Abstract
Microsporidia are obligate intracellular parasites with rigid spore walls that protect against various environmental pressures. Despite an extensive description of the spore wall, little is known regarding the mechanism by which it is deposited or the role it plays in cell adhesion and infection. In this study, we report the identification and characterization of two novel spore wall proteins, SWP7 and SWP9, in the microsporidian species Nosema bombycis. SWP7 and SWP9 are mainly localized to the exospore and endospore of mature spores and the cytoplasm of sporonts, respectively. In addition, a portion of SWP9 is targeted to the spore wall of sporoblasts earlier than SWP7 is. Both SWP7 and SWP9 are specifically colocalized to the spore wall in mature spores. Furthermore, immunoprecipitation, far-Western blotting, unreduced SDS-PAGE, and yeast two-hybrid data demonstrated that SWP7 interacted with SWP9. The chitin binding assay showed that, within the total spore protein, SWP9 and SWP7 can bind to the deproteinated chitin spore coats (DCSCs) of N. bombycis. However, binding of the recombinant protein rSWP7-His to the DCSCs is dependent on the combination of rSWP9–glutathione S-transferase (GST) with the DCSCs. Finally, rSWP9-GST, anti-SWP9, and anti-SWP7 antibodies decreased spore adhesion and infection of the host cell. In conclusion, SWP7 and SWP9 may have important structural capacities and play significant roles in modulating host cell adherence and infection in vitro. A possible major function of SWP9 is as a scaffolding protein that supports other proteins (such as SWP7) that form the integrated spore wall of N. bombycis.
INTRODUCTION
Microsporidia, which infect numerous vertebrate and invertebrate species, are a specific group of obligate spore-forming, intracellular, and unicellular eukaryotic protists (1). The microsporidian species Nosema bombycis causes pebrine, a disease that was common in Europe, America, and Asia during the mid-19th century and still causes heavy losses in silk-producing countries, such as China and India (1). It was first recognized in 1857 by Nägeli (2). Recently, phylogenetic analyses based on conserved proteins (3–5), ribosomal DNA (rDNA) sequences (6, 7), and complete sequencing of the Encephalitozoon cuniculi genome (8) have suggested that microsporidia, which undergo a highly reductive evolution, are closely related to fungi (9). However, the N. bombycis genome has greatly expanded due to transposable elements and gene duplications (10).
The microsporidian spore wall, which is usually comprised of an electron-dense proteinaceous outer layer (exospore) of 25 to 30 nm and an electron-transparent chitinous inner layer (endospore) of 30 to 35 nm, maintains the spore's morphology and helps the mature spore resist the outer environment (11–13). Although chitin is the major component of the spore wall, only one chitin-associated protein has been identified. The chitin deacetylase activity of E. cuniculi (EcCDA) has been confirmed to be expressed during sporogonic stages (14). Currently, the deproteinated chitin spore coats (DCSCs) of the N. bombycis endospore are isolated using a heated alkali solution (15). However, few studies have reported on the interactions between spore wall proteins (SWPs) and the DCSCs in microsporidia. At present, several spore wall proteins that are localized to the exospore or endospore have been identified in the genus Encephalitozoonidae (14, 16–20). For N. bombycis, only two exosporal proteins (21–23), three endosporal proteins (21, 24, 25), and a BAR-2 spore wall protein (26) have been characterized. In addition, several spore wall proteins that adhere to host cells via a heparin-binding motif (HBM) and interact with sulfated glycosaminoglycans (GAGs) of the host cell surface have been identified (20, 24, 27, 28). In recent years, several studies have found that the spores of N. bombycis not only infect the Bombyx mori ovarian cell line BmN-SWU1, but also adhere to and infect the B. mori embryonic cell line BmE-SWU1. SWP30 (SWP1), SWP25 (SWP2), and SWP32 (SWP3) are found in the spore walls of mature spores of N. bombycis cultured in BmE-SWU1 cells (21, 25).
Microsporidian spores contain a special invasion organelle (29), known as the polar tube, that is composed of three distinct polar tube proteins: PTP1 (30, 31), PTP2 (32), and PTP3 (33). Spores invade the host cell using two different mechanisms (34). Germination involves polar tube penetration into the host cell's cytoplasmic membrane to deliver the infectious sporoplasm to the host cell. The other process is phagocytosis, in which a spore is phagocytosed by a host cell, allowing germination to occur (34–36). However, little research has been performed to investigate the processes and mechanisms by which N. bombycis infects host cells.
Recently, 14 hypothetical N. bombycis spore wall proteins (including SWP7 and SWP9) were predicted using proteomics-based approaches (21). In this study, we identified two novel spore wall proteins (SWP7 and SWP9) and obtained immature spores. To understand the formation of the spore wall and the functions of its chitin and spore wall proteins, the interaction between SWP9 and SWP7 was studied for the first time. SWP9 in the chitin layer was also demonstrated to act as a scaffolding protein that supports SWP7 in the spore wall. More importantly, SWP9 and SWP7 mediate the infectious process by enabling spore adherence to host cells.
MATERIALS AND METHODS
Ethics statement.
All animal experiments, including animal care and procedures, were conducted in accordance with the guidelines of the China Council on Animal Care. This study was approved by the Laboratory Animal Welfare and Ethics Committee of the Third Military Medical University with the animal utilization protocol number SYXK-PLA-2007035.
Cell culture.
BmE-SWU1 cells were used for the cultivation of N. bombycis spores. The procedure for spore infection of BmE-SWU1 cells was performed as previously described (21, 25, 37, 38). Adherent and infectious cells were maintained in Grace's insect culture medium, manufactured by Gibco Co. (Carlsbad, CA), supplemented with 10% fetal bovine serum, manufactured by HyClone Co. (Logan, UT), at 28°C for the adherence and infection assays.
Production and purification of N. bombycis spores.
The N. bombycis isolate CQ1, originally isolated from infected silkworms in Chongqing, China, is conserved in the China Veterinary Culture Collection Center (CVCC no. 102059). The life cycle stages of N. bombycis were purified and harvested from laboratory-reared silkworm larvae as previously described (23, 39–41). The harvested spores were purified on a discontinuous Percoll gradient (GE Healthcare, Beijing, China; 30, 45, 60, 75, and 90% [vol/vol]) and centrifuged at 12,000 × g for 40 min. In addition, we obtained the N. bombycis spore coats by the two methods of glass bead agitation and germination. The purified spores and spore coats were washed and stored in phosphate-buffered saline (PBS) with antibiotics (100 U/ml streptomycin or 100 U/ml penicillin) at 4°C.
Protein extraction.
Total mature N. bombycis protein, spore coat protein, SDS-soluble protein, and alkali-soluble protein were extracted. Mature spores (109 spores) were broken in 400 μl PBS (pH 7.3) containing a protease inhibitor (phenylmethylsulfonyl fluoride [PMSF]) by vibration with 0.4 g glass beads (Sigma; 150 to 212 μm) at 4°C for 6 h. The supernatant protein was collected as total spore protein by centrifugation at 13,200 × g for 10 min and boiled with 3× loading buffer for Western blotting. The pellet was subjected to a discontinuous Percoll gradient (30, 45, 50, 75, and 90% [vol/vol]) and centrifuged at 12,000 × g for 30 min for the indirect immunofluorescence assay (IFA). The spore coat proteins of N. bombycis were extracted as previously reported (21, 42) with minor differences. In brief, in spores (109), germination was induced by 0.1 mol/liter K2CO3 (pH 8.0) at 28°C for 30 min. Then, the spores were collected by centrifugation at 10,000 × g for 15 min, purified on a discontinuous Percoll gradient (30, 45, 60, 75, and 90% [vol/vol]), and centrifuged at 12,000 × g for 40 min. The 75% Percoll fraction was spore coats and was extracted with 3× loading buffer for SDS-PAGE and Western blotting. Under the alkali and SDS conditions, the spore surface protein dropped from the wall at room temperature (42–44). In simple terms, 108 spores of N. bombycis were treated with 0.1 M NaOH or 2% SDS for 20 min at room temperature and centrifuged at 10,000 × g for 20 min. The supernatant contained alkali-soluble spore surface protein and SDS-soluble protein.
Sequence analysis of Nbswp9 and Nbswp7.
The signal peptide and transmembrane domains of N. bombycis swp9 (Nbswp9) and Nbswp7 and the N- and O-glycosylation and phosphorylation sites were predicted by the CBS prediction servers (http://www.cbs.dtu.dk/services/). The NCBI protein database (http://www.ncbi.nlm.nih.gov/protein/) was used to search for homologous proteins, and amino acid sequence alignments were performed using ClustalW (45).
Antiserum production and Western blotting.
Polyclonal antibody production and immunoblotting protocols to detect NbSWP9 and NbSWP7 in the total protein of N. bombycis have been previously described (21, 22, 24). In short, rabbits and mice were injected with recombinant rSWP9–glutathione S-transferase (GST) and rSWP7-His proteins, respectively. Meanwhile, one rabbit and one mouse were injected subcutaneously with PBS and adjuvant as a negative control. Finally, we placed the rabbit serum onto a desalting column, after purifying the serum by the hexylacetic acid-saturated ammonium sulfate method. The production of polyclonal antibody against polar tube protein 1 (PTP1) has been previously described (46).
For the immunoblot analysis, total mature N. bombycis protein, spore coat protein, and SDS-soluble and alkali-soluble spore surface proteins were transferred onto polyvinylidene difluoride (PVDF) membranes after SDS-PAGE and blocked overnight at 4°C in Tris-buffered saline–Tween 20 (TBST) with 5% (wt/vol) nonfat dry milk. Then, for primary-antibody incubation, the samples were incubated with either 10 μg anti-NbSWP9 or anti-NbSWP7 polyclonal antibody diluted at 1:6,000 or a negative-control serum. After washing six times, the membranes were reacted with horseradish peroxidase (HRP)-labeled goat anti-mouse or anti-rabbit IgG antiserum (Sigma-Aldrich, St. Louis, MO) at a 1:8,000 dilution, washed another six times, and developed with the ECL Western blot detection kit (Thermo Fisher Scientific, Rockford, IL).
IFA.
For the IFA, the above-mentioned purified spores and spore coats were fixed with 4% paraformaldehyde for 20 min at room temperature. Then, they were washed in PBS and blocked with a blocking solution consisting of 5% (wt/vol) bovine serum albumin (BSA) and 10% (vol/vol) goat serum in PBS overnight at 4°C or for 2 h at 37°C. The samples were incubated sequentially for 1 h with polyclonal anti-SWP9 or anti-SWP7 antibody (diluted 1:200 in blocking solution) or a negative-control serum (diluted 1:200 in blocking solution) and then washed and incubated for an additional 1 h with a 1:64 dilution of fluorescein isothiocyanate (FITC)-conjugated anti-mouse or anti-rabbit IgG (Sigma) in a moist chamber at 37°C. Nuclear material was stained with DAPI (4′6-diamidino-2-phenylindole) (Sigma) at 37°C for 10 min.
To analyze the interaction between SWP9 and SWP7 and the localization of SWP9 and SWP7 in the sporonts, sporoblasts, and mature spores, the life cycle stages of N. bombycis were acquired as described above. The purified life cycle stages of N. bombycis were fixed, washed, and blocked with blocking solution overnight at 4°C or for 2 h at 37°C. Samples were incubated for 1 h with polyclonal anti-SWP9 mouse and anti-SWP7 rabbit antibodies simultaneously and then washed and incubated for an additional 1 h with a 1:64 dilution of FITC-conjugated anti-mouse IgG and a 1:2,000 dilution of anti-rabbit Alexa Fluor 647 (Invitrogen, Carlsbad, CA) in a moist chamber at 37°C.
Transmission electron microscopy (TEM) immunolabeling.
The life cycle stages of N. bombycis spores were fixed in 3% paraformaldehyde (freshly prepared) in 0.1 M sodium cacodylate buffer (pH 7.2 to 7.4) containing 0.1% glutaraldehyde, 4% sucrose overnight at 4°C. The fixed pellets were rinsed four times with 0.1 M sodium cacodylate buffer containing 4% sucrose (pH 7.4) at 4°C for 15 min each time and then sequentially dehydrated with graded methanol. The dehydrated spores were then embedded and photopolymerized in Lowicryl K4M (Zhongjingkeyi Technology Co., Beijing, China) under UV light (360 nm) and placed 20 to 30 cm away from the UV source for 72 h at −35°C. Ultrathin sections were placed on 200-mesh nickel grids coated with Formvar and carbon. The nickel grids were incubated in blocking buffer (1% BSA [Sigma], 0.05% Triton X-100, and 0.05% Tween 20) at room temperature for 1 h, followed by incubation with a 1:300 dilution of primary rabbit anti-SWP9, anti-SWP7, and negative-control rabbit antibodies overnight at 4°C; rinsed 6 times in PBS; and then incubated with a 1:70 dilution of goat anti-rabbit IgG conjugated to 18-nm colloidal gold (Jackson ImmunoResearch, West Grove, PA) at room temperature for 1 h. The grids were rinsed with PBS, dried, stained with 3% uranyl acetate, and then examined and photographed with a Hitachi H-7650 transmission electron microscope at an accelerating voltage of 80 kV.
SDS-PAGE of unreduced, treated rSWP9-GST and rSWP7-His; immunoprecipitation; far-Western blotting; and yeast two-hybrid assay.
Colocalization; SDS-PAGE of unreduced, treated samples; immunoprecipitation; far-Western blotting; and yeast two-hybrid assays were used to demonstrate the interaction between SWP9 and SWP7 of N. bombycis as follows. (i) Unreduced, treated samples were subjected to SDS-PAGE. Briefly, rSWP7-His was incubated with rSWP9-GST for 2 h at 4°C, and then the sample was separated into two portions. Part of the sample was treated with normal 3× loading buffer (containing β-mercaptoethanol), and another part of the sample was prepared in a nonreducing sample buffer (without β-mercaptoethanol). The proteins were transferred to PVDF membranes and incubated with anti-SWP9 and anti-SWP7 antibodies to detect the complex and normal bands. (ii) To determine whether SWP9 coimmunoprecipitates with SWP7, we performed experiments on total mature-spore proteins. The immunoprecipitation procedure was as previously described (22, 47). In brief, total mature-spore proteins were agitated overnight at 4°C with 20 μg of rabbit anti-SWP9 or anti-SWP7 antibody with protein A plus G agarose beads (Beyotime, Chongqing, China). The next day, Western blotting was conducted using mouse anti-SWP9 or anti-SWP7 antibodies. Specifically, the total mature-spore protein was immunoprecipitated with antibodies specific for either SWP9 or SWP7 and then immunoblotted for SWP7 or SWP9, respectively. (iii) Far-Western blotting was then used to further identify the protein-protein interactions of SWP9 and SWP7. The rSWP7-His or rSWP9-GST protein and rGST as the prey proteins were separated by SDS-PAGE and then transferred to the surface of the PVDF membrane. After the prey proteins were transferred to the membrane, the membrane was blocked in TBST with 5% (wt/vol) nonfat dry milk. The PVDF membrane containing the prey protein rSWP7-His was probed with a purified rSWP9-GST bait protein, as well as purified rGST. Following binding of the bait protein with the prey protein, the anti-SWP9 or anti-GST detecting antibody was used to identify the corresponding bands. Furthermore, a PVDF membrane containing the prey proteins rSWP9-GST and rGST was incubated with the rSWP7-His bait protein. Following binding of the bait protein with the prey protein, the anti-SWP7 detecting antibodies were used to identify the corresponding bands. (iv) For further interaction study, the full lengths of Nbswp7 and Nbswp9 were amplified from N. bombycis DNA using the following primers: Nbswp7-Forward-EcoRI (CGGAATTCATGATAAAAGGTTTAATTTAC) and Nbswp7-Reverse-XhoI (CCCTCGAGTTATTTATTTATTAAGTGGTC), and Nbswp9-Forward-EcoRI (CGGAATTCATGTCAACACAACCAACAAAA) and Nbswp9-Reverse-SalI (GCGTCGACTTATTTAGATAATAATTCCTT). The procedure was described in detail previously (48). In brief, Nbswp7 and Nbswp9 were fused to the yeast two-hybrid vectors pGADT7 and pGBKT7. Yeast competent cells were transformed simultaneously with bait and prey constructs, pGBKT7-Nbswp9/pGADT7-Nbswp7. The yeast cells were plated on synthetic defined premix (SD) agar base plates that did not contain leucine, tryptophan, histidine, and adenine but contained 5-bromo-4-chloro-3-indoxyl-α-d-galactopyranoside (X-α-Gal) (Clontech Biosciences, San Jose, CA).
Chitin binding assay.
A chitin binding assay was designed to detect the combination of rSWP9-GST and rSWP7-His or native SWP9 and SWP7 (within the total spore protein) of N. bombycis with the DCSCs. We obtained DCSCs from 1 M heat- and NaOH-treated spores of N. bombycis. Briefly, 107 spores were suspended in 100 μl of 1 M NaOH and boiled for 1 h at 100°C. Then, insoluble spore wall material was obtained by centrifugation at 480 × g at 25°C for 10 min. The pellet was suspended in 0.75 M NaOH and incubated at 75°C for 1 h. The suspension was then cooled to 25°C, and the extraction was sustained for 24 h. The insoluble material was collected by centrifugation at 480 × g at room temperature for 10 min, and the sample was then washed 10 times with deionized water (15). Specifically, DCSCs of N. bombycis were incubated with 2 μg of total mature-spore proteins, rSWP9-GST or rSWP7-His, or both rSWP9-GST and rSWP7-His. All of the samples were centrifuged for 5 min at 4,320 × g, and the pellets were washed six times with PBS (0.01 M, pH 7.2). DCSC pellets, bound to proteins, were stained with Fluostain I {4,4-bis[4-anilino-6-di(hydroxyethyl)amino-triazine-2-ylamino]-2,2-stilbene disulfonic acid, disodium salt} to verify the chitin component of N. bombycis. Both IFA and Western blotting were used to detect the SWP9 and SWP7 proteins, which can bind to the chitin spore coats.
Host cell binding assay with total spore protein or rSWP9-GST and rSWP7-His.
To determine whether SWP9, SWP7, or both attach to the host cell surface, a host cell binding assay was designed with rSWP9-GST and rSWP7-His or the total mature-spore protein of N. bombycis and was performed as in previous studies (20, 24). The total spore protein of N. bombycis and purified rSWP9-GST and rSWP7-His were placed onto BmE cell monolayers in 12-well plates for 2 h at 28°C. Control wells were treated with PBS buffer, rGST, and total spore proteins. The BmE cell monolayers were washed six times with sterile PBS (pH 7.3). A portion of the BmE cell monolayers was solubilized in 3× loading buffer and boiled for 8 to 10 min, and another portion was fixed and then used to observe fluorescence with IFA.
Spore adherence and infectivity inhibition assays.
To further analyze the roles of SWP9 and SWP7 in spore adherence and infection, antibody-blocking and protein inhibition assays were designed. The procedures for these assays were similar to those previously reported, with some changes (20, 24, 27). For the antibody-blocking assay, the anti-SWP9 rabbit antibody (5 μg/ml), anti-SWP7 rabbit antibody (5 μg/ml), and negative-control rabbit antibody (5 μg/ml) were used to block N. bombycis spores (2.8 × 106) for 2 h at 28°C prior to placing the spores onto BmE cell monolayers (1 × 105 cells). In addition, both anti-SWP9 and anti-SWP7 antibodies were used together to block N. bombycis spores. For the protein inhibition assay, the rSWP9-GST (5 μg/ml) and the control rGST (5 μg/ml) proteins were incubated with BmE cell monolayers (1 × 105 cells) in 24-well plates for 2 h at 28°C. Subsequently, 2.8 × 106 N. bombycis spores were placed onto the BmE cell monolayers (1 × 105 cells) for 2 h at 28°C. The subsequent steps were as described above. Following fixation, the BmE host cells attached to by spores were stained with DAPI to detect adherence. This was evaluated here as BmE host cell adherence by spores of N. bombycis. Bound spores were quantified by counting bound microsporidia in at least 40 fields at ×600 magnification. The results are expressed as the percentages of adherent spores relative to control samples. Statistical significance was determined using Student's t test.
To estimate host cell infection following adherence of N. bombycis, the unbound spores were removed by washing, and incubation of host cells with attached spores was continued for 48 h, so that the host cells became infected. The interiors of BmE cells contained N. bombycis spores, signifying infected BmE cells. Following culture, the 24-well plates were fixed and then stained with DAPI. A fluorescence microscope was used to calculate the number of infected BmE cells per field at ×600 magnification. The results are expressed as the percentage of infected host cells per field of magnification.
The above-mentioned experiments were performed in triplicate and repeated at least three times, always with similar results. Each experiment was performed separately with its own negative control. The significances of the differences among the control and experimental assays were measured using the two-tailed Student t test in the Statistical Package for Social Science (SPSS) version 12.0 (SPSS Inc., Chicago, IL). P values of 0.05 or less were considered statistically significant; P values of 0.01 or less were considered highly significant.
Nucleotide sequence accession numbers.
The sequences of Nbswp7 and Nbswp9 have been deposited in GenBank under accession numbers EOB13707.1 and EOB13793.1, respectively.
RESULTS
Homologs and sequence characteristics of Nbswp7 and Nbswp9.
NbSWP7 has a theoretical molecular mass of 32.8 kDa and a predicted pI of 4.50. Amino acid sequence alignment indicated that NbSWP7 shares 27% and 40% identity with the ATP binding cassette (ABC)-type multidrug transport systems (ABCTs) of Enterocytozoon bieneusi (EbABCT; accession number EED44486.1) and Nosema apis (accession number EQB61147.1), respectively. NbSWP7 was predicted to have a signal peptide (Fig. 1A). The gene for NbSWP9 encodes 367 amino acids (theoretically, approximately 42.8 kDa) equipped with an N-terminal transmembrane domain (Fig. 1B). Multiple-sequence alignment showed that homologs of Nbswp7 and Nbswp9 were identified in other microsporidian species. Syntenic analysis showed that the gene loci of Nbswp7 and Nbswp9 are conserved in the microsporidia (data not shown). In addition, the reverse transcription (RT)-PCR results showed that Nbswp7 and Nbswp9 were transcribed in every developmental period of N. bombycis (data not shown).
FIG 1.
Alignment of multiple NbSWP7- and NbSWP9-homologous sequences in various microsporidian species. (A) Orthologs of NbSWP7 in various microsporidian species. The predicted sequence of the signal peptide of SWP7 is boxed. ABCT, ABC-type multidrug transport system, ATPase and permease component. (B) Multiple amino acid sequence alignment of NbSWP9 orthologs in other microsporidian species. There is a predicted transmembrane helix region (TMHMM) in the sequence of SWP9 (underlined). The asterisks indicate the many similar amino acid residues of these species. Black shading indicates identical amino acids. Grey shading represents amino acids with similar properties.
SWP9 and SWP7 are two novel spore wall proteins of N. bombycis and primarily localize to the spore wall.
To further characterize SWP9 and SWP7, the expressed recombinant proteins rSWP9-GST and rSWP7-His migrated as bands of approximately 64.7 kDa and 35.8 kDa in SDS-PAGE, respectively (data not shown). The antibodies to SWP9 and SWP7 recognize the corresponding bands of rSWP9-GST and rSWP7-His, respectively. These antibodies do not cross-react with each other (data not shown).
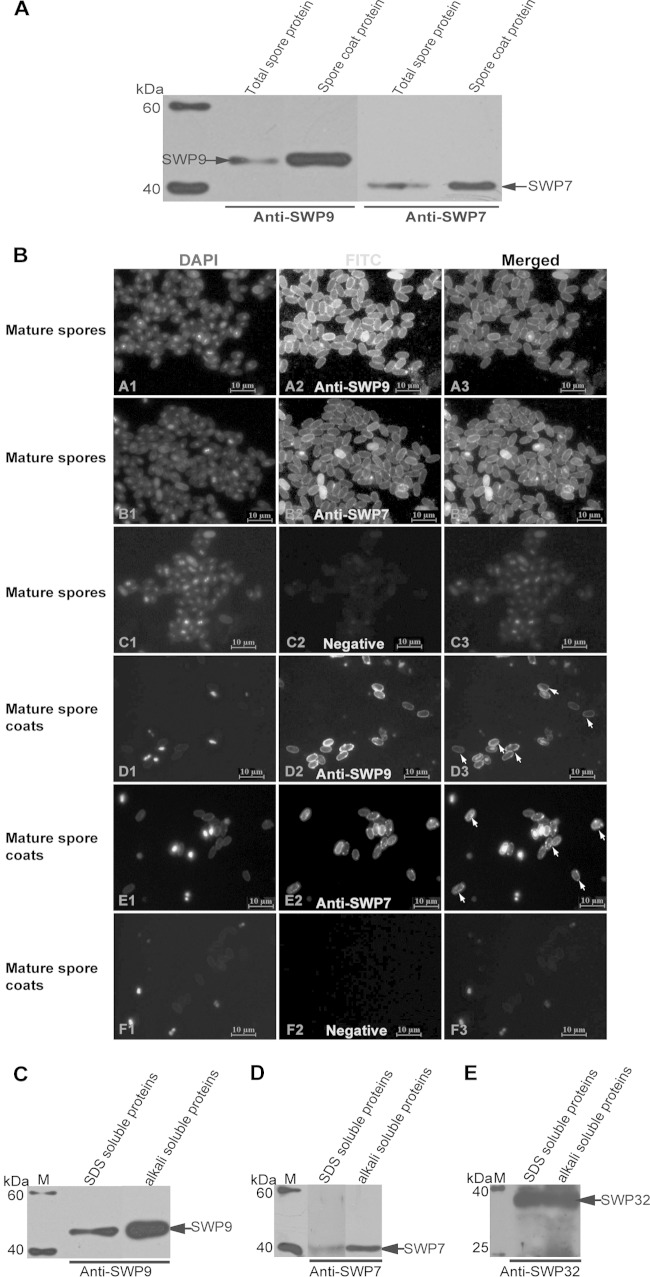
For Western blotting, the corresponding single SWP7 and SWP9 bands were detected from the total N. bombycis spore and spore coat proteins using mouse or rabbit anti-SWP9 antibodies (Fig. 2). The SWP9 band was in agreement with the size predicted by the SWP9 sequences. The SWP7 band was slightly larger than the predicted size. These data show that the antibodies do not cross-react with other spore proteins.
FIG 2.
Localization of SWP9 and SWP7 in N. bombycis mature spores and spore coats. (A) Western blotting was performed using anti-SWP9 and anti-SWP7 antibodies. A single 43-kDa band of SWP9 and 33-kDa band of SWP7 were detected on a Western blot of N. bombycis total mature-spore and spore coat protein using anti-SWP9 and anti-SWP7 antibodies. (B) IFA with rabbit anti-SWP9 and anti-SWP7 antibodies showing the spore wall localization of purified mature spores and spore coats. The purified mature spores (A2 and B2) and spore coats (D2 and E2) were visualized with a fluorescence microscope after incubation with the primary antibodies against SWP9 and SWP7. Negative-control mature spores (C2) and spore coats (F2) and DAPI staining (A1, B1, C1, D1, E1, and F1) were designed. The anti-SWP9 and anti-SWP7 antibodies were diluted 1:200. The secondary antibody was FITC-conjugated goat anti-rabbit IgG (Sigma) at a 1:64 dilution. All images, ×1,000 magnification. (C, D, and E) To further determine whether the exospores of N. bombycis contained SWP9 and SWP7, we analyzed the 0.1 M alkali- and 2% SDS-soluble proteins to detect SWP9 and SWP7 using the anti-SWP9 and anti-SWP7 antibodies. The arrows indicate SWP9 and SWP7. Furthermore, positive-control Western blotting was conducted to detect SWP32 using anti-SWP32 antibody. The corresponding band was recognized and demonstrated that the protein isolations worked properly. Lanes M, EasySee Western marker (Beyotime, Chongqing, China).
To investigate the localization of SWP9 and SWP7 in mature N. bombycis spores, anti-SWP9 and anti-SWP7 antibodies were used for IFA (Fig. 2B) and immunoelectron microscopy (IEM) (Fig. 3A). FITC labeling demonstrated that the anti-SWP9 and anti-SWP7 polyclonal antibodies reacted with the spore walls of the mature spores and spore coats with an intense emerald green fluorescence signal (Fig. 2B). Previous studies showed that surface antigens of mature spores could be removed by SDS and alkali extraction (42, 43, 49). The reports showed that the SWP32 protein of N. bombycis is an exospore protein (21). Therefore, positive-control Western blotting was also performed using anti-SWP32 antibody to confirm that the protein isolation had been performed properly (Fig. 2E). The SDS- and alkali-soluble spore surface proteins contained SWP9 and SWP7 (Fig. 2C and D). In summary, these results showed that SWP9 and SWP7 are localized in the spore wall and that the exospores contain SWP9 and SWP7.
FIG 3.
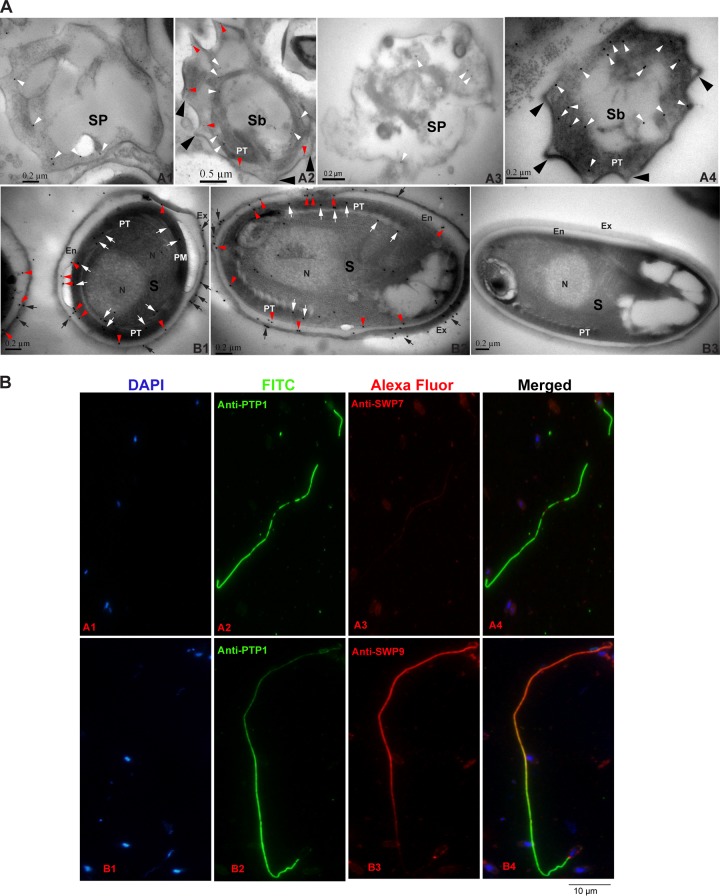
Immunolocalization and developmental expression of N. bombycis SWP9 and SWP7. (A) Immunoelectron microscopy localization and developmental expression of SWP9 and SWP7. (A1 and A3) Images of 30% and 45% Percoll gradient centrifugation fractions showing strong enrichment of sporonts with a visible cell coat and internal colloidal gold particles (18 nm) of SWP9 and SWP7 (white arrowheads). (A2 and A4) The 60% and 75% Percoll gradient centrifugation fractions are rich in sporoblasts, with internal and surface coat localization of SWP9 and SWP7. The sporoblast-containing portions have a scattered developing polar tube and a thick surface coat (large black arrowheads). A portion of the SWP9 gold particles are secreted and are present in the thickened surface coat (red arrowheads). The cytoplasm still has some diffuse gold particles of SWP9 in it (white arrowheads). However, the vast majority of the SWP7 colloidal gold particles are distributed in the cytoplasm. (B1 and B2) The 90% Percoll gradient centrifugation fraction shows nearly mature N. bombycis spores with localization of the exospore (black arrows), endospore (red arrowheads), and polar tube (white arrows). The surface of the mature N. bombycis spore plasmalemma is enclosed by a relatively thick electron-lucent endospore that is in turn covered by an electron-dense exospore coat. Colloidal gold particles of SWP9 and SWP7 are primarily dispersed in the exospore (black arrows), endospore (red arrowheads), and polar tube (white arrows) of the mature N. bombycis spores. (B3) Note the absence of gold particles in the control sections of the mature N. bombycis spore (using negative-control rabbit serum). Sp, sporonts; Sb, sporoblasts; S, mature spores; PT, polar tube; N, nucleus; Ex, exospores; En, endospore. (B) SWP9 and SWP7 of N. bombycis also localized at the polar tubes of germinated spores. N. bombycis spore germination was induced in 0.1 mol K2CO3 for 30 min at 28°C and then incubation with mouse anti-PTP1 (A2 and B2) and rabbit anti-SWP7 (A3) and anti-SWP9 (B3) antibodies. DAPI was used to stain the nucleus (A1 and B1). (A4 and B4) Merged images.
To more accurately determine the locations of SWP9 and SWP7 in mature N. bombycis spores, IEM was performed using anti-SWP9 and anti-SWP7 antibodies (Fig. 3A). As shown in Fig. 3A, B1 and B2, SWP9 and SWP7 are primarily localized in the spore walls and polar tubes of N. bombycis spores. SWP9 and SWP7 are expressed in both the endospore (red arrowheads) and the exposed exospore (black arrows) of the spore wall. Furthermore, SWP7 and SWP9 are found in the polar tube (white arrows) of the mature spore. In addition, to further confirm the polar tube localization of SWP7 and SWP9, N. bombycis spores were induced to germinate in 0.1 M K2CO3 solution. Then, the polar tubes were incubated with anti-PTP1, anti-SWP7, and anti-SWP9 antibodies. As shown in Fig. 3B, both anti-SWP7 and anti-SWP9 antibodies could recognize the polar tube. These results showed that SWP7 and SWP9 also localized in the polar tube of N. bombycis. Therefore, as demonstrated in Fig. 2 and 3, anti-SWP7 and anti-SWP9 primarily reacted with the spore walls of mature spores, suggesting that the genes encode two novel spore wall proteins and that SWP9 and SWP7 are primarily localized to the exospore and endospore.
SWP9 is secreted to the spore wall prior to SWP7 during the development process of N. bombycis, and both are colocalized at the spore walls of mature spores.
To detect the expression features of SWP7 and SWP9 during the development process of N. bombycis spores, we isolated the different life cycle stages of N. bombycis spores using Percoll gradient centrifugation. We also observed the morphologies of the different life cycle stages of the N. bombycis parasite by TEM (Fig. 3). The maturity of the parasite's stages can be assessed by the thickness of the plasmalemma. The sporont is characterized by uniform thickening of the surface coat, and the sporont eventually transitions into a sporoblast. Therefore, the fractions obtained by 30% and 45% Percoll gradient centrifugation were strongly enriched in sporonts with a visible cell coat (Fig. 3A, A1 and A3, and 4). The sporoblast is an irregularly shaped dense cell with a thick surface coat (Fig. 3A, A2 and A4) and a cytoplasm that contains portions of the developing polar tube and other organelles. Therefore, the 60% and 75% Percoll gradient centrifugation fractions are rich in sporoblasts (Fig. 3A, A2 and A4, and 4). The surface of the mature N. bombycis spore plasmalemma is abutted by a relatively thick electron-lucent endospore that is in turn covered by an electron-dense exospore coat. Thus, the 90% Percoll gradient centrifugation fraction contains nearly mature spores (Fig. 3A, B1, B2, and B3, and 4).
FIG 4.
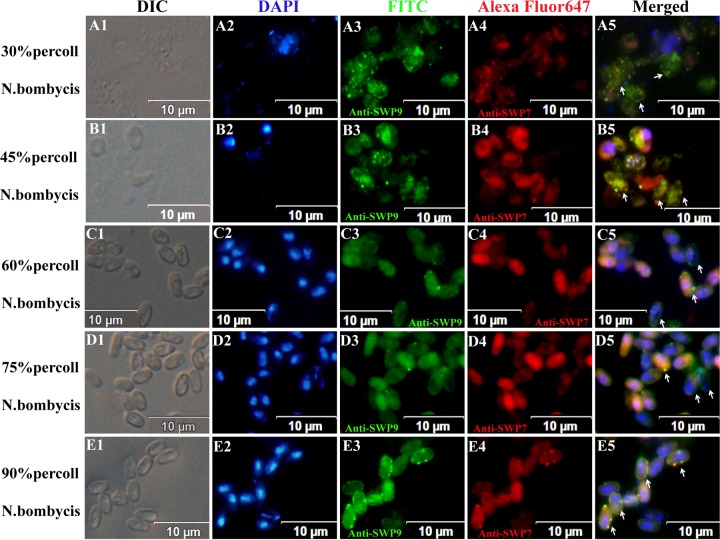
IFA micrographs of N. bombycis collected from different Percoll gradient fractions. N. bombycis was incubated with anti-SWP9 (A3, B3, C3, D3, and E3) labeled with FITC (green) and anti-SWP7 (A4, B4, C4, D4, and E4) labeled with Alexa Fluor 647 (red). Spores were visualized with a differential interference contrast (DIC) microscope (A1, B1, C1, D1, and E1) and stained with DAPI (A2, B2, C2, D2, and E2). The images of the 30% and 45% Percoll gradient centrifugation fractions show internal spot signals (arrows). (A2 and B2) At the same time, the nuclei of the sporonts were unstained with DAPI. (C5 and D5) Images from the 60% and 75% Percoll gradient centrifugation fractions show that these fractions are rich in early sporoblasts and late sporoblasts, with fluorescent ring signals and developing walls (arrows). The 90% Percoll gradient centrifugation fraction contains mature spores with intense fluorescent ring signals that are marked by a typical and intact thick wall. Note the presence of both red and green signals inside the spore and around the spore wall and areas of yellow signal where they overlap (arrows).
Spores at the different developmental stages were incubated with anti-SWP9 and anti-SWP7 antibodies. SWP9 and SWP7 are differently distributed during the life cycle stages of N. bombycis. Colloidal gold particles (Fig. 3A, A1 and A3, white arrowheads) and spot fluorescence signals (Fig. 4A5 and B5, white arrows) of SWP9 and SWP7 are distributed in the cytoplasm of sporonts, indicating the presence of SWP9 and SWP7 in the cytoplasm of the sporonts. However, there are fewer SWP7 gold particles than SWP9 particles. In addition, DAPI did not stain the nuclei of the sporonts (Fig. 4A2 and B2). The sporoblasts from the 60% and 75% Percoll gradient fraction, which possess the developing polar tube, had some SWP9 gold particles in their cytoplasm (white arrowheads) and a noticeable concentration of gold along the thickened plasmalemmal surfaces (Fig. 3A, A2, red arrowheads). However, the vast majority of the colloidal gold particles representing SWP7 were distributed in the cytoplasm (Fig. 3A, A4, white arrowheads). At the same time, IFA also showed that the anti-SWP9 antibody labeled the developing walls of sporoblasts with a fluorescent ring (Fig. 4C5 and D5, white arrows), while the anti-SWP7 antibody labeled the interiors of the sporoblasts. Therefore, a portion of SWP9 localizes to the spore wall, and SWP7 is distributed in the interiors of sporoblasts, suggesting that SWP9 is secreted to the spore wall before SWP7. Furthermore, IFA also showed that SWP9 and SWP7 were colocalized at the spore wall in mature spores (Fig. 4E5, white arrows). Overall, as demonstrated in Fig. 3 and 4, anti-SWP9 and SWP7 clearly reacted with the cytoplasm of sporonts, the cytoplasm (SWP7) and surface coats (SWP9) of sporoblasts, and the spore walls of mature spores, suggesting that these genes are expressed during the development process of N. bombycis and that SWP9 and SWP7 are colocalized to the spore wall.
FIG 5.
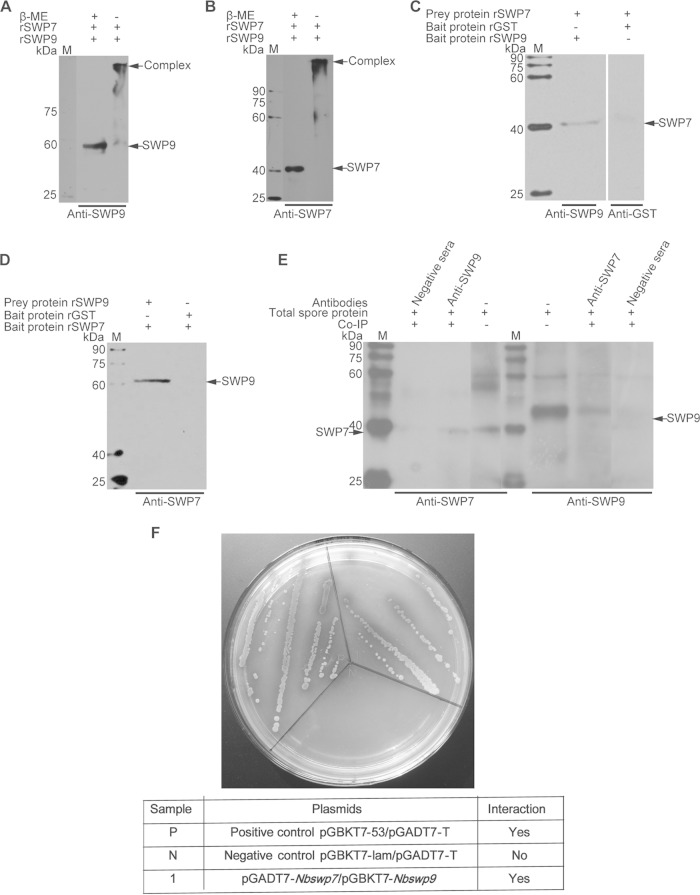
Immunoprecipitation, far-Western blotting, and SDS-PAGE of unreduced, treated samples and yeast two-hybrid assay, showing the interaction of SWP9 and SWP7. (A and B) SDS-PAGE of the unreduced, treated sample for analysis of the interaction between SWP9 and SWP7. Shown are SDS-PAGE gels of protein samples made from rSWP9-GST and rSWP7-His treated with (+) or without (−) β-mercaptoethanol. Mixed protein samples of rSWP9-GST and rSWP7-His, when treated without β-mercaptoethanol, formed a large complex (arrow) at the top of the gel, as demonstrated by SDS-PAGE. (C and D) Detection of prey protein rSWP7-His or rSWP9-GST by conventional far-Western blotting using rSWP9-GST or rSWP7-His and anti-SWP9 or anti-SWP7 antibody. In addition, rGST and monoclonal anti-GST antibodies were used as controls. (E) Immunoblots of SWP7 and SWP9 were coimmunoprecipitated with rabbit anti-SWP9 and anti-SWP7 antibodies, respectively. The immunoprecipitates were treated with loading buffer and run on SDS-12% PAGE, transferred to a PVDF membrane, and probed with 10 μg mouse anti-SWP7 (1:6,000) or anti-SWP9 (1:6,000). The samples were incubated with HRP-labeled anti-mouse IgG (1:8,000) and detected by enhanced chemiluminescence (ECL). The negative-control antibody did not precipitate SWP9 and SWP7. Lanes M, EasySee Western marker. (F) A yeast two-hybrid assay was used to determine the in vivo interaction between SWP9 and SWP7. Nbswp7 (prey) and Nbswp9 (bait) constructs were simultaneously transformed into yeast competent cells. Several independent blue colonies grew up on the Leu− Trp− His− Ade− SD plates that contained X-α-Gal. This demonstrates the interactions of the pGADT7-Nbswp7 and pGBKT7-Nbswp9 constructs. The positive-control pGBKT7-53/pGADT7-T and negative-control pGBKT7-lam/pGADT7-T reactions are also shown.
SWP9 interacts with SWP7 in the spore walls of mature N. bombycis spores.
The above-mentioned immunofluorescence assay showed that SWP9 and SWP7 were colocalized in the spore wall of mature N. bombycis (Fig. 4E5). Therefore, we inferred that SWP9 and SWP7 may interact in the spore wall. To determine whether SWP9 interacts with SWP7, a coimmunoprecipitation (co-IP) assay was employed. Briefly, protein lysates from N. bombycis were first immunoprecipitated with rabbit anti-SWP9 and then immunoblotted with mouse antibodies against SWP7. We found that SWP9 coprecipitated with SWP7. Furthermore, we observed that SWP7 also coprecipitated with SWP9 (Fig. 5E). The coimmunoprecipitation results showed that anti-SWP9 and anti-SWP7 antibodies precipitated SWP9 and SWP7, respectively, out of the completely mature spore proteins (data not shown). An SDS-PAGE assay of the unreduced, treated recombinant proteins rSWP9-GST and rSWP7-His was also used to determine whether these interactions occur in rSWP9-GST and rSWP7-His. When the rSWP9-GST and rSWP7-His mixture was treated with 3× loading buffer (containing β-mercaptoethanol), the Western blotting result showed that anti-SWP9 and anti-SWP7 could recognize the corresponding bands of SWP9 and SWP7, respectively (Fig. 5A and B). However, when the mixture was treated with an unreduced sample loading buffer (without β-mercaptoethanol), there was a larger band present in the PVDF membrane (Fig. 5A and B). This result shows that rSWP9-GST and rSWP7-His form a large protein complex. Far-Western blotting was also performed to further validate the interaction of SWP9 and SWP7. The prey protein rSWP7-His was able to bind the bait protein rSWP9-GST but not rGST (Fig. 5C), and the prey protein rSWP9-GST interacted with the bait protein rSWP7-His (Fig. 5D). The bait proteins rSWP7-His and rSWP9-GST could not attach to the PVDF membrane without the prey protein (data not shown). Finally, the possibility of an interaction between SWP7 and SWP9 was further investigated using yeast two-hybrid analysis. In this experiment, NbSWP9 was used as the bait and NbSWP7 as the prey to determine if the proteins interacted in vivo. Figure 5F demonstrates that the NbSWP9 protein interacts in vivo with NbSWP7. The above data indicate that SWP9 interacts with SWP7.
SWP7 binding to DCSCs depends upon the combination of SWP9 and DCSCs during the biosynthesis of the spore wall.
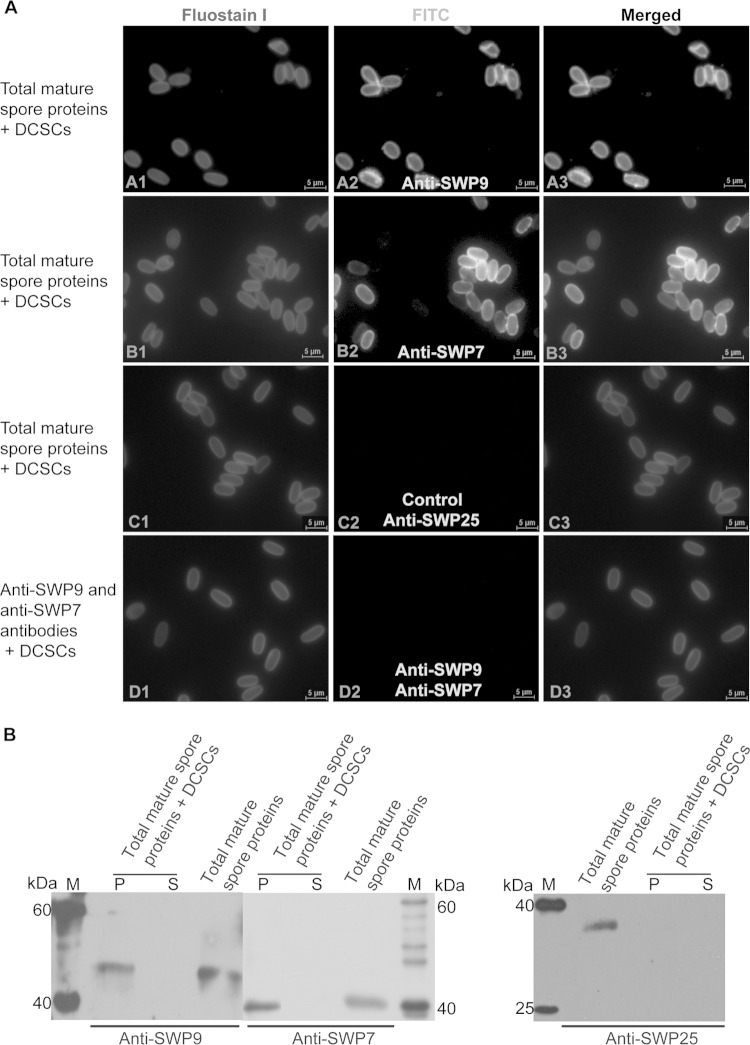
Given their localization in the developmental spores, it is likely that SWP7 and SWP9 are involved in the biosynthesis of the spore wall. To determine the roles of SWP7 and SWP9 in spore wall synthesis, mature spores were agitated with glass beads to obtain total mature-spore proteins, as described in Materials and Methods. In addition, DCSCs were isolated by boiling N. bombycis spores in 1 M NaOH (15). Anti-SWP9 and anti-SWP7 antibodies were employed to detect whether both native SWP9 and SWP7 could bind to the DCSCs. At the same time, the anti-SWP25 (SWP2) antibody was used as a control (Fig. 6A, C2, and B), because previous research demonstrated that SWP25 does not bind to DCSCs (15). The Western blotting results demonstrated that native SWP9 and SWP7 bound to the chitin spore coats, but SWP25 was unable to interact with the coats (Fig. 6B). Furthermore, IFA showed intense green fluorescence in the chitin spore coats that were bound by total mature-spore proteins, meaning anti-SWP9 and anti-SWP7 polyclonal antibodies recognized the chitin spore coats that were bound by proteins (Fig. 6A, A2 and B2). The IFA results are in agreement with the Western blotting results. These results indicate that native SWP9 and SWP7 from the total soluble protein fraction, which was obtained from glass bead agitation, could bind the N. bombycis DCSCs.
FIG 6.
Native SWP9 and SWP7 within the total spore protein fraction bound to DCSCs. (A) IFA of DCSCs bound by total mature-spore proteins. (A2, B2, and C2) DCSCs were visualized using a fluorescence microscope after incubating the DCSCs with total mature-spore proteins and primary antibodies against SWP9, SWP7, and SWP25 (SWP3). (C2) DCSCs were incubated with total mature-spore proteins, using anti-SWP25 antibody as a control, and showed no DCSC green fluorescence. (D2) At the same time, DCSCs were also incubated with both anti-SWP9 and SWP7 antibodies as a negative control and showed no DCSC green fluorescence. (A1, B1, C1, and D1) Fluostain I was used to stain the chitin component. The primary antibody was diluted 1:200. The secondary antibody was FITC-conjugated goat anti-mouse IgG (1:64 dilution; Sigma). All images, ×1,000 magnification. (B) Anti-SWP9 and SWP7 antibodies were used for Western blot analysis to detect native total mature-spore protein binding to DCSCs. In addition, anti-SWP25 antibody was used as a negative control. Lanes: S, supernatant after six washes, indicating DCSCs bound by total mature-spore proteins; P, DCSC pellet bound by total mature-spore proteins after six washes; M, EasySee Western marker.
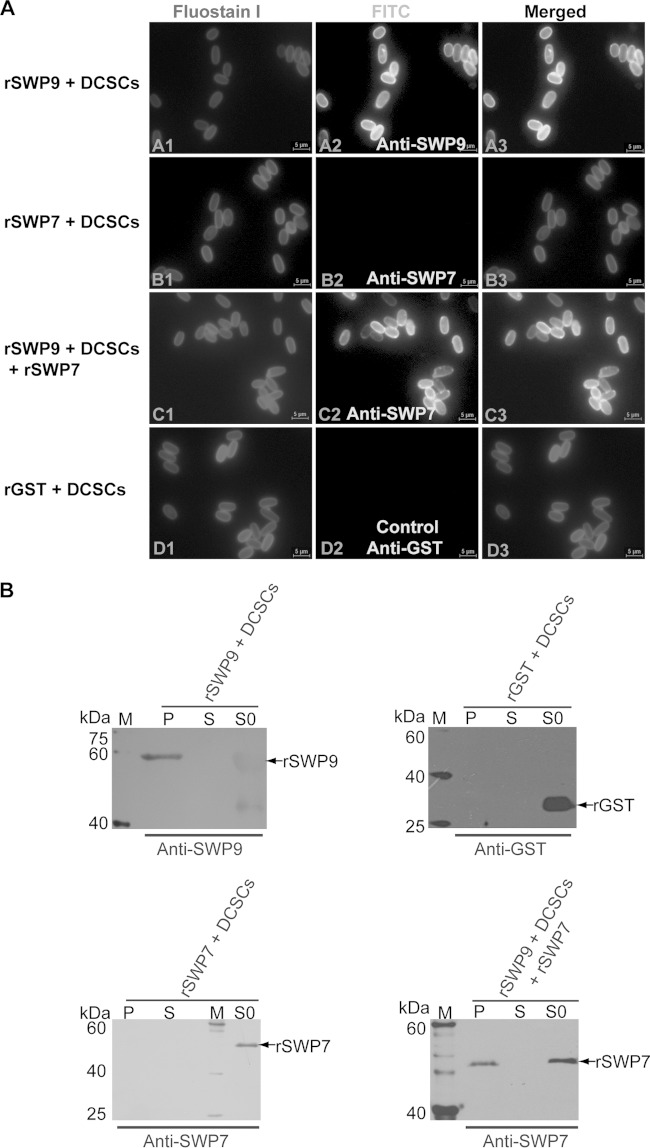
The above-described experiments showed that SWP9 interacts with SWP7 and that native SWP9 and SWP7 can bind to the DCSCs in the spore wall of N. bombycis. To demonstrate whether the SWP9 and SWP7 proteins directly or indirectly bind to the DCSCs or if one spore wall protein depends on another spore wall protein to bind to the chitin coats, the DCSCs were incubated with rSWP9-GST and rSWP7-His and then observed after silver staining, Western blotting, and IFA (Fig. 7A and B). Silver staining (data not shown) and Western blotting using anti-SWP9 and anti-SWP7 polyclonal antibodies as the primary antibodies showed that, after six washes, the DCSC pellets were bound to rSWP9-GST. Additionally, rSWP7-His could not bind the DCSC pellets. However, rSWP7-His could bind to the DCSCs when rSWP9-GST was preincubated with the DCSCs (Fig. 7B). Furthermore, IFA also demonstrated that the anti-SWP9 polyclonal antibody stained the chitin layer to which rSWP9-GST was bound with an intense emerald fluorescence signal, but rSWP7-His was unable to interact with the chitin coats (Fig. 7A, A2 and B2). In addition, IFA also demonstrated that the anti-SWP7 antibody stained the chitin layer to which both rSWP9-GST and rSWP7-His were bound with an intense emerald green fluorescence signal (Fig. 7A, C2). These results suggested that rSWP9-GST directly binds to the deproteinated chitin spore coats but rSWP7-His is unable to bind to the DCSCs. However, rSWP7-His could bind to the DCSCs after incubation of rSWP9-GST with the DCSCs. Therefore, the above results showed that rSWP7-His depends on rSWP9-GST being bound to the DCSCs. A possible explanation for this result is that SWP9 may bind to the chitin layer as a scaffolding protein for spore wall formation during development. Furthermore, SWP9 interacts with the SWP7 and SWP9 that are bound to the chitin spore coats to constitute and assemble the intact spore wall, which maintains the spore's morphology and protects the mature spore from the outer environment.
FIG 7.
Binding of rSWP7-His to DCSCs depends upon the combination of rSWP9-GST and DCSCs. (A) IFA of DCSCs bound by rSWP9-GST, rSWP7-His, and rGST proteins. (A2, B2, and D2) DCSCs were visualized using a fluorescence microscope after incubating the DCSCs with recombinant proteins and primary antibodies against SWP9, SWP7, and GST. (C2) DCSCs were incubated with rSWP7-His after incubating the DCSCs with rSWP9-GST, removing the unbound rSWP9-GST and rSWP7-His with PBS, and incubation with the anti-SWP7 primary antibody. (D2) DCSCs were incubated with rGST using anti-GST as a control. (A1, B1, C1, and D1) Fluostain I was used to stain the chitin component. (B) Immunoblotting analysis to detect whether rSWP9-GST and rSWP7-His proteins bind to DCSCs using anti-SWP9 and anti-SWP7 antibodies. In addition, rGST was used as a negative control. Lanes: S0, recombinant-protein supernatant after incubation with DCSCs; S, supernatant after six washes, indicating DCSCs bound by recombinant protein; P, DCSC pellet bound by recombinant protein after six washes; M, EasySee Western marker.
SWP9 and SWP7 help spores adhere to and infect the BmE host cell in vitro.
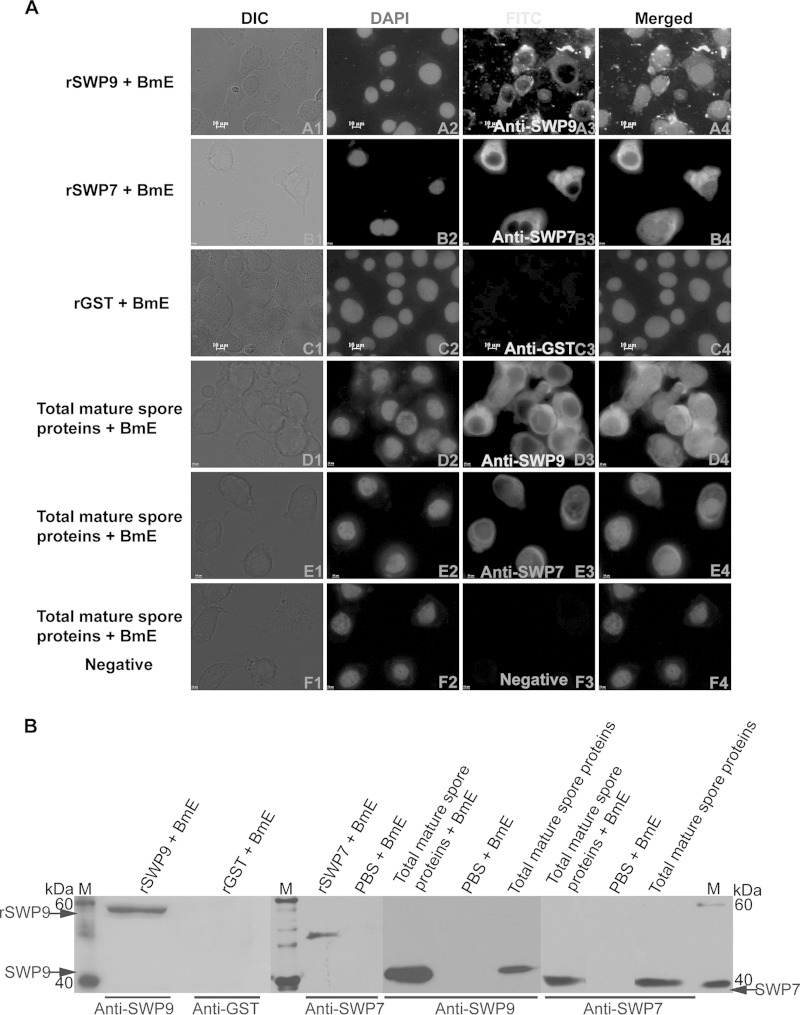
Because the N. bombycis exospore contains SWP9 and SWP7, the host cell binding assay was performed by incubating rSWP9-GST, rSWP7-His, or total mature-spore proteins with BmE cells to confirm whether the SWP9 and SWP7 proteins attach to the host cell surface. As shown in Fig. 8B, there are protein bands corresponding to rSWP9-GST, rSWP7-His, and native SWP9 and SWP7 within the total mature-spore proteins, demonstrating that rSWP9-GST, rSWP7-His, and native SWP9 and SWP7 can attach to the BmE host cell surface. When BmE cells were incubated with rSWP9-GST, rSWP7-His, and total mature-spore proteins, these proteins were detected as uniform circles on the cell surface with green FITC and red Alexa Fluor 647-phycoerythrin signals (Fig. 8A, A3, B3, D3, and E3). Furthermore, anti-SWP9 and anti-total BmE protein antibodies inhibited the ability of rSWP9-GST to bind to BmE cells in a dose-dependent manner (data not shown). These results showed that rSWP9-GST, rSWP7-His, and native SWP9 and SWP7 can attach to the surface of the BmE host cell and that anti-SWP9 and anti-BmE inhibit the binding of rSWP9-GST to BmE cells.
FIG 8.
SWP9 and SWP7 can attach to BmE host cells. (A) IFA of the BmE host cell bound by total mature-spore proteins and recombinant proteins. (A3, B3, C3, D3, and E3) BmE cells were visualized using a fluorescence microscope after incubation with native total mature-spore proteins or recombinant proteins and primary antibodies against SWP9, SWP7, and GST. (C3 and F3) BmE cells were incubated with rGST or total mature-spore proteins using anti-GST (C3) or negative-control antibodies (F3) as controls and showed no fluorescence at the cell membrane. (A2, B2, C2, D2, E2, and F2) DAPI was used to stain the nuclei. All images, ×1,000 magnification. (B) Anti-SWP9 and anti-SWP7 antibodies were used for Western blot analysis to detect rSWP9-GST and rSWP7-His and native SWP9 and SWP7 binding to BmE cells. BmE cells were incubated with rGST or PBS, using anti-GST or anti-SWP9 and SWP7 antibodies as negative controls. Lanes M, EasySee Western marker.
Based on the analysis of localization and the host cell binding assays of SWP9 and SWP7 in vitro, adherence and infection inhibition experiments with antibodies and recombinant proteins were performed to determine whether anti-SWP9 or anti-SWP7 inhibits spore adherence to and infection of host cells (data not shown). This was evaluated using adherence of spores of N. bombycis to BmE host cells. As expected, the anti-SWP9 and anti-SWP7 antibodies reduced spore adherence by approximately 20% and 10%, respectively, compared with the negative control (Fig. 9A). When both anti-SWP9 and anti-SWP7 antibodies were incubated with the spores, adherence was reduced by more than 35% (Fig. 9A). These data indicate that anti-SWP9 and anti-SWP7 antibodies reduce N. bombycis spore adherence to BmE cells.
FIG 9.
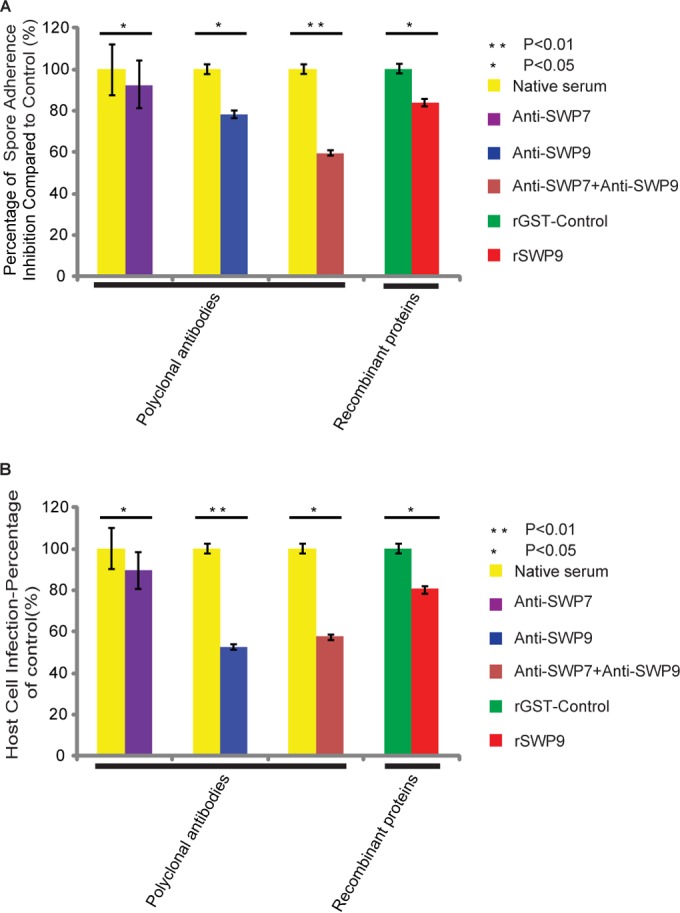
SWP9 and SWP7 contribute to N. bombycis adherence to and infection of the host cell. (A) Exogenous rSWP9-GST or anti-SWP9 and anti-SWP7 antibodies inhibited spore adherence to the host cell. SWP9- and SWP7-specific antibodies, individually and in combination, or a control preimmune antibody was incubated in an adherence assay with N. bombycis spores at a concentration of 5 μg/ml. Furthermore, rSWP9-GST and control rGST were also used as inhibitors of N. bombycis spore adherence at a concentration of 5 μg/ml. The data are shown as the percentage of adherence (compared to control samples) with preimmune antibody or rGST. (B) Exogenous rSWP9-GST or anti-SWP9 and SWP7 antibodies inhibited spore infection of the host cell. The data are shown as the percentages of infected host cells relative to control samples incubated at the same concentration (the infection percentage of control samples was treated as 100%). The experiments were repeated three times and produced similar results each time. At least 20 random fields were calculated for each data point. The error bars indicate standard deviations (SD).
Previous studies have shown that spore adherence to the host cell is necessary prior to spore activation and infection (20, 24). An infection assay was performed to survey the infection rate with anti-SWP9 and anti-SWP7 antibody treatment. The interiors of BmE cells contained N. bombycis spores, signifying infected BmE cells (data not shown). As shown in Fig. 9B, the anti-SWP9 antibody inhibited infection by more than 40% and the anti-SWP7 antibody inhibited infection by approximately 8%. Furthermore, the combination of anti-SWP9 and anti-SWP7 antibodies inhibited infection by more than 40%. These data show that antibodies against SWP9 and SWP7 not only inhibit adherence, but also decrease the efficiency of infection of BmE cells by spores of N. bombycis.
Furthermore, in vitro assays were also performed to assess the influence of rSWP9-GST on N. bombycis adherence and infection. The addition of rSWP9-GST (at a dose of 5 μg) to the spore adherence assay mixture decreased both spore adherence and BmE cell infection, while control rGST did not affect either adherence or infection. The inhibition of adherence (approximately 20%) and inhibition of infection (more than 20%) occurred when 5 μg of rSWP9-GST was used (Fig. 9A and B). The capability of rSWP9-GST to decrease spore adherence and infection in vitro confirms SWP9 localization to the accessible exospore region of N. bombycis and its ability to attach to the host cell. The above-mentioned results further support previous research showing that adherence to the host cell is a crucial step for infection.
DISCUSSION
The differentiation of unicellular eukaryotic parasites is necessarily related to transitions between complex life cycle stages (41). Among microsporidia, the complete life cycle occurs within the host cell. First, a proliferative growth stage (merogony) is followed by sporogony, when the meronts transform into sporonts. This stage is characterized by the deposition of an electron-dense material on the plasma membrane. Then, the sporonts divide into sporoblasts, and the formation of a thick wall is associated with the differentiation of specific cytoplasmic structures, such as the polar tube. After maturation, the rupture of host cells may lead to the release of resistant spores into the environment (13, 34). However, very few large-scale studies on the stage-specific expression of proteins in these parasites have been performed because it is difficult to acquire sufficient quantities of meronts, sporonts, and sporoblasts. We purified different life cycle stage spores using an improved method of discontinuous Percoll gradient separation (30, 45, 60, 75, and 90% [vol/vol]). The main cell stages isolated using this process were sporoblasts plus immature spores (60 and 75% [vol/vol]) and mature spores (90% [vol/vol]). Further centrifugation of the fraction at the 30% and 45% Percoll gradient generated a sporont-rich fraction. In addition, SWP9 and SWP7 were found to have different distributions during the different life cycle stages of the spores. IEM and IFA together showed that both SWP9 and SWP7 localized to the interiors of the sporonts. In sporoblasts, most of the SWP9 localized to the spore wall, while SWP7 was distributed mainly in the interior. Therefore, we inferred that SWP9 was secreted to the spore wall before SWP7. The change in protein localization from the cytoplasm to the cell surface corresponds to the life cycle of N. bombycis and illustrates the contributions of SWP7 and SWP9 to the formation of the thick spore wall during maturation. In contrast, the microsporidian spore wall is comprised of the exospore, endospore, and plasma membrane. This wall provides structural rigidity, maintains the osmotic pressure of the spore, and protects the mature spore from the environment (50, 51). At present, only a few spore wall proteins have been identified in microsporidia. Recently, a number of studies have shown that NbSWP5 localizes to the exospore and polar tube. NbSWP5 was demonstrated to protect spores from phagocytic uptake (23) and to interact with PTP2 and PTP3; it may also play an important role in supporting the structural integrity of the spore wall and modulating the process of N. bombycis infection (22). This study showed that SWP9 and SWP7 not only localized to the spore wall, but also localized to the polar tube. This result suggests that SWP9 and SWP7 may interact with polar tube proteins. We further described the interaction of SWP9 and SWP7 by performing SDS-PAGE of unreduced, treated rSWP9-GST and rSWP7-His; immunoprecipitation; far-Western blotting; and yeast two-hybrid analysis. However, the interaction motif that contributes to SWP9 interaction with SWP7 is still unknown. SWP7 and SWP9 contain 8 and 3 cysteine residues, respectively, that may form a disulfide bridge and mediate the interaction between SWP9 and SWP7.
A recent study extracted the N. bombycis DCSCs and the Giardia lamblia cyst wall β-1,3-linked N-acetylgalactosamine (GalNAc) homopolymer using hot NaOH. The DCSCs and GalNAc were much thicker and looser than the chitin layer of untreated spores and the intact G. lamblia cyst wall (15, 52). In addition, N. bombycis SWP26, SWP30 (SWP1), and SWP32 (SWP3) from the total soluble spore protein fraction could bind to the DCSCs, and recombinant NbSWP12 also adhered to the DCSCs (26, 52). These spore wall proteins may compress the N-acetylglucosamine homopolymer to form a narrow surface (26). Our data showed that both native SWP7 and SWP9 within the total spore protein and rSWP9-GST could bind to the DCSCs, but rSWP7-His could not. However, rSWP7-His could bind to the DCSCs after the DCSCs were incubated with rSWP9-GST. Thus, SWP7 binding to the DCSCs is an indirect action and is dependent upon the binding of SWP9 to the chitin layer of the DCSCs. Therefore, it has been hypothesized that SWP9 acts as a scaffolding protein for spore wall formation during development and contributes to the compression of the N-acetylglucosamine homopolymer to form a narrow surface.
The silkworm is an economically important lepidopteran insect and has a midgut containing a peritrophic membrane (PM) (53). The insect PM is the first barrier to pathogens that are ingested through oral feeding. The larval PM is primarily composed of chitin, proteoglycans, and proteins (54). Based on our results, we inferred that the N. bombycis spore may bind to and breach the silkworm's PM prior to infecting the epithelial cell. SWP9, SWP12, SWP26, SWP30 (SWP1), and SWP32 (SWP3) can bind to the DCSCs of N. bombycis (15, 26), suggesting that these spore wall proteins may bind to the chitin component of the PM. Once the spore binds to the PM, some spore wall proteins may be activated by this combination, inducing the expression of chitinases that degrade and breach the PM chitin. Then, the spores successfully infect the silkworm's epithelial cells. In summary, we infer that SWP9 may bind to the chitin component of the peritrophic membrane or that SWP9 and SWP7 may interact with the proteins of the PM and contribute to spore binding to the PM, suggesting that both SWP9 and SWP7 play roles in adherence and infection. Recently, some of the PM proteins have been identified by proteomic analysis, allowing convenient analysis of the interaction between N. bombycis and the silkworm (55).
The host cell binding assay showed that native SWP7 and SWP9 and rSWP9-GST can attach to the BmE host cell and that rSWP9-GST, anti-SWP9, and anti-SWP7 antibodies reduce N. bombycis spore adherence and infection of the BmE cells. Previous research found that some pathogens use a surface heparin-binding protein to bind the host cells (56–61). However, no HBM was identified in SWP7 or SWP9. SWP9 and SWP7 bind to host cells and may act as ligands for adherence; it is therefore possible that they could interact with receptors on the host cell. There are few in vivo reports of the process by which N. bombycis infects its host, the silkworm.
In conclusion, this study presents the first examination of the interaction of two newly identified spore wall proteins, SWP9 and SWP7, which are localized to the exospores, endospore, and polar tube of N. bombycis. The study provides the necessary data for a more detailed analysis of the formation of the spore wall. Furthermore, both SWP9 and SWP7 are localized to the interiors of the sporonts. However, in sporoblasts, part of SWP9 appears to be secreted to the spore wall earlier than SWP7, and SWP9 may bind to the chitin layer as a scaffolding protein for spore wall formation during development. Subsequently, SWP7 is secreted to the spore wall and interacts with SWP9 to play a common role in adherence and infection. Furthermore, both SWP9 and SWP7 exist and are conserved in other species of microsporidia. Therefore, research regarding SWP9 and SWP7 has profound implications for the future study of microsporidia. Finally, SWP9 appears to be a structural support protein for spore wall formation during development. It also plays a role in spore adherence and infection of the host cell. Therefore, determination of the structures of SWP9 and SWP7 may be useful for understanding how SWP9 and SWP7 interact to form the thick spore wall used by this widely disseminated parasitic protist group.
ACKNOWLEDGMENTS
We are grateful to all those who provided the means for us to acquire software free of charge, which we have used and cited in this article. We thank Hongjuan Cui and Liqun Yang (State Key Laboratory of Silkworm Genome Biology, Southwest University, Chongqing, China) for the fluorescence microscope and all of the people who assisted us in this study. In addition, we are very grateful to the Center of Electron Microscope, Zhejiang University (Life Science Division).
REFERENCES
- 1.Becnel JJ, Andreadis TG. 1999. Microsporidia in insects, p 447–501. In Wittner M, Weiss LM (ed), The microsporidia and microsporidiosis. American Society for Microbiology, Washington, DC. [Google Scholar]
- 2.Nägeli K. 1857. Uber die neue Krankheit der Seidenraupe und verwandte Organismen. Bot Z 15:760–761. [Google Scholar]
- 3.Keeling PJ, Luker MA, Palmer JD. 2000. Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Mol Biol Evol 17:23–31. doi: 10.1093/oxfordjournals.molbev.a026235. [DOI] [PubMed] [Google Scholar]
- 4.Thomarat F, Vivares CP, Gouy M. 2004. Phylogenetic analysis of the complete genome sequence of Encephalitozoon cuniculi supports the fungal origin of microsporidia and reveals a high frequency of fast-evolving genes. J Mol Evol 59:780–791. doi: 10.1007/s00239-004-2673-0. [DOI] [PubMed] [Google Scholar]
- 5.Gill EE, Fast NM. 2006. Assessing the microsporidia-fungi relationship: combined phylogenetic analysis of eight genes. Gene 375:103–109. doi: 10.1016/j.gene.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 6.Baker MD, Vossbrinck CR, Didier ES, Maddox JV, Shadduck JA. 1995. Small subunit ribosomal DNA phylogeny of various microsporidia with emphasis on AIDS related forms. J Eukaryot Microbiol 42:564–570. doi: 10.1111/j.1550-7408.1995.tb05906.x. [DOI] [PubMed] [Google Scholar]
- 7.Hartskeerl R, Schuitema A. 1993. Secondary structure of the small subunit ribosomal RNA sequence of the microsporidium Encephalitozoon cuniculi. Nucleic Acids Res 21:1489. doi: 10.1093/nar/21.6.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 9.Williams BAP, Keeling PJ. 2011. Microsporidia—highly reduced and derived relatives of fungi, p 25–36. In Pöggeler S, Wöstemeyer J (ed), The mycota, vol 14 Evolution of fungi and fungal-like organisms. Springer, Berlin, Germany. [Google Scholar]
- 10.Pan G, Xu J, Li T, Xia Q, Liu S-L, Zhang G, Li S, Li C, Liu H, Yang L. 2013. Comparative genomics of parasitic silkworm microsporidia reveal an association between genome expansion and host adaptation. BMC Genomics 14:186. doi: 10.1186/1471-2164-14-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vavra J. 1976. Structure of the microsporidia, p 1–86. In Bulla LA, Cheng TC (ed), Comparative pathology, vol 1 Biology of the microsporidia. Plenum Press, New York, NY. [Google Scholar]
- 12.Bigliardi ESMG, Lupetti P, Corona S, Gatti S, Scaglia MSL. 1996. Microsporidian spore wall: ultrastructural findings on Encephalitozoon hellem exospore. J Eukaryot Microbiol 43:181–186. doi: 10.1111/j.1550-7408.1996.tb01388.x. [DOI] [PubMed] [Google Scholar]
- 13.Vavra J, Larsson J. 1999. Structure of the microsporidia, p 7–84. In Wittner M, Weiss LM (ed), The microsporidia and microsporidiosis. American Society for Microbiology, Washington, DC. [Google Scholar]
- 14.Brosson D, Kuhn L, Prensier G, Vivarès CP, Texier C. 2005. The putative chitin deacetylase of Encephalitozoon cuniculi: a surface protein implicated in microsporidian spore wall formation. FEMS Microbiol Lett 247:81–90. doi: 10.1016/j.femsle.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 15.Yang D, Dang X, Tian R, Long M, Li C, Li T, Chen J, Li Z, Pan G, Zhou Z. 2014. Development of an approach to analyze the interaction between Nosema bombycis (microsporidia) deproteinated chitin spore coats and spore wall proteins. J Invertebr Pathol 115:1–7. doi: 10.1016/j.jip.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Hayman JR, Hayes SF, Amon J, Nash TE. 2001. Developmental expression of two spore wall proteins during maturation of the microsporidian Encephalitozoon intestinalis. Infect Immun 69:7057–7066. doi: 10.1128/IAI.69.11.7057-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohne W, Ferguson DJP, Kohler K, Gross U. 2000. Developmental expression of a tandemly repeated, glycine- and serine-rich spore wall protein in the microsporidian pathogen Encephalitozoon cuniculi. Infect Immun 68:2268–2275. doi: 10.1128/IAI.68.4.2268-2275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Takvorian P, Cali A, Wang F, Zhang H, Orr G, Weiss LM. 2006. Identification of a new spore wall protein from Encephalitozoon cuniculi. Infect Immun 74:239–247. doi: 10.1128/IAI.74.1.239-247.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peuvel-Fanget I, Polonais V, Brosson D, Texier C, Kuhn L, Peyret P, Vivares C, Delbac F. 2006. EnP1 and EnP2, two proteins associated with the Encephalitozoon cuniculi endospore, the chitin-rich inner layer of the microsporidian spore wall. Int J Parasitol 36:309–318. doi: 10.1016/j.ijpara.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Southern TR, Jolly CE, Lester ME, Hayman JR. 2007. EnP1, a microsporidian spore wall protein that enables spores to adhere to and infect host cells in vitro. Eukaryot Cell 6:1354–1362. doi: 10.1128/EC.00113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Z, Li Y, Pan G, Tan X, Hu J, Zhou Z, Xiang Z. 2008. Proteomic analysis of spore wall proteins and identification of two spore wall proteins from Nosema bombycis (Microsporidia). Proteomics 8:2447–2461. doi: 10.1002/pmic.200700584. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Pan G, Li T, Huang W, Chen J, Geng L, Yang D, Wang L, Zhou Z. 2012. SWP5, a spore wall protein, interacts with polar tube proteins in the parasitic microsporidian Nosema bombycis. Eukaryot Cell 11:229–237. doi: 10.1128/EC.05127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai S, Lu X, Qiu H, Li M, Feng Z. 2011. Identification of a Nosema bombycis (Microsporidia) spore wall protein corresponding to spore phagocytosis. Parasitology 138:1102–1109. doi: 10.1017/S0031182011000801. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Wu Z, Pan G, He W, Zhang R, Hu J, Zhou Z. 2009. Identification of a novel spore wall protein (SWP26) from microsporidia Nosema bombycis. Int J Parasitol 39:391–398. doi: 10.1016/j.ijpara.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Wu Z, Li Y, Pan G, Zhou Z, Xiang Z. 2009. SWP25, a novel protein associated with the Nosema bombycis endospore 1. J Eukaryot Microbiol 56:113–118. doi: 10.1111/j.1550-7408.2008.00375.x. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Geng L, Long M, Li T, Li Z, Yang D, Ma C, Wu H, Ma Z, Li C. 2013. Identification of a novel chitin-binding spore wall protein (NbSWP12) with a BAR-2 domain from Nosema bombycis (Microsporidia). Parasitology 140:1394–1402. doi: 10.1017/S0031182013000875. [DOI] [PubMed] [Google Scholar]
- 27.Hayman JR, Southern TR, Nash TE. 2005. Role of sulfated glycans in adherence of the microsporidian Encephalitozoon intestinalis to host cells in vitro. Infect Immun 73:841–848. doi: 10.1128/IAI.73.2.841-848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Southern TR, Jolly CE, Russell Hayman J. 2006. Augmentation of microsporidia adherence and host cell infection by divalent cations. FEMS Microbiol Lett 260:143–149. doi: 10.1111/j.1574-6968.2006.00288.x. [DOI] [PubMed] [Google Scholar]
- 29.Franzen C. 2005. How do microsporidia invade cells. Folia Parasitol 52:36–40. doi: 10.14411/fp.2005.005. [DOI] [PubMed] [Google Scholar]
- 30.Keohane EM, Orr GA, Zhang HS, Takvorian PM, Cali A, Tanowitz HB, Wittner M, Weiss LM. 1998. The molecular characterization of the major polar tube protein gene from Encephalitozoon hellem, a microsporidian parasite of humans. Mol Biochem Parasitol 94:227–236. doi: 10.1016/S0166-6851(98)00071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delbac F, Peyret P, Méténier G, David D, Danchin A, Vivarès CP. 1998. On proteins of the microsporidian invasive apparatus: complete sequence of a polar tube protein of Encephalitozoon cuniculi. Mol Microbiol 29:825–834. doi: 10.1046/j.1365-2958.1998.00975.x. [DOI] [PubMed] [Google Scholar]
- 32.Delbac F, Duffieux F, David D, Metenier G, Vivares CP. 1998. Immunocytochemical identification of spore proteins in two microsporidia, with emphasis on extrusion apparatus. J Eukaryot Microbiol 45:224–231. doi: 10.1111/j.1550-7408.1998.tb04529.x. [DOI] [PubMed] [Google Scholar]
- 33.Peuvel I, Peyret P, Méténier G, Vivarès CP, Delbac F. 2002. The microsporidian polar tube: evidence for a third polar tube protein (PTP3) in Encephalitozoon cuniculi. Mol Biochem Parasitol 122:69–80. doi: 10.1016/S0166-6851(02)00073-7. [DOI] [PubMed] [Google Scholar]
- 34.Franzen C. 2004. Microsporidia: how can they invade other cells? Trends Parasitol 20:275–279. doi: 10.1016/j.pt.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Magaud A, Achbarou A, Desportes-Livage I. 1997. Cell invasion by the microsporidium Encephalitozoon intestinalis. J Eukaryot Microbiol 44:81S. doi: 10.1111/j.1550-7408.1997.tb05795.x. [DOI] [PubMed] [Google Scholar]
- 36.Bigliardi E, Sacchi L. 2001. Cell biology and invasion of the microsporidia. Microbes Infect 3:373–379. doi: 10.1016/S1286-4579(01)01393-4. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Wu Z, Pan G, Pang M, Hu J, Zhang R, Zhou Z. 2008. Polar tube extrusion of Nosema bombycis in Bombyx mori-SWU1 embryonic cell line. Curr Zool 53:1107–1112. [Google Scholar]
- 38.Pan G, Li Y, Xie L, Pang M, Lu C, Zhou Z. 2005. Comparison of cytopathic effect (CPE) of Sf21 cell and embry cell of silkworm infected by Nosema bombycis. J Chongqing Normal University 22:25–28. [Google Scholar]
- 39.Seleznev KV, Issi IV, Dolgikh VV, Belostotskaya GB, Antonova OA, Sokolova JJ. 1995. Fractionation of different life cycle stages of microsporidia Nosema grylli from crickets Gryllus bimaculatus by centrifugation in Percoll density gradient for biochemical research. J Eukaryot Microbiol 42:288–292. doi: 10.1111/j.1550-7408.1995.tb01582.x. [DOI] [Google Scholar]
- 40.Gatehouse HS, Malone LA. 1998. The ribosomal RNA gene region of Nosema apis (Microspora): DNA sequence for small and large subunit rRNA genes and evidence of a large tandem repeat unit size. J Invertebr Pathol 71:97. doi: 10.1006/jipa.1997.4737. [DOI] [PubMed] [Google Scholar]
- 41.Taupin V, Méténier G, Vivarès CP, Prensier G. 2006. An improved procedure for Percoll gradient separation of sporogonial stages in Encephalitozoon cuniculi (Microsporidia). Parasitol Res 99:708–714. doi: 10.1007/s00436-006-0231-y. [DOI] [PubMed] [Google Scholar]
- 42.Wu Z, Tan X, Li Y, Pan G, Zhou Z. 2007. Extraction of spore wall proteins of Nosema bombycis with improved methods. Sci Sericulture 33:62–66. [Google Scholar]
- 43.Kawarabata T, Hayasaka S. 1987. An enzyme-linked immunosorbent assay to detect alkali-soluble spore surface antigens of strains of Nosema bombycis (Microspora: Nosematidae). J Invertebr Pathol 50:118–123. doi: 10.1016/0022-2011(87)90111-X. [DOI] [Google Scholar]
- 44.Wang JY, Chambon C, Lu CD, Huang KW, Vivares CP, Texier C. 2007. A proteomic-based approach for the characterization of some major structural proteins involved in host-parasite relationships from the silkworm parasite Nosema bombycis (Microsporidia). Proteomics 7:1461–1472. doi: 10.1002/pmic.200600825. [DOI] [PubMed] [Google Scholar]
- 45.Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, McWilliam H, Valentin F, Wallace I, Wilm A, Lopez R. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 46.Wu Y, Long M, Chen J, Zhi L, Li Z, Pan G, Li T, Zhou Z. 2014. Cloning and prokaryotic expression of Nosema bombycis polar tube protein 1 (PTP1). Sci Sericulture 40:258–264. [Google Scholar]
- 47.Schenk S, Horowitz JF. 2006. Coimmunoprecipitation of FAT/CD36 and CPT I in skeletal muscle increases proportionally with fat oxidation after endurance exercise training. Am J Physiol Endocrinol Metab 291:E254–E260. doi: 10.1152/ajpendo.00051.2006. [DOI] [PubMed] [Google Scholar]
- 48.Bouzahzah B, Nagajyothi F, Ghosh K, Takvorian PM, Cali A, Tanowitz HB, Weiss LM. 2010. Interactions of Encephalitozoon cuniculi polar tube proteins. Infect Immun 78:2745–2753. doi: 10.1128/IAI.01205-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chioralia G, Trammer T, Maier WA, Seitz HM. 1998. Morphologic changes in Nosema algerae (Microspora) during extrusion. Parasitol Res 84:123–131. [DOI] [PubMed] [Google Scholar]
- 50.Koudela B, Kucerova S, Hudcovic T. 1999. Effect of low and high temperatures on infectivity of Encephalitozoon cuniculi spores suspended in water. Folia Parasitol 46:171–174. [PubMed] [Google Scholar]
- 51.Frixione E, Ruiz L, Cerbon J, Uneddn AH. 2007. Germination of Nosema algerae (Microspora) spores: conditional inhibition by D2O, ethanol and Hg2+ suggests dependence of water influx upon membrane hydration and specific transmembrane pathways. J Eukaryot Microbiol 44:109–116. [DOI] [PubMed] [Google Scholar]
- 52.Chatterjee A, Carpentieri A, Ratner DM, Bullitt E, Costello CE, Robbins PW, Samuelson J. 2010. Giardia cyst wall protein 1 is a lectin that binds to curled fibrils of the GalNAc homopolymer. PLoS Pathog 6:e1001059. doi: 10.1371/journal.ppat.1001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, Zhao P, Zha X, Cheng T, Chai C. 2004. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 54.Peters W. 1992. Peritrophic membranes, p 238 In Bradshaw SD, Burggren W, Heller HC, Ishii S, Langer H, Neuweiler G, Randall D (ed), Zoophysiology, vol 30 Springer Verlag, Berlin, Germany. [Google Scholar]
- 55.Hu X, Chen L, Xiang X, Yang R, Yu S, Wu X. 2012. Proteomic analysis of peritrophic membrane (PM) from the midgut of fifth-instar larvae, Bombyx mori. Mol Biol Rep 39:3427–3434. doi: 10.1007/s11033-011-1114-6. [DOI] [PubMed] [Google Scholar]
- 56.Butcher BA, Sklar L, Seamer L, Glew R. 1992. Heparin enhances the interaction of infective Leishmania donovani promastigotes with mouse peritoneal macrophages. A fluorescence flow cytometric analysis. J Immunol 148:2879–2886. [PubMed] [Google Scholar]
- 57.Frevert U, Sinnis P, Cerami C, Shreffler W, Takacs B, Nussenzweig V. 1993. Malaria circumsporozoite protein binds to heparan sulfate proteoglycans associated with the surface membrane of hepatocytes. J Exp Med 177:1287–1298. doi: 10.1084/jem.177.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love DC, Esko JD, Mosser DM. 1993. A heparin-binding activity on Leishmania amastigotes which mediates adhesion to cellular proteoglycans. J Cell Biol 123:759–766. doi: 10.1083/jcb.123.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herrera EM, Ming M, Ortega-Barria E, Pereira MEA. 1994. Mediation of Trypanosoma cruzi invasion by heparan sulfate receptors on host cells and penetrin counter-receptors on the trypanosomes. Mol Biochem Parasitol 65:73–83. doi: 10.1016/0166-6851(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 60.Alvarez-Dominguez C, Vazquez-Boland JA, Carrasco-Marin E, Lopez-Mato P, Leyva-Cobián F. 1997. Host cell heparan sulfate proteoglycans mediate attachment and entry of Listeria monocytogenes, and the listerial surface protein ActA is involved in heparan sulfate receptor recognition. Infect Immun 65:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pethe K, Aumercier M, Fort E, Gatot C, Locht C, Menozzi FD. 2000. Characterization of the heparin-binding site of the mycobacterial heparin-binding hemagglutinin adhesin. J Biol Chem 275:14273–14280. doi: 10.1074/jbc.275.19.14273. [DOI] [PubMed] [Google Scholar]