid.
Deletion of epsL and pilD affects epsC and hemolysin gene expression in pPHDD1-harboring strains.
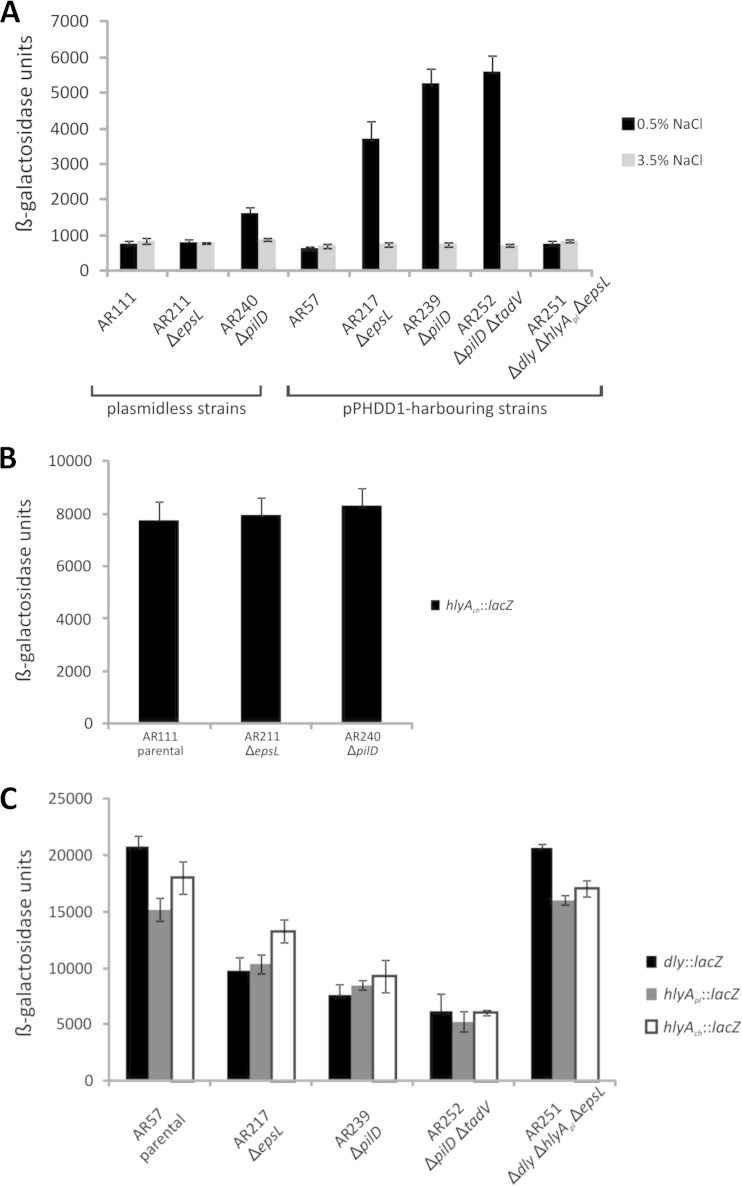
It is known that quorum sensing regulates T2SS genes, as well as the T2SS-secreted substrates in Pseudomonas aeruginosa (48). In contrast, eps secretion genes of V. cholerae were shown to be constitutive under laboratory conditions (49). In an attempt to gain some knowledge on conditions that might affect T2SS expression in P. damselae subsp. damselae, we evaluated changes in promoter expression of the first gene of the cluster, epsC, since P. damselae subsp. damselae T2SS genes all are transcribed from the same strand (Fig. 1B). The epsC expression levels in the parental strains AR111 and AR57 (ca. 800 Miller units) (Fig. 7A) were similar to those observed in other Gram-negative bacteria (50). Interestingly, we observed that under standard LB culture conditions, deletion of epsL in the pPHDD1-harboring strain caused a 6-fold increase in β-galactosidase levels of the epsC::lacZ fusion, but this effect was not observed in the plasmidless strain epsL mutant AR211 (Fig. 7A). Deletion of pilD caused a 2-fold increase in epsC promoter activity in plasmidless strains. In the pPHDD1 strain, an 8.5-fold increase was observed upon both pilD and pilD tadV deletions (Fig. 7A). It is interesting that pPHDD1-harboring derivatives express three hemolysins simultaneously, whereas plasmidless derivatives only express one. Thus, these observations suggest that the defect of the cell to secrete the hemolysins in epsL and pilD mutants leads to an increase in epsC expression (that would alleviate hemolysin accumulation in the cell), and this increase correlates with the predicted degree of hemolysin accumulation.
FIG 7.
Transcriptional activity (β-galactosidase units) of lacZ fusions to the epsC, dly, hlyApl, and hlyAch promoters under different parental and mutant backgrounds. (A) β-Galactosidase units of epsC promoter in strains grown in standard LB medium with 0.5% NaCl and in LB medium with 3.5% NaCl. (B) β-Galactosidase units of hlyAch promoter in plasmidless strain derivatives grown in standard LB medium with 0.5% NaCl. (C) β-Galactosidase units of each of the three hemolysin gene promoters, under different genetic backgrounds derived from the pPHDD1-harboring strain AR57, all grown under standard LB medium conditions. All experiments were carried out in triplicate. Mean values with standard deviations denoted by error bars are shown.
According to this hypothesis, we would expect that conditions that diminish hemolysin expression would also decrease epsC expression. We know from a previous study that NaCl concentration regulates hemolysin genes in P. damselae subsp. damselae in a manner that gene transcription is enhanced under standard LB culture conditions (0.5% NaCl) and diminished under growth at 3.5% NaCl (11). As expected, epsC expression levels at 3.5% NaCl were similar in the parental and mutant strains (Fig. 7A) and similar to the levels shown by the parental strains grown in 0.5% NaCl. These results demonstrate that NaCl concentration, a strong environmental regulator of P. damselae subsp. damselae hemolysins (11), is not a direct regulator of epsC and point to intracellular hemolysin accumulation (or RNA transcripts accumulation) being a signal that affects epsC expression. In accordance with this hypothesis, we observed that converting AR217 into a single hemolysin-producing strain (ΔepsL Δdly ΔhlyApl) (AR251) yielded the same epsC expression values observed both in the pPHDD1 and in the naturally plasmidless parental strains AR57 and AR111 (Fig. 7A). These results together suggest that intracellular hemolysin accumulation might modulate epsC expression.
The hemolysin accumulation hypothesis might fit well with a diminution of hemolysin gene expression in epsL or pilD mutants. To assess this possibility, we analyzed the promoter activity of hlyAch in the plasmidless strains and of dly, hlyApl and hlyAch in the pPHDD1-harboring strains. As a result, we did not observe changes in hlyAch promoter expression in the plasmidless AR211 (ΔepsL) and AR240 (ΔpilD) strains compared to AR111 (parental) (Fig. 7B). However, as expected, we did observe a decrease of promoter activity of hlyAch, hlyApl, and dly in the pPHDD1-harboring ΔepsL (AR217), ΔpilD (AR239), and ΔpilD ΔtadV (AR252) strains with respect to AR57 (parental) (Fig. 7C), confirming that preventing hemolysin secretion causes a diminution in their gene expression.
Most interestingly, in support of the hemolysin accumulation hypothesis, we found that conversion of the ΔepsL (AR217) mutant into a single hemolysin-producing strain by deleting dly and hlyApl (yielding AR251) caused this strain to behave as a parental strain in terms of expression levels of hemolysin genes (Fig. 7C). Thus, reducing the hemolysin gene number in a P. damselae subsp. damselae strain (via hemolysin gene deletion) diminishes the effect (positive in epsC and negative in the three hemolysin genes) that epsL or pilD mutations have on the promoter expression of epsC, dly, hlyApl, and hlyAch genes. Since these effects were observed only under conditions of laboratory-designed deletion mutants, the biological significance of the underlying mechanisms of epsC and hemolysin regulation influenced by epsL or pilD mutations awaits further studies. Recently, it was reported that expression of T2SS in Vibrio cholerae is positively regulated by the alternative sigma factor RpoE (49). The accumulation of hemolysins in P. damselae subsp. damselae mutants might thus induce a stress response governed by RpoE, which would result in the observed elevated transcription of epsC. Similarly, RpoE might be involved in the reduction in hemolysin gene expression we observed in the epsL or pilD mutants. In this regard, it has been recently discovered that inactivation of negative regulators (namely, rseB) of rpoE diminishes hemolytic activity in Vibrio harveyi (51).
Our results thus suggest that, under the conditions tested, T2SS expression in P. damselae subsp. damselae might be constitutive and the regulation of hemolysin production might be implemented by controlling hemolysin gene expression rather that regulating their secretion through T2SS. This constitutive expression of the T2SS can be beneficial for P. damselae subsp. damselae, a bacterium able to live either as a pathogen or as in a free-living style (3), that may change its secretome depending on the environmental requirements. This is the case of V. cholerae T2SS that secretes cholera toxin in the human intestine and chitinase in the aquatic environment (52).
epsL and pilD play a major role in virulence for mice.
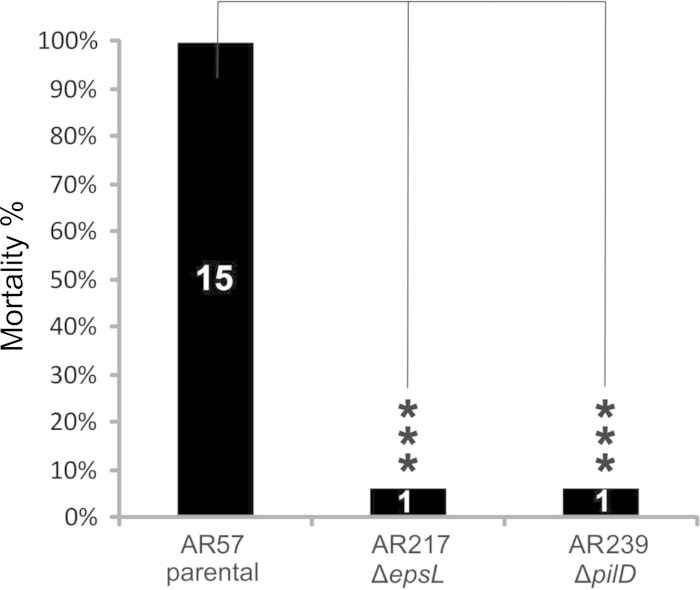
In a previous work we demonstrated that Dly, HlyApl, and HlyAch play a key role in the virulence of P. damselae subsp. damselae for mice (11). Since we have demonstrated that the T2SS is involved in the secretion of these toxins, we wanted to evaluate the contribution of epsL and pilD to P. damselae subsp. damselae virulence. For this purpose, we conducted virulence tests in mice inoculating the highly hemolytic parental strain AR57, as well as its epsL and pilD derivative mutants AR217 and AR239, respectively. Statistically significant differences were observed among the assayed strains (Fig. 8). The parental strain caused death in the 15 animals inoculated, whereas only one death was recorded among animals inoculated with the ΔepsL and ΔpilD single mutants.
FIG 8.
Results of mouse virulence assays with P. damselae subsp. damselae parental strain and epsL and pilD mutants. The results are expressed as mortality percentages. The numbers inside the bars denote the number of dead animals. The results from three independent groups of five animals each per strain tested were pooled and compared using a chi-square test. Asterisks are used to denote statistically significant differences between strains (***, P < 0.001).
Hemorrhage and tissue damage are hallmarks of infections caused by P. damselae subsp. damselae, a highly hemolytic bacterium (1, 5). Since the secretion mutants ΔepsL and ΔpilD are drastically reduced in hemolytic activity, it is thus conceivable that T2SS mutations cause a drastic descent in virulence. However, it is possible that this pathogen may also secrete as-yet-unknown T2SS-dependent virulence factors in addition to Dly, HlyApl, and HlyAch. We present here the first evidence of a secretion system in this pathogen and demonstrate that T2SS plays a major role in P. damselae subsp. damselae virulence.
ACKNOWLEDGMENTS
This study was supported by grant EM2012/043 from Xunta de Galicia, Spain, and by grants AGL2012-39274-C02-01 and AGL2013-48353-R from the Ministry of Economy and Competitiveness (MINECO) of Spain (both cofunded by the FEDER Programme from the European Union).
We acknowledge Manuel Noia (Department of Cellular Biology and Ecology, University of Santiago de Compostela) for support in the processing of samples for TEM and Martina Meyenburg for technical support in processing samples for Edman degradation.
REFERENCES
- 1.Fouz B, Larsen JL, Nielsen B, Barja JL, Toranzo AE. 1992. Characterization of Vibrio damsela strains isolated from turbot Scophthalmus maximus in Spain. Dis Aquat Org 12:155–166. doi: 10.3354/dao012155. [DOI] [Google Scholar]
- 2.Labella A, Sánchez-Montes N, Berbel C, Aparicio M, Castro D, Manchado M, Borrego JJ. 2010. Toxicity of Photobacterium damselae subsp. damselae strains isolated from new cultured marine fish. Dis Aquat Org 92:31–40. doi: 10.3354/dao02275. [DOI] [PubMed] [Google Scholar]
- 3.Rivas AJ, Lemos ML, Osorio CR. 2013. Photobacterium damselae subsp. damselae, a bacterium pathogenic for marine animals and humans. Front Microbiol 4:283. doi: 10.3389/fmicb.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarridge JE, Zighelboim-Daum S. 1985. Isolation and characterization of two hemolytic phenotypes of Vibrio damsela associated with a fatal wound infection. J Clin Microbiol 21:302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser SL, Purcell BK, Delgado B Jr, Baker AE, Whelen AC. 1997. Rapidly fatal infection due to Photobacterium (Vibrio) damsela. Clin Infect Dis 25:935–936. doi: 10.1086/597647. [DOI] [PubMed] [Google Scholar]
- 6.Morris JG, Wilson R, Hollis DG, Weaver RE, Miller HG, Tacket CO, Hickman FW, Blake PA. 1982. Illness caused by Vibrio damsela and Vibrio hollisae. Lancet i:1294–1297. [DOI] [PubMed] [Google Scholar]
- 7.Yamane K, Asato J, Kawade N, Takahashi H, Kimura B, Arakawa Y. 2004. Two cases of fatal necrotizing fasciitis caused by Photobacterium damsela in Japan. J Clin Microbiol 42:1370–1372. doi: 10.1128/JCM.42.3.1370-1372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hundenborn J, Thurig S, Kommerell M, Haag H, Nolte O. 2013. Severe wound infection with Photobacterium damselae ssp. damselae and Vibrio harveyi, following a laceration injury in marine environment: a case report and review of the literature. Case Rep Med 2013:610632. doi: 10.1155/2013/610632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim HR, Kim JW, Lee MK, Kim JG. 2009. Septicemia progressing to fatal hepatic dysfunction in a cirrhotic patient after oral ingestion of Photobacterium damselae: a case report. Infection 37:555–556. doi: 10.1007/s15010-009-9049-8. [DOI] [PubMed] [Google Scholar]
- 10.Rivas AJ, Balado M, Lemos ML, Osorio CR. 2011. The Photobacterium damselae subsp. damselae hemolysins damselysin and HlyA are encoded within a new virulence plasmid. Infect Immun 79:4617–4627. doi: 10.1128/IAI.05436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivas AJ, Balado M, Lemos ML, Osorio CR. 2013. Synergistic and additive effects of chromosomal and plasmid-encoded hemolysins contribute to hemolysis and virulence in Photobacterium damselae subsp. damselae. Infect Immun 81:3287–3299. doi: 10.1128/IAI.00155-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivas AJ, Labella AM, Borrego JJ, Lemos ML, Osorio CR. 2014. Evidence for horizontal gene transfer, gene duplication and genetic variation as driving forces of the diversity of haemolytic phenotypes in Photobacterium damselae subsp. damselae. FEMS Microbiol Lett 355:152–162. doi: 10.1111/1574-6968.12464. [DOI] [PubMed] [Google Scholar]
- 13.Kreger AS. 1984. Cytolytic activity and virulence of Vibrio damsela. Infect Immun 44:326–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kreger AS, Bernheimer AW, Etkin LA, Daniel LW. 1987. Phospholipase D activity of Vibrio damsela cytolysin and its interaction with sheep erythrocytes. Infect Immun 55:3209–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christie R, Atkins NE, Munch-Petersen E. 1944. A note on a lytic phenomenon shown by group B streptococci. Aust J Exp Biol Med Sci 22:197–200. doi: 10.1038/icb.1944.26. [DOI] [PubMed] [Google Scholar]
- 16.Sandkvist M. 2001. Type II secretion and pathogenesis. Infect Immun 69:3523–3535. doi: 10.1128/IAI.69.6.3523-3535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campos M, Cisneros DA, Nivaskumar M, Francetic O. 2013. The type II secretion system: a dynamic fiber assembly nanomachine. Res Microbiol 164:545–555. doi: 10.1016/j.resmic.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Nivaskumar M, Francetic O. 2014. Type II secretion system: a magic beanstalk or a protein escalator. Biochim Biophys Acta 1843:1568–1577. doi: 10.1016/j.bbamcr.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 19.Voulhoux R, Ball G, Ize B, Vasil ML, Lazdunski A, Wu LF, Filloux A. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J 20:6735–6741. doi: 10.1093/emboj/20.23.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korotkov KV, Sandkvist M, Hol WG. 2012. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol 10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrero M, de Lorenzo V, Timmis KN. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol 172:6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mouriño S, Osorio CR, Lemos ML. 2004. Characterization of heme uptake cluster genes in the fish pathogen Vibrio anguillarum. J Bacteriol 186:6159–6167. doi: 10.1128/JB.186.18.6159-6167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu PV. 1957. Survey of hemolysin production among species of pseudomonads. J Bacteriol 74:718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernheimer AW. 1988. Assay of hemolytic toxins. Methods Enzymol 165:213–217. doi: 10.1016/S0076-6879(88)65033-6. [DOI] [PubMed] [Google Scholar]
- 25.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 26.Parales RE, Harwood CS. 1993. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for Gram− bacteria. Gene 133:23–30. doi: 10.1016/0378-1119(93)90220-W. [DOI] [PubMed] [Google Scholar]
- 27.Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 28.Fouz B, Toranzo AE, Biosca EG, Mazoy R, Amaro C. 1994. Role of iron in the pathogenicity of Vibrio damsela for fish and mammals. FEMS Microbiol Lett 121:181–188. doi: 10.1111/j.1574-6968.1994.tb07097.x. [DOI] [PubMed] [Google Scholar]
- 29.Schulein R, Gentschev I, Schlor S, Gross R, Goebel W. 1994. Identification and characterization of two functional domains of the hemolysin translocator protein HlyD. Mol Gen Genet 245:203–211. [DOI] [PubMed] [Google Scholar]
- 30.Sikora AE, Zielke RA, Lawrence DA, Andrews PC, Sandkvist M. 2011. Proteomic analysis of the Vibrio cholerae type II secretome reveals new proteins, including three related serine proteases. J Biol Chem 286:16555–16566. doi: 10.1074/jbc.M110.211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandkvist M, Hough LP, Bagdasarian MM, Bagdasarian M. 1999. Direct interaction of the EpsL and EpsM proteins of the general secretion apparatus in Vibrio cholerae. J Bacteriol 181:3129–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray MD, Bagdasarian M, Hol WG, Sandkvist M. 2011. In vivo cross-linking of EpsG to EpsL suggests a role for EpsL as an ATPase-pseudopilin coupling protein in the type II secretion system of Vibrio cholerae. Mol Microbiol 79:786–798. doi: 10.1111/j.1365-2958.2010.07487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang F, Waterfield NR, Yang J, Yang G, Jin Q. 2014. A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe 15:600–610. doi: 10.1016/j.chom.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 34.Braun P, Tommassen J, Filloux A. 1996. Role of the propeptide in folding and secretion of elastase of Pseudomonas aeruginosa. Mol Microbiol 19:297–306. doi: 10.1046/j.1365-2958.1996.381908.x. [DOI] [PubMed] [Google Scholar]
- 35.Hase CC, Finkelstein RA. 1991. Cloning and nucleotide sequence of the Vibrio cholerae hemagglutinin/protease (HA/protease) gene and construction of an HA/protease-negative strain. J Bacteriol 173:3311–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto K, Ichinose Y, Shinagawa H, Makino K, Nakata A, Iwanaga M, Honda T, Miwatani T. 1990. Two-step processing for activation of the cytolysin/hemolysin of Vibrio cholerae O1 biotype El Tor: nucleotide sequence of the structural gene (hlyA) and characterization of the processed products. Infect Immun 58:4106–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valeva A, Walev I, Weis S, Boukhallouk F, Wassenaar TM, Endres K, Fahrenholz F, Bhakdi S, Zitzer A. 2004. A cellular metalloproteinase activates Vibrio cholerae pro-cytolysin. J Biol Chem 279:25143–25148. doi: 10.1074/jbc.M313913200. [DOI] [PubMed] [Google Scholar]
- 38.von Heijne G. 1986. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res 14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. 2004. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel 17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 40.Cristobal S, de Gier JW, Nielsen H, von Heijne G. 1999. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J 18:2982–2990. doi: 10.1093/emboj/18.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Natale P, Brüser T, Driessen AJM. 2008. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane-distinct translocases and mechanisms. Biochim Biophys Acta 1778:1735–1756. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 42.Marsh JW, Taylor RK. 1998. Identification of the Vibrio cholerae type 4 prepilin peptidase required for cholera toxin secretion and pilus formation. Mol Microbiol 29:1481–1492. doi: 10.1046/j.1365-2958.1998.01031.x. [DOI] [PubMed] [Google Scholar]
- 43.Paranjpye RN, Strom MS. 2005. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect Immun 73:1411–1422. doi: 10.1128/IAI.73.3.1411-1422.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clock SA, Planet PJ, Pérez BA, Figurski DH. 2008. Outer membrane components of the Tad (tight adherence) secreton of Aggregatibacter actinomycetemcomitans. J Bacteriol 190:980–990. doi: 10.1128/JB.01347-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomich M, Fine DH, Figurski DH. 2006. The TadV protein of Actinobacillus actinomycetemcomitans is a novel aspartic acid prepilin peptidase required for maturation of the Flp1 pilin and TadE and TadF pseudopilins. J Bacteriol 188:6899–6914. doi: 10.1128/JB.00690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bally M, Filloux A, Akrim M, Ball G, Lazdunski A, Tommassen J. 1992. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol Microbiol 6:1121–1131. doi: 10.1111/j.1365-2958.1992.tb01550.x. [DOI] [PubMed] [Google Scholar]
- 47.Sikora AE, Lybarger SR, Sandkvist M. 2007. Compromised outer membrane integrity in Vibrio cholerae type II secretion mutants. J Bacteriol 189:8484–8495. doi: 10.1128/JB.00583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chapon-Herve V, Akrim M, Latifi A, Williams P, Lazdunski Bally AM. 1997. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol Microbiol 24:1169–1178. doi: 10.1046/j.1365-2958.1997.4271794.x. [DOI] [PubMed] [Google Scholar]
- 49.Zielke RA, Simmons RS, Park BR, Nonogaki M, Emerson S, Sikora AE. 2014. The type II secretion pathway in Vibrio cholerae is characterized by growth-phase dependent expression of exoprotein genes and is positively regulated by sigmaE. Infect Immun 82:2788–2801. doi: 10.1128/IAI.01292-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Putker F, Tommassen-van Boxtel R, Stork M, Rodríguez-Herva JJ, Koster M, Tommassen J. 2013. The type II secretion system (Xcp) of Pseudomonas putida is active and involved in the secretion of phosphatases. Environ Microbiol 15:2658–2671. doi: 10.1111/1462-2920.12115. [DOI] [PubMed] [Google Scholar]
- 51.Rattanama P, Thompson JR, Kongkerd N, Srinitiwarawong K, Vuddhakul V, Mekalanos JJ. 2012. Sigma E regulators control hemolytic activity and virulence in a shrimp pathogenic Vibrio harveyi. PLoS One 7:e32523. doi: 10.1371/journal.pone.0032523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sikora AE. 2013. Proteins secreted via the type II secretion system: smart strategies of Vibrio cholerae to maintain fitness in different ecological niches. PLoS Pathog 9:e1003126. doi: 10.1371/journal.ppat.1003126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Le Roux F, Binesse J, Saulnier D, Mazel D. 2007. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol 73:777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vieira J, Messing J. 1987. Production of single-stranded plasmid DNA. Methods Enzymol 153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 55.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. doi: 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 56.Silva-Rocha R, Martínez-García E, Calles B, Chavarria M, Arce-Rodríguez A, de Las Heras A, Páez-Espino AD, Durante-Rodríguez G, Kim J, Nikel PI, Platero R, de Lorenzo V. 2013. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res 41:D666–D675. doi: 10.1093/nar/gks1119. [DOI] [PMC free article] [PubMed] [Google Scholar]