Abstract
Staphylococcus aureus is a Gram-positive bacterium that is carried by a quarter of the healthy human population and that can cause severe infections. This pathobiosis has been linked to a balance between Toll-like receptor 2 (TLR2)-dependent pro- and anti-inflammatory responses. The relationship between these two types of responses is unknown. Analysis of 16 nasal isolates of S. aureus showed heterogeneity in their capacity to induce pro- and anti-inflammatory responses, suggesting that these two responses are independent of each other. Uncoupling of these responses was corroborated by selective signaling through phosphoinositol 3-kinase (PI3K)-Akt-mTOR and extracellular signal-regulated kinase (ERK) for the anti-inflammatory response and through p38 for the proinflammatory response. Uncoupling was also observed at the level of phagocytosis and phagosomal processing of S. aureus, which were required solely for the proinflammatory response. Importantly, the anti-inflammatory properties of an S. aureus isolate correlated with its ability to modulate T cell immunity. Our results suggest the presence of anti-inflammatory TLR2 ligands in the staphylococcal cell wall, whose identification may provide templates for novel immunomodulatory drugs.
INTRODUCTION
Staphylococcus aureus is a Gram-positive bacterium that is frequently associated with localized soft tissue infections (e.g., impetigo and dermatitis) and also systemic complications (e.g., bacteremia, sepsis, and toxic shock syndrome [TSS]) (1–3). It is the most common microbe isolated from intrahospital microbiological samples and the second most common microbe isolated from outpatient samples (4). However, S. aureus is also part of the healthy human microbiome of the upper respiratory tract, being chronically carried by more than 25% of the general population with no long-term ill effects (5–7). Therefore, S. aureus can be classified as a pathobiont: an organism that is typically safe to its host but that can become pathogenic under certain circumstances other than immunosuppression.
One of the remarkable features of this state of pathobiosis is that commensal S. aureus isolates contain many, if not all, of the known virulence factors and microbe-associated molecular patterns (MAMPs) linked to disease (8–10). The pathogenic potential of these isolates is exemplified by the risk of staphylococcal nasal carriers to develop systemic infections caused by the endogenous S. aureus strain they carry (7, 11). How these highly pathogenic microbes can behave as commensals and only rarely cause disease remains unknown (12, 13).
Early recognition of S. aureus is initiated by pattern recognition receptors (PRRs) on epithelial cells and innate phagocytic cells. Toll-like receptor 2 (TLR2) has emerged as the most important of these PRRs in detecting extracellular S. aureus (14). It heterodimerizes with TLR1 or TLR6 to recognize lipopeptides and glycopolymers embedded in the staphylococcal cell envelope, triggering proinflammatory responses. Conventional proinflammatory TLR2 signaling begins with the recruitment of the adaptor proteins TIRAP and MyD88 and the Ser/Thr kinases IRAK-1 and -4. Distal TLR2 signaling activates the NF-κB and mitogen-activated protein kinase (MAPK) pathways to upregulate proinflammatory cytokines (i.e., interleukin 1β [IL-1β], IL-6, tumor necrosis factor alpha [TNF-α], and IL-12p70) and chemokine (i.e., IL-8, CCL2, CCL3, CCL4, and RANTES) production that will then coordinate microbial clearance (15). The importance of this pathway is highlighted by the susceptibility of MyD88/IRAK4-deficient patients to staphylococcal infections (16, 17).
TLR2 also cross talks with other PRRs, including NOD1/2 and TLR9, which recognize fragments of the peptidoglycan (PGN) backbone and CpG DNA, respectively (18). TLR9 activates a similar signaling pathway as TLR2 but without the need for TIRAP bridging, whereas NOD1/2 activate the NF-κB pathway through RIP-2. Signaling from these receptors requires phagocytosis and subsequent endosomal processing of S. aureus to liberate typically hidden ligands on the staphylococcal cell wall or in the DNA (19, 20). Digestion of S. aureus also releases additional TLR2 ligands that amplify the inflammatory response. Ultimately, cross talk between signaling from these receptors enhances the host's ability to clear infection and avoid disease.
It has been recently shown that in addition to the proinflammatory response described above, S. aureus is capable of inducing a robust anti-inflammatory response as measured by production of IL-10 (21–23). We and others have shown that this anti-inflammatory response results from TLR2 signaling upon recognition of staphylococcal PGN-embedded molecules and activation of PI3K-Akt signaling to stimulate IL-10 production (21, 24, 25). Moreover, downregulation of the costimulatory molecule CD86 and upregulation of the immunoregulatory molecule PD-L1 may provide complementary effects to limit the development of an adaptive immune response (23). Interestingly, monocytes and macrophages are more potent at activating this response than dendritic cells (22). Together, these studies have shown that anti-inflammatory TLR2 signaling may promote an environment of disease tolerance to S. aureus and support commensalism by this microbe (26).
It has been assumed that both pro- and anti-inflammatory responses to TLR2 engagement emanate coordinately and simultaneously from this receptor. If this paradigm is correct, then one would expect that both types of responses result at the same ratio upon receptor engagement. In contrast to this paradigm, we report here that the pro- and anti-inflammatory responses to S. aureus are uncoupled, i.e., independent of one another. Such an uncoupling can be observed in the analysis of responses to nasal isolates of S. aureus from community carriers of this microbe, suggesting an ongoing selective process for these properties. We show that the human anti-inflammatory response to these S. aureus isolates is mediated by the phosphoinositol 3-kinase (PI3K)-Akt-mTOR and extracellular signal-regulated kinase (ERK) pathways and does not require internalization of S. aureus, whereas the proinflammatory response utilizes the p38 pathway and is dependent on phagocytosis of this microbe. Moreover, the magnitude of the IL-10-inducing response translates into different regulation of adaptive T cell responses to S. aureus. Based on these data, we propose that the cell wall of S. aureus contains two sets of TLR2 ligands: one that induces predominantly proinflammatory responses and a second set that induces predominantly anti-inflammatory responses.
MATERIALS AND METHODS
Cells.
Human peripheral blood mononuclear cells (PBMCs) were isolated from venous blood of healthy volunteers by Ficoll-Hypaque density gradient centrifugation. Volunteers gave their informed consent in compliance with the Research Ethics Office at McGill University. PBMCs were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin, l-glutamine, nonessential amino acids, and sodium pyruvate. For experiments comparing clindamycin-treated and heat-killed S. aureus, clindamycin (1 μg ml−1) was substituted for penicillin-streptomycin in supplemented RPMI 1640. Peripheral blood neutrophils were isolated by red blood cell lysis of the pellet following Ficoll-Hypaque centrifugation.
Bacteria.
S. aureus isolates were obtained from the nostrils of individuals attending an ear, nose, and throat clinic. All the isolates were confirmed as S. aureus using PCR with primers specific for the 16S rRNA gene common to all bacteria (5′-AGAGTTTGATCATGGCTCAG-3′ and 5′-GGACTACCAGGGTATCTAAT-3′) and the S. aureus nuc gene (5′-GCGATTGATGGTGATACGGTT-3′ and 5′-ACGCAAGCCTTGACGAACTAAAGC-3′) (see Fig. S1 in the supplemental material). In addition, full-genome sequencing and multilocus sequence typing (MLST) of isolates further confirmed that they were S. aureus (27). The clonality of these isolates is representative of S. aureus strains in the community (see Table S1 in the supplemental material). Bacteria were grown overnight to stationary phase in tryptic soy broth (TSB), washed, and resuspended in sterile phosphate-buffered saline (PBS). Culture supernatants were collected, filtered through a 0.2-μm filter, and stored at −20°C. Bacteria (∼1010 CFU) were plate counted and heat killed for 1 h at 100°C in a heating block. Culture supernatants were diluted to 1% in RPMI complete media. For bacterial internalization experiments, S. aureus (109 CFU) was stained with 5 μg/ml of 6-carboxytetramethylrhodamine (6-TAMRA; Sigma-Aldrich) in 50 μl of PBS for 1 h at room temperature and washed and resuspended in sterile PBS. For bacterial fractionation experiments, S. aureus isolates were digested with 1 mg/ml of lysozyme and 0.1 mg/ml of mutanolysin for 1 h in TES buffer (10 mM Tris, 1 mM EDTA, 25% sucrose [pH 8]). Cell wall fractions were separated from protoplast fractions by differential centrifugation at 2,500 × g for 10 min, precipitated with 10% trichloroacetic acid, and resuspended in PBS. Protoplast fractions were resuspended in TE buffer (10 mM Tris, 1 mM EDTA, 2% SDS) and boiled for 5 min to reduce viscosity. The protein content of these subcellular fractions was determined by bicinchoninic acid (BCA) assay. For clindamycin treatment experiments, S. aureus was grown overnight in TSB, followed by culture in TSB-containing clindamycin (1 μg ml−1) for an additional 6 h.
Reagents.
Clindamycin, cytochalasin D, dynasore, PD-98059, rapamycin, SB-203580, staphylococcal peptidoglycan, and wortmannin were purchased from Sigma-Aldrich. The PI3K p110 isoform-specific inhibitors PIK-75, TGX-221, AS-604850, and IC-87114 were purchased from EMB-Millipore. BIRB-0796 was purchased from Cayman Chemicals. Antibodies to phosphorylated Akt at Ser473 (clone 193H12), pan-Akt (clone 11E7), phosphorylated p38 (Thr180/Tyr182; clone 12F8), pan-p38 (9212), phosphorylated ERK1/2 (Thr202/Tyr204; clone 197G2), and pan-ERK1/2 (clone 137F5) were purchased from Cell Signaling Technology. Conjugated antibodies used for flow cytometry were CD3-allophycocyanin (APC)-eF780 from eBioscience and CD14-peridinin chlorophyll protein (PerCP)-Cy5.5, CD19-APC, IL-10–phycoerythrin (PE), TNF-α–Alexa Fluor 700, phospho-Akt(S473)–Alexa Fluor 488, and pan-Akt-BV421 from BD Biosciences.
Functional assays.
PBMCs were seeded in 96-well plates (200,000 cells in a volume of 200 μl per well) and stimulated under the conditions indicated in the relevant figure legends. When inhibitors were used, cells were incubated for 1 h prior to stimulation, using 0.1% dimethyl sulfoxide (DMSO) as a control. Cell-free supernatants were collected and stored at −20°C until analyzed for accumulation of cytokines by enzyme-linked immunosorbent assay (ELISA) (eBioscience).
Flow cytometry.
PBMCs (1 × 106 cells per group) were stimulated with S. aureus isolates under the conditions indicated in the relevant figure legends. For intracellular cytokine staining, 3 μg/ml of brefeldin A (eBioscience) was added after 6 h of stimulation, and the stimulation was continued for an additional 12 h. Dead cells were excluded from the analysis using a Zombie Aqua fixable viability kit (BioLegend). Cells were washed in PBS containing 2% FBS and 2 mM EDTA, blocked with 10% normal human serum, and stained for CD3, CD14, and CD19. Cells were fixed and permeabilized using the Cytofix/Cytoperm kit (BD Biosciences) and stained for IL-10 and TNF-α. For phospho-flow, PBMCs were stimulated for 30 min, fixed with Fix Buffer I (BD Biosciences), stained for extracellular markers, permeabilized using Perm/Wash Buffer I (BD Biosciences), and stained for the intracellular molecules of interest. Events were acquired on a LSRII Fortessa (BD), and doublets were excluded based on forward side scatter-area (FSC-A)/forward side scatter-height (FSC-H). Data analysis was performed using FlowJo version 10.x (Tree Star Inc.).
Western blotting.
PBMCs (5 × 106 cells per group) were resuspended in 100 μl of media and rested for 5 min at 37°C. When used, inhibitors were added at a 1:1 (vol/vol) ratio to the cells at twice the concentration indicated in the figure legends for 1 h. Next, the stimulants were added for 30 min, and cell lysates were prepared, run on 10% acrylamide gels, and immunoblotted as described previously (21, 28).
Statistics.
Statistical analysis of intragroup differences was performed using the Student t test or analysis of variance (ANOVA) with post hoc Bonferroni test on Prism GraphPad. A P value of <0.05 was deemed significant.
RESULTS
Uncoupling of pro- and anti-inflammatory responses to nasal isolates of S. aureus.
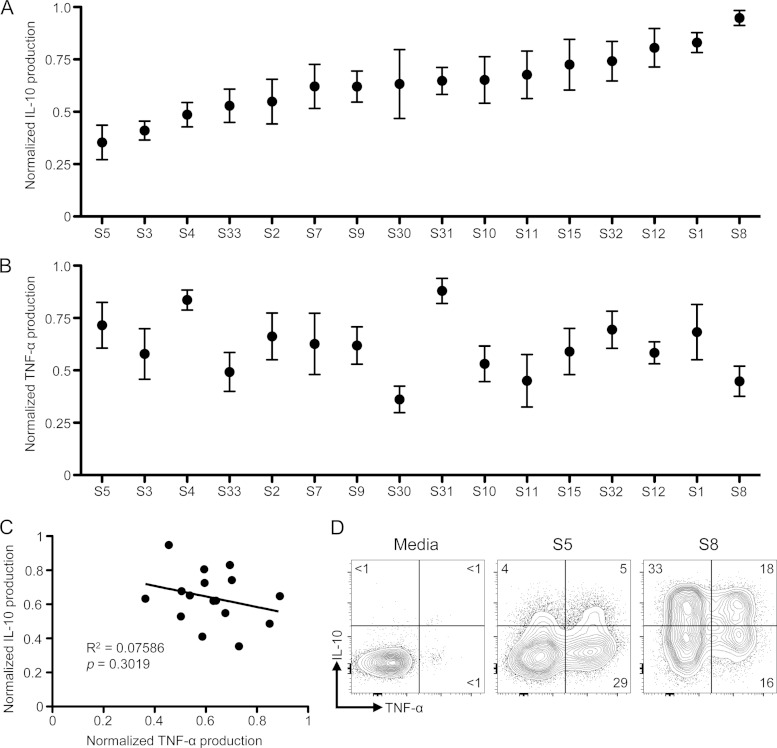
S. aureus recognition by host TLR2 induces both proinflammatory and anti-inflammatory cytokine production (14). However, the mechanisms governing these responses have been studied only in the context of laboratory isolates, crude staphylococcal preparations, or synthetic TLR2 ligands (21, 22). These experimental systems may not account for the diversity of interactions that the human immune system has with S. aureus and the subsequent heterogeneity of responses to this microbe. Indeed, S. aureus is capable of causing a spectrum of diseases, in addition to being a part of the human commensal flora. To determine the variation of the host responses to S. aureus, we stimulated human PBMCs with 16 S. aureus isolates obtained from the nostrils of human carriers. These isolates were representative of a cross-section of S. aureus found in the community as indicated by MLST (see Table S1 in the supplemental material) (29). We then measured TNF-α and IL-10 production to assess their pro- and anti-inflammatory properties, respectively. We found up to a 3-fold difference in the IL-10 production in response to these nasal S. aureus isolates by PBMCs (Fig. 1A). This response was reproducible and consistent for multiple PBMC donors and was largely determined by the bacterial isolate. A similar heterogeneity was seen in the capacity of these isolates to induce a proinflammatory TNF-α response (Fig. 1B). However, when we compared the capacities for an isolate of S. aureus to induce both IL-10 and TNF-α, we observed no correlation between these two responses (r2 = 0.07586; P = 0.3019 [Fig. 1C]). From these results, we postulated that the pro- and anti-inflammatory responses to S. aureus can be uncoupled.
FIG 1.
Uncoupling of pro- and anti-inflammatory PBMC responses to nasal S. aureus isolates. (A and B) Human PBMCs were stimulated with 16 nasal S. aureus isolates for 18 h, and accumulation of IL-10 (A) and TNF-α (B) in the supernatants was measured by ELISA. Normalized data are plotted as means ± SEMs of triplicates of 4 or 5 independent experiments involving 5 different donors. (C) Scattered plot of IL-10 versus TNF-α production by human PBMCs in response to nasal S. aureus isolates. (D) Intracellular cytokine staining of IL-10 and TNF-α in human monocytes stimulated with two representative S. aureus isolates inducing low-level (S5) or high-level (S8) IL-10 responses (MOI = 5) for 18 h. Stained PBMCs were gated on single, live CD14+ cells. Plots are representative of three independent experiments involving three different donors. Intracellular cytokine staining of T cells, B cells, and neutrophils is shown in Fig. S2 in the supplemental material.
Human PBMCs are a heterogeneous population consisting mostly of T cells, B cells, and monocytes, which can all produce IL-10 and TNF-α under different conditions (30). One possible explanation for uncoupling of the pro- and anti-inflammatory responses is a different cellular source of these cytokines. To address which population(s) was producing IL-10 and TNF-α, we stimulated human PBMCs with isolates that induced a high- or low-level IL-10 response and used intracellular flow cytometry to identify the PBMC population producing these cytokines. For simplicity and from here onward, we show the results obtained with isolates S8 and S5 as representative of the results obtained of high- and low-level IL-10 inducers. Monocytes mounted a robust TNF-α and IL-10 response to both S. aureus isolates (Fig. 1D). We detected monocytes producing only TNF-α or IL-10 or both TNF-α and IL-10. More monocytes responded to the S8 S. aureus isolate and produced more IL-10 on a per-cell basis than with the S5 S. aureus isolate, whereas levels of TNF-α production in response to each isolate were similar. We observed little to no contribution of T cells or B cells to either of these responses (see Fig. S2A and B in the supplemental material). In addition, human peripheral blood neutrophils did not produce IL-10 or TNF-α in response to S. aureus when mixed with autologous PBMCs (see Fig. S2C in the supplemental material). Thus, nasal S. aureus isolates have differential capacities to independently induce pro- and anti-inflammatory cytokine production by human monocytes.
Nasal S. aureus isolates contain qualitatively different IL-10-inducing TLR2 ligands in their cell walls.
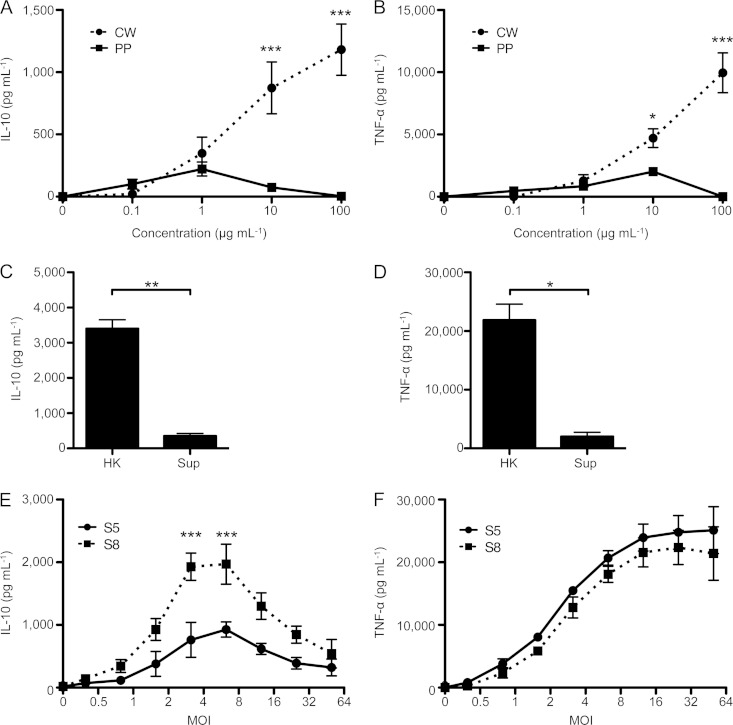
We have previously shown that the IL-10 response to S. aureus is primarily initiated by TLR2 engagement (21). These ligands could be membrane-bound, cell wall-anchored, or secreted molecules. To determine where the IL-10-inducing ligand(s) is located in nasal S. aureus isolates, we fractionated a high-level IL-10-inducing S. aureus isolate (S8) into its cell wall and protoplasm and tested the IL-10-inducing capacities of these fractions. We observed that the IL-10 and TNF-α responses were almost exclusively induced by the staphylococcal cell wall (Fig. 2A and B). Moreover, culture supernatants of S. aureus, which contain the secreted toxins and shedded components of the staphylococcal cell wall, minimally induced IL-10 and TNF-α production by PBMCs (<10% of the response to heat-killed S. aureus) (Fig. 2C and D). Together, these findings show that the pro- and anti-inflammatory TLR2 ligands are largely restricted to the staphylococcal cell wall. We therefore focused subsequent experiments on the pro- and anti-inflammatory properties of heat-killed S. aureus.
FIG 2.
Nasal S. aureus isolates have qualitatively different IL-10-inducing ligands embedded in their cell walls. (A and B) Human PBMCs were stimulated with protoplasm (PP) or cell wall (CW) fractions of the S8 S. aureus isolate for 18 h. (C and D) PBMC response to heat-killed bacteria (HK) (MOI = 5) or supernatants (Sup) (1% of overnight culture). (E and F) Human PBMCs were stimulated with increasing amounts of two representative S. aureus isolates inducing low-level (S5; circles and solid line) or high-level (S8; squares and dashed line) IL-10 responses for 18 h. Quantification of IL-10 (A, C, and E) and TNF-α (B, D, and F) in the supernatants was performed by ELISA. Data are plotted as means ± SEMs for 3 to 5 different donors in experiments performed in triplicate. Statistical analysis was performed using ANOVA with post hoc Bonferroni test (A, B, E, and F) or Student t test (C and D). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Next, we wanted to determine if high-level IL-10-inducing S. aureus isolates contain quantitatively more IL-10-inducing ligands or qualitatively different ligands than the low-level IL-10-inducing counterparts. To test this, we performed an extensive titration of S. aureus isolates representative of high and low IL-10 induction capacities (shown here for S8 and S5 isolates) to determine the multiplicity of infection (MOI) that induced maximal pro- and anti-inflammatory responses. IL-10 was already detectable at an MOI of 1 and peaked at an MOI of 6 for both isolates (Fig. 2E). At all MOIs tested, the IL-10 production in response to S. aureus S8 was at least 2-fold greater than the response to S5. Importantly, the levels of TNF-α production in response to the isolates did not differ (Fig. 2F). These results suggest that the anti-inflammatory response is due to a qualitatively different ligand(s) than those responsible for the proinflammatory response, and therefore, the anti-inflammatory response to S. aureus can be uncoupled from the proinflammatory response.
Pro- and anti-inflammatory responses to S. aureus are uncoupled at the signaling level.
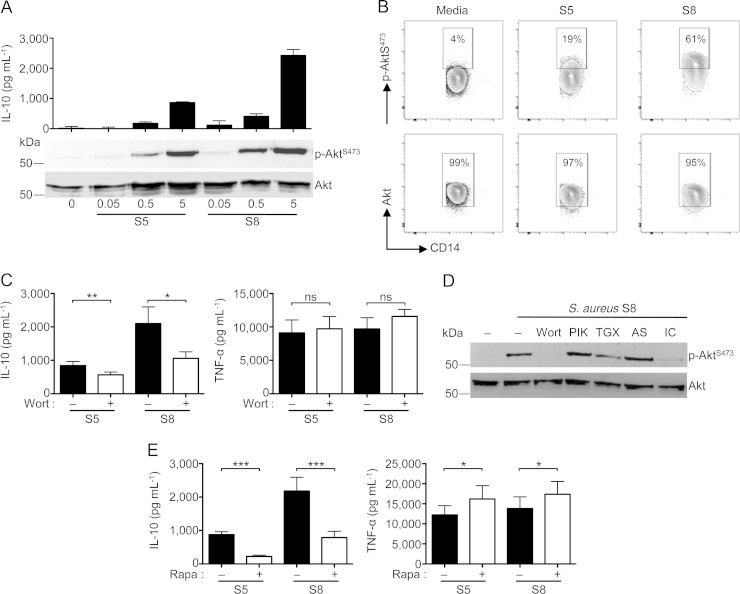
Given that S. aureus may contain multiple IL-10-inducing ligands that act on TLR2, we next asked if these ligands activated the same pathway(s). The PI3K-Akt pathway has previously been shown to be essential for the anti-inflammatory response to TLR2 (22) and TLR4 (31) ligands. Thus, we examined activation of this pathway in response to several nasal S. aureus isolates. As shown in Fig. 3A, for two representative isolates, we found that phosphorylation of Akt at S473 correlated with the IL-10-inducing capacity of nasal S. aureus isolates. Similar results were obtained using intracellular staining of phospho-AktS473 in monocytes (Fig. 3B). Interestingly, we also observed Akt phosphorylation in a subset of B cells (∼ 20%), but this did not differ between isolates inducing IL-10 at high and low levels (see Fig. S2 in the supplemental material). Inhibition of signaling through this pathway using the pan-PI3K inhibitor wortmannin significantly reduced IL-10 production in response to both S. aureus S5 and S8 but did not significantly affect the TNF-α response (Fig. 3C). Furthermore, using PI3K p110 isoform-specific inhibitors, we observed that p110δ was the dominant isoform mediating the IL-10 response, with a minor contribution from p110β (Fig. 3D).
FIG 3.
The PI3K/Akt/mTOR pathway mediates the IL-10 response to nasal S. aureus isolates. (A) PBMCs were stimulated with two S. aureus isolates at the indicated MOIs for 30 min for Western blot experiments or for 18 h in experiments looking at IL-10 accumulation by ELISA. (B) Flow cytometric analysis of Akt phosphorylation (S473) in CD14+ gated human PBMCs stimulated with two representative S. aureus isolates inducing low-level (S5) or high-level (S8) IL-10 responses. (C) PBMCs were pretreated with wortmannin (1 μM) and then stimulated with S. aureus (MOI = 5) for 18 h. Quantification of IL-10 and TNF-α accumulation in the supernatants was done by ELISA. (D) Western blot of PBMCs pretreated with the pan-PI3K-p110 inhibitor wortmannin (Wort) or p110 isoform inhibitors PIK-75 (PIK; p110α inhibitor; 100 nM), TGX-221 (TGX; p110β inhibitor; 500 nM), AS-604580 (AS; p110γ inhibitor; 10 μM), and IC-87114 (IC; p110δ inhibitor; 5 μM) for 1 h and then stimulated with S. aureus S8 (MOI = 5) for 30 min. (E) PBMCs were pretreated with rapamycin (10 nM) and then stimulated with S. aureus (MOI = 5) for 18 h. Quantification of IL-10 and TNF-α accumulation in the supernatants was performed by ELISA. Data in panels A, B, and D are representative of three independent experiments from three different donors. Data in panels C and E are plotted as means ± SEMs from eight individual donors in experiments performed in triplicate. Statistical analysis was performed using theStudent t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. ns, not significant.
To further corroborate the selective participation of the PI3K-Akt pathway in the anti-inflammatory response, we examined signaling steps further downstream in this cascade. We found that the mTOR inhibitor rapamycin significantly decreased the IL-10 response to S. aureus S5 and S8 (Fig. 3E). Interestingly, we observed a small but significant increase in TNF-α, which is most likely a reflection of the antagonistic properties IL-10 has on TNF-α production (32). Together, these results show that the PI3K-Akt-mTOR pathway is differentially activated by nasal S. aureus isolates, regulating the anti-inflammatory but not the proinflammatory response to this microbe.
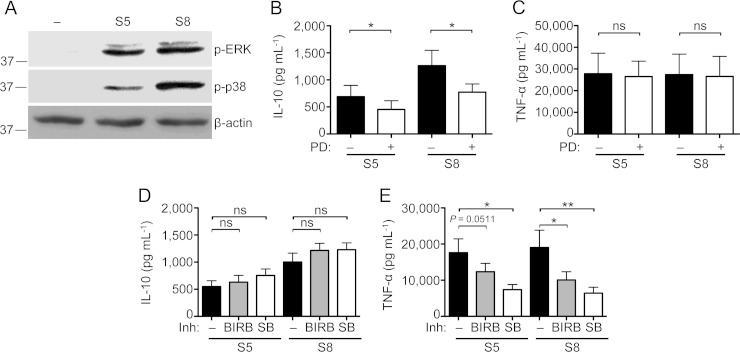
The importance of MAPK signaling in the cytokine response to MAMPs is well documented (33). Moreover, p38 and ERK have both been documented to regulate IL-10 production in response to various stimuli (30). Consistent with this, we observed that both p38 and ERK were activated in response to the nasal S. aureus isolates (Fig. 4A). To elucidate the specific roles of these members of the MAPK family in the response to S. aureus, we used the selective inhibitors PD-98059 for ERK1/2 and SB-203580 and BIRB-0796 for p38. PD-98059 slightly but significantly decreased the IL-10 response to S. aureus (Fig. 4B) and had no effect on the TNF-α response (Fig. 4C). In contrast, the p38 inhibitors SB-203580 and BIRB-0796 significantly decreased the TNF-α response without affecting the IL-10 response (Fig. 4D and E). These results further document the uncoupling of pro- and anti-inflammatory responses to S. aureus at the level of MAPK signaling by showing the selective dependence of the proinflammatory response on the p38 MAPK signaling pathway.
FIG 4.
The MAPK p38 mediates the proinflammatory response to nasal S. aureus isolates. (A) Western blot of phospho-ERK and -p38 from human PBMCs stimulated with two representative S. aureus isolates inducing low-level (S5) or high-level (S8) IL-10 responses (MOI = 5) for 30 min. Blots are representative of three independent experiments involving three different donors. (B to E) PBMCs were pretreated with the ERK inhibitor PD-98059 (B and C) or either of the p38 inhibitors SB-203580 and BIRB-0796 (D and E) for 1 h and then stimulated with two representative S. aureus isolates inducing low-level (S5) or high-level (S8) IL-10 responses (MOI = 5) for 18 h. Quantification of accumulation of IL-10 (B and D) and TNF-α (C and E) in the supernatants was done by ELISA. Bar graphs show means ± SEMs of data from five different donors in experiments performed in triplicate. Statistical analysis was performed using the Student t test. *, P < 0.05; **, P < 0.01.
Pro- and anti-inflammatory responses to S. aureus have differential requirements for microbial internalization and phagosome maturation.
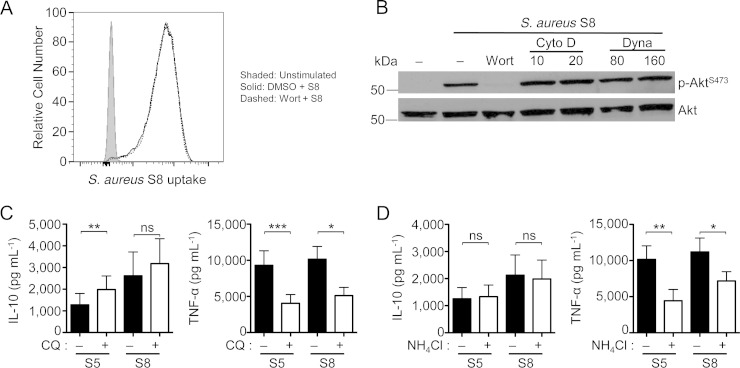
It has been recently shown that the anti-inflammatory IL-10 response to Gram-negative Escherichia coli lipopolysaccharide (LPS) requires PI3K-dependent internalization of TLR4 (31). Thus, we examined whether a similar mechanism could be involved in the anti-inflammatory response to S. aureus through TLR2. First, we tested if inhibition of PI3K could prevent internalization of S. aureus. As shown in Fig. 5A, this was not the case, as S. aureus was still internalized in the presence of wortmannin, suggesting that PI3K signaling is not required for S. aureus phagocytosis. Inhibition of phagocytosis with either the actin inhibitor cytochalasin D or the dynamin inhibitor dynasore did not prevent S. aureus-induced phospho-Akt (Fig. 5B), which is required for the anti-inflammatory response to S. aureus, even though inhibition of phagocytosis was confirmed by flow cytometry (data not shown). Unfortunately, due to cytotoxicity of these inhibitors, we were unable to examine their effects on cytokine production (data not shown). Together, these findings implied that the IL-10-induced signaling from S. aureus occurred at the cell surface and did not require S. aureus internalization. Consistent with this claim, we saw that the IL-10 response to S. aureus was not affected by inhibition of endophagosome acidification, whereas the proinflammatory response was significantly inhibited by chloroquine and NH4Cl (Fig. 5C and D). These results demonstrate a differential requirement for microbe internalization on pro- and anti-inflammatory responses, further documenting the spontaneous uncoupling of these properties in S. aureus.
FIG 5.
Pro- and anti-inflammatory responses to S. aureus have differential requirements for microbial internalization and processing. (A) Human PBMCs were pretreated with DMSO or wortmannin (1 μM) and cultured with TAMRA-labeled S. aureus (MOI = 5) for 30 min. Uptake of S. aureus by CD14+ monocytes was determined by flow cytometry. The plot is representative of three independent experiments. (B) Phospho-Akt Western blot of human PBMCs stimulated with S. aureus S8 for 30 min with or without pretreatment with wortmannin or either of the internalization inhibitors cytochalasin D and dynasore. Blots are representative of three independent experiments from three different donors. (C and D) PBMCs were pretreated for 1 h with chloroquine (CQ) (C) or NH4Cl (D) and then stimulated with two representative S. aureus isolates inducing low-level (S5) or high-level (S8) IL-10 responses (MOI = 5) for 18 h. Accumulation of IL-10 or TNF-α was quantified by ELISA. Bar graphs show means ± SEMs from five donors in experiments performed in triplicate. Statistical analysis was performed using the Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Uncoupling of pro- and anti-inflammatory properties of S. aureus translates into differential modulation of adaptive immunity to staphylococcal SAgs.
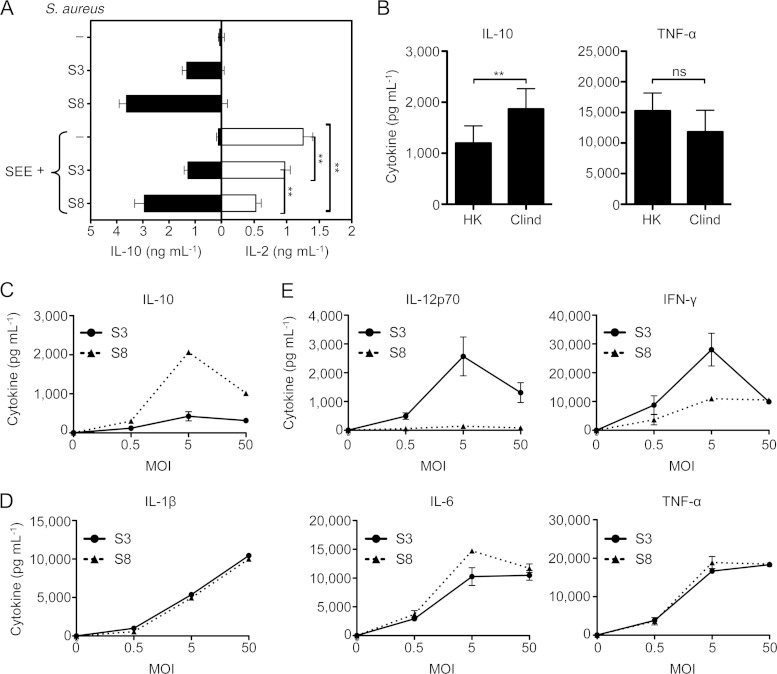
We have previously reported that the IL-10 response to S. aureus suppresses superantigen (SAg)-induced T cell activation and may be protective against staphylococcal TSS (21). We therefore predicted that those isolates able to induce a high-level IL-10 response are better at suppressing this T cell activation. We found that this was the case: even though all S. aureus isolates induced enough IL-10 to decrease the SAg-induced T cell activation as measured by IL-2 production (Fig. 6A), the high-level IL-10-inducing S. aureus isolates (e.g., S8) were significantly better suppressors than the low-level IL-10-inducing isolates (e.g., S3).
FIG 6.
The IL-10-inducing capacity of nasal S. aureus isolates correlates with the modulation of adaptive immunity. (A) PBMCs were stimulated with staphylococcal enterotoxin E (SEE) (10 ng ml−1) for 18 h in the presence or absence of representative S. aureus isolates inducing low-level (S3) or high-level (S8) IL-10 responses (MOI = 5). Quantification of IL-2 and IL-10 in the supernatants was done by ELISA. Data are plotted as means ± SDs and are representative of three independent experiments involving three different donors. (B) IL-10 and TNF-α responses by PBMCs to heat-killed (HK) or clindamycin-treated (Clind) S. aureus S8 (MOI = 5). Data are plotted as means ± SEMs from 5 donors in experiments performed in triplicate. Statistical analysis was performed using the Student t test. (C to E) Profiles of cytokine responses of PBMCs to S. aureus. PBMCs were stimulated for 18 h with S. aureus S3 or S8, and IL-10 (C), the proinflammatory cytokines IL-1β, IL-6, and TNF-α (D), and the Th1 cytokines IL-12p70 and IFN-γ (E) were measured. Data are plotted as means ± SDs and are representative of two independent experiments involving two different donors. **, P < 0.01.
Bacteriostatic antibiotics, such as clindamycin, have been recommended for the management of staphylococcal TSS (34). The effectiveness of clindamycin to treat staphylococcal TSS is attributed to blocking the translation of staphylococcal SAg (35). We postulated that in addition to this mechanism, clindamycin may also maintain the integrity of the staphylococcal cell wall to induce IL-10 production. To test this hypothesis, we cultured S. aureus in TSB containing clindamycin for 6 h and compared its immunostimulatory capacity to that of heat-killed S. aureus. We observed that clindamycin-treated S. aureus induced significantly more IL-10 than heat-killed S. aureus but did not significantly change TNF-α production (Fig. 6B). This result reveals a potential biological implication of the uncoupling of proinflammatory and anti-inflammatory properties and suggests that clindamycin may be effective for treating staphylococcal TSS by promoting an anti-inflammatory response to S. aureus, in addition to inhibiting SAg production.
To further characterize the modulatory effect of the community isolates of S. aureus on the adaptive immune response to staphylococcal superantigens, we performed a multiplex analysis of the cytokine response to high- and low-level IL-10-inducing nasal S. aureus isolates (Fig. 6C). We observed no difference in the production of proinflammatory IL-1β, IL-6, or TNF-α cytokines among these isolates (Fig. 6D), suggesting that these cytokines are similarly regulated in response to S. aureus. However, the Th1 cytokines IL-12p70 and gamma interferon (IFN-γ) were induced to a greater extent by S. aureus isolates that had less anti-inflammatory properties (Fig. 6E). Altogether, these results imply that the low-level IL-10-inducing capacity of an isolate of S. aureus imprints adaptive immunity to a Th1 profile and thus influences the development of protective proinflammatory responses in the context of staphylococcal diseases.
DISCUSSION
TLR signaling leading to the production of IL-10 and other anti-inflammatory mediators in response to MAMPs has been previously reported (21, 22, 31, 36). However, its qualitative and quantitative relationship to the proinflammatory cytokine response (i.e., IL-1β, IL-6, TNF-α, etc.) triggered by PRR signaling has not yet been studied. Specifically, it is not known whether the two types of responses occur in parallel and to similar extents or whether pro- and anti-inflammatory responses are the result of qualitatively different MAMP:PRR recognition events that can be uncoupled. On the basis of results obtained using multiple nasal isolates of S. aureus from chronic human carriers of this microbe, we report here that for TLR2, these responses are mechanistically distinct and are naturally uncoupled.
Such an uncoupling was seen in all the isolates of a representative sample of community S. aureus independently of the magnitude of IL-10 response they induced. Our findings support the idea that the pro- and anti-inflammatory properties of a given S. aureus isolate are the result of qualitatively different responses to recognition of MAMPs on the staphylococcal cell wall. In support of this conclusion, we found that IL-10 production in response to S. aureus was dependent on PI3K-Akt-mTOR and ERK signaling, whereas the TNF-α response was dependent on p38. Furthermore, internalization and phagosome maturation were required only for the proinflammatory response and not the anti-inflammatory response. Lastly, natural differences in the capacity of nasal S. aureus isolates to induce an IL-10 response in the host, independently of the proinflammatory response they can induce, differentially imprints the adaptive immune response to S. aureus.
Whether the uncoupling of pro- and anti-inflammatory properties of a given S. aureus isolate is the result of multiple ligands on the staphylococcal cell wall recognized by one PRR or by different PRRs is still unclear. Our previous data suggested that the proinflammatory response occurs from multiple PRRs, including TLR2, TLR9, and NOD1/2, whereas the anti-inflammatory response to S. aureus is predominantly secondary to TLR2 signaling (21). NOD1/2 were ruled out as players in the anti-inflammatory IL-10 response because ultrapure staphylococcal PGN (PGN-Sandi), which lacks TLR2-stimulating capacity and thus can only activate NOD1/2, did not induce IL-10 production, despite giving the same proinflammatory profile as crude staphylococcal PGN (22). Both NODs and TLR9 can also be excluded by the fact that IL-10 production in response to S. aureus is independent of phagocytosis. Thus, our data support the idea that the uncoupling occurs, in part, by several ligands acting on TLR2. Indeed, previous work from our laboratory has shown that the TLR2 accessory molecules CD14 and CD36 are only required for the proinflammatory response to staphylococcal PGN preparations (22). Moreover, CD36 is required for S. aureus internalization (37), and this may explain the dependency of this molecule only for the proinflammatory response (22). Together, these findings point to structural differences in pro- and anti-inflammatory TLR2 ligands. The molecular nature of these ligands is unknown at the moment, although it may be diverse and present in other microbial species (e.g., polysaccharide A in Bacillus fragilis [25]).
TLR2-based uncoupling of pro- and anti-inflammatory responses may stem from differential TLR2 oligomer formation. For example, TLR2 dimerization with TLR6 has been linked to anti-inflammatory responses, whereas TLR2/1 complexes promote proinflammatory responses (38–40). The molecular definition of the resulting signalosomes is uncertain (41, 42). It is plausible to suggest that in response to S. aureus, the availability of TLR2 on the cell surface is a limiting factor of the response. If so, the relative abundance of the pro- versus anti-inflammatory ligands would dictate the outcome of the response. We cannot rule out that under some circumstances (e.g., S. aureus isolates lacking TLR2-stimulatory capacity [43]) an IL-10 response may occur through an alternative, less efficient mechanism.
Phagocytosis is an important defense mechanism to mount an effective inflammatory response against S. aureus (19, 20). We have corroborated this fact using nasal S. aureus isolates. Interestingly, though, the anti-inflammatory response did not require phagocytosis by monocytes/macrophages. Since we used heat-killed bacteria as well as cell wall preparations, our findings indicate that the anti-inflammatory ligands are present in the staphylococcal cell wall in a recognizable conformation, unlike their proinflammatory counterparts. Such an arrangement may ensure an anti-inflammatory response that downplays the Th1 response to S. aureus and promotes a state of disease tolerance to this microbe (44).
The mechanism of TLR2-dependent anti-inflammatory response is different from that reported recently for the TLR4-dependent anti-inflammatory response. The anti-inflammatory response to TLR4 signaling by LPS requires activation of PI3K p110δ for endosomal translocation and a switch from the TIRAP-MyD88 pathway to TRAM-TRIF pathway for anti-inflammatory signaling to occur (31). In contrast, for TLR2 signaling, PI3K p110δ, although required for the IL-10 response, is not involved in endosomal trafficking of TLR2. We found that inhibition of PI3K did not affect S. aureus internalization by monocytes and the IL-10 response was independent of phagocytosis and phagosome maturation. Furthermore, although the switch to anti-inflammatory TLR4 signaling resulted in a diminished proinflammatory response (31), we did not observe such an effect with nasal S. aureus isolates. Together, our findings are consistent with those showing that the PI3K-Akt-mTOR pathway directly leads to IL-10 production (36, 45, 46).
We have previously shown that the IL-10 response to S. aureus is predominantly mounted by monocytes/macrophages. This is in contrast to the IL-10 response to other MAMPs (25, 47–49). Monocytes/macrophages are early responders to S. aureus, and their phenotype would be highly influential in establishing the microenvironment that sets up subsequent adaptive responses. Previous work from our laboratory has shown that the IL-10-producing monocytes/macrophages are classically activated and not alternatively activated macrophages, because of the robust inflammatory response simultaneously observed and the inability to show IL-4/IL-13 during the generation of this response (22). Moreover, a high proportion of these monocytes acquire a dual phenotype (i.e., IL-10+ TNF-α+), a phenotype not reported for M2 macrophages (reviewed in reference 50). The characterization of the different macrophage subsets responding to nasal S. aureus isolates based on their cytokine production profiles is a matter for future studies. It is likely that these cells show a phenotype similar to that of inflammatory monocytes seen in the gut during Toxoplasma gondii infection (51), in which simultaneous expression of proinflammatory and anti-inflammatory mediators is observed. Proper balance of this expression may determine pathogen elimination while limiting tissue damage and deleterious effects on commensals.
The findings reported here may have clinical implications. The balance between pro- and anti-inflammatory responses to a given S. aureus isolate may determine the outcome of the encounter, i.e., commensalism versus disease. S. aureus isolates with a high capacity to induce IL-10 would be better at colonizing the upper respiratory tract, as the heightened IL-10 levels may provide a tolerogenic environment and be less likely to cause staphylococcal TSS. Alternatively, a predominance of inflammatory cytokine production may exacerbate mucosal injury (52). Lastly, high-level IL-10 induction by a given isolate may be detrimental during staphylococcal bacteremia by dampening adaptive immunity (53). Formal proof of concept for this model will require in vivo testing.
In conclusion, our findings have revealed that the pro- and anti-inflammatory properties of nasal S. aureus isolates are independent of each other; i.e., they can be uncoupled. Such an uncoupling obeys different mechanistic requirements. The ability to naturally uncouple these two types of responses suggests that S. aureus contains two sets of MAMPs: one that preferentially induces a proinflammatory response and minimal anti-inflammatory mediators and a second set that induces a robust anti-inflammatory response, with few proinflammatory cytokines. Balance between the ensuing responses may determine colonization and disease tolerance versus pathogenicity and disease by S. aureus.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark Hancock and the McGill Multiplexing Facility (McGill University) for assistance with multiplexing and the Canadian Foundation for Innovation for infrastructure support, and we thank David E. Heinrichs and Martin McGavin (University of Western Ontario, London, ON, Canada) and members of the Madrenas Laboratory for helpful comments and criticisms.
This work was supported by the Canadian Institutes for Health Research (to J.M.). A.G.P. is a Fonds de la Recherche en Santé du Québec Research Scholar. J.M. holds a Tier I Canada Research Chair in Human Immunology.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02832-14.
REFERENCES
- 1.Kuehnert MJ, Hill HA, Kupronis BA, Tokars JI, Solomon SL, Jernigan DB. 2005. Methicillin-resistant-Staphylococcus aureus hospitalizations, United States. Emerg Infect Dis 11:868–872. doi: 10.3201/eid1106.040831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein E, Smith DL, Laxminarayan R. 2007. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg Infect Dis 13:1840–1846. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dantes R, Mu Y, Belflower R, Aragon D, Dumyati G, Harrison LH, Lessa FC, Lynfield R, Nadle J, Petit S, Ray SM, Schaffner W, Townes J, Fridkin S, Emerging Infections Program-Active Bacterial Core Surveillance MRSA Surveillance Investigators . 2013. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med 173:1970–1978. doi: 10.1001/jamainternmed.2013.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Styers D, Sheehan DJ, Hogan P, Sahm DF. 2006. Laboratory-based surveillance of current antimicrobial resistance patterns and trends among Staphylococcus aureus: 2005 status in the United States. Ann Clin Microbiol Antimicrob 5:2. doi: 10.1186/1476-0711-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eriksen NH, Espersen F, Rosdahl VT, Jensen K. 1995. Carriage of Staphylococcus aureus among 104 healthy persons during a 19-month period. Epidemiol Infect 115:51–60. doi: 10.1017/S0950268800058118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Belkum A, Verkaik NJ, de Vogel CP, Boelens HA, Verveer J, Nouwen JL, Verbrugh HA, Wertheim HF. 2009. Reclassification of Staphylococcus aureus nasal carriage types. J Infect Dis 199:1820–1826. doi: 10.1086/599119. [DOI] [PubMed] [Google Scholar]
- 7.Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, van Keulen PH, Vandenbroucke-Grauls CM, Meester MH, Verbrugh HA. 2004. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364:703–705. doi: 10.1016/S0140-6736(04)16897-9. [DOI] [PubMed] [Google Scholar]
- 8.Nashev D, Toshkova K, Salasia SI, Hassan AA, Lammler C, Zschock M. 2004. Distribution of virulence genes of Staphylococcus aureus isolated from stable nasal carriers. FEMS Microbiol Lett 233:45–52. doi: 10.1016/j.femsle.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 9.Lozano C, Gomez-Sanz E, Benito D, Aspiroz C, Zarazaga M, Torres C. 2011. Staphylococcus aureus nasal carriage, virulence traits, antibiotic resistance mechanisms, and genetic lineages in healthy humans in Spain, with detection of CC398 and CC97 strains. Int J Med Microbiol 301:500–505. doi: 10.1016/j.ijmm.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Netsvyetayeva I, Fraczek M, Piskorska K, Golas M, Sikora M, Mlynarczyk A, Swoboda-Kopec E, Marusza W, Palmieri B, Iannitti T. 2014. Staphylococcus aureus nasal carriage in Ukraine: antibacterial resistance and virulence factor encoding genes. BMC Infect Dis 14:128. doi: 10.1186/1471-2334-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall C, McBryde E. 2014. The role of Staphylococcus aureus carriage in the pathogenesis of bloodstream infection. BMC Res Notes 7:428. doi: 10.1186/1756-0500-7-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown AF, Leech JM, Rogers TR, McLoughlin RM. 2014. Staphylococcus aureus colonization: modulation of host immune response and impact on human vaccine design. Front Immunol 4:507. doi: 10.3389/fimmu.2013.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peres AG, Madrenas J. 2013. The broad landscape of immune interactions with Staphylococcus aureus: from commensalism to lethal infections. Burns 39:380–388. doi: 10.1016/j.burns.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Fournier B, Philpott DJ. 2005. Recognition of Staphylococcus aureus by the innate immune system. Clin Microbiol Rev 18:521–540. doi: 10.1128/CMR.18.3.521-540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 16.von Bernuth H, Picard C, Jin ZB, Pankla R, Xiao H, Ku CL, Chrabieh M, Ben Mustapha I, Ghandil P, Camcioglu Y, Vasconcelos J, Sirvent N, Guedes M, Vitor AB, Herrero-Mata MJ, Arostegui JI, Rodrigo C, Alsina L, Ruiz-Ortiz E, Juan M, Fortuny C, Yague J, Anton J, Pascal M, Chang HH, Janniere L, Rose Y, Garty BZ, Chapel H, Issekutz A, Marodi L, Rodriguez-Gallego C, Banchereau J, Abel L, Li XX, Chaussabel D, Puel A, Casanova JL. 2008. Pyogenic bacterial infections in humans with MyD88 deficiency. Science 321:691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Picard C, von Bernuth H, Ghandil P, Chrabieh M, Levy O, Arkwright PD, McDonald D, Geha RS, Takada H, Krause JC, Creech CB, Ku CL, Ehl S, Marodi L, Al-Muhsen S, Al-Hajjar S, Al-Ghonaium A, Day-Good NK, Holland SM, Gallin JI, Chapel H, Speert DP, Rodriguez-Gallego C, Colino E, Garty BZ, Roifman C, Hara T, Yoshikawa H, Nonoyama S, Domachowske J, Issekutz AC, Tang M, Smart J, Zitnik SE, Hoarau C, Kumararatne DS, Thrasher AJ, Davies EG, Bethune C, Sirvent N, de Ricaud D, Camcioglu Y, Vasconcelos J, Guedes M, Vitor AB, Rodrigo C, Almazan F, Mendez M, Arostegui JI, Alsina L, Fortuny C, Reichenbach J, Verbsky JW, Bossuyt X, Doffinger R, Abel L, Puel A, Casanova JL. 2010. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine (Baltimore) 89:403–425. doi: 10.1097/MD.0b013e3181fd8ec3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawai T, Akira S. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Ip WK, Sokolovska A, Charriere GM, Boyer L, Dejardin S, Cappillino MP, Yantosca LM, Takahashi K, Moore KJ, Lacy-Hulbert A, Stuart LM. 2010. Phagocytosis and phagosome acidification are required for pathogen processing and MyD88-dependent responses to Staphylococcus aureus. J Immunol 184:7071–7081. doi: 10.4049/jimmunol.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolf AJ, Arruda A, Reyes CN, Kaplan AT, Shimada T, Shimada K, Arditi M, Liu G, Underhill DM. 2011. Phagosomal degradation increases TLR access to bacterial ligands and enhances macrophage sensitivity to bacteria. J Immunol 187:6002–6010. doi: 10.4049/jimmunol.1100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chau TA, McCully ML, Brintnell W, An G, Kasper KJ, Vines ED, Kubes P, Haeryfar SM, McCormick JK, Cairns E, Heinrichs DE, Madrenas J. 2009. Toll-like receptor 2 ligands on the staphylococcal cell wall downregulate superantigen-induced T cell activation and prevent toxic shock syndrome. Nat Med 15:641–648. doi: 10.1038/nm.1965. [DOI] [PubMed] [Google Scholar]
- 22.Frodermann V, Chau TA, Sayedyahossein S, Toth JM, Heinrichs DE, Madrenas J. 2011. A modulatory interleukin-10 response to staphylococcal peptidoglycan prevents Th1/Th17 adaptive immunity to Staphylococcus aureus. J Infect Dis 204:253–262. doi: 10.1093/infdis/jir276. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Roderiquez G, Norcross MA. 2012. Control of adaptive immune responses by Staphylococcus aureus through IL-10, PD-L1, and TLR2. Sci Rep 2:606. doi: 10.1038/srep00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, Wu ZR, Hooper LV, Schmidt RR, von Aulock S, Radek KA, Huang CM, Ryan AF, Gallo RL. 2009. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med 15:1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. 2011. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mele T, Madrenas J. 2010. TLR2 signalling: at the crossroads of commensalism, invasive infections and toxic shock syndrome by Staphylococcus aureus. Int J Biochem Cell Biol 42:1066–1071. doi: 10.1016/j.biocel.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 27.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baroja ML, Cieslinski LB, Torphy TJ, Wange RL, Madrenas J. 1999. Specific CD3 epsilon association of a phosphodiesterase 4B isoform determines its selective tyrosine phosphorylation after CD3 ligation. J Immunol 162:2016–2023. [PubMed] [Google Scholar]
- 29.Feng Y, Chen CJ, Su LH, Hu S, Yu J, Chiu CH. 2008. Evolution and pathogenesis of Staphylococcus aureus: lessons learned from genotyping and comparative genomics. FEMS Microbiol Rev 32:23–37. doi: 10.1111/j.1574-6976.2007.00086.x. [DOI] [PubMed] [Google Scholar]
- 30.Saraiva M, O'Garra A. 2010. The regulation of IL-10 production by immune cells. Nat Rev Immunol 10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 31.Aksoy E, Taboubi S, Torres D, Delbauve S, Hachani A, Whitehead MA, Pearce WP, Berenjeno IM, Nock G, Filloux A, Beyaert R, Flamand V, Vanhaesebroeck B. 2012. The p110delta isoform of the kinase PI(3)K controls the subcellular compartmentalization of TLR4 signaling and protects from endotoxic shock. Nat Immunol 13:1045–1054. doi: 10.1038/ni.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P, Wu P, Siegel MI, Egan RW, Billah MM. 1994. IL-10 inhibits transcription of cytokine genes in human peripheral blood mononuclear cells. J Immunol 153:811–816. [PubMed] [Google Scholar]
- 33.Arthur JS, Ley SC. 2013. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol 13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 34.Annane D, Clair B, Salomon J. 2004. Managing toxic shock syndrome with antibiotics. Expert Opin Pharmacother 5:1701–1710. doi: 10.1517/14656566.5.8.1701. [DOI] [PubMed] [Google Scholar]
- 35.Schlievert PM, Kelly JA. 1984. Clindamycin-induced suppression of toxic-shock syndrome-associated exotoxin production. J Infect Dis 149:471. doi: 10.1093/infdis/149.3.471. [DOI] [PubMed] [Google Scholar]
- 36.Weichhart T, Costantino G, Poglitsch M, Rosner M, Zeyda M, Stuhlmeier KM, Kolbe T, Stulnig TM, Horl WH, Hengstschlager M, Muller M, Saemann MD. 2008. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity 29:565–577. doi: 10.1016/j.immuni.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Stuart LM, Deng J, Silver JM, Takahashi K, Tseng AA, Hennessy EJ, Ezekowitz RA, Moore KJ. 2005. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J Cell Biol 170:477–485. doi: 10.1083/jcb.200501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Depaolo RW, Tang F, Kim I, Han M, Levin N, Ciletti N, Lin A, Anderson D, Schneewind O, Jabri B. 2008. Toll-like receptor 6 drives differentiation of tolerogenic dendritic cells and contributes to LcrV-mediated plague pathogenesis. Cell Host Microbe 4:350–361. doi: 10.1016/j.chom.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DePaolo RW, Kamdar K, Khakpour S, Sugiura Y, Wang W, Jabri B. 2012. A specific role for TLR1 in protective T(H)17 immunity during mucosal infection. J Exp Med 209:1437–1444. doi: 10.1084/jem.20112339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skabytska Y, Wolbing F, Gunther C, Koberle M, Kaesler S, Chen KM, Guenova E, Demircioglu D, Kempf WE, Volz T, Rammensee HG, Schaller M, Rocken M, Gotz F, Biedermann T. 2014. Cutaneous innate immune sensing of Toll-like receptor 2-6 ligands suppresses T cell immunity by inducing myeloid-derived suppressor cells. Immunity 41:762–775. doi: 10.1016/j.immuni.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Kenny EF, Talbot S, Gong M, Golenbock DT, Bryant CE, O'Neill LA. 2009. MyD88 adaptor-like is not essential for TLR2 signaling and inhibits signaling by TLR3. J Immunol 183:3642–3651. doi: 10.4049/jimmunol.0901140. [DOI] [PubMed] [Google Scholar]
- 42.Santos-Sierra S, Deshmukh SD, Kalnitski J, Kuenzi P, Wymann MP, Golenbock DT, Henneke P. 2009. Mal connects TLR2 to PI3Kinase activation and phagocyte polarization. EMBO J 28:2018–2027. doi: 10.1038/emboj.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hilmi D, Parcina M, Stollewerk D, Ostrop J, Josten M, Meilaender A, Zaehringer U, Wichelhaus TA, Bierbaum G, Heeg K, Wolz C, Bekeredjian-Ding I. 2014. Heterogeneity of host TLR2 stimulation by Staphylocoocus aureus isolates. PLoS One 9:e96416. doi: 10.1371/journal.pone.0096416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medzhitov R, Schneider DS, Soares MP. 2012. Disease tolerance as a defense strategy. Science 335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Over B, Ziegler S, Foermer S, Weber AN, Bode KA, Heeg K, Bekeredjian-Ding I. 2013. IRAK4 turns IL-10+ phospho-FOXO+ monocytes into pro-inflammatory cells by suppression of protein kinase B. Eur J Immunol 43:1630–1642. doi: 10.1002/eji.201243217. [DOI] [PubMed] [Google Scholar]
- 46.Katholnig K, Kaltenecker CC, Hayakawa H, Rosner M, Lassnig C, Zlabinger GJ, Gaestel M, Muller M, Hengstschlager M, Horl WH, Park JM, Saemann MD, Weichhart T. 2013. p38alpha senses environmental stress to control innate immune responses via mechanistic target of rapamycin. J Immunol 190:1519–1527. doi: 10.4049/jimmunol.1202683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zielinski CE, Mele F, Aschenbrenner D, Jarrossay D, Ronchi F, Gattorno M, Monticelli S, Lanzavecchia A, Sallusto F. 2012. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature 484:514–518. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
- 48.Ziegler S, Gartner K, Scheuermann U, Zoeller T, Hantzschmann J, Over B, Foermer S, Heeg K, Bekeredjian-Ding I. 2014. Ca(2+)-related signaling events influence TLR9-induced IL-10 secretion in human B cells. Eur J Immunol 44:1285–1298. doi: 10.1002/eji.201343994. [DOI] [PubMed] [Google Scholar]
- 49.Parcina M, Miranda-Garcia MA, Durlanik S, Ziegler S, Over B, Georg P, Foermer S, Ammann S, Hilmi D, Weber KJ, Schiller M, Heeg K, Schneider-Brachert W, Gotz F, Bekeredjian-Ding I. 2013. Pathogen-triggered activation of plasmacytoid dendritic cells induces IL-10-producing B cells in response to Staphylococcus aureus. J Immunol 190:1591–1602. doi: 10.4049/jimmunol.1201222. [DOI] [PubMed] [Google Scholar]
- 50.Van Dyken SJ, Locksley RM. 2013. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu Rev Immunol 31:317–343. doi: 10.1146/annurev-immunol-032712-095906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grainger JR, Wohlfert EA, Fuss IJ, Bouladoux N, Askenase MH, Legrand F, Koo LY, Brenchley JM, Fraser ID, Belkaid Y. 2013. Inflammatory monocytes regulate pathologic responses to commensals during acute gastrointestinal infection. Nat Med 19:713–721. doi: 10.1038/nm.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szkaradkiewicz A, Karpinski TM, Zeidler A, Szkaradkiewicz AK, Masiuk H, Giedrys-Kalemba S. 2012. Cytokine response in patients with chronic infections caused by Staphylococcus aureus strains and diversification of their Agr system classes. Eur J Clin Microbiol Infect Dis 31:2809–2815. doi: 10.1007/s10096-012-1633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rose WE, Eickhoff JC, Shukla SK, Pantrangi M, Rooijakkers S, Cosgrove SE, Nizet V, Sakoulas G. 2012. Elevated serum interleukin-10 at time of hospital admission is predictive of mortality in patients with Staphylococcus aureus bacteremia. J Infect Dis 206:1604–1611. doi: 10.1093/infdis/jis552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.