Abstract
To discern virulent from innocuous microbes, the innate immune system senses events associated with bacterial access to immunoprivileged sites such as the host cell cytosol. One such pathway is triggered by the cytosolic delivery of flagellin, the major subunit of the flagellum, by bacterial secretion systems. This leads to inflammasome activation and subsequent proinflammatory cell death (pyroptosis) of the infected phagocyte. In this study, we demonstrate that the causative agent of typhoid fever, Salmonella enterica serovar Typhi, can partially subvert this critical innate immune recognition event. The transcriptional regulator TviA, which is absent from Salmonella serovars associated with human gastroenteritis, repressed the expression of flagellin during infection of human macrophage-like (THP-1) cells. This mechanism allowed S. Typhi to dampen inflammasome activation, leading to reduced interleukin-1β (IL-1β) secretion and diminished cell death. Likewise, the introduction of the tviA gene in nontyphoidal Salmonella enterica serovar Typhimurium reduced flagellin-induced pyroptosis. These data suggest that gene regulation of virulence factors enables S. Typhi to evade innate immune recognition by concealing a pathogen-induced process from being sensed by the inflammasome.
INTRODUCTION
To distinguish between self and nonself, the host employs a multitude of innate pattern recognition receptors (PRRs) to initiate adequate host responses aimed at eradicating invading microbes. Activation of PRRs can be achieved by two conceptually different types of events: recognition of conserved microbial structures (pathogen-associated molecular patterns [PAMPs]) (1) and recognition of virulence-associated strategies aimed at manipulating host cells (patterns of pathogenesis) (2, 3). PAMPs are specific for certain groups of microbes regardless of their pathogenic potential, while host cell manipulation is a trait found exclusively in pathogens (2, 3).
PAMPs are typically detected by Toll-like receptors (TLRs), resulting in the activation of the NF-κB pathway (4). For example, flagellin, the major component of the flagellum, is recognized by TLR5, which enables the innate immune system to detect flagellated bacteria, regardless of their pathogenic potential (5, 6). In contrast, detection of host cell manipulation events allows the host to discern virulent from commensal microbes and scale the host defenses commensurate with the threat (2, 3). A prime example is the translocation of bacterial flagellin into the host cell cytosol through virulence-associated type III secretion system or type IV secretion system, which triggers a proinflammatory form of cell death termed pyroptosis (7–9). Pyroptotic cell death is characterized by the assembly and activation of the inflammasome (10, 11). In the center of this multiprotein complex, active caspase-1 processes pro-interleukin-1β (IL-1β) as well as pro-IL-18 into the biologically active forms by proteolytic cleavage. IL-1β and IL-18 are highly proinflammatory cytokines that are critical for coordinating host responses during bacterial infections (10, 11). In addition to providing a proinflammatory signal, pyroptotic macrophage cell death releases intracellular bacteria from their protected intracellular niche, thus exposing the microbes to uptake and killing by neutrophils (12).
Not surprisingly, a subset of bacterial pathogens has evolved to evade immune recognition to establish infection. One well-documented strategy to avoid detection is to alter the structure of a PAMP, thus decreasing its affinity for the cognate PRR. For example, the flagellin expressed by Campylobacter jejuni lacks a TLR5 binding site and is a poor activator of this signaling pathway (13, 14). Likewise, Yersinia pestis can evade TLR4 signaling by producing a tetra-acylated form of lipid A when grown at a temperature that resembles that of the mammalian host (15, 16). Although strategies designed to avoid recognition of PAMPs by their respective PRRs have been described extensively, it is unclear how bacterial pathogens can evade inflammasome activation, a critical host defense pathways poised to detect manipulation of host cells by bacterial pathogens.
Human infection with nontyphoidal serovars of Salmonella enterica, e.g., serovar Typhimurium, results in an acute, self-limiting gastroenteritis with a quick onset of symptoms (<24 h on average) (17). Invasion and replication inside the intestinal mucosa ignite a vigorous host response characterized by a massive infiltration of neutrophils (18). In contrast, ingestion of the human-restricted pathogen Salmonella enterica serovar Typhi results in typhoid fever, a severe febrile illness due to the dissemination of S. Typhi to systemic sites (19, 20). After a median incubation period of 2 weeks, only a fraction of patients develop intestinal complications, with the infiltrates being dominated by mononuclear cells (21). The absence of an initial severe inflammatory response in the mucosa suggests that unlike Salmonella serovars associated with gastroenteritis, S. Typhi evades detection by innate PRRs in the gut (19, 20). Interestingly, the genome sequences of S. Typhi and S. Typhimurium are highly syntenic (22), thus raising the question of which genetic determinants are involved in escaping innate immune detection. Here we investigated whether and by which mechanisms S. Typhi is able to evade the activation of the inflammasome in human macrophage-like THP-1 cells. Our results demonstrate that reduced flagellin expression enables S. Typhi to avoid inflammasome activation. Tight regulation of the flagellar operon in S. Typhi is mediated by TviA, a regulatory protein that is absent from Salmonella serovars causing gastroenteritis in humans.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All plasmids and bacterial strains used in this study are listed in Table 1. Salmonella strains were routinely cultured in LB broth (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl) or on LB agar plates (15 g/liter agar) at 37°C. To induce optimal expression of TviA and capsule biosynthesis, strains were cultured aerobically in tryptone yeast extract (TYE) broth (10 g/liter tryptone, 5 g/liter yeast extract) or in Dulbecco's modified Eagle's medium (DMEM), as indicated. Antibiotics were added, when appropriate, at the following concentrations: 100 mg/liter carbenicillin, 30 mg/liter chloramphenicol, 50 mg/liter kanamycin, and 50 mg/liter nalidixic acid. Acidic phosphatase (PhoN) activity was determined on agar plates containing the chromogenic substrate 5-bromo-4-chloro-3-indolyl phosphate (X-phos) at a concentration of 40 mg/liter.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or characteristic(s) | Source or referencea |
|---|---|---|
| Strains | ||
| S. Typhi | ||
| Ty2 | Wild type; Vi+ | ATCC 19430 |
| SW74 | Ty2 ΔtviB-vexE::Cmr; Vi− | 37 |
| SW347 | Ty2 ΔviaB; Vi− | 25 |
| SW359 | Ty2 ΔfliC; Vi+ | 25 |
| SW483 | Ty2 ΔviaB::Kanr ΔfliC | 25 |
| S. Typhimurium | ||
| IR715 | Wild type; Nalr derivative of ATCC 14028 | 54 |
| SL1344 | Wild type | 55 |
| AJB715 | IR715 phoN::Kanr | 56 |
| SW473 | IR715 ΔfliC fljB5001::MudJ | 23 |
| SW474 | IR715 phoN::tviA-Cmr | 25 |
| SW681 | IR715 phoN::Kanr ΔfliC fljB5001::MudCm | 26 |
| SW718 | IR715 phoN::tviA-Cmr ΔfliC fljB5001::MudJ | This study |
| Plasmids | ||
| pWSK29 | ori(pSC101); Ampr | 57 |
| pTVIA1 | S. Typhi tviA under the control of its native promoter in pWSK29 | 58 |
ATCC, American Type Culture Collection.
Differentiation of THP-1 cells.
THP-1 cells were routinely cultured as a suspension in RPMI 1640 containing 1 mM sodium pyruvate, 10 mM HEPES, 2 mM glutamine, and 10% FBS (fetal bovine serum; Gibco) (complete medium) for a maximum of ∼20 passages. Cells were seeded at a density of 5 × 105 cells per well in 0.5 ml complete medium containing 50 ng/ml phorbol 12-myristate 13-acetate (PMA; Sigma). After 48 h, cells were washed twice with Dulbecco's phosphate-buffered saline (DPBS), and fresh complete medium was added. The medium was replaced daily for another 4 days. On the last day, cells were switched to complete medium containing 2% FBS.
Generation of bone marrow-derived macrophages.
Bone marrow-derived macrophages (BMMs) were isolated from C57BL/6 mice as reported previously (23). Briefly, bone marrow cells were flushed from femora with 10 ml of cold RPMI medium. Cells were seeded in 5 petri dishes containing 20 ml RPMI medium supplemented with 20% heat-inactivated FBS, L929 cell-conditioned medium, 1 mM glutamine, 1% nonessential amino acids, and 1% antibiotic-antimycotic mix (Invitrogen) (BMM medium). On day 3, 10 ml of fresh BMM medium was added, and cells were incubated for an additional 5 days. Harvested BMMs were resuspended in RPMI supplemented with 10% heat-inactivated FBS and 1 mM glutamine, seeded at a density of 5 × 105 cells per well in a 24-well plate, and incubated for 24 h. The medium was then replaced with RPMI medium supplemented with 2% FBS and 1 mM glutamine, and cells were incubated for another 24 h prior to infection.
Immortalized bone marrow-derived macrophages.
v-raf/v-myc immortalized bone marrow macrophages (24) derived from C57BL/6 and isogenic, Nlrc4-deficient mice were a kind gift from K. Fitzgerald. Cells were routinely cultured in DMEM supplemented with 10% FBS and 1 mM glutamine. For experiments, cells were seeded at a density of 1 × 105 cells per well in a 24-well plate and incubated at 37°C in 5% CO2 for 24 h prior to infection.
Macrophage infection experiments.
Bacterial strains for macrophage infection studies were grown in TYE broth for 9 h at 30°C, as described previously (9). Macrophages were treated with 100 ng/ml lipopolysaccharide (Escherichia coli O55:B5; Sigma) 4 h prior to infection. If applicable, the inhibitor Z-WEHD-FMK (R&D Systems) (dissolved in dimethyl sulfoxide [DMSO]), specific for caspase-1, -4, and -5, was added 4 h prior to infection at a final concentration of 25 μM (as indicated in the figure legends). Mock treatment consisted of treatment with DMSO at a final concentration of 0.05% or 0.125%. Cells were infected with a multiplicity of infection (MOI) of 5 bacteria to 1 macrophage. Bacterial uptake was synchronized by centrifugation at 1,000 × g for 5 min at room temperature. Cells were incubated for 25 min at 37°C in 5% CO2. The medium was replaced with fresh medium containing 25 μg/ml gentamicin. At the indicated time points after infection (4 h, 8 h, or 16 h), plates were centrifuged at 1,000 × g for 5 min at room temperature to sediment particles. Supernatants were collected and stored at −80°C for further analysis. To determine the bacterial load at the end of the experiment, cells were lysed in 0.5 ml 1% Triton X-100 in phosphate-buffered saline (PBS), the wells were washed with 0.5 ml PBS, and both suspensions were combined. Serial dilutions of the cell lysates were spread onto LB agar plates, and CFU were enumerated.
Quantification of IL-1β secretion.
The total concentration of IL-1β, including pro-IL-1β and IL-1β, in culture supernatants was determined by using mouse-specific (eBioscience) and human-specific (Biolegend) IL-1β enzyme-linked immunosorbent assay kits according to the manufacturers' instructions.
Measurement of macrophage cell death.
To determine macrophage cell death, the relative amount of lactate dehydrogenase (LDH) released into the supernatant was measured with a coupled enzymatic assay (CytoTox96; Promega) according to the instructions provided by the manufacturer. The percentage of cell death was calculated by dividing the LDH activity of the sample (minus the LDH activity of mock-treated cells) by the activity of cells lysed by Triton X-100 treatment (minus the LDH activity of mock-treated cells) and multiplying this value by 100. All raw absorbance values were corrected by measuring cell death in control reaction mixtures with medium only added.
Quantification of mRNA levels from bacterial cultures.
Bacterial gene expression was quantified as described previously (25, 26). Briefly, S. Typhi strains were cultured in TYE broth for 9 h at 30°C. RNA was isolated by using the Aurum Total RNA minikit (Bio-Rad) and treated with DNase (DNAfree DNase treatment and removal reagents; Ambion). RNA was reverse transcribed according to the recommendations of the manufacturer (TaqMan reverse transcription reagents; Applied Biosystems). SYBR green-based real-time PCR (Applied Biosystems) was performed by using the primers listed in Table 2. Data were analyzed by using the comparative threshold cycle (CT) method, and the transcription level of flagellar genes was normalized to that of the housekeeping gene gmk, encoding guanylate kinase.
TABLE 2.
Oligonucleotides used in this study
| Target gene | Oligonucleotide sequence | Reference |
|---|---|---|
| fliC | 5′-GTAACGCTAACGACGGTATC-3′ | 25 |
| 5′-ATTTCAGCCTGGATGGAGTC-3′ | ||
| flhC | 5′-CTGTAACTGCTGCGGTGGGAAC-3′ | 25 |
| 5′-GGATGGCGGCTGGCATAAACTAC-3′ | ||
| gmk | 5′-TTGGCAGGGAGGCGTTT-3′ | 59 |
| 5′-GCGCGAAGTGCCGTAGTAAT-3′ | ||
| 16S rRNA | 5′-TGTTGTGGTTAATAACCGCA-3′ | 60 |
| 5′-GACTACCAGGGTATCTAATCC-3′ |
Detection of bacterial gene expression in macrophages.
PMA-differentiated THP-1 cells and immortalized bone marrow-derived macrophages were infected with Salmonella strains at an MOI of 10, as described above. Three hours after infection, RNA was extracted from ∼3 × 106 macrophage-like cells by the Tri reagent method (Molecular Research Center), further purified by using the RNeasy MinElute cleanup kit (Qiagen), and treated with DNase (DNAfree DNase treatment and removal reagents). One microgram of total RNA was used in a 50-μl reverse transcription-PCR (RT-PCR) reaction mixture (TaqMan reverse transcription reagents). Two microliters of cDNA was used as a template for SYBR-based (Applied Biosystems) real-time PCR in a 12.5-μl volume with a final concentration of 250 nM each primer (Table 2). Data analysis was performed by using the comparative CT method. The expression level of the bacterial fliC gene was normalized to Salmonella 16S rRNA levels.
Analysis of bacterial protein expression by Western blotting.
For the experiment shown in Fig. 1B, strains were cultured at 37°C for 16 h in LB broth, while for experiments shown in Fig. S2 in the supplemental material, bacteria were grown at 30°C for 9 h in TYE broth. The turbidity of the culture (optical density at 600 nm [OD600]) was measured, and cells were lysed in loading buffer (25 mM Tris-HCl [pH 6.8], 8% glycerol, 0.8% SDS, 0.04% bromophenol blue, 10% 2-mercaptoethanol). A fraction of this boiled solution, corresponding to ∼5 × 107 CFU, was used for SDS-polyacrylamide gel electrophoresis (PAGE). To determine bacterial protein levels in infected macrophages (Fig. 2D), PMA-differentiated THP-1 cells were infected with bacterial strains precultured in TYE medium at an MOI of 5. The bacterial infection was synchronized by mild centrifugation, as described above. Cells were then incubated for 25 min at 37°C. The medium was replaced with fresh medium containing 25 μg/ml gentamicin to kill extracellular bacteria. Two hours thirty minutes later, ∼3 × 106 THP-1 cells were lysed in 50 μl of loading buffer. Samples were boiled for 3 min and resolved by SDS-PAGE. Proteins were transferred from gels onto a polyvinylidene fluoride membrane (Millipore) by using a semidry transfer method (Bio-Rad Laboratories). Expression of FliC, GroEL, and RpoA was detected by using rabbit Salmonella H antiserum d (Difco) and Salmonella H antiserum i (Difco), anti-GroEL antiserum (Sigma), and anti-RpoA antiserum (4RA2; Santa Cruz Biotechnology), respectively, as the primary antibodies. A horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse antibody (Bio-Rad) was used as the secondary antibody. Chemiluminescence (Western Lightning Plus ECL chemiluminescent substrate; PerkinElmer) was visualized by using a BioSpectrum (UVP) or a G:Box (Syngene) imaging system. Raw images were processed with Photoshop CS2 (Adobe Systems) to uniformly adjust brightness levels of the entire image.
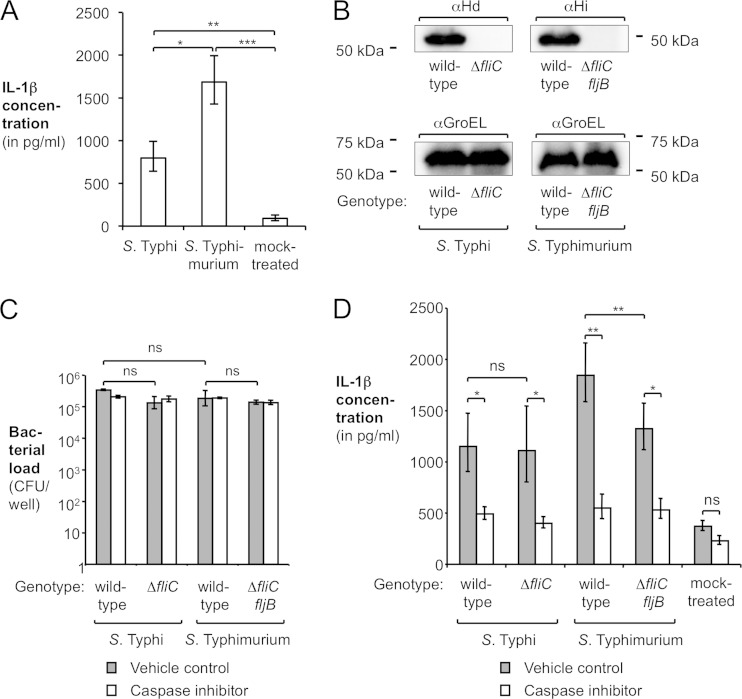
FIG 1.
Effect of flagellin on the interaction of S. Typhi and S. Typhimurium with human macrophage-like cells. (A) PMA-differentiated THP-1 cells were infected with the S. Typhi wild-type strain (Ty2) or the S. Typhimurium wild-type strain (IR715) or mock treated, and the concentration of IL-1β in the supernatant was determined by an enzyme-linked immunosorbent assay (n = 3). (B) In vitro expression of flagellin by the S. Typhi wild-type strain, an isogenic ΔfliC mutant, the S. Typhimurium wild-type strain, or an isogenic ΔfliC fljB mutant. S. Typhi and S. Typhimurium flagellins were detected by Western blotting using Salmonella H antiserum d (αHd) and antiserum i (αHi), respectively. As a loading control, the amount of the chaperone GroEL (αGroEL) was determined. The approximate locations of standard proteins are indicated. (C and D) PMA-differentiated THP-1 cells were treated with DMSO (vehicle control) or 25 μM caspase inhibitor Z-WEHD-FMK and subsequently infected with the S. Typhi wild-type strain (Ty2), an isogenic ΔfliC mutant (SW359), the S. Typhimurium wild-type strain (IR715), or an isogenic ΔfliC fljB mutant (SW473) or mock treated. After 8 h, the intracellular bacterial load (C) and the amount of IL-1β released into the supernatant (D) were determined (n = 4). Bars represent geometric means ± standard errors. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not statistically significant.
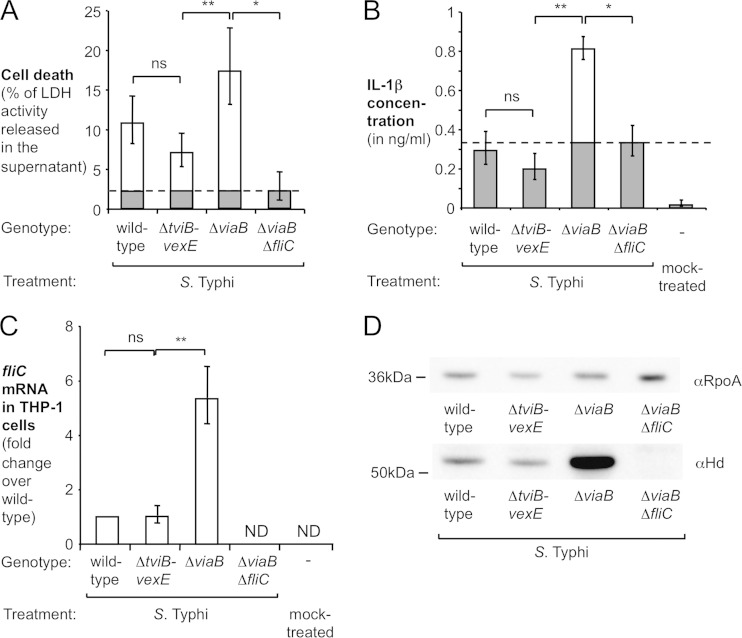
FIG 2.
Effect of TviA-mediated flagellin repression on cell death and IL-1β secretion induced by S. Typhi. (A and B) PMA-differentiated THP-1 cells were infected for 8 h with the indicated S. Typhi strains precultured in TYE broth. (A) Cell death was determined by using an LDH release assay. (B) IL-1β secretion into the supernatant was measured by an enzyme-linked immunosorbent assay. FliC-dependent cell death and IL-1β release are indicated by white bars, while LDH release and IL-1β secretion independent of FliC (i.e., amount induced by a ΔviaB ΔfliC mutant [dashed lines]) are indicated by gray bars (n = 6). (C and D) Quantification of S. Typhi fliC levels inside macrophage-like cells. PMA-differentiated THP-1 cells were infected with the S. Typhi wild-type strain, a ΔviaB mutant, a ΔtviB-vexE mutant, and a ΔviaB ΔfliC mutant for 3 h. (C) Total RNA was extracted, and relative fliC mRNA levels, normalized to the amount of 16S rRNA, were determined by quantitative RT-PCR (n = 4). (D) Lysates of infected THP-1 cells were analyzed by Western blotting for expression of flagellin (αHd antiserum). As a loading control, the alpha subunit of the bacterial RNA polymerase (RpoA) was quantified. The approximate locations of marker proteins are indicated on the left. Bars represent geometric means ± standard errors. *, P < 0.001; **, P < 0.01; ns, not statistically significant; ND, none detected.
Proteolytic cleavage of caspase-1.
Immortalized BMMs were infected, as described above, at an MOI of 5 with S. Typhimurium strains precultured in TYE broth. After 16 h, cells were lysed in loading buffer, and the lysate as well as the culture supernatant were analyzed by Western blotting, as described above. As a primary antibody, a rat polyclonal antibody against caspase-1 p20 (kindly provided by Genentech) was used.
Mouse experiments.
This study was performed in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals (27). The protocol was approved by the Institutional Animal Care and Use Committee of the University of California, Davis.
Female C57BL/6 mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and housed under specific-pathogen-free conditions. Access to food and water was provided ad libitum. S. Typhimurium strains, cultured in TYE broth as described above, were washed once with DPBS. Groups of 4 animals were intraperitoneally infected with a total of 1 × 104 CFU of S. Typhimurium (5 × 103 CFU of each strain; competitive infection design) in 0.1 ml DPBS. The ratio of the indicated strains in the inoculum was determined by culturing on selective agar plates. Animals were euthanized 48 h after infection. The spleen was removed, weighed, and homogenized in 5 ml DPBS. Homogenates were serially diluted in PBS and plated onto selective, chromogenic medium (X-phos agar plates containing nalidixic acid). The competitive index for each animal was calculated by dividing the bacterial load of the tviA-expressing strain by the bacterial load of the strain lacking the tviA gene, normalized by the ratio of the strains in the inoculum. One group of animals was infected with the S. Typhimurium wild type and the phoN mutant, with both strains being recovered in similar numbers (data not shown).
Statistics.
Raw data were log transformed for descriptive and inferential statistical analysis. The Student t test was used to assess statistical significance. P values of <0.05 were considered to be statistically significant. For each figure panel, the number of biological repeats (n) and statistical significance for relevant comparisons are indicated.
RESULTS
Absence of flagellin-dependent IL-1β secretion by S. Typhi-infected macrophages.
The mechanisms of inflammasome activation during S. Typhimurium infection have been studied extensively by the use of cultured mouse macrophages and murine disease models. In contrast, S. Typhi is a human-restricted pathogen and does not infect rodents. To investigate the interaction of S. Typhi and S. Typhimurium with phagocytes, we used differentiated THP-1 cells, a human macrophage-like cell line (28). THP-1 cells were infected with the S. Typhi wild-type strain (Ty2) or the S. Typhimurium wild-type strain (IR715). After 8 h, the total concentration of IL-1β in the supernatant was determined by an enzyme-linked immunosorbent assay. The IL-1β detected in this assay comprised both immature, pro-IL-1β leaking from damaged cells as well as the biologically active, mature form of IL-1β and thus serves as an indirect readout for inflammasome activation. Infection of THP-1 cells with S. Typhimurium or S. Typhi induced IL-1β secretion compared to mock-treated cells (Fig. 1A), which was reported previously (29–31). Curiously, the S. Typhi wild-type strain elicited significantly less IL-1β secretion than did the S. Typhimurium wild-type strain, suggesting that S. Typhi induces less inflammasome activation than does S. Typhimurium.
Previous reports demonstrated that S. Typhimurium induces IL-1β secretion through type III secretion system-mediated translocation of flagellin (FliC) into the cytosol of murine macrophages (7, 9, 32). We therefore sought to determine whether flagellin contributes to inflammasome activation by S. Typhi in human macrophages. The genome of biphasic S. Typhimurium contains two flagellin genes (fliC and fljB), while the chromosome of monophasic S. Typhi possesses only one flagellin gene (fliC). We thus utilized flagellin-deficient mutants (Fig. 1B) of S. Typhi (ΔfliC mutant [SW359]) and S. Typhimurium (ΔfliC fljB mutant [SW473]) and infected THP-1 cells with these strains. THP-1 cells were treated with either Z-WEHD-FMK, an inhibitor specific for caspase-1, -4, and -5 (dissolved in DMSO), or vehicle (DMSO treatment) (Fig. 1C and D). The number of intracellular bacteria (Fig. 1C) and IL-1β secretion (Fig. 1D) were determined 8 h after infection. The intracellular bacterial loads of the flagellin-deficient S. Typhi ΔfliC mutant as well as the S. Typhimurium ΔfliC fljB mutant were found to be similar to those of the respective wild-type strains, indicating that the absence of flagellin did not affect uptake and intracellular replication in this model system (Fig. 1C). Consistent with the concept that flagellin translocation into human macrophage-like cells results in inflammasome activation, inactivation of fliC and fljB in S. Typhimurium (ΔfliC fljB mutant) significantly reduced IL-1β secretion (P < 0.01) (Fig. 1D). In contrast, inactivation of fliC in S. Typhi (ΔfliC mutant) did not further reduce the amount of IL-1β secreted by infected macrophage-like cells (Fig. 1D), and the levels of IL-1β production induced by the S. Typhi strains were similar to those induced by the flagellin-deficient S. Typhimurium ΔfliC fljB mutant. Interestingly, the nonflagellated S. Typhi ΔfliC mutant and S. Typhimurium ΔfliC fljB mutant still induced a significant amount of IL-1β secretion compared to that in mock-treated cells, suggesting that other flagellin-independent pathways contribute to inflammasome activation, as reported previously (33). IL-1β secretion was dependent on caspase-1, since the IL-1β concentration in supernatants from inhibitor-treated cells was comparable to that in uninfected cells. Taken together, these results suggested that S. Typhi triggered little flagellin-dependent inflammasome activation during infection of human macrophage-like cells.
S. Typhi reduces IL-1β secretion and macrophage cell death by repressing FliC expression.
One DNA region that is present in the S. Typhi chromosome but absent from S. Typhimurium is Salmonella pathogenicity island 7 (SPI-7), a 134-kb insertion that was likely acquired by horizontal gene transfer (34). Situated within SPI-7 is the viaB locus (see Fig. S1A in the supplemental material). This operon contains the genes for the regulation (tviA), biosynthesis (tviBCDE), and export (vexABCDE) of the virulence-associated (Vi) capsular polysaccharide (19). TviA is a transcriptional activator of the viaB operon (35, 36) and regulates virulence factors outside SPI-7, including the flagellar regulon (25). TviA controls the expression of FliC (see Fig. S1B and S1C in the supplemental material) by repressing the transcription of the flagellar master regulator FlhDC (see Fig. S1D in the supplemental material) in response to osmolarity (25, 37). TviA-mediated repression of flagellin expression occurs under conditions that are similar to those of the osmolarity encountered in tissue (plasma osmolarity) (26). We hypothesized that TviA might repress the transcription of the flagellar regulon, thereby concealing flagellin from inflammasome sensing in macrophages. To test this hypothesis, we infected THP-1 cells with an S. Typhi wild-type strain (Ty2), a nonencapsulated ΔtviB-vexE mutant (SW74) that expresses tviA, a ΔviaB mutant (SW347) that lacks the capsule biosynthesis genes as well as the regulator TviA (deletion of tviABCDE vexABCDE), and a ΔviaB ΔfliC mutant (SW483). Cell death (lactate dehydrogenase [LDH] release assay) and IL-1β secretion were monitored at various time points (Fig. 2A and B; see also Fig. S2 in the supplemental material). We reasoned that this comparison would allow us to disentangle the contribution of the Vi capsular polysaccharide from the contribution of TviA, since a simple deletion of tviA is pleiotropic (35) and would affect Vi capsular polysaccharide expression and fliC expression simultaneously. The S. Typhi wild-type strain and the nonencapsulated, tviA-expressing ΔtviB-vexE mutant induced similar levels of LDH release (Fig. 2A; see also Fig. S2A and S2B in the supplemental material) and IL-1β secretion (Fig. 2B; see also Fig. S2C and S2D in the supplemental material) into the supernatant, suggesting that the Vi capsular polysaccharide did not alter inflammasome activation. In contrast, cells infected with the nonencapsulated ΔviaB mutant lacking the tviA gene secreted significantly larger amounts of LDH and IL-1β. Inactivation of the fliC gene in the absence of the negative regulator tviA (ΔviaB ΔfliC mutant) significantly diminished cell death and IL-1β production (Fig. 2A and B; see also Fig. S2 in the supplemental material). This result suggested that in the absence of TviA, FliC of S. Typhi triggers inflammasome activation, which was similar to the flagellin-mediated inflammasome activation observed in nontyphoidal S. Typhimurium (Fig. 1D). Collectively, these results suggested that TviA enables S. Typhi to evade host detection through the inflammasome by negatively regulating the expression of FliC.
To directly test whether fliC transcription was indeed repressed when bacteria were residing intracellularly, we infected differentiated THP-1 cells with the S. Typhi wild-type strain (Ty2), a ΔtviB-vexE mutant (SW74), a ΔviaB mutant (SW347), and a ΔviaB ΔfliC mutant (SW483) precultured in TYE medium (Fig. 2C) or in tissue culture medium (see Fig. S3A in the supplemental material). Total RNA was isolated, and the abundance of bacterial fliC mRNA (standardized to 16S rRNA) was determined by quantitative reverse transcription-PCR (qRT-PCR) (Fig. 2C; see also Fig. S3A in the supplemental material). Inactivation of capsule biosynthesis genes (ΔtviB-vexE mutant) did not affect fliC transcription; however, a significant increase in the level of fliC mRNA was observed in the ΔviaB mutant compared to the ΔtviB-vexE mutant (P < 0.01). Similarly, flagellin protein levels were significantly increased in the viaB mutant in comparison with the wild-type strain and the ΔtviB-vexE mutant during infection of THP-1 cells (Fig. 2D).
Introduction of TviA into S. Typhimurium decreases activation of the NLRC4 inflammasome.
To further study the role of TviA in evading inflammasome activation, we utilized an S. Typhimurium strain in which the phoN gene was replaced with the S. Typhi tviA promoter and coding sequence. The phoN gene encodes an acidic phosphatase that does not contribute to pathogenesis and is considered a neutral locus (26). Paralleling the results from S. Typhi (see Fig. S1B in the supplemental material), the expression of tviA reduced protein levels of FliC in S. Typhimurium in vitro (see Fig. S1E in the supplemental material). No significant expression of the second-phase flagellin FljB was detected (data not shown).
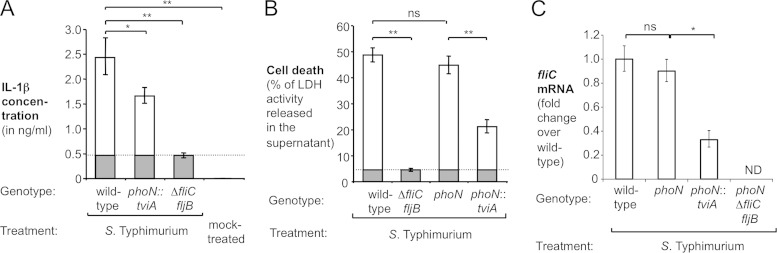
We next investigated whether TviA-mediated changes in gene transcription altered the interaction of S. Typhimurium with mouse macrophages. To this end, murine bone marrow-derived macrophages (BMMs) were infected with the S. Typhimurium wild-type strain (IR715), the tviA-expressing phoN::tviA mutant (SW474), and a flagellin-deficient ΔfliC fljB mutant (SW473). Neither the presence of tviA nor the lack of flagellin expression altered the amount of bacteria recovered at the end of the experiment (see Fig. S3C in the supplemental material). As demonstrated in previous studies (7, 9), the ΔfliC fljB mutant elicited significantly (P < 0.01) less IL-1β secretion than did the wild-type strain (Fig. 3A), indicating that in this model system, flagellin is a major factor contributing to inflammasome activation. Most importantly, the expression of tviA significantly (P < 0.05) reduced IL-1β production in BMMs (Fig. 3A). Similar results were obtained when human THP-1 cells were infected with S. Typhimurium strains (see Fig. S3B in the supplemental material). In a second set of experiments, BMMs were infected with the S. Typhimurium wild-type strain, the tviA-expressing phoN::tviA mutant, the flagellin-deficient ΔfliC fljB mutant, and a phoN mutant (AJB715), and LDH release after 16 h was determined, to study the effect of tviA on cell death (Fig. 3B). Inactivation of phoN did not alter cell death induced by S. Typhimurium, while the flagellin-deficient ΔfliC fljB mutant as well as the strain carrying the tviA gene (phoN::tviA mutant) induced significantly less cell death than did their isogenic parents (Fig. 3B). Mirroring the findings with S. Typhi (Fig. 2C), the expression of tviA in S. Typhimurium reduced fliC mRNA levels during infection of immortalized murine bone marrow-derived macrophages (Fig. 3C). Collectively, these data supported the concept that tviA reduces flagellin-dependent pyroptosis in murine macrophages.
FIG 3.
Heterologous expression of TviA in S. Typhimurium reduces IL-1β secretion. The indicated S. Typhimurium strains were cultured in TYE broth prior to infections. (A and B) Murine BMMs were infected with the indicated S. Typhimurium strains, and IL-1β secretion (A) (n = 3) as well as cell death (B) (n = 4) were measured after 16 h. FliC-dependent IL-1β release and cell death are indicated by white bars, while LDH release and IL-1β secretion independent of FliC (i.e., amount induced by a ΔfliC fljB mutant [dashed lines]) are indicated by gray bars. (C) Immortalized BMMs isolated from wild-type animals were infected with the indicated S. Typhimurium strains for 3 h. Total RNA was extracted from infected cells, and relative fliC mRNA levels, normalized to 16S rRNA levels, were determined by quantitative RT-PCR (n = 4). Bars represent geometric means ± standard errors. *, P < 0.001; **, P < 0.01; ns, not statistically significant.
One critical step during the activation of the inflammasome is the proteolytic cleavage of procaspase-1 into two fragments, p10 and p20 (11, 38). To test the hypothesis that TviA reduces inflammasome activation, we analyzed lysates of infected BMMs by Western blotting by probing membranes with an antibody specific for the cleaved p20 subunit. Both the S. Typhimurium wild type and the phoN mutant induced processing of caspase-1, while no caspase-1 cleavage was detected in cells infected with the tviA-expressing phoN::tviA mutant or a phoN ΔfliC fljB mutant (SW681) (Fig. 4A), thus providing direct support for the hypothesis that TviA-mediated repression of flagellin expression can mediate inflammasome evasion.
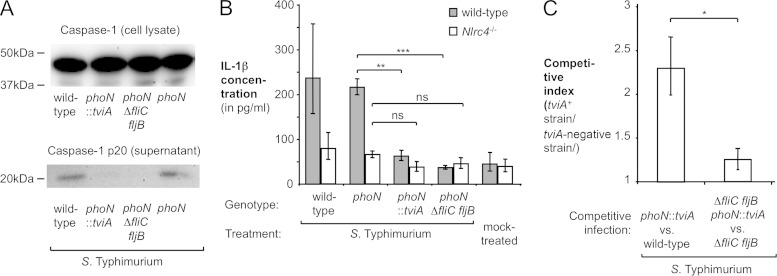
FIG 4.
Effect of TviA on inflammasome activation. S. Typhimurium strains were cultured in TYE broth prior to infections. (A and B) Immortalized murine BMMs were infected with the indicated S. Typhimurium strains for 16 h. (A) Cell lysates and supernatants were analyzed by Western blotting using caspase-1-specific antiserum. The approximate locations of marker proteins are indicated. (B) Immortalized BMMs isolated from wild-type animals or Nlrc4-deficient animals were infected with S. Typhimurium strains or mock treated, and IL-1β secretion was quantified (n = 3). (C) Groups of C57BL/6 animals (n = 4) were intraperitoneally infected with an equal mixture of the wild type (IR715) and the phoN::tviA mutant (SW474) or the ΔfliC fljB mutant (SW473) and the ΔfliC fljB phoN::tviA mutant (SW718). Bacterial numbers in the spleen were determined at 48 h postinfection, and the ratio of tviA-expressing strain over the strain lacking the tviA gene (competitive index) was calculated. Bars represent geometric means ± standard errors. *, P < 0.001; **, P < 0.01; ns, not statistically significant.
The inflammasome is activated upon the assembly of a multiprotein complex, a process that is mediated by a NOD-like receptor (NLR) and that is enhanced by the adaptor protein apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) (10, 11). In S. Typhimurium-infected murine macrophages, inflammasome activation in response to flagellin translocation requires the NLR protein NLRC4 (7, 9, 39). To test the hypothesis that the TviA-mediated repression of flagellar genes diminishes the activation of the NLRC4 inflammasome, we infected immortalized BMMs generated from wild-type (C57BL/6) mice and isogenic NLRC4-deficient mice with S. Typhimurium strains and measured the production of IL-1β (Fig. 4B; see also Fig. S3D in the supplemental material). While the wild-type strain and the phoN mutant induced IL-1β secretion into the supernatant by wild-type BMMs, significantly less cytokine production was observed for cells infected with the phoN::tviA mutant or the phoN ΔfliC fljB mutant. Significantly less IL-1β was detected in supernatants from NLRC4-deficient macrophages infected with the wild type and the phoN mutant, demonstrating that inflammasome inactivation is in part dependent on this NLR. Most importantly, no effect of TviA on IL-1β secretion was observed in the absence of NLRC4 (Fig. 4B).
Due to the host restriction of S. Typhi, animal models available to study typhoid fever pathogenesis are very limited. Since the transfer of the tviA gene into S. Typhimurium recapitulated aspects of S. Typhi inflammasome evasion in vitro, we investigated potential consequences of TviA-mediated flagellin repression in a murine model of S. Typhimurium infection. In a model for the systemic phase of infection, it was recently demonstrated that flagellin-overexpressing S. Typhimurium strains are cleared in a manner independent of IL-1β and IL-18. Infected macrophages that undergo NLRC4-depdendent pyroptosis release bacteria into the extracellular milieu. This removal of S. Typhimurium from its intracellular location makes the pathogen susceptible to uptake and killing by neutrophils, thus enhancing tissue clearance (12). To test whether tviA-mediated repression of flagellin expression could enhance bacterial survival in this model of the systemic phase of infection, groups of C57BL/6 mice were intraperitoneally infected with an equal mixture of the S. Typhimurium wild-type strain and a phoN::tviA mutant. The bacterial loads of both strains in the spleen were determined 48 h after infection (Fig. 4C). The wild type was recovered in lower numbers than was the tviA-expressing strain. Most importantly, the expression of tviA in S. Typhimurium did not impact bacterial numbers in the absence of flagellin (fliC fljB mutant background) (Fig. 4C). Taken together, these experiments suggested that TviA-mediated flagellin repression enhances the fitness of S. Typhimurium in this model of the systemic phase of infection.
DISCUSSION
The innate immune system deploys a multitude of PRRs to detect the presence of bacteria in the tissue, with the aim of clearing invading microbes. Conversely, pathogenic microbes have evolved mechanisms to overcome antibacterial host defenses. For example, virulence-associated type III secretion systems in S. Typhimurium accidentally translocate flagellin into the cytosol of murine host cells in vitro, an event that triggers the activation of the NLRC4 inflammasome and pyroptotic cell death (7, 9). S. Typhimurium averts this course of events during infection of the murine host by limiting the expression of flagellin. FliC is expressed in the lumen of the intestinal tract as well as in Peyer's patches, while the S. Typhimurium populations colonizing the mesenteric lymph nodes and the spleen lack FliC expression (40–42). Tissue-specific regulation of the flagellar regulon in the mouse model is critical for colonization of systemic sites, since S. Typhimurium mutants engineered to constitutively express FliC induce pyroptosis of their macrophage host, with the extracellular bacteria being rapidly cleared by neutrophils (12). Downregulation of FliC expression in S. Typhimurium is mediated by PhoPQ (41), a two-component system that is conserved between S. Typhi and S. Typhimurium. It is conceivable that S. Typhi restricts FliC expression in a PhoPQ-dependent manner and thus limits inflammasome activation once the bacteria have reached the mesenteric lymph nodes or systemic sites of the human host.
In comparison to S. Typhimurium, S. Typhi exhibits an additional, tighter layer of flagellin regulation. Repression of flagellin expression further reduces inflammatory cell death and cytokine production by S. Typhi-infected macrophages. Downmodulation of flagellin expression is coordinated by TviA, a transcriptional regulator encoded within the S. Typhi-specific pathogenicity island 7. Expression of TviA itself is regulated by the EnvZ/OmpR system in response to osmolarity (43). Under conditions encountered in the gut lumen, TviA expression is repressed (26, 44), thus permitting the production of flagella to assist in invasion through motility. Once the bacteria reach the mucosa, the decrease in osmolarity leads to the expression of TviA and a rapid repression of flagellin expression (26). This swift downregulation of flagellin production occurring in the mucosa has been shown to be critical for reducing pattern recognition by TLR5 (26, 37) and limiting adaptive immune responses (45). In addition to the recognition of extracellular flagellin by TLR5 and subsequent NF-κB activation, the mammalian host also senses the delivery of flagellin to the cytosol of mononuclear cells such as macrophages or dendritic cells, ultimately leading to the activation of the NLRC4 inflammasome. While flagellation is a property shared by commensal and pathogenic bacteria, cytosolic access is a typical signature of bacterial pathogens and thus a “pattern of pathogenesis” (2, 3). The ability to differentiate between the localization of flagellin (intracellular versus extracellular) allows the mammalian host to discriminate pathogens from microbes with lower pathogenic potential. Here we provide evidence that S. Typhi dampens inflammasome activation in human macrophage-like cells in a TviA-dependent manner. The presence of the tviA gene reduced the expression of the bacterial stimulant flagellin. Additionally, tviA represses the expression of the invasion-associated type III secretion system (46), which is involved in the translocation of bacterial flagellin into cultured macrophages (7, 9). The concomitant downregulation of the translocated bacterial flagellin as well as the secretion apparatus by TviA may allow S. Typhi to mask the cytosolic translocation of flagellin, a pattern of pathogenesis, in the intestinal mucosa.
Upon reaching the intestinal mucosa, extracellular bacteria are phagocytosed by mononuclear cells (47). Evasion of inflammasome activation during early phases of the infection process by S. Typhi may have multiple potential consequences. Inflammasome activation results in the release of IL-1β and IL-18, proinflammatory cytokines critical for orchestrating innate immune responses against S. Typhimurium (48–50). In addition to coordinating cytokine release, inflammasome activation also results in the death of infected macrophages (12). Upon initial invasion, bacteria are thought to disseminate from the gastrointestinal tract to deeper tissues inside CD18-expressing phagocytes (51), and survival within macrophages is thought to be critical for replication within tissues (52, 53). Here we demonstrate that the expression of the S. Typhi regulator TviA in S. Typhimurium can reduce flagellin-dependent cell death of murine macrophages and augments fitness in a systemic model of infection. These findings suggest that evasion of inflammasome activation by S. Typhi by concealing flagellin expression might be an important virulence strategy aimed to aid in enhancing fitness within the human host.
Taken together, our data provide evidence that S. Typhi can obscure a pattern of pathogenesis through stringent gene regulation of the immunostimulant flagellin. This mechanism might allow the organism to avoid death of the infected host cell and aid the pathogen in establishing a protected niche.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI044170. E.L.R. was supported by a scholarship from the Brazilian Science without Borders program (CNPq).
We thank Genentech for providing the caspase-1 antibody.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02803-14.
REFERENCES
- 1.Janeway CA., Jr 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol 54(Part 1):1–13. [DOI] [PubMed] [Google Scholar]
- 2.Vance RE, Isberg RR, Portnoy DA. 2009. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe 6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blander JM, Sander LE. 2012. Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat Rev Immunol 12:215–225. doi: 10.1038/nri3167. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, Akira S. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng J, Akira KS, Underhill DM, Aderem A. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 6.Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SL, Cookson BT, Aderem A. 2003. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol 4:1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 7.Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol 7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 8.Fink SL, Cookson BT. 2005. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun 73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, Jagirdar R, Inohara N, Vandenabeele P, Bertin J, Coyle A, Grant EP, Nunez G. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol 7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 10.Rathinam VA, Vanaja SK, Fitzgerald KA. 2012. Regulation of inflammasome signaling. Nat Immunol 13:333–332. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamkanfi M, Dixit VM. 2012. Inflammasomes and their roles in health and disease. Annu Rev Cell Dev Biol 28:137–161. doi: 10.1146/annurev-cellbio-101011-155745. [DOI] [PubMed] [Google Scholar]
- 12.Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, Warren SE, Wewers MD, Aderem A. 2010. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol 11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watson RO, Galan JE. 2005. Signal transduction in Campylobacter jejuni-induced cytokine production. Cell Microbiol 7:655–665. doi: 10.1111/j.1462-5822.2004.00498.x. [DOI] [PubMed] [Google Scholar]
- 14.de Zoete MR, Keestra AM, Wagenaar JA, van Putten JP. 2010. Reconstitution of a functional Toll-like receptor 5 binding site in Campylobacter jejuni flagellin. J Biol Chem 285:12149–12158. doi: 10.1074/jbc.M109.070227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montminy SW, Khan N, McGrath S, Walkowicz MJ, Sharp F, Conlon JE, Fukase K, Kusumoto S, Sweet C, Miyake K, Akira S, Cotter RJ, Goguen JD, Lien E. 2006. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat Immunol 7:1066–1073. doi: 10.1038/ni1386. [DOI] [PubMed] [Google Scholar]
- 16.Robinson RT, Khader SA, Locksley RM, Lien E, Smiley ST, Cooper AM. 2008. Yersinia pestis evades TLR4-dependent induction of IL-12(p40)2 by dendritic cells and subsequent cell migration. J Immunol 181:5560–5567. doi: 10.4049/jimmunol.181.8.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glynn JR, Palmer SR. 1992. Incubation period, severity of disease, and infecting dose: evidence from a Salmonella outbreak. Am J Epidemiol 136:1369–1377. [DOI] [PubMed] [Google Scholar]
- 18.McGovern VJ, Slavutin LJ. 1979. Pathology of salmonella colitis. Am J Surg Pathol 3:483–490. doi: 10.1097/00000478-197912000-00001. [DOI] [PubMed] [Google Scholar]
- 19.Wangdi T, Winter SE, Baumler AJ. 2012. Typhoid fever: “you can't hit what you can't see.” Gut Microbes 3:88–92. doi: 10.4161/gmic.18602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsolis RM, Young GM, Solnick JV, Baumler AJ. 2008. From bench to bedside: stealth of enteroinvasive pathogens. Nat Rev Microbiol 6:883–892. doi: 10.1038/nrmicro2012. [DOI] [PubMed] [Google Scholar]
- 21.Olsen SJ, Bleasdale SC, Magnano AR, Landrigan C, Holland BH, Tauxe RV, Mintz ED, Luby S. 2003. Outbreaks of typhoid fever in the United States, 1960-99. Epidemiol Infect 130:13–21. doi: 10.1017/S0950268802007598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 23.Winter SE, Thiennimitr P, Nuccio SP, Haneda T, Winter MG, Wilson RP, Russell JM, Henry T, Tran QT, Lawhon SD, Gomez G, Bevins CL, Russmann H, Monack DM, Adams LG, Baumler AJ. 2009. Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype Typhimurium infection. Infect Immun 77:1904–1916. doi: 10.1128/IAI.01341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blasi E, Mathieson BJ, Varesio L, Cleveland JL, Borchert PA, Rapp UR. 1985. Selective immortalization of murine macrophages from fresh bone marrow by a raf/myc recombinant murine retrovirus. Nature 318:667–670. doi: 10.1038/318667a0. [DOI] [PubMed] [Google Scholar]
- 25.Winter SE, Winter MG, Thiennimitr P, Gerriets VA, Nuccio SP, Russmann H, Baumler AJ. 2009. The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Mol Microbiol 74:175–193. doi: 10.1111/j.1365-2958.2009.06859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winter SE, Winter MG, Godinez I, Yang HJ, Russmann H, Andrews-Polymenis HL, Baumler AJ. 2010. A rapid change in virulence gene expression during the transition from the intestinal lumen into tissue promotes systemic dissemination of Salmonella. PLoS Pathog 6:e1001060. doi: 10.1371/journal.ppat.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 28.Tsuchiya S, Kobayashi Y, Goto Y, Okumura H, Nakae S, Konno T, Tada K. 1982. Induction of maturation in cultured human monocytic leukemia cells by a phorbol diester. Cancer Res 42:1530–1536. [PubMed] [Google Scholar]
- 29.Monack DM, Raupach B, Hromockyj AE, Falkow S. 1996. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci U S A 93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen LM, Kaniga K, Galan JE. 1996. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol 21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 31.Monack DM, Detweiler CS, Falkow S. 2001. Salmonella pathogenicity island 2-dependent macrophage death is mediated in part by the host cysteine protease caspase-1. Cell Microbiol 3:825–837. doi: 10.1046/j.1462-5822.2001.00162.x. [DOI] [PubMed] [Google Scholar]
- 32.Sun YH, Rolan HG, Tsolis RM. 2007. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J Biol Chem 282:33897–33901. doi: 10.1074/jbc.C700181200. [DOI] [PubMed] [Google Scholar]
- 33.Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM. 2010. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med 207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O'Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 35.Virlogeux I, Waxin H, Ecobichon C, Popoff MY. 1995. Role of the viaB locus in synthesis, transport and expression of Salmonella typhi Vi antigen. Microbiology 141(Part 12):3039–3047. [DOI] [PubMed] [Google Scholar]
- 36.Virlogeux I, Waxin H, Ecobichon C, Lee JO, Popoff MY. 1996. Characterization of the rcsA and rcsB genes from Salmonella typhi: rcsB through tviA is involved in regulation of Vi antigen synthesis. J Bacteriol 178:1691–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winter SE, Raffatellu M, Wilson RP, Russmann H, Baumler AJ. 2008. The Salmonella enterica serotype Typhi regulator TviA reduces interleukin-8 production in intestinal epithelial cells by repressing flagellin secretion. Cell Microbiol 10:247–261. doi: 10.1111/j.1462-5822.2007.01037.x. [DOI] [PubMed] [Google Scholar]
- 38.Molineaux SM, Casano FJ, Rolando AM, Peterson EP, Limjuco G, Chin J, Griffin PR, Calaycay JR, Ding GJ, Yamin TT, Palyha OC, Luell S, Fletcher D, Miller DK, Howard AD, Thornberry AD, Kostura MJ. 1993. Interleukin 1 beta (IL-1 beta) processing in murine macrophages requires a structurally conserved homologue of human IL-1 beta converting enzyme. Proc Natl Acad Sci U S A 90:1809–1813. doi: 10.1073/pnas.90.5.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kofoed EM, Vance RE. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cummings LA, Barrett SL, Wilkerson WD, Fellnerova I, Cookson BT. 2005. FliC-specific CD4+ T cell responses are restricted by bacterial regulation of antigen expression. J Immunol 174:7929–7938. doi: 10.4049/jimmunol.174.12.7929. [DOI] [PubMed] [Google Scholar]
- 41.Cummings LA, Wilkerson WD, Bergsbaken T, Cookson BT. 2006. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol Microbiol 61:795–809. doi: 10.1111/j.1365-2958.2006.05271.x. [DOI] [PubMed] [Google Scholar]
- 42.Lai MA, Quarles EK, Lopez-Yglesias AH, Zhao X, Hajjar AM, Smith KD. 2013. Innate immune detection of flagellin positively and negatively regulates salmonella infection. PLoS One 8:e72047. doi: 10.1371/journal.pone.0072047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickard D, Li J, Roberts M, Maskell D, Hone D, Levine M, Dougan G, Chatfield S. 1994. Characterization of defined ompR mutants of Salmonella typhi: ompR is involved in the regulation of Vi polysaccharide expression. Infect Immun 62:3984–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran QT, Gomez G, Khare S, Lawhon SD, Raffatellu M, Baumler AJ, Ajithdoss D, Dhavala S, Adams LG. 2010. The Salmonella enterica serotype Typhi Vi capsular antigen is expressed after the bacterium enters the ileal mucosa. Infect Immun 78:527–535. doi: 10.1128/IAI.00972-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atif SM, Winter SE, Winter MG, McSorley SJ, Baumler AJ. 2014. Salmonella enterica serovar Typhi impairs CD4 T cell responses by reducing antigen availability. Infect Immun 82:2247–2254. doi: 10.1128/IAI.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winter SE, Winter MG, Poon V, Keestra AM, Sterzenbach T, Faber F, Costa LF, Cassou F, Costa EA, Alves GE, Paixao TA, Santos RL, Baumler AJ. 2014. Salmonella enterica serovar Typhi conceals the invasion-associated type three secretion system from the innate immune system by gene regulation. PLoS Pathog 10:e1004207. doi: 10.1371/journal.ppat.1004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santos RL, Zhang S, Tsolis RM, Baumler AJ, Adams LG. 2002. Morphologic and molecular characterization of Salmonella typhimurium infection in neonatal calves. Vet Pathol 39:200–215. doi: 10.1354/vp.39-2-200. [DOI] [PubMed] [Google Scholar]
- 48.Monack DM, Hersh D, Ghori N, Bouley D, Zychlinsky A, Falkow S. 2000. Salmonella exploits caspase-1 to colonize Peyer's patches in a murine typhoid model. J Exp Med 192:249–258. doi: 10.1084/jem.192.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mastroeni P, Clare S, Khan S, Harrison JA, Hormaeche CE, Okamura H, Kurimoto M, Dougan G. 1999. Interleukin 18 contributes to host resistance and gamma interferon production in mice infected with virulent Salmonella typhimurium. Infect Immun 67:478–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raupach B, Peuschel SK, Monack DM, Zychlinsky A. 2006. Caspase-1-mediated activation of interleukin-1beta (IL-1beta) and IL-18 contributes to innate immune defenses against Salmonella enterica serovar Typhimurium infection. Infect Immun 74:4922–4926. doi: 10.1128/IAI.00417-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vazquez-Torres A, Jones-Carson J, Baumler AJ, Falkow S, Valdivia R, Brown W, Le M, Berggren R, Parks WT, Fang FC. 1999. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 52.Shea JE, Hensel M, Gleeson C, Holden DW. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci U S A 93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ochman H, Soncini FC, Solomon F, Groisman EA. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci U S A 93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stojiljkovic I, Baumler AJ, Heffron F. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol 177:1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 56.Kingsley RA, Humphries AD, Weening EH, De Zoete MR, Winter S, Papaconstantinopoulou A, Dougan G, Baumler AJ. 2003. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype Typhimurium: identification of intestinal colonization and persistence determinants. Infect Immun 71:629–640. doi: 10.1128/IAI.71.2.629-640.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. doi: 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 58.Raffatellu M, Santos RL, Chessa D, Wilson RP, Winter SE, Rossetti CA, Lawhon SD, Chu H, Lau T, Bevins CL, Adams LG, Baumler AJ. 2007. The capsule encoding the viaB locus reduces interleukin-17 expression and mucosal innate responses in the bovine intestinal mucosa during infection with Salmonella enterica serotype Typhi. Infect Immun 75:4342–4350. doi: 10.1128/IAI.01571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bohez L, Ducatelle R, Pasmans F, Botteldoorn N, Haesebrouck F, Van Immerseel F. 2006. Salmonella enterica serovar Enteritidis colonization of the chicken caecum requires the HilA regulatory protein. Vet Microbiol 116:202–210. doi: 10.1016/j.vetmic.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 60.Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. 2008. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun 76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.