Abstract
A critical step in the life cycle of all organisms is the duplication of the genetic material during cell division. Ribonucleotide reductases (RNRs) are essential enzymes for this step because they control the de novo production of the deoxyribonucleotides required for DNA synthesis and repair. Enterobacteriaceae have three functional classes of RNRs (Ia, Ib, and III), which are transcribed from separate operons and encoded by the genes nrdAB, nrdHIEF, and nrdDG, respectively. Here, we investigated the role of RNRs in the virulence of adherent-invasive Escherichia coli (AIEC) isolated from Crohn's disease (CD) patients. Interestingly, the LF82 strain of AIEC harbors four different RNRs (two class Ia, one class Ib, and one class III). Although the E. coli RNR enzymes have been extensively characterized both biochemically and enzymatically, little is known about their roles during bacterial infection. We found that RNR expression was modified in AIEC LF82 bacteria during cell infection, suggesting that RNRs play an important role in AIEC virulence. Knockout of the nrdR and nrdD genes, which encode a transcriptional regulator of RNRs and class III anaerobic RNR, respectively, decreased AIEC LF82's ability to colonize the gut mucosa of transgenic mice that express human CEACAM6 (carcinoembryonic antigen-related cell adhesion molecule 6). Microarray experiments demonstrated that NrdR plays an indirect role in AIEC virulence by interfering with bacterial motility and chemotaxis. Thus, the development of drugs targeting RNR classes, in particular NrdR and NrdD, could be a promising new strategy to control gut colonization by AIEC bacteria in CD patients.
INTRODUCTION
Ribonucleotide reductase (RNR) is an essential enzyme in all living organisms. It catalyzes the reduction of ribonucleotides (nucleoside triphosphates [NTPs]) to their corresponding 2′-deoxyribonucleotides (deoxynucleoside triphosphates [dNTPs]) and therefore plays an essential role in DNA synthesis and repair. Three RNR classes (classes I, II, and III) exist; these classes exhibit different primary structures, subunit cofactor requirements, and quaternary three-dimensional (3D) structures, but they all are allosterically regulated and share similar catalytic mechanisms (1, 2). Class I RNRs are oxygen-dependent enzymes that occur in eubacteria, eukaryotes, and some viruses. This class comprises two main subgroups (Ia and Ib). Class Ia RNRs are encoded by an operon containing nrdA and nrdB genes. These genes encode the NrdA subunit, which is catalytically and allosterically regulated, and the NrdB subunit, which possesses radical-generating activity. Class Ib RNRs are encoded by an operon containing the nrdH, nrdI, nrdE, and nrdF genes, which encode the corresponding specific redoxin NrdH, the activating subunit NrdI, the catalytic subunit NrdE, and the radical-generating subunit NrdF. Class III RNRs are present in facultative anaerobic and strict anaerobic microorganisms and use S-adenosylmethionine and iron-sulfur clusters in the NrdG accessory protein to create a stable glycyl radical. nrdD and nrdG form an operon and encode the catalytic NrdD subunit and the activating protein NrdG, respectively. This system works only under strict anaerobic conditions because oxygen is toxic to these enzymes (1, 2). Notably, the distribution of the different RNR classes is difficult to elucidate, as the different bacterial phylogenetic groups do not display any common RNR combinations. One possible RNR distribution appears to correlate with the bacterial growth conditions. We identified bacteria that encode one RNR class, whereas others encode more than one RNR class, and many RNR combinations exist in nature (3, 4).
Escherichia coli and all Enterobacteriaceae encode three RNR classes: classes Ia, Ib, and III (5, 6). In E. coli, class Ia RNRs are active during aerobic growth, class III RNRs are active under anaerobic growth conditions, and class Ib RNRs are active under some special circumstances, such as growth in an iron-deficient medium or biofilm formation (6, 7). However, the mechanisms that govern different RNR activities in pathogenic E. coli are not fully understood. An additional protein, termed NrdR, was described as a novel transcriptional regulator capable of modulating the expression of all RNR present in one organism (8). NrdR proteins are composed of 140 to 200 amino acids, with two differentiated domains: a zinc ribbon DNA-binding domain and an ATP-cone domain with the capacity to bind nucleotides. NrdR changes its conformation and binds to its cognate DNA recognition sequences to repress RNR gene expression depending on which nucleotide it is bound to (9).
A role for E. coli has been demonstrated in Crohn's disease (CD), an inflammatory bowel disease that occurs in individuals with a genetic predisposition to environmental or infectious triggers that cause an abnormal immune response (10–13). Most E. coli strains isolated from the ileal mucosa of CD patients are able to adhere to and invade intestinal epithelial cells (14–16) and belong to the pathogenic group of adherent invasive E. coli (AIEC) (17). AIEC is highly associated with the ileal mucosa in CD patients (14–16, 18–23). CD-associated AIEC cells adhere to the brush border of primary ileal enterocytes isolated from CD patients but not to enterocytes isolated from controls without inflammatory bowel disease. This adhesion is involved in the recognition between the variant FimH adhesin motifs located on the top of type 1 pili expressed on the AIEC bacterial surface and the carcinoembryonic antigen-related cell-adhesion molecule 6 (CEACAM6) abnormally expressed in the ileal epithelial cells of CD patients (24, 25). AIEC strain LF82 colonizes and induces strong gut inflammation in CEABAC10 transgenic mice, which express human CEACAMs and mimic CD susceptibility (25, 26). In the present study, we gained insight into the roles of the different RNR classes during the course of infection with AIEC LF82 using intestinal cells and an in vivo model of infection. Interestingly, we demonstrated that the transcriptional regulator NrdR governs the virulence of CD-associated E. coli by regulating AIEC LF82 motility and chemotaxis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Adherent invasive Escherichia coli (AIEC) was used throughout this study and for constructing gene knockouts. The bacteria and plasmids used are listed in Table 1. The bacterial cells were routinely grown in LB medium at 37°C supplemented with 50 μg ampicillin ml−1, 50 μg kanamycin ml−1, 30 μg chloramphenicol ml−1, or 30 μg X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) ml−1. Bacterial growth was measured as optical density at 550 nm (OD550).
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Descriptiona | Source |
|---|---|---|
| Plasmids | ||
| pGEM-T easy | A/T cloning vector (Ampr) | Promega |
| pJET1.2 | Positive selection cloning vector (Ampr) | Fermentas |
| pBAD18 | Arabinose-inducible expression vector (Ampr) | 47 |
| pETS174 | Chromosomal nrdAB operon cloned into EcoRI and XbaI sites of pBAD18 (Ampr) | This work |
| pETS175 | Plasmid nrdAB operon cloned into EcoRI and XbaI sites of pBAD18 (Ampr) | This work |
| pETS179 | nrdR (with native promoter) cloned into ApaI-SacI sites of pBBR1mcs5 (Gmr) | This work |
| pETS188 | nrdDG coding region cloned into EcoRI-XbaI sites of pJN105 (Gmr) | This work |
| pBBR1mcs5 | Broad-host-range plasmid vector (pBBR) (Gmr) | 48 |
| pJN105 | araBAD promoter broad-host-range vector (Gmr) | 49 |
| pKD4 | Source of kanamycin-resistant cassette | 28 |
| pKD46 | Encode red protein from phage λ under l-arabinose promoter control | 28 |
| E. coli strains | ||
| DH5α | recA1 endA1 hsdR17 supE44 thi-1 relA1 Δ(lacZYA-argF)U169 deoR ϕ80ΔlacZM15 | Laboratory stock |
| LF82 | Wild-type adherent-invasive E. coli LF82 | 36 |
| LF82 ΔnrdR | E. coli LF82 ΔnrdR::kan (Kanr) | This work |
| LF82 ΔnrdE | E. coli LF82 ΔnrdE::kan (Kanr) | This work |
| LF82 ΔnrdD | E. coli LF82 ΔnrdD::kan (Kanr) | This work |
| LF82 ΔfliC | E. coli LF82 ΔfliC::kan (Kanr) | 43 |
| E101 | thr1 leuB6 fhuA21 lacY1 glnV44 rfbC1 nrdA(Ts) thyA6 rpsL67 thi1 deoC1 deoB37 | 50 |
Amp, ampicillin; Gm, gentamicin; Kan, kanamycin.
DNA manipulations.
Restriction endonucleases, T4 DNA ligase, alkaline phosphatase, DNA polymerase (Klenow fragment), and DNA-modifying enzymes were purchased from Fermentas and used according to the manufacturer's instructions. Plasmid DNA was isolated using the QIAprep spin Miniprep kit (Qiagen) as described by the manufacturer.
PCR amplifications were performed using high-fidelity polymerase (Fermentas). PCR master mix (2×) was used for screening assays according to the manufacturer's specifications (Fermentas) with the primers described in Table 2. Other molecular assays and manipulations were performed according to standard procedures (27).
TABLE 2.
Primers used in this study
| Name | Sequence (5′→3′) | Application |
|---|---|---|
| LF82 R up | AATGCGGGTAAAGGGTCATT | Cloning |
| LF82 R lw | CGCGCCATGTAATACTCGT | Cloning |
| LF82up-DGpBAD | AAAGAATTCATGACACCGCATGTGAA | Cloning |
| LF82lw-DGpBAD | AAATCTAGAGCAACGCCTATCACCAGA | Cloning |
| M13 dir | GTTTTCCCAGTCACGAC | Verification-cloning |
| M13 rev | CAGGAAACAGCTATGAC | Verification-cloning |
| EcE-up | CCGATGAACGCATTCGCAA | cDNA amplification |
| EcE-lw | AAAGTGCACAGGAGACGCAG | Reverse transcription-cDNA amplification |
| EcGAP-up | GGCCAGGACATCGTTTCCA | cDNA amplification |
| EcGAP-lw | GATGATGTTCTGGGAAGCGC | Reverse transcription-cDNA amplification |
| LF82 fliS RT up | GCAAGGCAAAGGCGTCTCTTTGTC | cDNA amplification |
| LF82 fliS RT lw | GACATCGTTGCGTAAATTGGCTTGC | Reverse transcription-cDNA amplification |
| LF82 motA RT up | CAATACCAAACGCCGGAAGTGAGTC | cDNA amplification |
| LF82 motA RT lw | GATAGCGTCATGCTTGAATTTATCGTCG | Reverse transcription-cDNA amplification |
| LF82 cheR RT up | GCTCCTGACGCACGCGTAC | cDNA amplification |
| LF82 cheR RT lw | CAGCATAGCGATGACGCTGGC | Reverse transcription-cDNA amplification |
| LF82 kdgK RT up | CGAAGCCGCCGCCAAATTCTGG | cDNA amplification |
| LF82 kdgK RT lw | GTCTCTTCTTTGCTGGCCCACAG | Reverse transcription-cDNA amplification |
| LF82 tsr RT up | GCTGGCGGAGCTGATACAACTG | cDNA amplification |
| LF82 tsr RT lw | GCCTACCAGAATCCACATCGCC | Reverse transcription-cDNA amplification |
| LF82 fliC RT up | CGGCAAATACCGCCTGATACGTC | cDNA amplification |
| LF82 fliC RT lw | GACTGCATCAGTCACGATGGGG | Reverse transcription-cDNA amplification |
| ABc-pBADup | AAGAATTCGGAGTGAAAGCATGAATCAGAATCTGCTG | pBAD cloning |
| ABc-pBADlow | AATCTAGAGGGCCATTCAGAGCTGGAAG | pBAD cloning |
| ABp-pBADup | AAGAATTCGGAGTGTACATGATAAGCATCGTAAAACGTAAC | pBAD cloning |
| ABp-pBADlow | AATCTAGATCGGTTTACAGGTCAGCAAAC | pBAD cloning |
| MInrdR(F) | TCAGTTCAGGGTAAAATAGATTTCTGTTAACCACCTGGTCAGGACGCCGTGTAGGCTGGAGCTGCTTCG | Construction of isogenic nrdR mutant |
| MInrdR(R) | CGTTGCGCCAGCTTTAGCGCCCGCGCCATGTAATACTCGTCCTGCACGGCCATATGAATATCCTCCTTAG | Construction of isogenic nrdR mutant |
| MInrdAc(F) | CTCACTACAGGTAGTCTGCATGAAGCTATTGAAAAACAGGTACGACATACGTAGGCTGGAGCTGCTTCG | Construction of isogenic nrdAc mutant |
| MInrdAc(R) | TGGACCCGTAGGCCGGGTAGGGCGCTCACGCCGCATCCGGCATCACAATACATATGAATATCCTCCTTAG | Construction of isogenic nrdAc mutant |
| MInrdAp(F) | TCTATTAACATGGGCCACTATATTTAGTGGCCCTCTTTATTTGGTGATACGTAGGCTGGAGCTGCTTCG | Construction of isogenic nrdAp mutant |
| MInrdAp(R) | AAACAAGGCTTACAAATCAAAGAGAAGGTGGGGATATCCCCCACCATATGCATATGAATATCCTCCTTAG | Construction of isogenic nrdAp mutant |
| MInrdD(F) | GCCTTTCCCAATTTCTGTGGATAACCTGTTCTTAAAAATATGGAGCGATCGTAGGCTGGAGCTGCTTCG | Construction of isogenic nrdD mutant |
| MInrdD(R) | GGATAAGGCGTTTACGCCGCATCCGGCATTTGCTTAACACAGAGTGAAGACATATGAATATCCTCCTTAG | Construction of isogenic nrdD mutant |
| MInrdE(F) | GTAACCGAATTTTGGCAACGACAACCGCAGAACGCCTGACGCAGGAAACGGTAGGCTGGAGCTGCTTCG | Construction of isogenic nrdE mutant |
| MInrdE(R) | ATCTTGTTCCAGTTGATGGCGCTGATACGTGAGAGTTTCATAGATATTCCCATATGAATATCCTCCTTAG | Construction of isogenic nrdE mutant |
Construction of E. coli LF82 nrd mutants and complementation plasmids.
Isogenic mutants were generated using PCR products, as described by Datsenko and Wanner (28) and modified by Chaveroche et al. (29). Briefly, we replaced a chromosomal sequence with a selectable antibiotic resistance gene (kanamycin or chloramphenicol) generated by PCR. For the nrd mutants, this PCR product was generated using primers with 50-nucleotide (nt) extensions that are homologous to regions adjacent to the target gene and the E. coli template strain harboring the kanamycin resistance gene on the pKD4 plasmid. In parallel, the E. coli AIEC strain LF82 was transformed with the pKD46 plasmid, which encodes Red proteins from phage λ synthesized under the control of an l-arabinose-inducible promoter. This plasmid was maintained in bacteria at 30°C with 25 μg/ml chloramphenicol and was eliminated at 37°C. Strain LF82/pKD46 was grown at 30°C with 1 mM l-arabinose to induce Red expression. When the OD620 reached 0.6, the bacterial culture was incubated for 20 min at 42°C to eliminate the plasmid. The bacteria were washed three times with 10% glycerol and then electroporated with the PCR products. Isogenic mutants were selected on LB agar containing 50 μg/ml kanamycin. Gene replacement by the kanamycin resistance cassette in mutants was confirmed by PCR (primers are listed in Table 2).
Complementation plasmids (pETS179 and pETS188) were constructed by cloning the nrdR gene (970 bp) with its promoter region and the nrdDG (class III RNR; 3,298-bp) coding region into plasmids pBBR1MCS-5 and pJN105, respectively, using the primer pairs LF82 R up/LF82 R low and LF82 DG up/LF82 DG low, respectively (Table 2). Sequencing and restriction analysis confirmed the correct cloning and orientation of the inserts.
Motility and swim plate chemotaxis assays.
Bacterial motility was evaluated on soft LB agar (Difco). Four different chemoattractants (glucose, maltose, l-serine, and l-fucose) and one chemorepellent (l-valine) (Sigma) were added to 0.25% LB agar plates to a final concentration of 2 mM. The LF82 and LF82 ΔnrdR overnight cultures were adjusted to an OD550 of 1.0, and 5 μl of the cultures was spotted onto the plate surface. The motility plates were incubated for 12 h at 30°C, after which the diameter of the bacterial movement was measured. Three independent experiments were performed, and significant differences in the motility diameter were determined and plotted.
The swim plates for chemotaxis assays were prepared as described previously (30). The chemoattractants were added at a final concentration of 2 μM for l-serine, 1 μM for thymine, and 100 μM for maltose. Similar to the procedure used for the motility plates, the overnight cultures of the LF82 and the LF82 ΔnrdR strains were adjusted to an OD550 of 1.0 and spotted (5 μl) onto the plate surfaces. The plates were incubated at 30°C, and the chemotaxis expansion halos were measured at 4 h, 8 h, 12 h, and 24 h of incubation. For statistical validation, three independent experiments were performed.
RNA isolation.
RNA was isolated from three independent E. coli cultures of the wild-type LF82 strain and the LF82 ΔnrdR strain, which were grown aerobically at 37°C to an OD550 of 0.5 using the RNeasy kit (Qiagen). The cultures were treated with the RNAprotect bacteria reagent (Qiagen) to stabilize and preserve the RNA before total RNA extraction and concentration, as described by the manufacturer. The RNA's integrity, quality, and quantity were assessed using a Bioanalyzer (Bio-Rad) and a NanoDrop spectrophotometer (ND-1000; NanoDrop).
RT-PCR and real-time PCR.
One microgram of total RNA was mixed with 1 pmol of each reverse primer (listed in Table 2) and 2 μl of 10 mM dNTPs, and the mixture was brought to a volume of 12 μl with sterile distilled water. The mixture was heated to 65°C and iced for 1 min. Four microliters of 5× First-Strand buffer, 1 μl of 0.1 M dithiothreitol (DTT), 1 μl RNaseOUT (Invitrogen), 1 μl of SuperScript reverse transcriptase (RT; 200 units/μl; Invitrogen), and 1 μl of DEPC (diethyl pyrocarbonate)-treated water (Invitrogen) were added to the mix. The PCR was performed according to the manufacturer's recommendations. One microliter of each reverse transcriptase reaction was used as a template for amplifying cDNA using a 2× PCR master mix (Fermentas) according to the manufacturer's instructions. The primers used for cDNA amplification are listed in Table 2.
For quantification of gene expression by real-time PCR, the RNAs were reverse transcribed and amplified using specific primers to nrdR, nrdAp, nrdAc, nrdD, nrdE, or 16S rRNA (Table 2). Results are expressed as the number of mRNA-encoding nrd genes for 107 copies of mRNA-encoding 16S. The amplification of a single expected PCR product was confirmed by electrophoresis on a 2% agarose gel. Real-time PCR was performed using an Eppendorf Realplex, and the RNA levels were quantified using the RNA master SYBR green I (Roche Diagnostic) with 0.25 mg of total RNA.
Microarray studies and data analysis.
For the nrdR microarray study, we used the Affymetrix GeneChip E. coli Genome 2.0 Array (Affymetrix). The microarray samples were analyzed to determine the nrdR expression profile in strain LF82. The data were normalized and compared to calculate the significance of the expression log ratios and standard deviations for open reading frames (ORFs) that exhibited at least a 2-fold change and P values of <0.05. Genes with expression ratios in the ΔnrdR strain that changed more than 2-fold were considered up- or downregulated compared with the wild-type strain.
Ribonucleotide reductase complementation assay.
The entire chromosomally carried E. coli LF82 wild-type nrdAB (nrdABc) and plasmid nrdAB genes (nrdABp) were amplified from genomic DNA by PCR using the primer combinations ABc-pBADup/ABc-pBADlow and ABp-pBADup/ABp-pBADlow, respectively. The respective primers were designed to generate an EcoRI and XbaI restriction site at the start and end of the resulting amplified fragment, respectively. All amplified fragments were cloned into the pGEM-T Easy vector as described by the manufacturer (Promega). The EcoRI/XbaI-digested DNA fragments ABc (3,606 bp) and ABp (3,573 bp) were cloned into the pBAD18 vector, thus generating pETS174 and pETS175, respectively. Both plasmids were sequenced in both directions and transformed into E. coli strain E101. This recipient strain contains conditional lethal mutations in nrdA(Ts), which result in a temperature-sensitive phenotype. As a result, E101 can grow at 42°C only if complementary NrdAB activity is supplied in trans (31). The complementation assay was previously used to verify the activity of atypical RNRs (31–33).
Complementation was determined by plating serial dilutions of liquid cultures on LB agar plates supplemented with thymine (50 μg/ml) and either 0.2% l-arabinose or 0.2% d-glucose to induce or repress the Para promoter, respectively, at both 30 and 42°C for 24 to 48 h. As a positive control, the LF82 nrdAB chromosomal copy cloned into the pBAD18 vector (plasmid pETS174) was used, and the pBAD18 vector was used as a negative control; both were transformed into strain E101.
Cell culture and infections.
T-84 cells were purchased from Flow Laboratories, Inc., McLean, VA. The cultured cells were maintained in 5% CO2 at 37°C in Dulbecco's modified Eagle medium (DMEM)-Ham's F12 medium (Seromed, Biochrom KG, Berlin, Germany) supplemented with 10% (vol/vol) fetal calf serum (FCS; Seromed), 1% nonessential amino acids (Life Technologies, Cergy-Pontoise, France), 1% l-glutamine (Life Technologies), 200 U of penicillin, 50 mg of streptomycin and 0.25 mg of amphotericin B per liter, and 1% minimal essential medium (EMEM) vitamin mix X-100 (Life Technologies). Monolayers were seeded in 24-well tissue culture plates (Polylabo, Strasbourg, France) with 4 × 105 cells/well and incubated for 48 h. The monolayers were then infected with 1 ml of the cell culture medium without antibiotics and with heat-inactivated fetal bovine serum at a multiplicity of infection (MOI) of 10 bacteria per epithelial cell. After a 3-h incubation period at 37°C, the monolayers were washed five times in phosphate-buffered saline (PBS; pH 7.2). The epithelial cells were then lysed with 1% Triton X-100 (Sigma Chemical Company, St. Louis, MO) in deionized water. The samples were diluted and plated onto Mueller-Hinton agar plates to determine the number of CFU corresponding to the total number of cell-associated bacteria (adherent and intracellular bacteria). The bacterial invasion was measured using the gentamicin protection assay (17). After a 3-h incubation period at 37°C, the monolayers were washed three times in PBS (pH 7.2). To determine the number of intracellular bacteria, fresh cell culture medium containing 100 mg/ml gentamicin (Sigma) was added for 1 h to kill extracellular bacteria. The monolayers were then lysed with 1% Triton X-100, and the intracellular bacteria were quantified as described above. When needed, the infected monolayers were centrifuged for 10 min at 1,000 × g before the 3-h infection period.
Animal model of infection.
Twelve-week-old FVB/N CEABAC10 transgenic male mice (body weight, 26 to 28 g) that expressed human CEACAM6 were first pretreated orally with the broad-spectrum antibiotic streptomycin (5 mg per mouse administered intragastrically) to disrupt the normal resident bacterial flora in the intestinal tract (34) and then orally challenged with 109 bacteria 24 h later. The animals received an extremely low dose of 0.25% (wt/vol) dextran sulfate sodium (DSS; molecular mass, 36,000 to 50,000 Da; MP Biomedicals) in their drinking water starting 3 days before infection to increase the bacteria's accessibility to the surface of the epithelial layer.
One, 2, 3, and 6 days after bacterial infection, fresh fecal pellets (100 to 200 mg) were collected from individual mice and resuspended in PBS. After serial dilution, the bacteria were enumerated by plating them on LB agar medium containing ampicillin (50 mg/μl) and erythromycin (20 mg/μl) to isolate AIEC LF82 bacteria and isogenic mutants and then incubating them at 37°C overnight.
Bacterial interactions were also studied using mouse colonic loops, as previously described (35). The mice were starved for 12 h before surgery, with water available ad libitum. They were anesthetized, and their intestines were exteriorized through a midline incision. Two colonic segments (approximately 1 cm) were ligated and inoculated with approximately 5 × 107 bacteria. After 4 h, the mice were anesthetized with isoflurane and then euthanized by cervical dislocation. Colonic loops were removed, and the associated bacteria were seeded onto agar plates containing the appropriate antibiotics.
Transmission electron microscopy.
The bacteria were grown overnight at 37°C without shaking in LB broth, placed for 1 min on carbon-Formvar copper grids (Electron Microscopy Sciences, Hatfield, England), and negatively stained for 1 min with phosphotungstic acid, pH 6.0. The grids were examined with a Hitachi H-7650 transmission electron microscope.
Statistical analysis.
Values are expressed as the means ± standard errors of the means (SEM) or medians. Data were compared using Student's t test analysis or nonparametric one-way analysis of variance Mann-Whitney test when appropriate. Statistical analyses were performed using the GraphPad Prism 4.00 (GraphPad Software, San Diego, CA, USA) software package for PC. Single comparisons were performed with the unpaired Mann-Whitney test. Correlation analyses were performed using a nonparametric correlation Spearman test. P values of <0.05 were considered statistically significant.
Ethics statement.
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the University of Clermont-Ferrand, Clermont-Ferrand, France. The animal protocol was approved by the Committee for Research and Ethical Issues, Auvergne department (CEMEA Auvergne), according to international directive 86/609/CEE (n°CE16-09).
Microarray data accession number.
Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-2288.
RESULTS
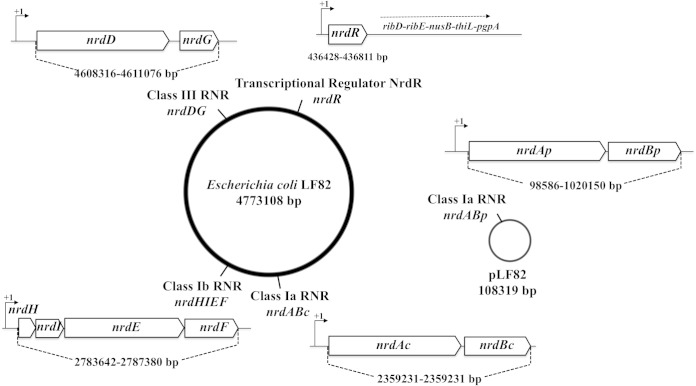
AIEC LF82 strain harbors four RNR classes in the genome.
Like all Enterobacteriaceae, the AIEC strain LF82 encodes three classes of RNRs (classes Ia, Ib, and III). Interestingly, this E. coli strain also harbors the pLF82 plasmid, with an additional operon encoding class Ia proteins (here denoted nrdABp). The positions of the different ribonucleotide reductase genes on the LF82 chromosome and plasmid are shown in Fig. 1. An alignment comparison between E. coli LF82 chromosomal and plasmid NrdAB proteins (see Fig. S1 in the supplemental material) revealed only 59% identity and 75% similarity. In contrast, a comparison between the LF82 chromosomal NrdAB proteins and chromosomal NrdABs from other Enterobacteriaceae revealed 97% identity and 98% similarity. Only extremely important amino acid residues located in the active site and those with allosteric specificity in the active site are present in the E. coli LF82 NrdAp. The residues that are important for iron ligand binding, R1 interaction, and hydrophobic pocket integrity in the NrdB proteins are also conserved in the E. coli LF82 NrdBp. Concerning class Ib and III RNRs, the proteins encoded by the chromosome exhibited 99 to 100% identity/similarity to other similar RNR classes in Enterobacteriaceae.
FIG 1.
Four different RNR classes in E. coli strain LF82. The localizations of the different ribonucleotide reductase classes and the transcriptional factor NrdR are illustrated in the representation of the adherent-invasive E. coli genome and plasmid. The genetic organizations of the different RNR classes are also presented, and their positions and lengths are drawn to scale.
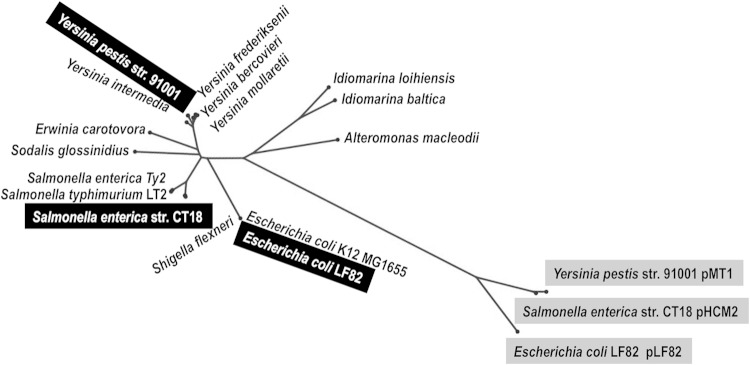
Plasmid-encoded NrdAB proteins have been detected only in the Salmonella enterica serovar Typhi strain CT18 and several Yersinia pestis biovars and in no other members of the E. coli species that have been sequenced to date. These sequences are located on several plasmids (pLF82, pHCM2, and pMT1) that share a plasmid structure, sequence, and genes (3, 4, 36). Phylogenetic reconstructions demonstrated that the plasmid-encoded NrdABp drastically differs from the chromosomal copy (NrdABc) (Fig. 2). The latter is much closer to other enterobacterial and gammaproteobacterial sequences. This suggests that plasmid copies evolve at a different rate from that of their corresponding chromosomal copies and that the acquisition of such copies could occur via horizontal gene transfer.
FIG 2.
Plasmid NrdAB genes are acquired by horizontal gene transfer. Unrooted phylogenetic tree of representative class Ia (NrdA) catalytic RNR subunit. The NeighborNet phylogenetic network was generated from a representative collection of the 26 closest BLAST neighbors in RNRdb to the class Ia catalytic subunit (NrdA) from E. coli LF82 and S. enterica CT18 that were encoded chromosomally and by the plasmid. Yersinia pestis strain 91001, Salmonella enterica strain CT18, and E. coli strain LF82 are highlighted in black when the class Ia RNR copy is encoded chromosomally and in light gray when encoded by a plasmid. All bootstrap values are higher than 950.
To assess whether the pLF82-encoded NrdA and NrdB were functional, an enzymatic complementation assay was performed. The complete nrdAB sequences from the chromosome and plasmid copy were cloned into an arabinose-inducible pBAD18 vector, as described in Materials and Methods. As expected, the positive-control pETS174 complemented the nrdA(Ts) deficiency, which indicates that these proteins are fully functional and active (Table 3). The absence of bacterial growth using the pETS175 construction at 42°C in arabinose LB plates revealed an absence of complementation of the temperature-sensitive phenotype. This indicates that the plasmid class Ia RNR (nrdABp) is not enzymatically functional under the conditions tested in our experiment.
TABLE 3.
Low enzymatic activity of plasmid NrdAB proteins in a heterologous complementation assay
| Plasmid | Activity (CFU/ml)a |
|||
|---|---|---|---|---|
| Arabinose (0.2%) |
Glucose (0.2%) |
|||
| 30°C | 42°C | 30°C | 42°C | |
| E101(pETS174) | 3.1 × 109 ± 1.2 × 107 | 2.9 × 109 ± 3.2 × 107 | 2.9 × 109 ± 4.2 × 106 | <10 |
| E101(pETS175) | 5.2 × 109 ± 7.2 × 106 | <10 | 3.1 × 109 ± 1.2 × 107 | <10 |
| E101(pBAD18) | 2.1 × 109 ± 1.2 × 106 | <10 | 3.1 × 109 ± 1.2 × 107 | <10 |
Values are based on the results of at least six independent experiments.
Expression of RNR genes during intestinal cell infection.
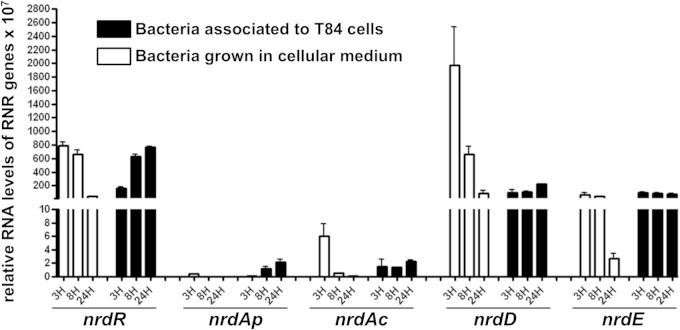
To explore if the different RNR proteins have a role in promoting the adherence of AIEC bacteria to intestinal epithelial cells, we measured the levels of nrdR, nrdAp, nrdAc, nrdD, and nrdE mRNA in AIEC LF82 bacteria using real-time PCR. This measurement was performed after different infection periods using T-84 intestinal epithelial cells or after bacterial growth in DMEM during similar periods (Fig. 3). The levels of nrdR mRNA increased during the bacterium-cell interaction, whereas the level of nrdR mRNA significantly decreased when the bacteria were grown in DMEM during a similar period. nrdAp was not expressed during bacterial growth in DMEM, but its expression increased after contact with T-84 cells. Finally, we observed decreased nrdAc, nrdD, and nrdE expression when the bacteria were grown in DMEM, whereas similar mRNA levels for these three genes were detected during the bacterium-cell interaction. These results indicate that ribonucleotide reductases may play crucial roles during the infection of intestinal epithelial cells by AIEC bacteria.
FIG 3.
Expression of the different nrd genes depends on cell interaction. mRNA levels of the nrdR, nrdAp, nrdAc, nrdD, and nrdE genes in associated bacteria (after a 3-h, 8-h, or 24-h infection period) relative to those of AIEC LF82 bacteria grown during similar periods in MEM cell culture medium were compared using quantitative RT-PCR. As controls, 16S rRNA levels were measured. Results are expressed as the number of mRNA-encoding nrd genes for 107 copies of mRNA-encoding 16S rRNA. Only experiments that demonstrated the same levels of 16S rRNA for each sample were taken into account. The data represent the means from at least three separate experiments, and the bars represent the SEM.
nrdR is required for AIEC adhesion to intestinal epithelial T-84 cells and the ileal mucosa.
To evaluate the roles of the different RNRs in promoting the adherence of the AIEC strain LF82 to T-84 cells, we constructed insertion mutants with interruptions in several nrd genes (nrdAp, nrdR, nrdD, nrdE). We could not create a chromosomal nrdAB mutant since class Ia RNR mutants are nonviable under laboratory growing conditions (5, 6).
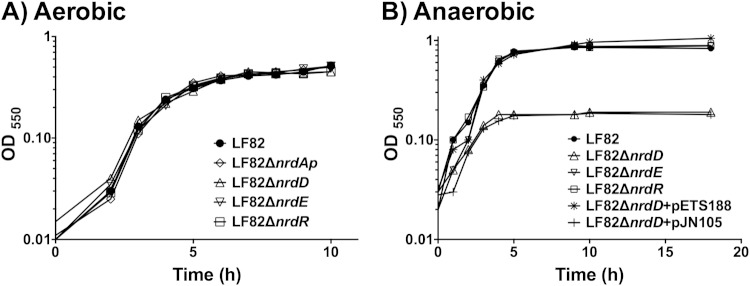
The mutants lacking nrd (nrdR, nrdD, and nrdE mutants) displayed growth curves in DMEM growth medium that were similar to those of the wild-type LF82 strain (Fig. 4A) over an 8-h period. Interestingly, under anaerobic conditions, the nrdD mutants grew more slowly than the wild-type strain and other nrd mutants did (Fig. 4B), indicating that this ribonucleotide reductase class is important under anaerobiosis. To correlate loss of function of the nrdD to anaerobic growth, this mutation was complemented with the corresponding wild-type nrdDG genes (pETS188), restoring the wild-type phenotype (Fig. 4B). The wild-type strain transformed with pJN105 did not show any change in the anaerobic growth curve (data not shown).
FIG 4.
Growth curves of different nrd mutants under aerobic (A) and anaerobic (B) conditions.
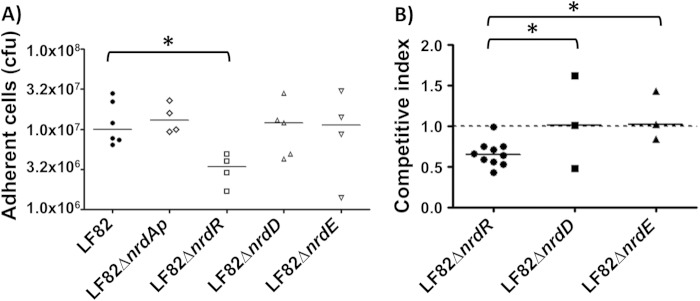
A significant decrease in adhesion was observed only for the LF82 ΔnrdR mutant (Fig. 5A). This mutant's decreased ability to adhere was type 1 pilus independent, as the LF82 ΔnrdR mutant still expressed functional type 1 pili on the bacterial surface, as determined by the ability of bacteria to aggregate yeast cells via binding to d-mannose residues located on the yeast surface (data not shown). The role of nrdR gene in adhesion ability of AIEC LF82 bacteria was confirmed by a transcomplementation assay restoring the wild-type phenotype (see Fig. S3 in the supplemental material). The bacterial interaction of LF82, LF82 ΔnrdR, LF82 ΔnrdD, and LF82 ΔnrdE was investigated using ex vivo mouse ileal loop assays. The mean competitive index for LF82 ΔnrdD and ΔnrdE isogenic mutants compared to wild-type LF82 bacteria was close to 1. In contrast, for LF82 ΔnrdR, the mean competitive index was 0.65 ± 0.15 (Fig. 5B). These indexes suggest that nrdR is important for the ileal mucosal interaction and that the loss of NrdR expression reduced LF82's ability to interact with the ileal mucosa.
FIG 5.
Adhesion of LF82 Δnrd mutants using T-84 cells or ileal loops. (A) Cell-associated bacteria were quantified using undifferentiated T-84 cells after a 3-h infection. (B) Competitive indexes of LF82 with LF82 ΔnrdR, LF82 ΔnrdD, and LF82 ΔnrdE. Intestinal ileal loops were inoculated with a mixed inoculum comprising equivalent numbers of two bacterial strains. After 5 h, the bacterial presence was compared using competitive index analysis, which provides a sensitive measurement of the relative degree of attenuation. *, P < 0.05.
Importance of nrdD and nrdR to AIEC persistence in the gut of CEABAC10 mice.
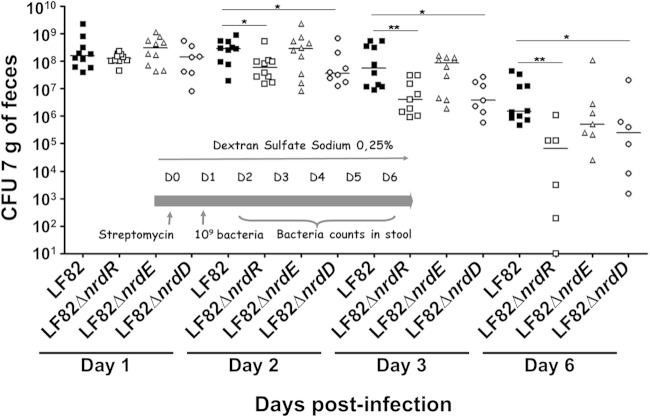
To investigate the role of Nrd proteins in gut colonization by AIEC LF82, CEABAC10 transgenic mice expressing human CEACAM molecules (CEACAM3, CEACAM5, CEACAM6, and CEACAM7) were challenged with LF82, LF82 ΔnrdR, LF82 ΔnrdD, or LF82 ΔnrdE bacteria. The quantification of bacteria in stool samples at day 2 postinfection revealed an 80.0-fold decrease for LF82 ΔnrdR (5.0 × 107 CFU/g of feces) and an 87.8-fold decrease for LF82 ΔnrdD (3.6 × 107 CFU/g of feces) compared with wild-type AIEC LF82 (3.0 × 108 CFU/g of feces) (Fig. 6). The significantly lower concentration of LF82 ΔnrdR and LF82 ΔnrdD mutants in the stool was confirmed at day 3 postinfection (4.0 × 106 for LF82 ΔnrdR; 3.9 × 106 for LF82 ΔnrdR versus 5.7 × 107 CFU/g of feces, respectively; P = 0.01 and P = 0.05) and at day 6 postinfection (6.7 × 104 for LF82 ΔnrdR; 2.5 × 106 for LF82 ΔnrdR versus 1.5 × 106 CFU/g of feces, respectively; P = 0.01 and P = 0.05). No significant difference in bacterial persistence in the gut of CEABAC10 mice was observed for LF82 ΔnrdE compared with wild-type LF82 (Fig. 6).
FIG 6.
Bacterial persistence in the gut of CEABAC10 mice. Quantification of LF82, LF82 ΔnrdR, LF82 ΔnrdE, and LF82 ΔnrdD bacteria in the feces of CEABAC10 mice that received 0.25% DSS in their drinking water after oral infection with 109 bacteria at day 0. *, P < 0.05; **, P < 0.01.
These results suggest that the deletion of nrdR and nrdD genes in the AIEC LF82 genome dramatically decreased the ability of bacteria to persist in the gut of CEABAC10 mice.
Interference of nrdR regulon in AIEC virulence.
We demonstrated that NrdR is involved in LF82 adhesion and persistence. Thus, we pursued an investigation of the mechanisms leading to these phenotypes. Total RNA from aerobic LF82 and LF82 ΔnrdR cultures grown to mid-exponential phase was extracted to obtain an nrdR expression profile in the AIEC LF82 strain. The microarray data from this transcriptomic analysis demonstrated that 33 genes were differentially expressed between the LF82 ΔnrdR mutant and a wild-type strain. Of those genes, only 7 were upregulated in the absence of nrdR, whereas 26 genes were downregulated in the LF82 ΔnrdR mutant (Table 4). As we expected from previous studies (8), the operon for the class Ib RNR expression, nrdHIEF, was induced in the nrdR mutant strain. This RNR is encoded by genes that were identified along with two genes involved in cell division, dicB and ydfD, among the few upregulated genes obtained in the LF82 ΔnrdR microarray.
TABLE 4.
NrdR-dependent expression of genes identified by transcriptomic analysis
| Gene | Operon arrangement | Log fold changea | Gene product |
|---|---|---|---|
| nrdH | nrdHIEF | 2.29 | Glutaredoxin-like protein |
| nrdI | 2.32 | Ribonucleotide reductase stimulatory protein: flavodoxin | |
| nrdE | 2.28 | Ribonucleotide-diphosphate reductase subunit alpha | |
| nrdF | 1.92 | Ribonucleotide-diphosphate reductase subunit beta | |
| dicB | dicB ydfDE insD intQ | 1.15 | Cell division inhibition protein |
| ydfD | 1.44 | Hypothetical protein | |
| yieI | yieIJ | 1.13 | Putative inner membrane protein |
| ycgR | ycgR | −1.08 | Flagellar function protein |
| fliD | fliDST | −1.21 | Flagellar filament capping protein |
| fliS | −1.34 | Flagellar biosynthesis protein FliS | |
| fliT | −1.39 | Flagellar biosynthesis protein FliT | |
| aer | aer | −1.47 | Aerotaxis receptor |
| fliC | fliC | −1.56 | Flagelline |
| motA | motAB cheAW | −1.63 | Flagellar motor protein; proton conduct |
| motB | −1.75 | Flagellar motor protein; motor rotation | |
| cheA | −1.82 | Chemotaxis protein; chemotactic sensory histidine kinase | |
| cheW | −2.06 | Purine-binding chemotaxis protein | |
| tar | tar tap cheRBYZ | −2.26 | Methyl-accepting chemotaxis protein II |
| tap | −2.36 | Methyl-accepting chemotaxis protein IV | |
| cheR | −1.96 | Chemotaxis methyltransferase; regulator | |
| cheB | −2.01 | Chemotaxis-specific methylesterase; chemotaxis regulator | |
| cheY | −2.18 | Chemotaxis regulator transmitting signal | |
| cheZ | −2.01 | Chemotaxis protein phosphatase for CheY; chemotaxis regulator | |
| tsr | tsr | −2.35 | Methyl-accepting chemotaxis protein I |
| yhjH | yhjH | −2.26 | EAL domain-containing protein |
| ymdA | ymdA | −1.31 | Hypothetical protein |
| yjcZ | yjdAZ | −1.88 | Hypothetical protein |
| yhjG | yhjG | −1.48 | Hypothetical protein |
| kdgK | kdgK | −2.79 | 2-Dehydro-3-deoxygluconokinase |
Log fold change was determined by comparing the transcription (ratio of mRNA) of Pseudomonas aeruginosa ΔnrdR to its wild-type strain PAO1 (ΔnrdR mRNA/wild-type mRNA).
Interestingly, 8 fully transcriptional units corresponding to 18 genes (ycgR, fliDST, aer, tsr, motAB, cheAW, tar, tap, cheRBYZ, fliC, and yhjH) from the 26 repressed genes have functions in bacterial motility and chemotaxis (Table 4). Four genes (aer, tsr, tar, and tap), encoding the chemotaxis receptors known as methyl-accepting chemotaxis proteins (MCPs), and the 6 che loci (cheA, cheB, cheR, cheZ, cheY, and cheW) responsible for the chemotactic movements of E. coli were downregulated by an approximately 2-log-fold change (FC) in an LF82 ΔnrdR mutant. The genes are involved in flagellar biosynthesis (fliDST) and the flagelline (fliC) and flagellar (ycgR) functions, and they were downregulated in the LF82 ΔnrdR mutant, demonstrating the possible participation of this transcriptional regulator in LF82 motility. The EAL domain-containing phosphodiesterase YhjH-encoding gene, yhjH, was also downregulated by 2.26 log FC in the LF82 ΔnrdR mutant compared with the wild-type strain. The results of the transcriptomic experiment were confirmed via quantitative PCR of several of the genes described in Table 4 (Fig. 7).
FIG 7.
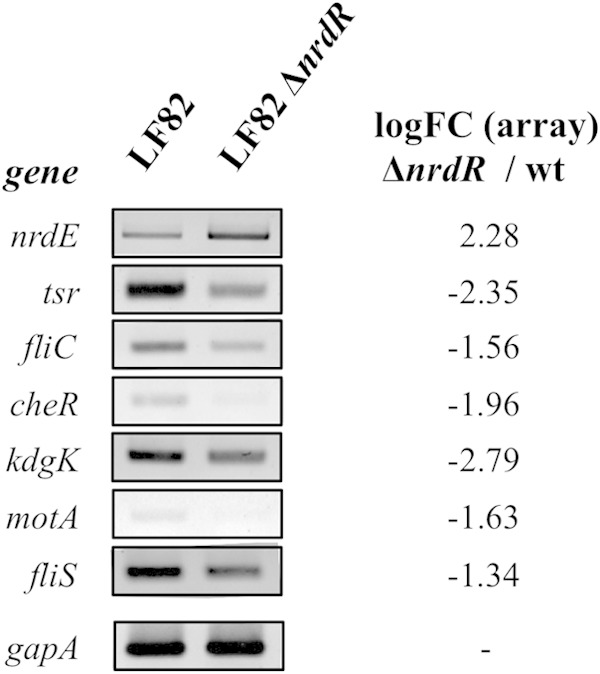
RT-PCR analysis of the transcription levels of the nrdE, tsr, fliC, cheR, kdgK, motA, and fliS genes from the AIEC LF82 (wt) and LF82 ΔnrdR isogenic mutant strains. The gapA gene was used as a control to ensure that equivalent quantities of templates were loaded. Log FC values from the array are shown. Twenty-five PCR cycles were performed in all cases.
Regulation of LF82 motility and chemotaxis by NrdR.
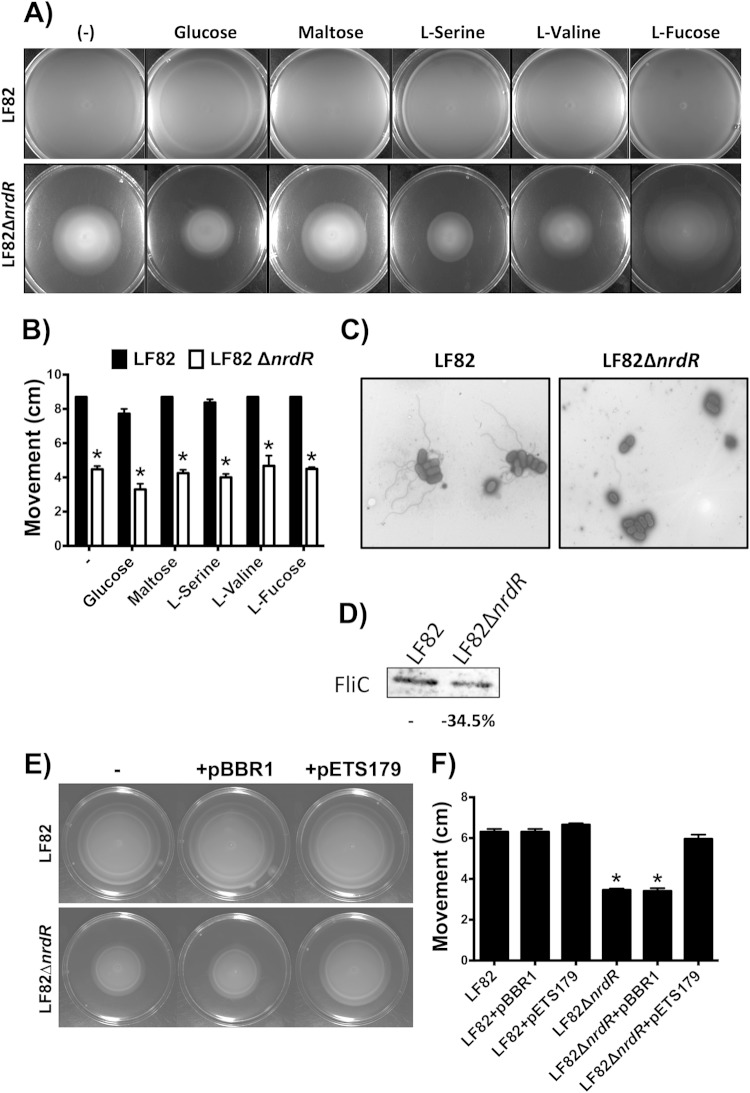
To phenotypically corroborate our microarray results, which indicated a possible role of NrdR in motility and chemotaxis, we evaluated the behavior of LF82 and LF82 ΔnrdR on soft agar supplemented with four different chemoattractants (d-glucose, maltose, l-serine, and l-fucose) and one chemorepellent (l-valine). After a 12-h incubation period at 30°C for all conditions, the LF82 ΔnrdR mutant exhibited reduced colony motility compared with its corresponding wild-type strain (Fig. 8A). The diameters of the motility halos in the LF82 ΔnrdR were one-half of those of the halos produced by the wild-type strain (Fig. 8B), indicating the importance of NrdR in E. coli LF82 motility. Experiments to evaluate chemotaxis using specific soft-agar plates supplemented with serine, maltose, and thymine yielded results similar to those of the motility assays, with reduced chemotactic expansion of the LF82 ΔnrdR mutant compared with the wild-type strain (see Fig. S2 in the supplemental material).
FIG 8.
Effect of ΔnrdR on LF82 motility. (A and B) Motility on soft LB agar of wild-type and nrdR mutant LF82 strains after 12 h of incubation at 30°C. The errors bars in panel B represent the standard deviations from three independent experiments. (C) Electron microscopy of AIEC LF82 and the nrdR mutants (magnification, ×10,000). (D) Western blot analysis of FliC expression in wild-type and LF82 ΔnrdR mutant strains. (E and F) Motility on soft LB agar of complemented ΔnrdR mutant strains with cloned nrdR gene (pETS179) and the empty vector (pBBR1). The error bars in panel F represent the standard deviations from three independent experiments. Significant differences in motility were determined using a paired Student t test; *, P < 0.01.
Electron microscopy indicated reduced expression of flagella at the surface of the LF82 ΔnrdR mutant bacteria compared with the wild-type strain (Fig. 8C). LF82 ΔnrdD, ΔnrdE, and ΔnrdABp mutants did not display any alterations in flagellum production compared with the wild-type strain (data not shown). In addition, Western blot analysis indicated reduced FliC expression in the LF82 ΔnrdR mutant compared with the wild-type strain, reinforcing our previous results of reduced motility and demonstrating a reduced FliC protein level (Fig. 8D). In addition, complementation of LF82 ΔnrdR mutant with a cloned nrdR gene restored the motility phenotype of the ΔnrdR mutant to a level similar to the wild-type level (Fig. 8E and F), confirming the effect of NrdR in the regulation of LF82 motility.
DISCUSSION
Ribonucleotide reductases operate under a range of environmental conditions, which suggests that although the propensity to synthesize deoxyribonucleotides is an essential function, the type of RNR present will have an impact on the microorganism's ability to adapt to different environments. In addition, when more than one RNR class is encoded by the bacterial genome, it is crucial to understand which RNR is important during the adhesion, colonization, and persistence of pathogenic bacteria. Adherent-invasive E. coli (AIEC), especially the reference strain LF82, is of particular interest because AIEC virulence evolution involves the selection of amino acid mutations in common bacterial proteins, such as the FimH protein, and leads to the development of chronic inflammatory bowel disease in a genetically susceptible host (25). The AIEC reference strain LF82 is of great interest among the different E. coli species and Enterobacteriaceae, as it provides an additional RNR class of plasmid origin. This bacterium harbors four different RNR classes, three of which are chromosomally encoded (classes Ia, Ib, and III) and an additional copy (class Ia) that is encoded by the pLF82 plasmid. In the present study, we demonstrated that this extra class Ia RNR copy encoded by pLF82 was acquired by horizontal gene transfer. Several other examples of plasmid- and prophage-encoded RNR proteins have been previously reported (for an example, see reference 3), providing clear evidence of horizontal gene transfer events that play an important role in the evolution of the RNR distribution for a great variety of RNR genes from all three domains of life.
However, for suspected horizontal gene transfer cases, we must assess whether the transferred copy is enzymatically active. In the AIEC LF82 strain, this is not the case because plasmid NrdAp was inactive in our experiments. Surprisingly, all-important residues located at the active and allosteric sites, iron ligand and NrdA, and the NrdA-NrdB interaction were also conserved in the E. coli LF82 NrdABp. Thus, the loss of E. coli LF82 NrdAB activity may involve residues that are important for protein folding, activation, and subunit interaction.
Bacteria need tight gene regulation to maintain the balanced dNTP pools important for DNA replication. For this reason, tight transcriptional regulation must be achieved when a bacterium encodes more than one class of RNR. Therefore, it is important to understand which is the role of each RNR class during AIEC LF82 virulence processes, such as adhesion to intestinal epithelial cells and AIEC LF82 persistence in the gut mucosa of a mouse model mimicking CD susceptibility. An expression analysis of the different RNR classes present when AIEC LF82 was grown in cells associated with T-84 cells compared with cells grown in cellular medium revealed important differences. nrdR and nrdD (class III) mRNA levels increased during growth when AIEC LF82 bacteria were associated with T-84 cells, whereas the corresponding mRNA levels decreased significantly when bacteria were grown in cellular medium during a similar period, suggesting that NrdR and NrdD protein levels in AIEC LF82 bacteria are crucial for controlling bacterial virulence.
The importance of each RNR class during infection was investigated by assessing the bacterium's ability to attach to T-84 cells and to the ileal loop and to persist in the gut mucosa of CEABAC10 mice. The adhesion assays demonstrated that the nrdR mutant exhibited an impaired ability to interact with intestinal epithelial cells compared with the wild type. The other nrd-lacking mutants used in this study did not display alterations in adhesion. Conversely, the results indicate that only the nrdR and nrdD gene products reduced the ability of AIEC LF82 to persist in the gut and thus impaired pathogenesis. Several authors have previously described the anaerobic environments in the gut of mammals (37), and under these conditions, anaerobic enzymes are transcriptionally activated. Class III enzymes are enzymatically active only under anaerobic conditions, and their importance has been well established (5). The nrdD mutant (class III) has a markedly reduced capacity to infect the gut, which is correlated with the capacity for DNA replication under anaerobic conditions. Other RNR classes cannot compensate for nrdD deficiency. A comparison of the LF82 nrdD promoter region with the E. coli MG1655 nrdD promoter region revealed two FNR boxes (TTGATGTAGCTCAA and TTGATGCAAAGCAC) located at −211 and −242 bp upstream of the translation start point. The almost-identical positions in the two promoter region strains demonstrate that this RNR class is transcriptionally activated during anaerobic metabolism and is important for the specific dNTP supply in the AIEC LF82 strain facing anaerobic conditions during gut infection.
We also assessed why the AIEC transcriptional regulator NrdR is important for AIEC's ability to adhere to T-84 cells, interact with intestinal mucosa in the ileal loop model, and persist in the gut mucosa of mice that mimic CD susceptibility. To answer this question, we investigated NrdR's ability to regulate global gene expression. In our transcriptomic analysis, the genes that were transcriptionally altered were ones that regulate motility and chemotaxis. Genes involved in chemotaxis, such as MotA and MotB, together apply the required force to FliG for flagellar rotation (38). MotA interaction with FliG is essential for switching the flagellar direction from clockwise (CW) movement and stimulating cell tumble to the counterclockwise (CCW) state that promotes the forward movement of the bacteria (39). The effect of deletion of motA and motB, which promotes CCW movement and reduces bacterial chemotaxis, is well known (40–42) and corroborates our array results indicating reduced chemotaxis for the AIEC LF82 strain (see Fig. S2 in the supplemental material). In addition, the AIEC LF82 ΔnrdR mutant exhibited reduced motility capacity compared to the wild-type strain (Fig. 8). Flagella of the AIEC LF82 strain have established roles in the adhesion to and invasion of intestinal epithelial cells (43, 44). Electronic microscopy analysis of the wild-type and LF82 ΔnrdR isogenic mutant demonstrated reduced flagellar expression when the nrdR gene was attenuated. This was confirmed by Western blot analysis of the expression of the major component of the flagella, FliC (Fig. 8). It is well described that flagella contribute to AIEC virulence. Indeed, the nonmotile aflagellar LF82 mutants (ΔfliC or ΔfliA mutants) exhibit a drastic reduction in adherence and invasion ability (43, 44), and the flagellar sigma factor FliA regulates the adhesion and invasion of Crohn's disease-associated Escherichia coli via a cyclic dimeric GMP-dependent pathway (44). In addition, Crohn's disease-associated virulent AIEC LF82 bacteria, via flagellar expression, are able to potentiate an inflammatory mucosal immune response in the dextran sulfate sodium (DSS)-injured colons of BALBc/J mice via increased expression of Toll-like receptor 5 (TLR5) and IPAF flagellin receptors (45). In addition, we reported that CEABAC10 mice infected with LF82 ΔfliC did not loss body weight compared to mice infected with LF82 bacteria (see Fig. S4 in the supplemental material). Thus, the decreased virulence ability of the LF82 mutant lacking nrdR could be explained in part by impaired flagellar expression. In addition, flagellin receptor TLR5-deficient mice (T5KO) display elevated intestinal proinflammatory gene expression and colitis. The Crohn's disease-associated E. coli LF82 strain induced chronic colitis in T5KO, which persisted well after the exogenously introduced bacterial species had been eliminated (46).
An extended understanding of the pathogenicity and physiology of AIEC could aid in the development of novel drugs active against these pathogens. In the present study, we investigated the role of RNRs in the virulence of AIEC isolated from Crohn's disease patients and demonstrated that knockout of the nrdR gene, which encodes a transcriptional regulator of RNR, decreased the ability of AIEC LF82 to adhere to intestinal epithelial T-84 cells, interact with the ileal mucosa using the ileal loop model, and colonize the gut mucosa of transgenic mice that express human CEACAM6. NrdR plays a crucial role in AIEC virulence by interfering with bacterial motility and chemotaxis. Thus, the development of new drugs targeting RNR classes, particularly NrdR, could be a promising new strategy for controlling AIEC flagellum expression and motility and subsequently controlling gut colonization and AIEC-induced inflammation in CD patients.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Ministerio de Economia y Competitividad with grants BFU2011-24066, CSD2008-00013, and ERA-NET PathoGenoMics (BIO2008-04362-E) to E.T. This work was also supported by the Generalitat de Catalunya SGR2014-01260. E.T. was supported by the Ramón y Cajal and I3 programs from the Ministerio de Ciencia e Innovación. This study was supported by INSERM (UMR1071) and INRA (USC-2018) and by grants from the Association F. Aupetit (AFA), ANR, in the frame of ERA-NET PathoGenomics (grant no. 0315443).
The funders did not participate in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02772-14.
REFERENCES
- 1.Nordlund P, Reichard P. 2006. Ribonucleotide reductases. Annu Rev Biochem 75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 2.Torrents E, Sahlin M, Sjöberg B-M. 2008. The ribonucleotide reductase family—genetics and genomics, p 17–77. In Anderson KK. (ed), Ribonucleotide reductases. Nova Science Publishers, Hauppauge, NY. [Google Scholar]
- 3.Lundin D, Gribaldo S, Torrents E, Sjoberg BM, Poole AM. 2010. Ribonucleotide reduction—horizontal transfer of a required function spans all three domains. BMC Evol Biol 10:383. doi: 10.1186/1471-2148-10-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lundin D, Torrents E, Poole AM, Sjöberg B-M. 2009. RNRdb, a curated database of the universal enzyme family ribonucleotide reductase, reveals a high level of misannotation in sequences deposited to Genbank. BMC Genomics 10:589. doi: 10.1186/1471-2164-10-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garriga X, Eliasson R, Torrents E, Jordan A, Barbe J, Gibert I, Reichard P. 1996. nrdD and nrdG genes are essential for strict anaerobic growth of Escherichia coli. Biochem Biophys Res Commun 229:189–192. doi: 10.1006/bbrc.1996.1778. [DOI] [PubMed] [Google Scholar]
- 6.Cendra M, Juarez A, Torrents E. 2012. Biofilm modifies expression of ribonucleotide reductase genes in Escherichia coli. PLoS One 7:e46350. doi: 10.1371/journal.pone.0046350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin JE, Imlay JA. 2011. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol Microbiol 80:319–334. doi: 10.1111/j.1365-2958.2011.07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torrents E, Grinberg I, Gorovitz-Harris B, Lundstrom H, Borovok I, Aharonowitz Y, Sjöberg BM, Cohen G. 2007. NrdR controls differential expression of the Escherichia coli ribonucleotide reductase genes. J Bacteriol 189:5012–5021. doi: 10.1128/JB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torrents E. 2014. Ribonucleotide reductases: essential enzymes for bacterial life. Front Cell Infect Microbiol 4:52. doi: 10.3389/fcimb.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strober W, Fuss I, Mannon P. 2007. The fundamental basis of inflammatory bowel disease. J Clin Invest 117:514–521. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xavier RJ, Podolsky DK. 2007. Unravelling the pathogenesis of inflammatory bowel disease. Nature 448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 12.Kaser A, Zeissig S, Blumberg RS. 2010. Inflammatory bowel disease. Annu Rev Immunol 28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chassaing B, Darfeuille-Michaud A. 2011. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology 140:1720–1728. doi: 10.1053/j.gastro.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 14.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel JF. 1998. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology 115:1405–1413. doi: 10.1016/S0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 15.Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, Englyst H, Williams HF, Rhodes JM. 2004. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology 127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 16.Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U, Schreiber S, Dietel M, Lochs H. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 17.Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. 1999. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect Immun 67:4499–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conte MP, Schippa S, Zamboni I, Penta M, Chiarini F, Seganti L, Osborn J, Falconieri P, Borrelli O, Cucchiara S. 2006. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut 55:1760–1767. doi: 10.1136/gut.2005.078824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotlowski R, Bernstein CN, Sepehri S, Krause DO. 2007. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 56:669–675. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neut C, Bulois P, Desreumaux P, Membre JM, Lederman E, Gambiez L, Cortot A, Quandalle P, van Kruiningen H, Colombel JF. 2002. Changes in the bacterial flora of the neoterminal ileum after ileocolonic resection for Crohn's disease. Am J Gastroenterol 97:939–946. doi: 10.1111/j.1572-0241.2002.05613.x. [DOI] [PubMed] [Google Scholar]
- 21.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, Orsi RH, Wiedmann M, McDonough P, Kim SG, Berg D, Schukken Y, Scherl E, Simpson KW. 2007. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J 1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, Garrett N, Alpern JA, Akyildiz A, Theiss AL, Nusrat A, Klapproth JM. 2007. Invasive Escherichia coli are a feature of Crohn's disease. Lab Invest 87:1042–1054. doi: 10.1038/labinvest.3700661. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Medina M, Aldeguer X, Lopez-Siles M, Gonzalez-Huix F, Lopez-Oliu C, Dahbi G, Blanco JE, Blanco J, Garcia-Gil LJ, Darfeuille-Michaud A. 2009. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm Bowel Dis 15:872–882. doi: 10.1002/ibd.20860. [DOI] [PubMed] [Google Scholar]
- 24.Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, Peeters H, Bommelaer G, Desreumaux P, Colombel JF, Darfeuille-Michaud A. 2007. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest 117:1566–1574. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dreux N, Denizot J, Martinez-Medina M, Mellmann A, Billig M, Kisiela D, Chattopadhyay S, Sokurenko E, Neut C, Gower-Rousseau C, Colombel JF, Bonnet R, Darfeuille-Michaud A, Barnich N. 2013. Point mutations in FimH adhesin of Crohn's disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog 9:e1003141. doi: 10.1371/journal.ppat.1003141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carvalho FA, Barnich N, Sivignon A, Darcha C, Chan CH, Stanners CP, Darfeuille-Michaud A. 2009. Crohn's disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J Exp Med 206:2179–2189. doi: 10.1084/jem.20090741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 28.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaveroche MK, Ghigo JM, d'Enfert C. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res 28:E97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adase CA, Draheim RR, Manson MD. 2012. The residue composition of the aromatic anchor of the second transmembrane helix determines the signaling properties of the aspartate/maltose chemoreceptor Tar of Escherichia coli. Biochemistry 51:1925–1932. doi: 10.1021/bi201555x. [DOI] [PubMed] [Google Scholar]
- 31.Roca I, Torrents E, Sahlin M, Gibert I, Sjoberg BM. 2008. NrdI essentiality for class Ib ribonucleotide reduction in Streptococcus pyogenes. J Bacteriol 190:4849–4858. doi: 10.1128/JB.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jordan A, Aragall E, Gibert I, Barbe J. 1996. Promoter identification and expression analysis of Salmonella typhimurium and Escherichia coli nrdEF operons encoding one of two class I ribonucleotide reductases present in both bacteria. Mol Microbiol 19:777–790. doi: 10.1046/j.1365-2958.1996.424950.x. [DOI] [PubMed] [Google Scholar]
- 33.Ekberg M, Birgander P, Sjoberg BM. 2003. In vivo assay for low-activity mutant forms of Escherichia coli ribonucleotide reductase. J Bacteriol 185:1167–1173. doi: 10.1128/JB.185.4.1167-1173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. 2007. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chassaing B, Etienne-Mesmin L, Bonnet R, Darfeuille-Michaud A. 2013. Bile salts induce long polar fimbriae expression favouring Crohn's disease-associated adherent-invasive Escherichia coli interaction with Peyer's patches. Environ Microbiol 15:355–371. doi: 10.1111/j.1462-2920.2012.02824.x. [DOI] [PubMed] [Google Scholar]
- 36.Miquel S, Peyretaillade E, Claret L, de Vallee A, Dossat C, Vacherie B, Zineb EH, Segurens B, Barbe V, Sauvanet P, Neut C, Colombel JF, Medigue C, Mojica FJ, Peyret P, Bonnet R, Darfeuille-Michaud A. 2010. Complete genome sequence of Crohn's disease-associated adherent-invasive E. coli strain LF82. PLoS One 5(9):pii:e12714. doi: 10.1371/journal.pone.0012714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones SA, Gibson T, Maltby RC, Chowdhury FZ, Stewart V, Cohen PS, Conway T. 2011. Anaerobic respiration of Escherichia coli in the mouse intestine. Infect Immun 79:4218–4226. doi: 10.1128/IAI.05395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kojima S, Blair DF. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- 39.Berg HC. 2003. The rotary motor of bacterial flagella. Annu Rev Biochem 72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 40.Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol 180:2729–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morimoto YV, Che YS, Minamino T, Namba K. 2010. Proton-conductivity assay of plugged and unplugged MotA/B proton channel by cytoplasmic pHluorin expressed in Salmonella. FEBS Lett 584:1268–1272. doi: 10.1016/j.febslet.2010.02.051. [DOI] [PubMed] [Google Scholar]
- 42.Parkinson JS, Houts SE. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol 151:106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnich N, Boudeau J, Claret L, Darfeuille-Michaud A. 2003. Regulatory and functional co-operation of flagella and type 1 pili in adhesive and invasive abilities of AIEC strain LF82 isolated from a patient with Crohn's disease. Mol Microbiol 48:781–794. doi: 10.1046/j.1365-2958.2003.03468.x. [DOI] [PubMed] [Google Scholar]
- 44.Claret L, Miquel S, Vieille N, Ryjenkov DA, Gomelsky M, Darfeuille-Michaud A. 2007. The flagellar sigma factor FliA regulates adhesion and invasion of Crohn disease-associated Escherichia coli via a cyclic dimeric GMP-dependent pathway. J Biol Chem 282:33275–33283. doi: 10.1074/jbc.M702800200. [DOI] [PubMed] [Google Scholar]
- 45.Carvalho FA, Barnich N, Sauvanet P, Darcha C, Gelot A, Darfeuille-Michaud A. 2008. Crohn's disease-associated Escherichia coli LF82 aggravates colitis in injured mouse colon via signaling by flagellin. Inflamm Bowel Dis 14:1051–1060. doi: 10.1002/ibd.20423. [DOI] [PubMed] [Google Scholar]
- 46.Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, Gonzalez A, Clemente JC, Cullender TC, Barnich N, Darfeuille-Michaud A, Vijay-Kumar M, Knight R, Ley RE, Gewirtz AT. 2012. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 49.Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203. doi: 10.1016/S0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- 50.Wechsler JA, Gross JD. 1971. Escherichia coli mutants temperature-sensitive for DNA synthesis. Mol Gen Genet 113:273–284. doi: 10.1007/BF00339547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.