Abstract
By producing toxins, Clostridium perfringens causes devastating diseases of both humans and animals. C. perfringens beta toxin (CPB) is the major virulence determinant for type C infections and is also implicated in type B infections, but little is known about the CPB structure-function relationship. Amino acid sequence comparisons of the CPBs made by 8 randomly selected isolates identified two natural variant toxins with four conserved amino acid changes, including a switch of E to K at position 168 (E168K) that introduces a potential trypsin cleavage site into the CPB protein of strain JGS1076. To investigate whether this potential trypsin cleavage site affects sensitivity to trypsin, a primary host defense against this toxin, the two CPB variants were assayed for their trypsin sensitivity. The results demonstrated a significant difference in trypsin sensitivity, which was linked to the E168K switch by using site-directed recombinant CPB (rCPB) mutants. The natural CPB variants also displayed significant differences in their cytotoxicity to human endothelial cells. This cytotoxicity difference was mainly attributable to increased host cell binding rather than the ability to oligomerize or form functional pores. Using rCPB site-directed mutants, differences in cytotoxicity and host cell binding were linked to an A300V amino acid substitution in the strain JGS1076 CPB variant that possessed more cytotoxic activity. Mapping of sequence variations on a CPB structure modeled using related toxins suggests that the E168K substitution is surface localized and so can interact with trypsin and that the A300V substitution is located in a putative binding domain of the CPB toxin.
INTRODUCTION
Clostridium perfringens is the etiological agent of numerous devastating diseases of both humans and animals (1). Illness caused by this Gram-positive, spore-forming bacterium is mediated by its extensive toxin arsenal, consisting of ∼17 toxins. Four of these toxins are used to classify individual strains into one of five types, A to E, depending on which toxins are encoded. By definition, both type B and C strains must produce alpha toxin (CPA) and beta toxin (CPB); type B strains are distinguished from type C strains by also producing epsilon toxin (2, 3).
Type B infection is typically confined to animals, including domestic livestock species, where it causes hemorrhagic enteritis and diarrhea, which may be accompanied by severe acute neurologic signs or sudden death (1, 4). Type C strains also cause animal diseases, typically in neonatal animals, which manifest as necrohemorrhagic enteritis and toxemia, resulting in rapid death of the animal (2, 4, 5). In addition to animal diseases, type C strains can infect people, causing enteritis necroticans (known regionally as Darmbrand or pig-bel) (6–9). These human infections, which are endemic in Southeast Asia, result in segmental necrotizing enteritis, which may lead to toxemia and are rapidly fatal without prompt medical intervention. Several predisposing factors for type C infection have been identified, including diets rich in trypsin inhibitors, such as sweet potato or colostrum (6–9). This association suggests that trypsin is a natural host defense against this disease, in large part because CPB is highly trypsin sensitive (10, 11). This sensitivity is consistent with the results of experiments using rabbits with ligated small intestinal loops, where purified CPB or C. perfringens type C cultures only caused intestinal lesions in the presence of trypsin inhibitor (12, 13).
Strong evidence supports CPB as the major factor for type C disease. First, type C culture supernatants treated with neutralizing anti-CPB antibodies lacked lethal properties in a mouse intravenous-injection lethality model, whereas supernatants treated with CPA or perfringolysin O (PFO) antibodies retained full lethality (14). Second, treatment of rabbit intestinal loops with highly purified beta toxin and trypsin inhibitor caused intestinal lesions consistent with those observed during challenge with type C strain CN3685 (13, 15). Finally, molecular Koch's postulates have been fulfilled using type C strain CN3685, which demonstrated sharply attenuated or no virulence for an isogenic CPB null mutant in both a rabbit small intestinal loop model of necrotic enteritis and a mouse oral challenge model of enterotoxemic disease (12, 13). In contrast, mutation-induced loss of both CPA and PFO production had little effect on the virulence of CN3685 in these animal models (12, 13).
CPB is the second-most-potent toxin produced by C. perfringens, with an intravenous 50% lethal dose (LD50) of ∼400 ng/kg of body weight in mice (16, 17). CPB is expressed as a 336-amino-acid single polypeptide that is processed to remove a 27-amino-acid signal sequence before secretion (3). Sequence homology exists between mature CPB and other toxins, including C. perfringens delta toxin (43% identity) (18), C. perfringens necrotic enteritis beta (NetB) toxin (38% identity) (19), and several Staphylococcus aureus toxins, including alpha toxin (28% identity) (20) and leukotoxin LukD (30% identity) (21). Based on this sequence homology, CPB is considered a member of the beta-pore-forming toxin family. Consistent with this view, CPB-induced cell death occurs through the generation of ion channels, which allow the unregulated efflux of K+ and influx of Na+, Ca2+, and Cl− ions (22, 23). Pore formation begins with the binding of CPB monomers to a currently unknown receptor that is believed to localize in lipid rafts on susceptible cells (22). Binding is then followed by rapid oligomerization of CPB monomers into heptameric or hexameric prepores on the cell surface. Once oligomerized, insertion of the prepore into the cellular membrane occurs, resulting in the generation of an active pore with an ion channel of approximately 12 Å (24). Recent studies suggest that the ion disruption mediated by CPB results in cellular death via programmed necrosis (25).
Little information about the CPB structure-function relationship is available. Since the alteration of a single amino acid can affect the function of clostridial pore-forming toxins (21, 23, 26–28), our current work identified natural CPB sequence variations and assessed their effects on the stability of these CPB variants in the presence of trypsin and on their CPB cytotoxicity in vitro.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains used in this study are described in Table 1. Clostridium perfringens strains were routinely cultured in fluid thioglycolate (FTG) broth (Difco). Toxin expression was carried out using TGY (3% tryptic soy broth [Becton Dickinson], 2% glucose [Sigma], 1% yeast extract [Becton Dickinson], and 0.1% thioglycolate [Sigma]) broth. Escherichia coli strains were routinely cultured in ATCC medium 1133 (1% tryptone [Becton Dickinson], 0.5% yeast extract [Becton Dickinson], 0.5% NaCl [Fisher Scientific], and 0.1% glucose [Sigma]) broth supplemented with 100 mg/liter of diaminopimelic acid (Alfa Aesar) and 10 mg/liter of thymidine (Sigma). As needed, E. coli was plated on ATCC medium 1133 agar plates supplemented with 100 μg/ml ampicillin (Fisher Scientific). All strains were incubated at 37°C.
TABLE 1.
Strains used in this study
| Strain | Type | Origin | Reference or source |
|---|---|---|---|
| CN1886 | B | Ovine isolate | 40 |
| CN3425 | B | Ovine isolate | 40 |
| NCTC8533 | B | Ovine isolatea | NCTC |
| JGS1984 | B | Unknown origin | 41 |
| CN3685 | C | Ovine isolatea | 14 |
| CN1797 | C | Animal origin, species unknown | 14 |
| JGS1495 | C | Porcine isolate | 41 |
| JGS1076 | C | Porcine isolatea | 41 |
Isolated from animal known to be experiencing disease.
Alignment of CPB sequences.
C. perfringens strains were grown overnight in FTG broth. Genomic DNA was harvested from cultures using the MasterPure Gram-positive DNA purification kit (Epicenter), following the manufacturer's instructions. The cpb gene was PCR amplified from the genomic preparations using primers JRT201201F and JRT201201R (Table 2). PCR products were confirmed by electrophoresis on a 1% agarose gel. Sequencing of PCR products was performed at the University of Pittsburgh Genomic Core Facility. Sequences were translated and aligned using CLC Workbench version 6.7.1 software.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| JRT201201F | TTCTCAAGTGTTGAAAAGAGCAA |
| JRT201201R | AAATTACATACGAAGGACTAATTTGAA |
| JRT201305F | TTGGATCCATGATATAGGTAAAACTA |
| JRT201205R | AAGAATTCCTAAATAGCTGTTAC |
| JT2014K40NF | CTAGAAATAATACATCAGATGGCTATAC |
| JT2014K40NR | TTATAGTAGTAGTTTTACCTATATC |
| JT2014E168KF | AGGTTCAATTAAAATAGAAGAAAATAAACC |
| JT2014E168KR | CCTATTTTGTATCCCATTGTATTG |
| JT2014V191IF | AATAGAATATATCCAACCTGATTTTTC |
| JT2014V191R | GTAGATGATTCAGCATATTCG |
| JT2014A300VF | TGGAATGGTGTTAACTGGGTAG |
| JT2014A300VR | ATTAAGATAATAACAGTTTCTTTCAC |
CPB purification.
Purification of CPB from C. perfringens strains CN3685 and JGS1076 was performed as described previously (13).
rCPB construction.
CPB was recombinantly expressed in chi-1776 E. coli (ATCC) using the pGEX-2T expression system (GE Healthcare), as described previously (28). Briefly, the cpb gene was PCR amplified from genomic DNA using primers JRT201305F and JRT201205R. The PCR inserts and pGEX-2T vector were digested with BamHI and EcoRI (NEB), ligated with T4 DNA ligase (NEB), and cloned into chi-1776 E. coli. DNA sequencing confirmed proper vector construction. The resultant construct allowed the expression of a glutathione S-transferase (GST)-CPB fusion protein, with thrombin cleavage allowing complete removal of the GST tag to generate free recombinant CPB (rCPB). Constructs for the CPB sequences of both strain CN3685 and strain JGS1076 were derived in this manner.
Site-directed mutagenesis of rCPB.
Site-directed rCPB mutants were generated in the CN3685 variant rCPB expression vector by individually switching each of the four amino acid substitutions to those present in the JGS1076 variant CPB by using the Q5 site-directed mutagenesis kit (NEB) according to the manufacturer's instructions. The following primer pairs were used to construct four point mutants: for the change of K to N at position 40 (K40N), JT2014K40NF and JT2014K40NR; for E168K, JT2014E168KF and JT2014E168KR; for V191I, JT2014V191IF and JT2014V191IR; and for A300V, JT2014A300VF and JT2014A300VR (Table 2). Sequencing of the constructs was performed to confirm successful site-directed mutagenesis and ensure that there were no secondary mutations in the coding region.
rCPB expression.
A 1-liter culture of ATCC 1133 medium was inoculated to an optical density at 600 nm (OD600) of 0.1 and then grown with aeration at 37°C until an OD600 of 0.5 to 0.6 was obtained. Isopropyl-β-d-thiogalactopyranoside (IPTG) (Gold Biotechnology) was added to a final concentration of 0.6 μM, and the culture was then incubated with aeration at room temperature for 3 h. Cells were harvested by centrifugation and frozen at −80°C overnight. Frozen pellets were thawed on ice, resuspended in phosphate-buffered saline (PBS) (pH 8.0), and disrupted by sonication. Insoluble material was removed from the lysates by centrifugation. A 1-ml volume of glutathione resin (GenScript) was added to the clarified lysate, and the mixture incubated at room temperature for 3 h. The sample was then transferred to a gravity column, the unbound material drained, and the resin rinsed with 20 ml of PBS. A 1-ml aliquot of PBS containing 3 U of thrombin (Novagen) was added to the resin, and the mixture was then incubated overnight at 4°C. Following incubation, the column was drained and rinsed (rCPB-containing fractions) with 5 ml of PBS. The final purity of rCPB was determined by SDS-PAGE, and its concentration was determined by Western blotting and densitometry using Image J software and known concentrations of a purified native CPB standard.
Trypsin digestion of CPB.
Purified CPB was diluted to 40 μg/ml in Hanks balanced salt solution (HBSS; Mediatech). Purified trypsin from porcine pancreas (Sigma) was added to a final concentration of 5 μg/ml (0.034 IU), and the reaction mixture was then incubated at 37°C. Samples were removed at 5-min intervals, mixed with 6× SDS-PAGE loading buffer, and boiled for 10 min to stop trypsin digestion. The boiled samples were then subjected to SDS-PAGE and Western blotting; the amount of CPB remaining relative to the starting amount was determined by densitometry for each time point, using Image J software. As a control for protease specificity, trypsin was pretreated with trypsin inhibitor (MP Biomedicals) for 30 min at room temperature prior to combining that mixture with CPB. The digestion was then performed as described above. rCPB was trypsin digested in the same fashion except that the starting concentrations were raised to 50 μg/ml CPB and the final trypsin concentration was reduced to 2.5 μg/ml (0.017 IU). Trypsin digestion of CPB was also performed in culture supernatants as follows: a CN3685 CPB null mutant, described elsewhere (13), was grown in TGY broth overnight at 37°C. Bacteria were removed by centrifugation at 10,000 × g for 5 min, followed by filtration of the supernatant through a 0.2-μM filter (Millipore). The cell-free supernatant was spiked with CPB to a final concentration of 40 μg/ml, and this sample was digested with 60 μg/ml (0.41 IU) of trypsin at 37°C for 60 min. Samples were removed at regular time intervals and processed as described above.
Cell culture.
Human umbilical vein endothelial cells (HUVEC; Life Technologies) were routinely cultured in medium 200 (Life Technologies) supplemented with low-serum growth supplement (Life Technologies) in a humidified chamber at 37°C with 5% CO2. To facilitate cell adhesion, all plates and flasks were pretreated with 60 μg/ml bovine collagen (Advanced Biomatrix) for 3 h prior to the addition of cells.
Cytotoxicity assays.
CPB cytotoxicity for HUVEC cells was measured using the CellTiter 96 one solution cell proliferation assay (Promega). HUVEC cells were seeded in 96-well plates and grown to confluent monolayers. Monolayers were treated for 1 h at 37°C with 2-fold dilutions (range, 2,000 to 31.3 ng/ml) of CPB diluted in HBSS. Positive- and negative-control wells received HBSS with or without 1% Triton X-100 (Sigma). After incubation, MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] reagent was added (20 μl/well), and incubation continued for an additional 2 h at 37°C. Following this incubation, the absorbance was determined with a plate reader set to 490 nm. For rCPB, HUVEC cell monolayers were grown in 96-well plates and then treated with 2-fold dilutions (range, 1,000 to 62.5 ng/ml) of rCPB for 6 h at 37°C. After this treatment, MTT reagent (20 μl/well) was added, and incubation continued for an additional 3 h. At that time, the absorbance was quantified with a plate reader set to 490 nm.
CPB cell binding and oligomerization assays.
Host cell binding and oligomerization assays were performed using the method of Steinthorsdottir et al. (23). Briefly, HUVEC cells were seeded in 6-well plates and grown to confluent monolayers. Those monolayers were then treated with 1 ml of HBSS containing 2 μg/ml CPB for 30 min at 37°C. Following this treatment, the cells were collected and rinsed 3 times with HBSS. After the final rinse, the cells were resuspended in 50 μl of HBSS containing 0.5% NP-40 (Fluka) and 250 U/ml Benzonase (Novagen) and incubated on ice for 10 min. Following this incubation, 6× SDS-PAGE loading buffer was added and the cells were boiled for 10 min. Samples were then subjected to SDS-PAGE and Western blotting. The relative levels of CPB binding were determined by densitometry using Image J software. rCPB binding assays were performed using the same method except that the initial incubation time for rCPB and HUVEC cells was increased from 30 min to 1 h.
Rubidium 86 release.
The formation of a functional CPB pore was assessed by measuring 86Rb release from host cells, as described previously (27). Briefly, 24-well plates were seeded with HUVEC cells and grown to confluent monolayers. These monolayers were radiolabeled with 1 ml of culture medium containing 2 μCi/ml 86Rb (specific activity of 1 Ci/g; PerkinElmer) for 4 h at 37°C. Following radiolabeling, cells were rinsed 2 times with HBSS and treated with 2-fold dilutions (range, 2,000 to 125 ng/ml) of purified CPB for 15 min at 37°C. After this toxin treatment, culture supernatants containing the released 86Rb were collected and quantified using a Cobra Quantum gamma counter (PerkinElmer). The maximum release of 86Rb was determined by treating cells with HBSS containing 1% Triton X-100 (Sigma); spontaneous release was determined by treating cells with HBSS alone. Data were converted and plotted as percentages of maximal release using the following calculation: [(experimental release − spontaneous release)/(maximal release − spontaneous release)] × 100.
Preparation of goat intestinal fluid.
The jejunal contents from a healthy adult Anglo Nubian goat euthanized by an overdose of sodium barbiturate (Beuthanasia; Schering-Plough Animal Health) were harvested and stored at −80°C until use. The intestinal contents were thawed at room temperature, and insoluble material was separated from the fluid fraction by two centrifugations, each at 15,000 × g. Total protease activity of the intestinal fluid was found to be ∼1 μg/μl using a colorimetric protease assay kit (Pierce). The goat intestinal fluid was stored at −80°C until needed. This animal work was approved by the University of California—Davis IACUC under permit number 16383.
Limited proteolytic digestion of CPB complex on the surface of HUVEC cells.
The protease sensitivity of oligomerized CPB complex present on the surface of HUVEC cells was assayed using a modification of previously described methods (27). Briefly, HUVEC cells were cultured in 6-well plates until confluent monolayers were achieved. These monolayers were treated for 30 min at 37°C with 1 ml of HBSS containing 2 μg/ml of CPB. Following this incubation, the cells were collected and rinsed 2 times with HBSS. After being rinsed, cells were treated with goat intestinal fluid (neat [full strength] or 1:3 and 1:6 dilutions) for 6 or 60 min at room temperature. Following this treatment, the cells were rinsed 3 times with HBSS. After the final rinse, the cells were resuspended in 50 μl of HBSS containing 0.5% NP-40 (Fluka) and 250 U/ml benzonase (Novagen) and incubated on ice for 10 min. Following this incubation, 6× SDS-PAGE loading buffer was added and the cells were boiled for 10 min. Samples were then subjected to SDS-PAGE and Western blotting. Binding levels relative to the binding of an undigested control were determined by densitometry using Image J software.
SDS-PAGE and Western blotting.
Samples for SDS-PAGE were mixed with 6× SDS-PAGE loading buffer and boiled for 10 min. Boiled samples were cooled to room temperature, loaded into a 10% acrylamide gel, and then electrophoresed. To identify proteins, gels were stained with Bio-Safe Coomassie G-250 stain (Bio-Rad) for 30 min and then destained with a 33% methanol (Fisher Scientific) and 7% acetic acid (Fisher Scientific) solution. Western blotting was performed as described elsewhere (13).
Modeling of the CPB structure.
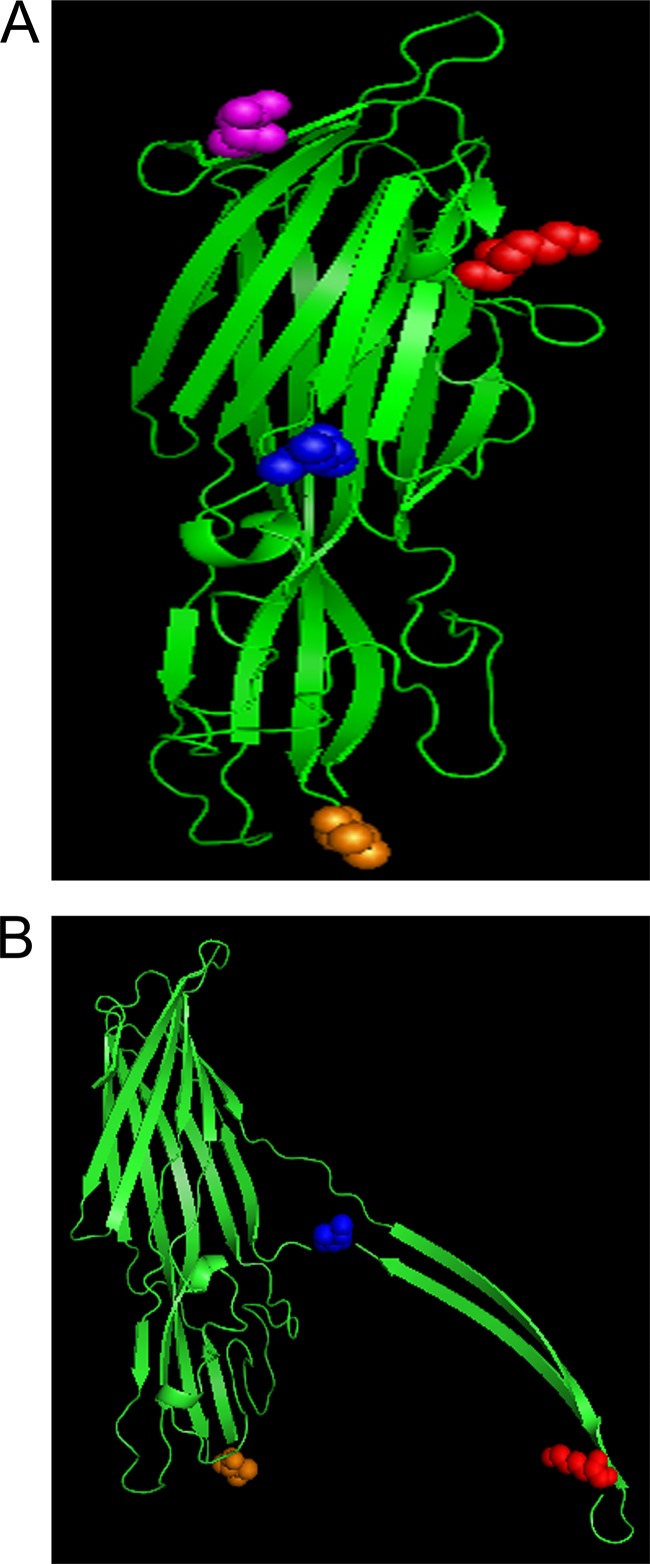
The CPB amino acid sequence was uploaded to the PHYRE2 program (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) for sequence analysis and threading. Based on the PHYRE2 results, two known structures were selected as the templates for threading of the CPB amino acid sequence. To model the secreted form of CPB, the known structure of C. perfringens delta toxin was selected as the template (29). For modeling the active form of CPB, the known structure of the membrane-active form of C. perfringens necrotic enteritis beta toxin (NetB) was selected as the template (30). Once modeled, further analysis and labeling of the model were performed using PyMOL software.
Statistical analysis.
For statistical analysis, each experiment was performed three to four times using two independently purified batches of CPB or rCPB. For single comparisons, significance was determined using the paired t test. For multiple comparisons, significance was determined using repeated measure one-way analysis of variance (ANOVA) with Dunnett's post hoc multiple-comparison test. All statistical analysis was performed using GraphPad Prism, version 6.
Nucleotide sequence accession numbers.
Nucleotide sequences are available in GenBank under accession numbers ABDU01000064.2 (strain JGS1495), KP064410 (strain CN1886), KP064409 (strain CN3425), KP064408 (strain NCTC8533), KP064407 (strain JGS1984), KP064405 (strain CN3685), KP064406 (strain CN1797), and KP064404 (strain JGS1076).
RESULTS
CPB sequence variations among isolates.
Eight CPB-positive strains (4 type B and 4 type C) were randomly chosen for nucleic acid sequencing of their cpb genes. The resultant sequences were then translated into amino acid sequences, and an alignment generated using CLC Workbench software (Fig. 1). This alignment indicated that 2 of the type C strains, i.e., JGS1495 and JGS1076, harbor the same 4 amino acid substitutions in their CPBs compared to the CPBs made by the other 6 sequenced strains, resulting in two natural variant toxins.
FIG 1.
Natural CPB sequence variations among eight surveyed type B and C strains. Nucleic acid sequencing of the cpb gene was performed on a randomly chosen sample set of C. perfringens type B and C isolates. Sequences were translated and aligned; the abridged results shown indicate the location of 4 amino acid substitutions (boxes). All other residues were identical between the CPBs made by these strains.
Trypsin digestion of CPB variants.
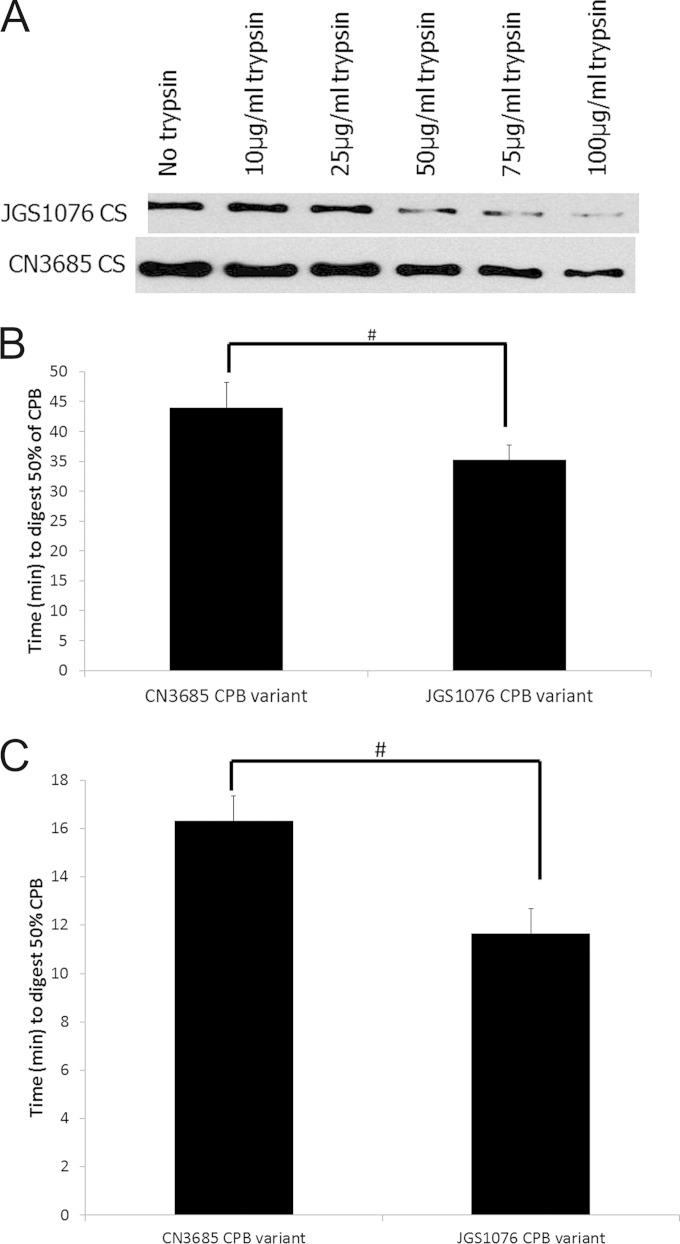
Since trypsin is an important host defense against CPB-mediated disease, the amino acid sequences for both CPB variants were analyzed for putative trypsin cleavage sites using the ExPASy Peptide Cutter (http://web.expasy.org/peptide_cutter/) program. This analysis predicted 34 trypsin cleavage sites in each CPB variant, with the amino acid differences between the two CPB variants noted in Fig. 1 creating one potential trypsin cleavage site unique to each variant. Therefore, the biological relevance of these unique putative trypsin cleavage sites was evaluated experimentally. Two C. perfringens strains, CN3685 and JGS1076 (one strain representing each CPB sequence variant identified), were selected for analysis of the trypsin sensitivity of their CPBs. The CPB of CN3685 has a putative unique trypsin cleavage site at residue 40 that is not present in the JGS1076 CPB variant, while the CPB of JGS1076 has a unique putative trypsin cleavage site at residue 168 that is not present in the CN3685 CPB variant. After a 30-min digestion of cell-free TGY culture supernatants generated from overnight cultures of CN3685 and JGS1076 with various amounts of trypsin (Fig. 2A), the results strongly suggested that the CPB variant produced by JGS1076 is more trypsin sensitive than the CN3685 CPB variant.
FIG 2.
Trypsin sensitivity of natural CPB variants. (A) Overnight cell-free TGY culture supernatants were produced for strains CN3685 and JGS1076 (representing each CPB variant) and subjected to digestion with various doses of trypsin at 37°C for 30 min. SDS-PAGE and Western blotting were performed to detect undigested CPB. A representative blot of CPB remaining after digestion is shown for each trypsin dose. Each digestion was performed twice. (B and C) An overnight TGY culture of a CN3685 CPB null mutant was used to generate a cell-free culture supernatant. Purified CPB (40 μg/ml) from both variants was used to spike culture supernatant (B) or HBSS (C), followed by digestion with trypsin. Western blotting and densitometry were performed to determine the amounts of CPB remaining relative to starting amounts of the toxin. The times for 50% digestion from four replicate digests are presented. Error bars indicate standard errors of the means. #, there was a significant (P < 0.05) difference in digestion times.
Variations in CPB production levels (CN3685 produces higher levels of CPB than JGS1076) or differences in supernatant backgrounds could conceivably affect the relative CPB trypsin sensitivities shown by the results presented in Fig. 2A. To control for these issues, each CPB variant was purified and the same amount of each purified CPB was added to a common background, i.e., a cell-free culture supernatant from a CN3685 CPB null mutant (13). When this spiked sample was digested with trypsin to determine the time needed for 50% digestion of CPB in the sample (Fig. 2B), a significant (P < 0.05) difference in trypsin sensitivity was again observed, with the JGS1076 CPB variant possessing greater sensitivity to trypsin than the CN3685 CPB variant.
Finally, to evaluate the effects of the unique trypsin cleavage sites on CPB trypsin sensitivity in the absence of any supernatant effects, an equal concentration of each purified CPB variant in HBSS was digested with trypsin to determine the time needed for 50% digestion of each CPB variant (Fig. 2C). As with the culture supernatant results, the JGS1076 CPB variant was significantly (P < 0.05) more sensitive to trypsin digestion than the CN3685 CPB variant. As a control for the specificity of the trypsin preparation used in this experiment, it was determined that treatment of this trypsin with trypsin inhibitor prior to its addition to samples blocked CPB digestion for both variants (data not shown).
The E168K amino acid substitution is responsible for the increased trypsin sensitivity of the JGS1076 CPB variant.
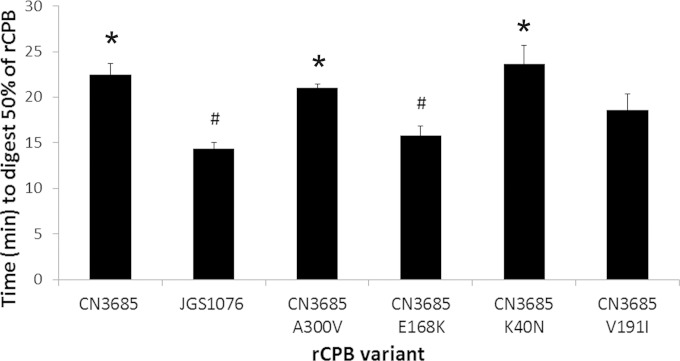
The results presented in Fig. 2 show that the JGS1076 CPB variant is more sensitive to trypsin than is the CN3685 CPB variant. Bioinformatic analysis of the sequences of the two CPB variants (Fig. 1) suggested that this difference could be due to the presence of a unique potential trypsin cleavage site that is predicted at residue 168 of the JGS1076 CPB variant but is absent from the CN3685 variant CPB. To test this hypothesis, both the JGS1076 and CN3685 CPB variants were recombinantly expressed in E. coli. Trypsin digestion of these rCPB variants in HBSS (Fig. 3) showed that the JGS1076 rCPB variant was significantly (P < 0.05) more trypsin sensitive than the CN3685 rCPB variant, in agreement with the native CPB results (Fig. 2). Amino acid substitutions were then introduced into the CN3685 rCPB variant by site-directed mutagenesis, individually switching each of the four variant amino acid residues to those present in the JGS1076 CPB variant sequence. Trypsin digestion of these site-directed rCPB variants showed that only the CN3685 E168K rCPB variant displayed a significant (P < 0.05) difference in trypsin sensitivity compared to that of the CN3685 rCPB. The trypsin sensitivity of the CN3685 E168K rCPB variant did not differ significantly from that of the JGS1076 rCPB (Fig. 3). The CN3685 V191I rCPB variant displayed intermediate trypsin sensitivity that was not statistically different from that of the CN3685 rCPB variant or the JGS1076 rCPB variant. This weak difference, if real, may be caused by a minor change in folding that has exposed another trypsin cleavage site.
FIG 3.
Trypsin digestion of rCPB variants. Recombinant CPBs containing JGS1076 or CN3685 sequences were constructed in pGEX-2T as GST fusion proteins. Site-directed mutagenesis was performed on the CN3685 rCPB variant, individually switching each of the 4 amino acids that differ between CN3685 CPB and JGS1076 CPB to the amino acids present in the JGS1076 sequence. Purified rCPB (50 μg/ml) from each of those variant rCPBs was then digested with trypsin in HBSS. Western blotting and densitometry were performed to determine the amounts of rCPB remaining relative to the starting amounts. The times for 50% digestion from four replicate digests are presented. Error bars indicate standard errors of the means. #, there was a significant (P < 0.05) difference from the results for the CN3685 variant rCPB. *, there was a significant (P < 0.05) difference from the results for the JGS1076 variant rCPB.
Effects of CPB sequence variations on cytotoxicity for cultured HUVEC cells.
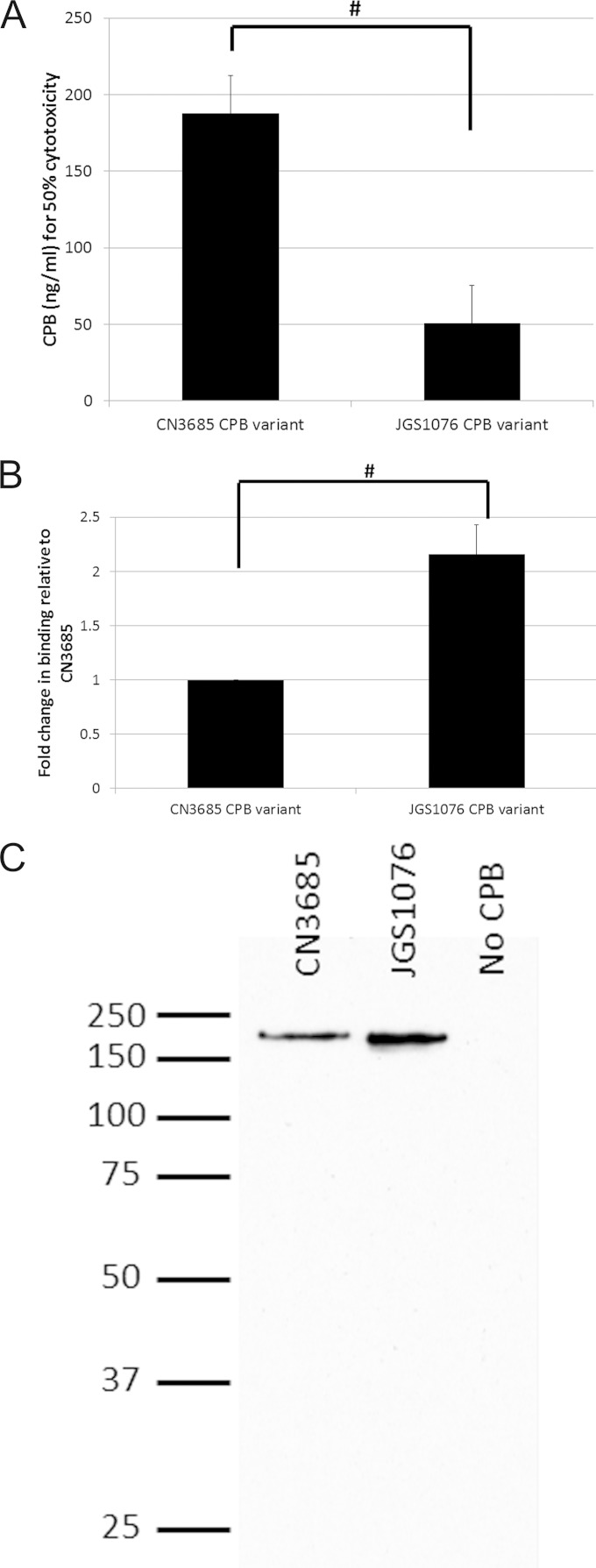
To further examine the phenotypic effects of CPB sequence variation, the cytotoxic activities of both purified CPB variants were compared using HUVEC monolayers and quantified with an MTT-based viable cell assay. As reported previously in the literature (23, 31–33), endothelial cells, such as HUVEC cells, are among the most CPB-susceptible cell lines known, and endothelial cells appear to be natural targets of CPB action in vivo. The results obtained here (Fig. 4A) indicate that both CPB variants exert cytotoxic effects on HUVEC cells. However, the JGS1076 CPB variant demonstrated a significant (P < 0.05) reduction of ∼3-fold in the amount of toxin needed to kill 50% of the cells in culture compared to the results for the CN3685 CPB variant.
Fig 4.
CPB sequence variation effects on host cell cytotoxicity and toxin binding. (A) CPB cytotoxicity for HUVEC cells was determined after a 3-h treatment using an MTT cell viability assay for both CPB variants. The mean amounts of CPB found to be necessary for causing 50% cytotoxicity from four replicate experiments are presented. (B) The ability of both CPB variants to bind to and oligomerize on HUVEC cells was assayed. Relative amounts of CPB binding/oligomerization were determined by Western blotting and densitometry. The mean results of four independent assays are presented. (C) A representative Western blot of the host cell binding is shown. Note the absence of any free (monomeric) CPB on these gels. #, there was a significant (P < 0.05) difference between the results for CPB variants. Error bars indicate standard errors of the means.
Differences in CPB variant binding to cultured HUVEC cells.
The early steps of CPB cytotoxicity involve binding of this toxin to susceptible host cells, followed immediately thereafter by oligomerization (22, 23). To begin determining the mechanism for the increase in cytotoxic activity of the JGS1076 CPB variant identified in the experiment whose results are shown in Fig. 4A, binding and oligomerization assays were performed with both CPB variants. The assays (Fig. 4B) identified a significant (P < 0.05), >2-fold increase in the amount of oligomerized CPB complex formed by the JGS1076 CPB variant compared to the amount formed by the CN3685 CPB variant. This difference was due to toxin binding and not to the ability of the CPB to oligomerize, since no bound monomeric CPB was detected for either CPB variant by Western blotting (Fig. 4C).
Comparison of insertion abilities of CPB variant complexes.
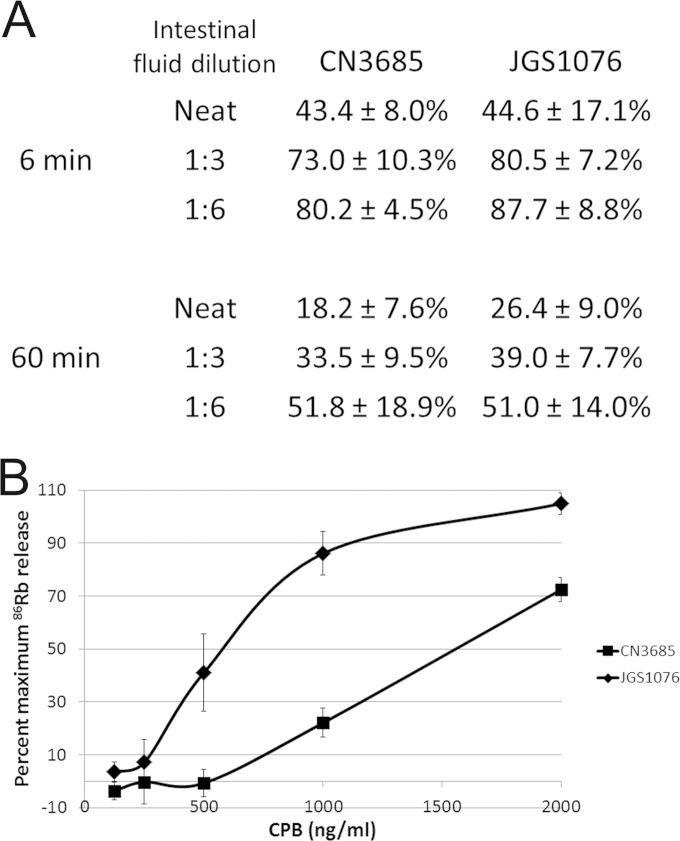
After its formation, the CPB complex is thought to insert into the host cell membrane, a process that should impart to the complex some protection from proteolytic digestion. To monitor for differences between the abilities of CPB variants to mediate insertion of their complexes into membranes, CPB complexes in HUVEC cells were subjected to digestion with goat intestinal fluid for 6 or 60 min, followed by Western blotting and densitometry. The results of this analysis, summarized in Fig. 5A, revealed similar digestion kinetics for the complexes formed by either CPB variant.
FIG 5.
Effects of CPB sequence variants on complex insertion and pore function. (A) HUVEC monolayers were treated with 2 μg/ml purified CPB variants for 1 h, allowing for complex formation and insertion. After rinsing away unbound CPB, on-cell proteolytic digestion of complexes was performed for 6 min or 1 h with goat intestinal fluid, diluted as indicated. Western blotting and densitometry were used to determine the amounts of CPB complexes remaining relative to the starting amount of each complex. The mean results from three independent experiments are shown, along with the standard errors of the means. (B) HUVEC cell monolayers were loaded with 86Rb and then treated with various CPB doses for 15 min. 86Rb released into the culture supernatant was quantified using a Cobra Quantum gamma counter. Averaged results from three independent experiments are presented as percentages of maximum 86Rb release. Error bars indicate standard errors of the means.
Proteolytic digestions are more typically performed with purified enzymes, such as pronase or trypsin, rather than intestinal fluid (34, 35). However, when this experiment was repeated using the complex formed by either CPB variant and 600 μg/ml trypsin or pronase, no digestion of the complex was observed (not shown). This absence of digestion could be due to complete insertion or internalization of the complex or to the intrinsic resistance of CPB complexes to these proteases. To clarify this situation, complexes were extracted from the cells prior to their digestion with 600 μg/ml either of trypsin or pronase. These assays still detected no complex digestion, indicating that the CPB complex is intrinsically insensitive to these proteases (data not shown).
Comparison of CPB pore formation.
Once the oligomerized CPB complex has inserted into the membrane, host cell death is elicited by increased ion fluxes, including efflux of K+ and influx of Na+, Ca2+, and Cl−, through an unregulated pore (22, 23). To assess whether their amino acid differences affect the ability of CPB variants to form functional ion pores, 86Rb release from labeled HUVECs was determined. These results indicated that complexes made with either CPB variant can form functional ion channels (Fig. 5B). However, the CN3685 CPB variant requires ∼3-fold more CPB than the JGS1076 CPB variant to achieve 50% maximal ion release.
Fine mapping of CPB variants suggests that the A300V substitution is critical for toxin binding and cytotoxicity.
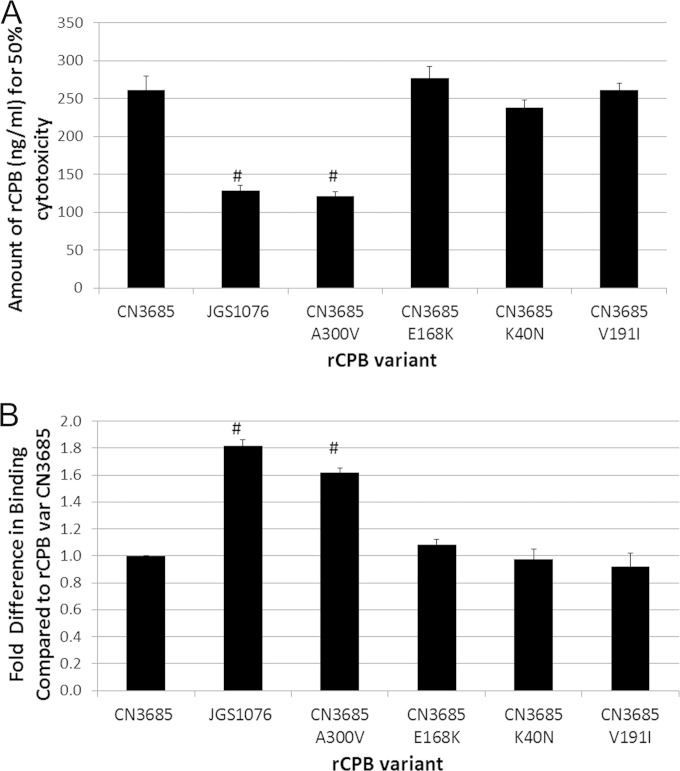
The results obtained with native CPB variants presented in Fig. 5 indicated that the increased cytotoxicity of the JGS1076 CPB variant in comparison to the cytotoxicity of the CN3685 CPB variant was primarily due to changes in binding to host cells. To fine map which of the four amino acid substitutions are critical for this binding difference, the rCPB site-directed mutants described earlier were utilized in cytotoxicity and binding experiments. Cytotoxicity testing with the CN3685 and JGS1076 variant rCPBs demonstrated a significant (P < 0.05) difference between these two rCPB variants, in agreement with the results for native CPBs. The results with each site-directed-mutant rCPB demonstrated that only the CN3685 A300V variant rCPB was significantly (P < 0.05) different from the CN3685 rCPB variant in its cytotoxic activity (Fig. 6A). The CN3685 A300V rCPB variant did not differ significantly from the JGS1076 rCPB variant. Similarly, when these rCPB site-directed mutants were assayed for binding and oligomerization ability on HUVECs, only the CN3685 A300V rCPB variant differed significantly (P < 0.05) from the CN3685 rCPB variant (Fig. 6B). No significant difference was observed between the JGS1076 rCPB variant and the CN3685 A300V rCPB variant.
FIG 6.
Testing individual site-directed rCPB mutants for binding and cytotoxicity phenotypes. (A) Recombinant CPBs containing JGS1076 and CN3685 sequences were constructed in pGEX-2T as GST fusion proteins. Site-directed mutagenesis was performed on the CN3685 rCPB variant, switching each of the 4 amino acids that differ between CN3685 CPB and JGS1076 CPB to the amino acid present in the JGS1076 sequence. Cytotoxicity of purified rCPBs for HUVEC cells was determined after a 9-h treatment using an MTT cell viability assay. Mean amounts of rCPB necessary to cause 50% cytotoxicity in three replicate experiments are presented. (B) Purified rCPB from the site-directed mutants was added to HUVEC cell monolayers for 1 h, allowing for binding/complex formation. After rinsing to remove unbound rCPB, Western blotting and densitometry were used to determine the relative amounts of rCPB variants bound to host cells. Mean results from three independent experiments are presented as fold changes in binding compared to that of the CN3685 variant rCPB. #, there was a significant (P < 0.05) difference compared to the results for the CN3685 variant rCPB.
DISCUSSION
Clostridium perfringens beta toxin, which is produced by type B and C strains, is the primary toxin responsible for disease caused by type C strains and likely also contributes to disease caused by type B strains (12, 13, 36). The work presented in this paper sought to identify whether natural CPB variations in a sampling of type B and C isolates have any effect on toxin function and trypsin sensitivity. Sequencing of 8 isolates identified CPB sequence variations among these strains, with the CPBs in two of the sequenced type C strains having 4 conserved amino acid changes compared to the CPBs made by the other six strains. Note that this limited sampling of strains does not preclude the possible existence of other CPB variants. Consistent with that possibility, the cpb genes from two additional porcine isolates were sequenced during revision of the manuscript, which identified a hybrid CPB variant possessing the V191I residue of JGS1076 but the K40N, E168K and A300V residues of CN3685 CPB (data not shown). This additional result indicates that the four observed CPB sequence variations in JGS1076 are not shared by all porcine isolates.
Trypsin is a key innate defense against disease mediated by CPB (1, 5–8, 13), rapidly inactivating the toxin and limiting the susceptible hosts to trypsin-deficient hosts, such as neonatal animals, people with diets that include foods rich in trypsin inhibitors, and individuals with pancreatic dysfunction (37, 38). No data exist on the actual trypsin levels in the intestine of animals with type C necrotizing enteritis or human patients with pig-bel. However, an older study (39) found that 10 of 17 young children living in the highlands of Papua New Guinea, a location where pig-bel was endemic, had fecal trypsin levels lower than the assay detection limit of 2 IU per ml of fecal material. This level of trypsin was significantly lower than the trypsin level measured in feces of children of the same age group living in Papua New Guinea cities, explaining why the highlands children with reduced trypsin levels were at increased risk for pig-bel. The analysis of cpb sequences in the current study suggested the possibility of differing trypsin sensitivities between CPB variants due to the presence or absence of a unique predicted novel trypsin cleavage site at residue 168 of the JGS1076 CPB, and this was established in the current study using a low, disease-relevant trypsin challenge of 0.41 IU. The importance of the trypsin cleavage site at residue 168 of the JGS1076 CPB variant was confirmed in vitro through trypsin digestion assays and by introducing an E186K amino acid substitution into the CN3685 rCPB variant.
This observation is interesting for several reasons. First, it demonstrates increased persistence of a CPB variant in the presence of trypsin levels that could be found in susceptible hosts (Fig. 2B). Second, it could suggest that CPB variants vary in their ability to cause disease in the host. However, it should be noted that the JGS1076 strain, which produces a CPB variant possessing the E168K amino acid switch, was isolated from an animal experiencing enteritis. Therefore, it is possible that some animals or humans affected by strains producing the more trypsin-sensitive CPB variant have sufficiently depressed trypsin levels to mask the increased trypsin sensitivity of the JGS1076 CPB variant, i.e., the trypsin levels of these hosts could be low due to diets rich in foods high in trypsin inhibitor, such as sweet potato or colostrum, or because of pancreatic dysfunction (1, 5–8, 13, 37, 38). Alternatively, it is possible that the sequence differences between the two CPB variants impart variations in activity (as discussed below) that may compensate for their increased trypsin sensitivity.
This work also identified a difference between the in vitro cytotoxicity of the two CPB variants for cultured HUVEC cells. Interestingly, although the JGS1076 CPB variant displayed increased trypsin sensitivity, it was more cytotoxic in vitro. This increased cytotoxicity was largely attributable to better host cell binding than the CN3685 CPB variant had. This increased cytotoxicity largely compensates for the higher trypsin sensitivity of the CN3685 CPB variant, and this may help to explain why strains producing a JGS1076-like CPB variant are still able to cause disease despite the increased trypsin sensitivity of their CPB variant. Conversely, strain CN3685 expresses ∼3 times more CPB than the JGS1076 strain (data not shown). During disease, this higher CPB expression, in combination with the increased trypsin resistance of its CPB variant, could help strains like CN3685 compensate for the decreased activity of their CPB variant.
Binding of CPB to host cells is rapidly followed by oligomerization of the toxin into either hexameric or heptameric complexes on the host cell (22, 23). Neither CPB variant was deficient for binding or oligomerization, since the CPB oligomer was detectable for both variants and no monomeric CPB was found bound to host cells. However, significantly more JGS1076 CPB bound and, thus, oligomerized to HUVEC cells than was seen for an equal dose of CN3685 CPB, suggesting that one of the amino acid differences in the JGS1076 variant CPB confers an enhanced binding phenotype for the currently unknown CPB receptor (further discussed below).
After the CPB complex forms, it inserts into host cell membranes, creating a pore. The insertion of toxins into membranes is often assayed by challenging a toxin complex present in host cell membranes with a purified protease, such as pronase, and then monitoring the degradation of the complex over time in order to detect partial or complete protection from digestion due to membrane insertion (27, 34, 35). Interestingly, when this assay was performed with CPB complexes in HUVEC cells, no digestion of the complex was observed using either pronase or trypsin for either CPB variant. This is apparently not due either to protection of the complex through its complete insertion into membranes or to the complex being internalized into the cytoplasm, since no CPB complex degradation was observed even if the complex was extracted from host cells (data not shown). This finding may suggest how CPB persists and causes disease in the intestine, i.e., although CPB is trypsin labile in the monomeric form, rapid oligomerization on host cells may provide CPB complexes with sufficient trypsin resistance to persist and induce cell death. In addition, the resistance of the CPB complex to trypsin or pronase provides further evidence of the stability of these complexes, which includes resistance to boiling, 2% SDS, and 8 M urea treatments (23).
Cell death by CPB is brought about by the formation of pores with a diameter of ∼12Å (24). These pores facilitate K+ efflux and Ca2+, Na+, and Cl− influx, resulting in cellular swelling and death via necrosis (22, 25). Testing CPB variants with an 86Rb release assay demonstrated that both variants can form a functional pore in HUVEC cells. Consistent with the cytotoxicity differences detected between the two CPB variants, the JGS1076 variant CPB was more efficient at 86Rb release than the CN3685 variant CPB; this enhanced pore formation correlates with the increased binding phenotype of the JGS1076 CPB variant, which should result in the release of more 86Rb using an equal dose of the two CPB variants.
To identify which amino acid substitution(s) were responsible for the binding and cytotoxicity phenotypes observed, we utilized recombinantly expressed CPB, assaying rCPBs containing the CN3685 or JGS1076 CPB sequences for their cytotoxicity and binding on cultured HUVECs. Consistent with the native CPB results, the JGS1076 rCPB variant displayed increased binding to and cytotoxicity of HUVEC cells compared to the cell binding and cytotoxicity of the CN3685 rCPB variant. Next, we generated a series of point mutations in the CN3685 variant rCPB sequence, thus converting individual residues to those present in the JGS1076 variant CPB sequence. The results of this work indicated that the A300V substitution is responsible for the observed phenotypic binding change, suggesting that this amino acid may lie in the binding or rim domain of CPB.
To better understand the effects of the sequence variations on CPB trypsin sensitivity and cytotoxic activity, CPB was modeled in both the secreted and membrane-active forms by threading the CPB sequence onto the published (29, 30) structures of C. perfringens delta and necrotic enteritis beta (NetB) toxins, respectively (Fig. 7). These toxins were chosen because delta toxin and NetB are also pore-forming toxins and they both share partial sequence identity with CPB. Based on this CPB modeling, the putative trypsin cleavage site at residue 168 should be surface exposed on monomeric CPB, allowing trypsin access to the cleavage site. This initial cleavage likely exposes additional trypsin sites, facilitating more rapid degradation of the JGS1076 CPB variant. Interestingly, based on the modeling of CPB in the secreted form, the putative trypsin site at residue 40 should also be surface exposed, although this variant was less sensitive to trypsin in this work. It is possible that this residue 40 cleavage site is not accessible to the active site of the trypsin enzyme or that cleavage of CPB close to the unstructured N terminus of the protein has a limited effect in exposing additional trypsin cleavage sites. Once CPB oligomerization on host cells has occurred, most of the CPB trypsin sites (including the residue 168 trypsin cleavage site of the JGS1076 CPB variant) will be buried in the complex, thereby offering protection from trypsin degradation.
FIG 7.
Modeling CPB to the known structures of C. perfringens delta and NetB toxin. The secreted (A) and membrane-active (B) structure of CPB was predicted by modeling CPB to the resolved structure of C. perfringens delta and NetB toxin, respectively (29, 30). The CN3685 CPB amino acid sequence was threaded onto the model toxin using the PHYRE2 program. Once modeled, the locations of the 4 amino acid substitutions were identified and labeled with PyMOL software as follows: purple, K40N; red, E168K; blue, V191I; orange, A300V. Note that the K40N substitution was not modeled on Net B due to size differences between the toxins.
To address the effect of the A300V amino acid substitution on CPB binding, the predicted membrane-active structure of CPB, derived from the known structure of NetB (30), was utilized (Fig. 7B). Using this threading technique, 3 of the 4 amino acid substitutions can be modeled; CPB is slightly larger than NetB (309 versus 293 amino acids, respectively) so the K40N substitution could not be modeled, as it is absent from NetB. This model predicts that the A300V switch occurs on one of 4 loops located near the bottom of the toxin, correlating with the predicted binding or rim domain of NetB (26, 30). It appears likely that, based on the homology with NetB and our result, these 4 loops create the binding domain of CPB, although more work is needed to confirm this hypothesis.
In summary, this work showed that CPB sequence variations directly affect toxin sensitivity to trypsin, as well as host cell binding and cytotoxicity. Furthermore, through identification of the A300V substitution as being responsible for the increased host cell binding and cytotoxicity and modeling of CPB using NetB as a scaffold, we can predict the rim domain of CPB, setting up future studies to characterize this critical CPB domain.
ACKNOWLEDGMENTS
This work was generously supported by National Institute of Allergy and Infectious Diseases (NIAID) grants T32 AI060525 and R01 AI056177.
REFERENCES
- 1.McClane BA, Uzal FA, Miyakawa MF, Lyerly D, Wilkins TD. 2006. The enterotoxic clostridia, p 688–752. In Dworkin M, Falkow S, Rosenburg E, Schleifer H, Stackebrandt E (ed), The prokaryotes, 3rd ed. Springer, New York, NY. [Google Scholar]
- 2.McClane BA, Lyerly DM, Wilkins TD. 2006. Enterotoxic clostridia: Clostridium perfringens type A and Clostridium difficile, p703–714. In Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood J (ed), Gram-positive pathogens, 3rd ed. ASM Press, Washington, DC. [Google Scholar]
- 3.Freedman JC, Theoret JR, Wisniewski JA, Uzal FA, Rood JI, McClane BA. 2 October 2014. Clostridium perfringens type A-E toxin plasmids. Res Microbiol doi: 10.1016/j.resmic.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Songer JG. 1996. Clostridial enteric diseases of domestic animals. Clin Microbiol Rev 9:216–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uzal FA, Vidal JE, McClane BA, Gurjar AA. 2010. Toxins involved in mammalian veterinary diseases. Open Toxinology J 2:24–42. [PMC free article] [PubMed] [Google Scholar]
- 6.Lawrence GW. 1997. The pathogenesis of enteritis necroticans, p 198–207. In Rood JI, McClane BA, Songer JG, Titball RW (ed), The clostridia: molecular genetics and pathogenesis. Academic Press, London, United Kingdom. [Google Scholar]
- 7.Johnson S, Gerding DN. 1997. Enterotoxemic Infections, p 117–140. In Rood JI, McClane BA, Songer JG, Titball RW (ed), The clostridia: molecular biology and pathogenesis. Academic Press, London, United Kingdom. [Google Scholar]
- 8.Walker PD. 1985. Pig-bel, p 93–115. In Borriello SP. (ed), Clostrida in gastrointestinal disease. CRC Press, Boca Raton, FL. [Google Scholar]
- 9.Ma M, Li J, McClane BA. 2012. Genotypic and phenotypic characterization of Clostridium perfringens isolates from Darmbrand cases in post-World War II Germany. Infect Immun 80:4354–4363. doi: 10.1128/IAI.00818-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakurai J, Duncan CL. 1978. Some properties of beta-toxin produced by Clostridium perfringens type C. Infect Immun 21:678–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macias Rioseco M, Beingesser J, Uzal FA. 2012. Freezing or adding trypsin inhibitor to equine intestinal contents extends the lifespan of Clostridium perfringens beta toxin for diagnostic purposes. Anaerobe 18:357–360. doi: 10.1016/j.anaerobe.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Uzal FA, Saputo J, Sayeed S, Vidal JE, Fisher DJ, Poon R, Adams V, Fernandez-Miyakawa ME, Rood JI, McClane BA. 2009. Development and application of new mouse models to study the pathogenesis of Clostridium perfringens type C enterotoxemias. Infect Immun 77:5291–5299. doi: 10.1128/IAI.00825-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sayeed S, Uzal FA, Fisher DJ, Saputo J, Vidal JE, Chen Y, Gupta P, Rood JI, McClane BA. 2008. Beta toxin is essential for the intestinal virulence of Clostridium perfringens type C disease isolate CN3685 in a rabbit ileal loop model. Mol Microbiol 67:15–30. doi: 10.1111/j.1365-2958.2007.06007.x. [DOI] [PubMed] [Google Scholar]
- 14.Fisher DJ, Fernandez-Miyakawa ME, Sayeed S, Poon R, Adams V, Rood JI, Uzal FA, McClane BA. 2006. Dissecting the contributions of Clostridium perfringens type C toxins to lethality in the mouse intravenous injection model. Infect Immun 74:5200–5210. doi: 10.1128/IAI.00534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vidal JE, McClane BA, Saputo J, Parker J, Uzal FA. 2008. Effects of Clostridium perfringens beta-toxin on the rabbit small intestine and colon. Infect Immun 76:4396–4404. doi: 10.1128/IAI.00547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill DM. 1982. Bacterial toxins: a table of lethal amounts. Microbiol Rev 46:86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakurai J, Fujii Y. 1987. Purification and characterization of Clostridium perfringens beta toxin. Toxicon 25:1301–1310. doi: 10.1016/0041-0101(87)90008-0. [DOI] [PubMed] [Google Scholar]
- 18.Manich M, Knapp O, Gibert M, Maier E, Jolivet-Reynaud C, Geny B, Benz R, Popoff MR. 2008. Clostridium perfringens delta toxin is sequence related to beta toxin, NetB, and Staphylococcus pore-forming toxins, but shows functional differences. PLoS One 3:e3764. doi: 10.1371/journal.pone.0003764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keyburn AL, Boyce JD, Vaz P, Bannam TL, Ford ME, Parker D, Di Rubbo A, Rood JI, Moore RJ. 2008. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog 4:e26. doi: 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter SE, Brown JE, Oyston PC, Sakurai J, Titball RW. 1993. Molecular genetic analysis of beta-toxin of Clostridium perfringens reveals sequence homology with alpha-toxin, gamma-toxin, and leukocidin of Staphylococcus aureus. Infect Immun 61:3958–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinthorsdottir V, Fridriksdottir V, Gunnarsson E, Andresson OS. 1998. Site-directed mutagenesis of Clostridium perfringens beta-toxin: expression of wild-type and mutant toxins in Bacillus subtilis. FEMS Microbiol Lett 158:17–23. doi: 10.1111/j.1574-6968.1998.tb12794.x. [DOI] [PubMed] [Google Scholar]
- 22.Nagahama M, Hayashi S, Morimitsu S, Sakurai J. 2003. Biological activities and pore formation of Clostridium perfringens beta toxin in HL 60 cells. J Biol Chem 278:36934–36941. doi: 10.1074/jbc.M306562200. [DOI] [PubMed] [Google Scholar]
- 23.Steinthorsdottir V, Halldorsson H, Andresson OS. 2000. Clostridium perfringens beta-toxin forms multimeric transmembrane pores in human endothelial cells. Microb Pathog 28:45–50. doi: 10.1006/mpat.1999.0323. [DOI] [PubMed] [Google Scholar]
- 24.Shatursky O, Bayles R, Rogers M, Jost BH, Songer JG, Tweten RK. 2000. Clostridium perfringens beta-toxin forms potential-dependent, cation-selective channels in lipid bilayers. Infect Immun 68:5546–5551. doi: 10.1128/IAI.68.10.5546-5551.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Autheman D, Wyder M, Popoff M, D'Herde K, Christen S, Posthaus H. 2013. Clostridium perfringens beta-toxin induces necrostatin-inhibitable, calpain-dependent necrosis in primary porcine endothelial cells. PLoS One 8:e64644. doi: 10.1371/journal.pone.0064644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan XX, Porter CJ, Hardy SP, Steer D, Smith AI, Quinsey NS, Hughes V, Cheung JK, Keyburn AL, Kaldhusdal M, Moore RJ, Bannam TL, Whisstock JC, Rood JI. 2013. Structural and functional analysis of the pore-forming toxin NetB from Clostridium perfringens. mBio 4(1):e00019-13. doi: 10.1128/mBio.00019-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Theoret JR, Shrestha A, Smedley JG III, McClane BA. 2012. Cysteine-scanning mutagenesis supports the importance of Clostridium perfringens enterotoxin amino acids 80 to 106 for membrane insertion and pore formation. Infect Immun 80:4078–4088. doi: 10.1128/IAI.00069-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagahama M, Kihara A, Miyawaki T, Mukai M, Sakaguchi Y, Ochi S, Sakurai J. 1999. Clostridium perfringens beta-toxin is sensitive to thiol-group modification but does not require a thiol group for lethal activity. Biochim Biophys Acta 1454:97–105. doi: 10.1016/S0925-4439(99)00026-5. [DOI] [PubMed] [Google Scholar]
- 29.Huyet J, Gilbert M, Popoff MR, Basak A. 2011. Crystallization and preliminary X-ray diffraction studies of delta-toxin from Clostridium perfringens. Acta Crystallogr Sect F Struct Biol Cryst Commun 67(Pt 3):369–371. doi: 10.1107/S1744309110054187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savva CG, Fernandes da Costa SP, Bokori-Brown M, Naylor CE, Cole AR, Moss DS, Titball RW, Basak AK. 2013. Molecular architecture and functional analysis of NetB, a pore-forming toxin from Clostridium perfringens. J Biol Chem 288:3512–3522. doi: 10.1074/jbc.M112.430223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurtner C, Popescu F, Wyder M, Sutter E, Zeeh F, Frey J, von Schubert C, Posthaus H. 2010. Rapid cytopathic effects of Clostridium perfringens beta-toxin on porcine endothelial cells. Infect Immun 78:2966–2973. doi: 10.1128/IAI.01284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miclard J, Jaggi M, Sutter E, Wyder M, Grabscheid B, Posthaus H. 2009. Clostridium perfringens beta-toxin targets endothelial cells in necrotizing enteritis in piglets. Vet Microbiol 137:320–325. doi: 10.1016/j.vetmic.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 33.Miclard J, van Baarlen J, Wyder M, Grabscheid B, Posthaus H. 2009. Clostridium perfringens beta-toxin binding to vascular endothelial cells in a human case of enteritis necroticans. J Med Microbiol 58:826–828. doi: 10.1099/jmm.0.008060-0. [DOI] [PubMed] [Google Scholar]
- 34.Smedley JG III, Uzal FA, McClane BA. 2007. Identification of a prepore large-complex stage in the mechanism of action of Clostridium perfringens enterotoxin. Infect Immun 75:2381–2390. doi: 10.1128/IAI.01737-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wieckowski EU, Kokai-Kun JF, McClane BA. 1998. Characterization of membrane-associated Clostridium perfringens enterotoxin following pronase treatment. Infect Immun 66:5897–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandez-Miyakawa ME, Fisher DJ, Poon R, Sayeed S, Adams V, Rood JI, McClane BA, Uzal FA. 2007. Both epsilon-toxin and beta-toxin are important for the lethal properties of Clostridium perfringens type B isolates in the mouse intravenous injection model. Infect Immun 75:1443–1452. doi: 10.1128/IAI.01672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrillo TM, Beck-Sague CM, Songer JG, Abramowsky C, Fortenberry JD, Meacham L, Dean AG, Lee H, Bueschel DM, Nesheim SR. 2000. Enteritis necroticans (pigbel) in a diabetic child. N Engl J Med 342:1250–1253. doi: 10.1056/NEJM200004273421704. [DOI] [PubMed] [Google Scholar]
- 38.Gui L, Subramony C, Fratkin J, Hughson MD. 2002. Fatal enteritis necroticans (pigbel) in a diabetic adult. Mod Pathol 15:66–70. doi: 10.1038/modpathol.3880491. [DOI] [PubMed] [Google Scholar]
- 39.Davis MW. 1984. Intestinal trypsin levels and susceptibility to pigbel, p 35-38. In Davis MW. (ed), Pigbel—necrotising enteritis in Papua New Guinea: proceedings of a workshop held in Goroka, Papua New Guinea, September 2-5, 1980. Monograph series (Papua New Guinea Institute of Medical Research), no. 6. PNG IMR, Goroka, Papua New Guinea. [Google Scholar]
- 40.Sayeed S, Li J, McClane BA. 2010. Characterization of virulence plasmid diversity among Clostridium perfringens type B isolates. Infect Immun 78:495–504. doi: 10.1128/IAI.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawires YS, Songer JG. 2005. Multiple-locus variable-number tandem repeat analysis for strain typing of Clostridium perfringens. Anaerobe 11:262–272. doi: 10.1016/j.anaerobe.2005.03.004. [DOI] [PubMed] [Google Scholar]