Abstract
Infection and lysogenic conversion with Shiga toxin-encoding bacteriophages (Stx phages) drive the emergence of new Shiga toxin-producing Escherichia coli strains. Phage attachment to the bacterial surface is the first stage of phage infection. Envelope perturbation causes activation of envelope stress responses in bacterial cells. Although many external factors are known to activate envelope stress responses, the role of these responses in the phage-bacterium interaction remains unexplored. Here, we investigate the link between three envelope signaling systems in E. coli (RcsBC, CpxAR, and BaeSR) and Stx2 phage infection by determining the success of bacterial lysogenic conversion. For this purpose, E. coli DH5α wild-type (WT) and mutant strains lacking RcsBC, CpxAR, or BaeSR signaling systems were incubated with a recombinant Stx2 phage (933W). Notably, the number of lysogens obtained with the BaeSR mutant was 5 log10 units higher than with the WT, and the same differences were observed when using 7 different Stx2 phages. To assess whether the membrane receptor used by Stx phages, BamA, was involved in the differences observed, bamA gene expression was monitored by reverse transcription-quantitative PCR (RT-qPCR) in all host strains. A 4-fold-higher bamA expression level was observed in the BaeSR mutant than in the WT strain, suggesting that differential expression of the receptor used by Stx phages accounted for the increase in the number of lysogenization events. Establishing the link between the role of stress responses and phage infection has important implications for understanding the factors affecting lysogenic conversion, which drives the emergence of new pathogenic clones.
INTRODUCTION
Temperate phages are responsible for the conversion of nonpathogenic strains of bacteria to pathogenic strains by transduction of virulence genes (1). Well-known examples are Shiga toxin (Stx) phages, which carry the genes for Shiga toxin (stx). Stx phages can cause the conversion of Escherichia coli and bacteria of other genera to Shiga toxin-producing E. coli (STEC) and other Shiga-toxigenic pathogens (2). A recent example is the STEC outbreak in Germany in May 2011, where an enteroaggregative E. coli O104:H4 strain incorporated an Stx2 phage (3, 4). Because the lysogeny of E. coli by Stx phages has relevant implications for their pathogenicity, a better understanding of the molecular mechanisms underlying phage-host interaction is a challenge for researchers and could help in designing strategies to avoid lysogenization by Stx phages.
The first step in phage infection is attachment to the specific receptor on the bacterial surface. The maintenance, adaptation, and protection of the bacterial envelope are essential for bacteria. Stresses that damage and alter the bacterial envelope can lead to activation of stress responses in bacteria by means of various phosphorelay signaling systems (5). In E. coli, five different envelope stress responses have been identified: BaeSR, CpxAR, RcsBC, Psp, and σE (5, 6). Each response triggers a signaling cascade that leads to the regulation of factors needed to combat envelope damage or to respond to the stress (5). Some envelope stress response signaling has been linked to virulence in Gram-negative bacteria (7, 8). For instance, the RcsBC signaling system enhances the locus of enterocyte effacement (LEE) and the adherence phenotype of the STEC O157:H7 isolate TW14359, the isolate responsible for a 2006 outbreak in the United States (8). CpxAR is involved in bacterial adhesion (6, 7) and is activated during prophage induction in E. coli (9). Finally, BaeSR, which mediates increased resistance to toxic compounds (10), is related to envelope biogenesis (10) and could regulate the expression of surface elements (10). Moreover, BaeSR activates the promoter of the casA gene, which is part of the CRISPR/Cas response, conferring resistance against phages in bacteria and archaea (11, 12). BaeSR consists of a histidine sensor kinase (BaeS) residing in the inner membrane and a response regulator (BaeR) in the cytoplasm (6, 10).
We hypothesized that by affecting the phage-bacterium interaction, envelope stress responses might impact the phage lysogenic conversion, but this possibility has not been well explored. This study evaluates three different membrane stress responses and their involvement in lysogenic conversion with Stx phages, which have been directly implicated in the emergence of new pathogenic STEC strains.
MATERIALS AND METHODS
Phages, plasmids, and bacterial strains.
933W is a reference phage isolated from a STEC strain (13), and the other seven Stx2 phages were induced from STEC strains isolated from cattle (14). All the phages were labeled with a cat gene (from plasmid pKD3) (Table 1) inserted into the stx2 gene (15) (stx2::cat phages) (Table 1) and were used to assess lysogenic conversion of E. coli DH5α (Table 1), a recA-negative E. coli K-12 derivative. The plasmids used to generate the mutant strains and the recombinant phages or to clone the baeSR operon for the complementation assay are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | Laboratory strain K-12, recA negative | Gibco-BRL |
| E. coli DH5αΔbaeSR::tet | Strain where BaeSR has been replaced by tet | 19 |
| E. coli DH5αΔbaeSR::tet(pGEM::baeSR) | DH5αΔbaeSR complemented with baeSR | This study |
| E. coli DH5αΔbaeSR::tet(pGEM) | DH5αΔbaeSR complemented with pGEM-T Easy without baeSR | This study |
| E. coli C600 (933W) | Lysogen with phage 933W | 13 |
| E. coli C600 (933WΔstx2::cat) | Lysogen with recombinant phage 933W | 15 |
| E. coli C600 (ΦA9Δstx2::cat) | Lysogen with recombinant phage ΦA9 | 15 |
| E. coli DH5α (ΦVTB55Δstx2::cat) | Lysogen with recombinant phage ΦVTB55 | 15 |
| E. coli DH5α (ΦA75Δstx2::cat) | Lysogen with recombinant phage ΦA75 | 15 |
| E. coli DH5α (ΦA312Δstx2::cat) | Lysogen with recombinant phage ΦA312 | 15 |
| E. coli DH5α (ΦA534Δstx2::cat) | Lysogen with recombinant phage ΦA534 | 15 |
| E. coli DH5α (ΦA549Δstx2::cat) | Lysogen with recombinant phage ΦA549 | 15 |
| E. coli DH5α (ΦA557Δstx2::cat) | Lysogen with recombinant phage ΦA557 | 15 |
| Plasmids | ||
| pKD46 | Containing Red recombinase system | GenBank AY048746; 18 |
| pKD3 | Source of chloramphenicol resistance gene (cat) | GenBank AY048742; 18 |
| pACYC184 | Source of tetracycline resistance gene (tet) | GenBank X06403; 38 |
| pGEM-T Easy | High-copy-number plasmid | Promega |
| pGEM-T Easy::baeSR | Containing baeSR gene | This study |
The bacterial strains were grown in Luria-Bertani (LB) broth and on LB agar. When necessary, media were supplemented with chloramphenicol (Cm) (15 μg/ml), tetracycline (Tc) (10 μg/ml), and ampicillin (Ap) (100 μg/ml) (Sigma-Aldrich, Steinheim, Germany).
Preparation of phage lysate.
LB cultures of the lysogens containing recombinant phages with the stx2::cat construct were grown at an optical density at 600 nm (OD600) of 0.5. Mitomycin C was added to a final concentration of 0.5 μg/ml, and the cultures were incubated for 7 h at 37°C in the dark. After incubation, the cultures were filtered through low-protein-binding 0.22-μm-pore-size membrane filters (Millex-GP; Millipore, Bedford, MA). Infectious recombinant phages were counted by the double-agar-layer method using the E. coli DH5α strain as the host, followed by hybridization with the cat-Dig probe as described previously (15). The nonrecombinant reference phage 933W (containing the intact stx2 gene) was also used in phage induction experiments.
Lysogenization by Stx2 phages.
Suspensions of eight recombinant Stx2 phages containing 106 PFU/ml, obtained and enumerated as described above, were used to lysogenize the host strains (Table 1). One milliliter of the phage suspension was incubated with 1 ml of mid-exponential-phase cultures (OD600 = 0.3) of each host strain and incubated at 37°C for different time intervals. After incubation, the mixture was decimally diluted, plated in LB agar plus Cm, and incubated at 37°C for 18 h. The colonies obtained were confirmed as lysogens by PCR using the primer combination UP 378/Cm-3 (see Table S1 in the supplemental material).
Evaluation of bacterial growth rates.
The growth rates of all the strains were monitored by optical density (OD600) and by colony counts on LB agar supplemented with the appropriate antibiotics at different time intervals. The exponential growth rate (μ) was calculated in three independent experiments by the following formula: μ = (ln Nt − ln N0)/(t − t0), where Nt is the number of cells (CFU) at a given time, N0 is the initial number of cells (CFU), and t is the time in hours.
PCR techniques.
PCRs were performed with a GeneAmp PCR system 2400 (Life Technologies, Barcelona, Spain). Phage and bacterial DNA was extracted as previously described (14) and used for the endpoint PCR amplification or for the stx2 quantitative PCR (qPCR) (16) with the oligonucleotides listed in Table S1 in the supplemental material.
RNA isolation and real-time quantitative PCR.
Total RNA was extracted from 1 ml cells grown to log phase (OD600 = 0.6) at 37°C using the SV Total RNA Isolation System (Promega) following the manufacturer's instructions. An 8.5-μl volume of the RNA extracted was used for each qPCR assay (E. coli 16S rRNA genes or bamA) (see Table S1 in the supplemental material); reverse transcription (RT) to cDNA and DNA amplification were carried out in a one-step reaction using the Power SYBR green RNA-to-CT 1-Step Kit (Life Technologies), following the manufacturer's protocol. Amplification was done under standard conditions in a StepOne Real Time PCR System (Life Technologies) in a final volume of 20 μl per reaction. Relative quantification was evaluated using the comparative threshold cycle (CT) method (17), for which the CT obtained with the qPCR assay of the 16S rRNA genes of E. coli (see Table S1 in the supplemental material) was used as an internal control for normalization of the data. The relative fold changes in transcript levels and standard deviations were calculated as the means of the results of three independent experiments.
Gene knockouts and complementation assay for the three envelope stress responses.
E. coli strain DH5α was used to generate three mutants in BaeSR, RcsBC, or CpxAR envelope stress responses by replacing the genes that encode those proteins with a tet cassette from plasmid pACYC184 (Table 1), using the Red recombinase method (15, 18, 19).
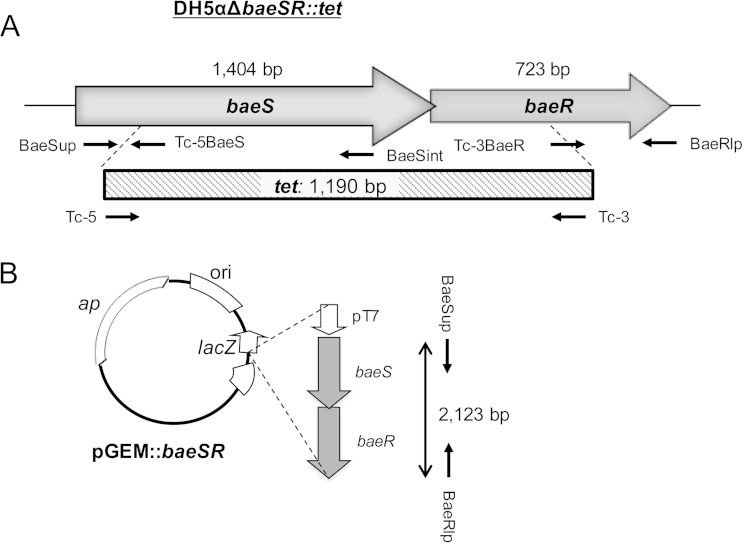
For complementation assays, E. coli DH5αΔbaeSΔbaeR::tet (here referred to as DH5αΔbaeSR) (Fig. 1) was complemented with the pGEM-T Easy vector containing baeSR, a 2,123-bp fragment obtained with primers BaeSup/BaeRlp (Fig. 1). DH5αΔbaeSR transformed with the pGEM-T Easy vector without a BaeSR fragment was used as a control. Insertion of the fragment and cloning in DH5α electrocompetent cells were performed following the manufacturer's instructions. The presence of the construct in colonies grown in LB agar plus Tc was confirmed by conventional PCR and sequencing.
FIG 1.
Generation of the BaeSR mutant in E. coli DH5α. (A) Positions and directions of transcription of the genes making up the BaeSR pathway in the host strains and the deletions used to generate the mutant DH5αΔbaeSΔbaeR::tet. (B) The pGEM plasmid construct containing the baeSR fragment used for complementation experiments is cloned under the T7 RNA polymerase promoter (pT7).
Statistical analyses.
Data and statistical tests used the Statistical Package for Social Science software (SPSS). The differences between the numbers of lysogens obtained with the different strains and under different conditions were evaluated by one-way analysis of variance (ANOVA).
RESULTS
Role of envelope stress responses in the generation of Stx2 lysogens.
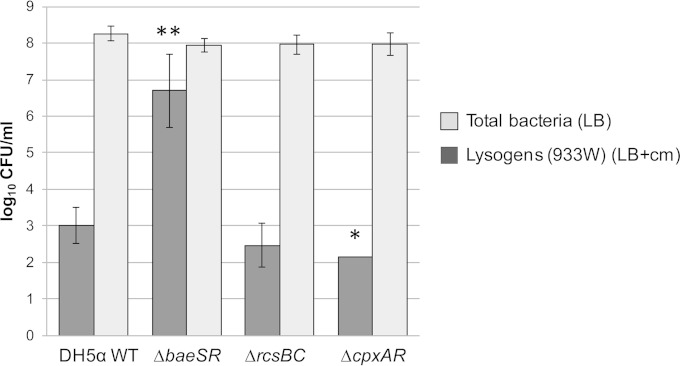
To evaluate the role of envelope stress responses in the generation of Stx2 lysogens, DH5α mutant strains lacking the envelope stress response signaling system BaeSR, RcsBC, or CpxAR were used as hosts for phages. The number of lysogens obtained with phage 933WΔstx2::cat was compared with the WT DH5α number. Mutations on RcsBC did not significantly (P = 0.319) alter the number of lysogens compared with the WT (Fig. 2). Deletion of CpxAR caused a significant (P = 0.049) reduction in the number of Stx2 lysogens compared with the WT strain. In contrast, a significant (P = 0.004) increase in the generation of lysogens was observed when using DH5αΔbaeSR (Fig. 2). The total counts for each mutant strain obtained in LB agar without antibiotic indicated that these differences were not caused by differential growth rates of the strains (Fig. 2). Between 10 and 20% of the lysogens of 933WΔstx2::cat phage grown on LB-plus-Cm agar plates were further confirmed by PCR.
FIG 2.
Roles of BaeSR, RcsBC, and CpxAR membrane stress responses in the generation of lysogens. Generation of lysogens of phage 933WΔstx2::cat was compared in the WT strain E. coli DH5α, DH5αΔbaeSR, DH5αΔrcsBC, and DH5αΔcpxAR. Total bacterial counts of each strain were obtained in LB agar without antibiotic selection. Lysogens of the 933WΔstx2::cat recombinant phage in each strain were obtained in LB agar plus Cm. *, P < 0.05; **, P < 0.01 (significant differences from the WT). The error bars indicate standard deviations.
BaeSR protects against lysogeny with the Stx2 phage 933WΔstx2::cat.
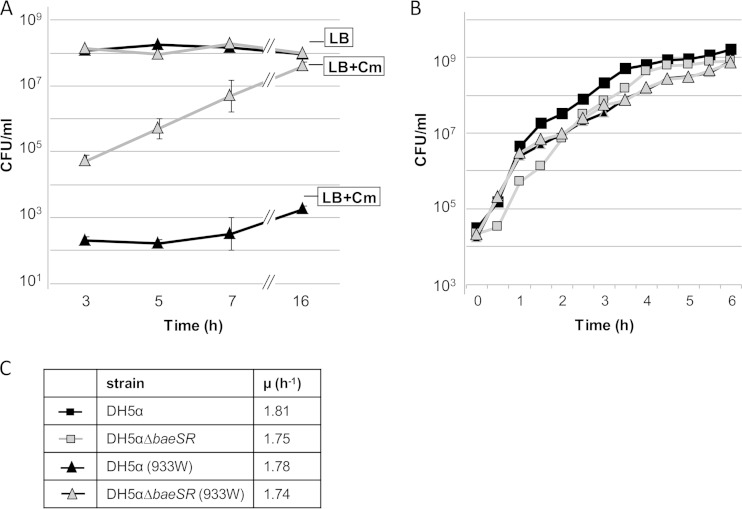
Lysogenic conversion obtained with phage 933WΔstx2::cat (from a phage lysate containing 5 × 106 PFU/ml) was further evaluated at different times of phage-host bacterium contact (Fig. 3A). Lysogens were detected 3 h after DH5αΔbaeSR was incubated with Stx2 phages. An increase in lysogens was observed over time and was most likely due to duplication in the lysogenic population, which was also augmented with new lysogenic cells. The total numbers of cells (measured without antibiotic selection) in WT and DH5αΔbaeSR cultures were the same, which confirms that the effect observed was not due to lysis of the host strain (Fig. 3B).
FIG 3.
(A) Effect of baeSR deletion on lysogenization of recombinant Stx2 phage 933WΔstx2::cat. In every period, bacterial cells were plated in LB agar plus Cm for detection of lysogenic strains or in LB agar without antibiotic for detection of the total number of cells (CFU/ml). (B) Representative experiment on the growing curves of the E. coli DH5α and BaeSR mutant host strains and in lysogens DH5α (933WΔstx2::cat) and DH5αΔbaeSR (933WΔstx2::cat). The symbols are defined in panel C. (C) Growth rates of all the strains calculated with data from panel B. The error bars indicate standard deviations.
To compare the effects of the mutations on bacterial fitness, we calculated the exponential growth rate (μ) of the WT and DH5αΔbaeSR strains and their respective 933W lysogens. Strains lacking the BaeSR envelope stress response had slightly reduced fitness compared to the WT (Fig. 3C). However, differences in the growth rates could not account for the lower number of lysogens obtained with the WT, because the WT showed growth equal to or slightly faster than that of the mutant.
Induction of phage 933W from DH5αΔbaeSR and DH5α lysogens.
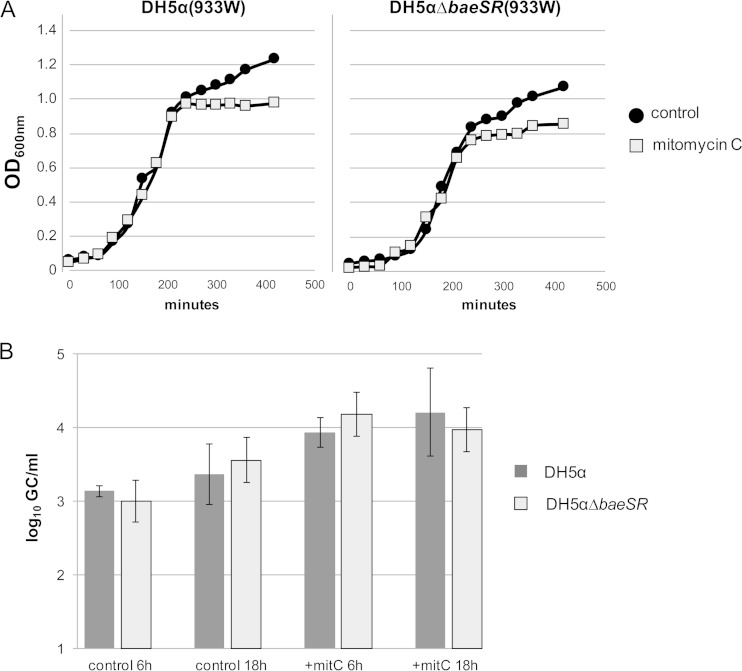
One plausible explanation for the results obtained could be that the WT lysogens and the BaeSR mutant lysogens had different phage induction abilities that could cause differential lysis of the lysogenic cells, thereby influencing the final numbers. To monitor the induction abilities of the lysogens, 933W phage lysogens were obtained after 18 h of incubation with DH5α and DH5αΔbaeSR. In these experiments, a nonrecombinant 933W phage was used, in order to apply the stx2 qPCR-based protocol for phage counting (16). Evaluation of the densities (OD600) of both cultures after mitomycin C treatment (time zero) showed similar patterns for the mutant and the WT strains (Fig. 4A). At 6 and 18 h and in the presence/absence of mitomycin C, no significant differences (under all conditions, P values were >0.05) in the number of phages produced by the mutant and WT strains were observed (Fig. 4B). The numbers of phages produced were the same for both strains. Our results show that differences in phage induction, and hence in the lysis of the lysogens, were not the cause of the higher number of lysogens obtained with the mutant strain.
FIG 4.
Evaluation of 933W phage induction in lysogens DH5α (933W) and DH5αΔbaeSR (933W) with and without mitomycin C induction. (A) Densities of the cultures monitored by OD600. (B) qPCR evaluation of 933W phage particles (no. of gene copies/ml) at 6 and 18 h with or without mitomycin C (mitC) treatment. The error bars indicate standard deviations.
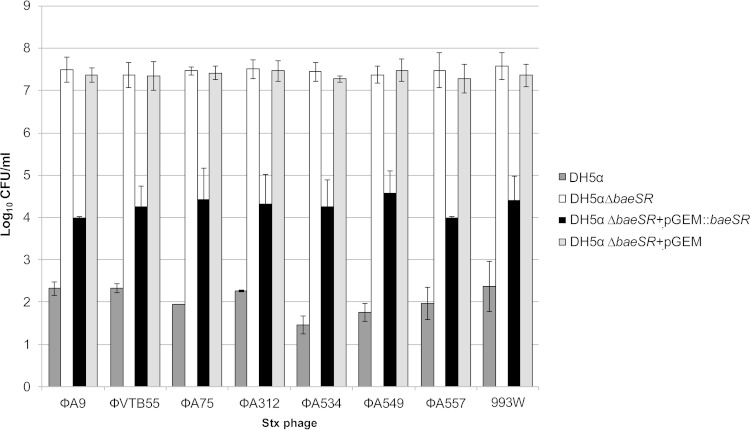
Role of BaeSR in lysogenic conversion with different Stx2 phages.
Stx phages are a very heterogeneous group defined by the presence of stx in their genomes (2). For this reason, we wanted to assess whether the effect of BaeSR on the lysogenic conversion observed for phage 933W could also occur with other Stx2 phages. To do this, seven distinct recombinant Stx2 phages having the stx2 gene truncated by a cat gene (Table 1) were used. The Stx phages were used to obtain lysogens in the host strains DH5α and DH5αΔbaeSR. To confirm the role of BaeSR, DH5αΔbaeSR complemented with pGEM::baeSR was also included as a host strain. DH5αΔbaeSR with a pGEM vector without an insert was used as a negative control. The Stx2 phages used in this study differed genetically from each other, and some also had different insertion sites (14, 20). Phage lysates containing 107 PFU/ml were used for at least three to five independent assays.
The effect of BaeSR on the generation of lysogens was consistent for all Stx2 phages. The DH5αΔbaeSR strain showed at least 5 log10 units more lysogens than the WT (Fig. 5). The role of the baeSR deletion in the number of lysogens was confirmed by baeSR complementation, with the resultant strain showing significantly (P = 0.0001) fewer lysogens, 2.8 to 3.5 log10 units less than the mutant strain (Fig. 5). The number of lysogens obtained with DH5αΔbaeSR containing the pGEM-T Easy vector without an insert (an average of 7.3 log10 lysogens/ml for all phages) did not reveal significant differences (P = 0.394) from the noncomplemented DH5αΔbaeSR strain.
FIG 5.
Generation of lysogens with various Stx recombinant phages in DH5α and DH5αΔbaeSR and complementation of DH5αΔbaeSR pGEM::baeSR. Lysogens of DH5α WT and DH5αΔbaeSR were grown in LB agar plus Cm. Lysogens of DH5αΔbaeSR complemented with pGEM::baeSR and control DH5αΔbaeSR with pGEM were grown in LB agar plus Cm plus Ap. The error bars indicate standard deviations.
We confirmed that no changes in the prophage insertion site could account for the differences observed. The insertion sites of the phages used in this study (wrbA for all phages except ϕA9, which inserts in yecE) (20) were confirmed to be the same in the WT and in DH5αΔbaeSR.
Reverse transcription-quantitative PCR assay of bamA expression.
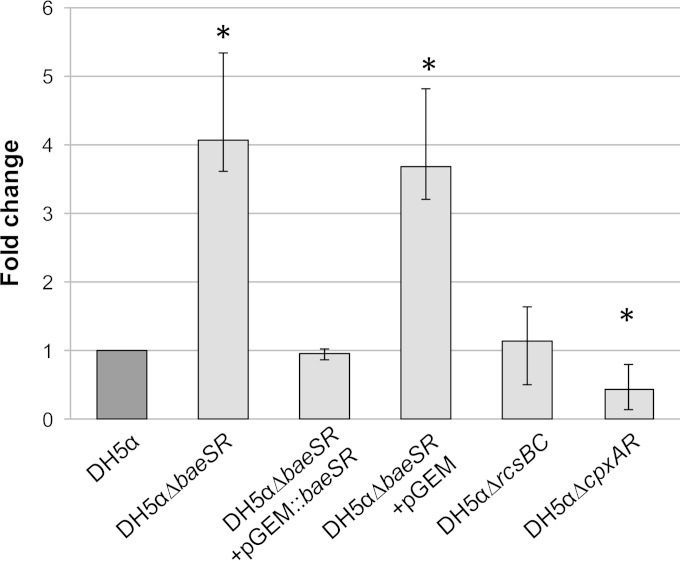
BaeSR is involved in cell envelope stress responses (10). These responses often result in the alteration of proteins in the cell envelope that may be involved in phage attachment to the bacterial surface. BamA (also called YaeT) has been reported to be the receptor for short-tailed Stx phages (21, 22). If BaeSR affects the expression of BamA, this may have affected phage attachment and hence lysogenic conversion, thus providing an explanation for the difference in the numbers of Stx phage lysogens observed between the WT and DH5αΔbaeSR.
To explore this possibility, we determined the expression levels of bamA mRNA in the different host strains by RT-qPCR. The results obtained for each strain were normalized by expression of the 16S rRNA genes of all the strains. The relative expression was recorded as fold changes with respect to the WT. The bamA gene expression in DH5αΔbaeSR was 4-fold higher than in the WT (Fig. 6), representing a significant increase (P = 0.005). bamA expression dropped again to WT levels (P = 0.552) when the strain was complemented with intact baeSR (Fig. 6). Significant changes (P = 0.002) in bamA expression were again observed in DH5αΔbaeSR containing the pGEM-T Easy plasmid without an insert (Fig. 6). Unlike DH5αΔbaeSR, the strains containing deletions of the cpxAR and rcsBC pathways showed no increase in bamA expression. While no significant (P = 0.372) changes in bamA expression were observed for the DH5αΔrcsBC mutant, the levels of expression in the DH5αΔcpxAR mutant were significantly (P = 0.002) lower than in the WT (Fig. 6).
FIG 6.
Relative fold change of bamA expression in the mutant strains (light gray) relative to the WT strain (dark gray) using quantitative real-time RT-PCR analysis. Quantification of bamA RNA transcripts was performed in triplicate, and the expression of E. coli 16S rRNA genes was used as a reference to normalize the results for each strain. The fold change is shown on a logarithmic scale. *, P > 0.01 (significant differences from the WT). The error bars indicate standard deviations.
DISCUSSION
The role of Stx phages in the pathogenicity of E. coli has been widely demonstrated (23, 24). Lysogenization of a susceptible host strain can result in the emergence of new, dangerous strains, as recently occurred in Germany (25, 26). Stx is not the only virulence factor in STEC, but it does seem, in addition to the colonization abilities of a given strain, to play a key role in the successful development of disease.
Signal transduction pathways control the adaptive response to envelope stress in E. coli (10) and could influence the virulence phenotypes. We explored in this study how three different envelope stress responses in STEC affect Stx phage lysogenization. Among the three pathways examined, only BaeSR enhanced Stx2 phage lysogenic conversion. RcsBC failed to show a clear influence, while the CpxAR mutation showed a slight but significant reduction in the number of lysogens obtained. Based on these results, a deeper exploration of the CpxAR pathway should be conducted in future studies.
The differences observed using the BaeSR mutant strain were not attributable to the fitness cost paid by the bacteria because of the mutation or the incorporation of the phage. We did not observe spontaneous lysis of the lysogens, either, which we attributed to the use of E. coli DH5α, which displays a low level of spontaneous induction with Stx phages, as the host (19). Therefore, the number of lysogens was not expected to be reduced by spontaneous activation of the phage lytic pathway. Actually, the BaeSR mutant showed the same level of phage induction as its WT counterparts, confirming that phage lysis could not be the cause of the observed differences. Our observations suggest that the lysogens, once generated, coexist with the nonlysogenic bacterial cells in the culture. The effect, with some variations, appears to be common to all the Stx2 phages studied here. This group of phages show some variability in their sequences (14) but also share common traits.
Some factors, such as indole, sodium tungstate, or zinc (27–29), induce BaeSR response and should cause interference with the acquisition of Stx phages by their potential host strains. For instance, zinc has been reported to inhibit virulence traits in pathogenic E. coli and, in particular, Stx phage induction (30). However, assays with indole, zinc, or sodium tungstate were not possible in this study, as various concentrations of these compounds were found to affect the growth of the strains (data not shown). This reduction in the number of cells of the host strains prevented evaluation of the impact of the factors on phage lysogenization.
BaeSR expression is related to drug and metal resistance (31, 32) and to zinc toxicity response (33). Together with CpxAR, it activates spy, which encodes a periplasmic chaperone of proteins involved in envelope biogenesis (10, 34). The role of BaeSR in envelope biogenesis could be linked to the up/down regulation of elements of the bacterial surface. Phages recognize and attach to receptors in the bacterial surface to initiate infection (35). Phage receptors are mainly represented by surface proteins, polysaccharides, and lipopolysaccharides (35). Short-tailed Stx phages, like the ones in this study (14), specifically recognize BamA (21, 22). BamA (or YaeT) is a conserved protein essential for outer membrane protein assembly in Gram-negative bacteria (21).
Increased BamA expression in the absence of BaeSR should improve the efficiency of infection by Stx phages and, hence, the frequency of lysogenization. This observation provides an explanation for the greater number of DH5αΔbaeSR lysogens observed in this study. The association between the BaeSR pathway and BamA could be an indirect or a synergistic effect or the consequence of the activation of an integrated and complex response to extracytoplasmic signals (5).
Perez-Rodriguez et al. (12) described the relationship between the BaeSR pathway and the activation of the CRISPR-Cas system. The reduction of lysogens observed in our study could be caused by CRISPR defense mechanisms that are not activated when BaeSR is absent. However, the CRISPR-Cas I-E system appears to be inactive in E. coli under normal laboratory growth conditions (36). Additionally, the host needs to acquire the appropriate spacers for recognizing foreign DNA (36). The acquisition of these spacers is not likely to take place very quickly, even under the prolonged incubation times used in this study.
Bacteria have developed mechanisms to avoid adsorption and/or lysogenization by a given phage (37). Pathogenic STEC strains pose a risk to human health, and therefore, identification of the mechanisms for Stx phage acquisition and maintenance, as well as determining the signal pathways activated in the recipient strains, could be a first step to reducing and controlling the emergence of new pathogenic clones.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Spanish Ministerio de Ciencia y Tecnología (AGL2009-07576 and AGL2012-30880), the Generalitat de Catalunya (2009SGR1043), and the Centre de Referència en Biotecnologia (XeRBa). A. Martínez-Castillo and L. Imamovic received grants from the Spanish Ministry of Education and Science.
We thank M. C. Rodriguez de Evgrafov for useful comments on the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02916-14.
REFERENCES
- 1.Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H. 2003. Phage as agents of lateral gene transfer. Curr Opin Microbiol 6:417–424. doi: 10.1016/S1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- 2.Muniesa M, Schmidt H. 2014. Shiga toxin-encoding phages: multifunctional gene ferries, p 57–77. . In Morabito S. (ed), Pathogenic Escherichia coli: molecular and cellular microbiology. Horizon Press, Linton, United Kingdom. [Google Scholar]
- 3.Muniesa M, Hammerl J, Hertwig S, Appel B, Brüssow H. 2012. Shiga toxin-producing Escherichia coli O104:H4: a new challenge for microbiology. Appl Environ Microbiol 78:4065–4073. doi: 10.1128/AEM.00217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laing CR, Zhang Y, Gilmour MW, Allen V, Johnson R, Thomas JE, Gannon VPJ. 2012. A comparison of Shiga-toxin 2 bacteriophage from classical enterohemorrhagic Escherichia coli serotypes and the German E. coli O104:H4 outbreak strain. PLoS One 7:e37362. doi: 10.1371/journal.pone.0037362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bury-Moné S, Nomane Y, Reymond N, Barbet R, Jacquet E, Imbeaud S, Jacq A, Bouloc P. 2009. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet 5:e1000651. doi: 10.1371/journal.pgen.1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz N, Silhavy TJ. 2005. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr Opin Microbiol 8:122–126. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Humphreys S, Rowley G, Stevenson A, Anjum MF, Woodward MJ, Gilbert S, Kormanec J, Roberts M. 2004. Role of the two-component regulator CpxAR in the virulence of Salmonella enterica serotype Typhimurium. Infect Immun 72:4654–4661. doi: 10.1128/IAI.72.8.4654-4661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan JK, Vendura KW, Stevens SM, Riordan JT. 2013. RcsB determines the locus of enterocyte effacement (LEE) expression and adherence phenotype of Escherichia coli O157:H7 spinach outbreak strain TW14359 and coordinates bicarbonate-dependent LEE activation with repression of motility. Microbiology 159:2342–2353. doi: 10.1099/mic.0.070201-0. [DOI] [PubMed] [Google Scholar]
- 9.Osterhout RE, Figueroa IA, Keasling JD, Arkin AP. 2007. Global analysis of host response to induction of a latent bacteriophage. BMC Microbiol 7:82. doi: 10.1186/1471-2180-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raffa RG, Raivio TL. 2002. A third envelope stress signal transduction pathway in Escherichia coli. Mol Microbiol 45:1599–1611. doi: 10.1046/j.1365-2958.2002.03112.x. [DOI] [PubMed] [Google Scholar]
- 11.Raivio T. 2011. Identifying your enemies—could envelope stress trigger microbial immunity? Mol Microbiol 79:557–561. doi: 10.1111/j.1365-2958.2010.07485.x. [DOI] [PubMed] [Google Scholar]
- 12.Perez-Rodriguez R, Haitjema C, Huang Q, Nam KH, Bernardis S, Ke A, DeLisa MP. 2011. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol Microbiol 79:584–599. doi: 10.1111/j.1365-2958.2010.07482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Brien AD, Newland JW, Miller SF, Holmes RK, Smith HW, Formal SB. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694–696. doi: 10.1126/science.6387911. [DOI] [PubMed] [Google Scholar]
- 14.Muniesa M, Blanco JE, De Simón M, Serra-Moreno R, Blanch AR, Jofre J. 2004. Diversity of stx2 converting bacteriophages induced from Shiga-toxin-producing Escherichia coli strains isolated from cattle. Microbiology 150:2959–2971. doi: 10.1099/mic.0.27188-0. [DOI] [PubMed] [Google Scholar]
- 15.Serra-Moreno R, Acosta S, Hernalsteens JP, Jofre J, Muniesa M. 2006. Use of the lambda Red recombinase system to produce recombinant prophages carrying antibiotic resistance genes. BMC Mol Biol 7:31. doi: 10.1186/1471-2199-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imamovic L, Serra-Moreno R, Jofre J, Muniesa M. 2010. Quantification of Shiga toxin 2-encoding bacteriophages, by real-time PCR and correlation with phage infectivity. J Appl Microbiol 108:1105–1114. doi: 10.1111/j.1365-2672.2010.04664.x. [DOI] [PubMed] [Google Scholar]
- 17.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 18.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamovic L, Muniesa M. 2012. Characterizing RecA-independent induction of Shiga toxin2-encoding phages by EDTA treatment. PLoS One 7:e32393. doi: 10.1371/journal.pone.0032393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serra-Moreno R, Jofre J, Muniesa M. 2007. Insertion site occupancy by stx2 bacteriophages depends on the locus availability of the host strain chromosome. J Bacteriol 189:6645–6654. doi: 10.1128/JB.00466-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith DL, James CE, Sergeant MJ, Yaxian Y, Saunders JR, McCarthy AJ, Allison HE. 2007. Short-tailed stx phages exploit the conserved YaeT protein to disseminate Shiga toxin genes among enterobacteria. J Bacteriol 189:7223–7233. doi: 10.1128/JB.00824-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Islam MR, Ogura Y, Asadulghani M, Ooka T, Murase K, Gotoh Y, Hayashi T. 2012. A sensitive and simple plaque formation method for the Stx2 phage of Escherichia coli O157:H7, which does not form plaques in the standard plating procedure. Plasmid 67:227–235. doi: 10.1016/j.plasmid.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Neely MN, Friedman DI. 1998. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of Shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol Microbiol 28:1255–1267. doi: 10.1046/j.1365-2958.1998.00890.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Z, Li X, Liu B, Beutin L, Xu J, Ren Y, Feng L, Lan R, Reeves PR, Wang L. 2010. Derivation of Escherichia coli O157:H7 from its O55:H7 precursor. PLoS One 5:e8700. doi: 10.1371/journal.pone.0008700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchholz U, Bernard H, Werber D, Böhmer MM, Remschmidt C, Wilking H, Deleré Y, an der Heiden M, Adlhoch C, Dreesman J, Ehlers J, Ethelberg S, Faber M, Frank C, Fricke G, Greiner M, Höhle M, Ivarsson S, Jark U, Kirchner M, Koch J, Krause G, Luber P, Rosner B, Stark K, Kühne M. 2011. German outbreak of Escherichia coli O104:H4 associated with sprouts. N Engl J Med 365:1763–1770. doi: 10.1056/NEJMoa1106482. [DOI] [PubMed] [Google Scholar]
- 26.King LA, Nogareda F, Weill F-X, Mariani-Kurkdjian P, Loukiadis E, Gault G, Jourdan-DaSilva N, Bingen E, Macé M, Thevenot D, Ong N, Castor C, Noël H, Van Cauteren D, Charron M, Vaillant V, Aldabe B, Goulet V, Delmas G, Couturier E, Le Strat Y, Combe C, Delmas Y, Terrier F, Vendrely B, Rolland P, de Valk H. 2012. Outbreak of Shiga toxin-producing Escherichia coli O104:H4 associated with organic fenugreek sprouts, France, June 2011. Clin Infect Dis 54:1588–1594. doi: 10.1093/cid/cis255. [DOI] [PubMed] [Google Scholar]
- 27.Leblanc SKD, Oates CW, Raivio TL. 2011. Characterization of the induction and cellular role of the BaeSR two-component envelope stress response of Escherichia coli. J Bacteriol 193:3367–3375. doi: 10.1128/JB.01534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Appia-Ayme C, Patrick E, Sullivan MJ, Alston MJ, Field SJ, AbuOun M, Anjum MF, Rowley G. 2011. Novel inducers of the envelope stress response BaeSR in Salmonella Typhimurium: BaeR is critically required for tungstate waste disposal. PLoS One 6:e23713. doi: 10.1371/journal.pone.0023713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D, Ding X, Rather PN. 2001. Indole can act as an extracellular signal in Escherichia coli. J Bacteriol 183:4210–4216. doi: 10.1128/JB.183.14.4210-4216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crane JK, Byrd IW, Boedeker EC. 2011. Virulence inhibition by zinc in Shiga-toxigenic Escherichia coli. Infect Immun 79:1696–1705. doi: 10.1128/IAI.01099-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baranova N, Nikaido H. 2002. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J Bacteriol 184:4168–4176. doi: 10.1128/JB.184.15.4168-4176.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto K, Ogasawara H, Ishihama A. 2008. Involvement of multiple transcription factors for metal-induced spy gene expression in Escherichia coli. J Biotechnol 133:196–200. doi: 10.1016/j.jbiotec.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Wang D, Fierke CA. 2013. The BaeSR regulon is involved in defense against zinc toxicity in E. coli. Metallomics 5:372–383. doi: 10.1039/c3mt20217h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosner JL, Martin RG. 2013. Reduction of cellular stress by TolC-dependent efflux pumps in Escherichia coli indicated by BaeSR and CpxARP activation of spy in efflux mutants. J Bacteriol 195:1042–1050. doi: 10.1128/JB.01996-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samson JE, Magadán AH, Sabri M, Moineau S. 2013. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 36.Almendros C, Mojica FJM, Díez-Villaseñor C, Guzmán NM, García-Martínez J. 2014. CRISPR-Cas functional module exchange in Escherichia coli. mBio 5:e00767-13. doi: 10.1128/mBio.00767-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 38.Rose RE. 1988. The nucleotide sequence of pACYC184. Nucleic Acids Res 16:355. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.